Abstract
Canonical Wnt/β-catenin signaling is crucial during embryonic development. Upon Wnt stimulation, Dishevelled proteins relay the signal from upstream Frizzled receptors to downstream effectors. By using affinity purification followed by ion-trap mass spectrometry we identified K-homology splicing regulator protein (KSRP) as a novel Dishevelled-interacting protein. We show that KSRP negatively regulates Wnt/β-catenin signaling at the level of post-transcriptional CTNNB1 (β-catenin) mRNA stability. Thus, Dishevelled-KSRP complex operates in Wnt regulation of β-catenin, functioning post-transcriptionally upon CTNNB1 mRNA stability.
Keywords: Wnt, β-catenin, Frizzled, Dvl, KSRP, RNA decay, Canonical pathway, Lef/Tcf, Primitive endoderm, K-Homology splicing regulatory protein
Introduction
Wnt signaling is crucial during normal embryonic development and cellular homeostasis (Logan and Nusse, 2004; Malbon, 2005; Moon et al., 2004; Polakis, 2007; Reya and Clevers, 2005). Wnt3a acts via the Frizzled1-LRP5/6-Dishevelled pathway to suppress phosphorylation and ubiquitin-mediated degradation of β-catenin and thereby increases the stability and accumulation of intracellular β-catenin. Nuclear accumulation of β-catenin provokes activation of Lymphoid-enhancer factor/T-cell factor (Lef/Tcf)-sensitive transcription of developmentally related genes (Behrens et al., 1996; Molenaar et al., 1996). Dishevelled (Dsh/Dvl) has a key role in the canonical Wnt/β-catenin pathway (Axelrod et al., 1998; Boutros et al., 1998; Heisenberg et al., 2000; Sokol, 1996), but its mode of action is poorly defined.
To fully appreciate the functional role of Dvl in the regulation of Wnt-dependent pathways, we sought to identify Dvl-associated proteins by using proteomics, with the atypical C-terminus of Dvl3 (residues 497-716) as ‘bait’. Studies in F9 teratocarcinoma cells and genetically modified mice demonstrate that Dvl3, one of the three homologues of mammalian Dvl proteins, is a prime candidate for interrogation (Etheridge et al., 2008; Lee et al., 2008). In addition, the primary sequence of Dvl3 also displays greater variance from Dvl1 and Dvl2 in the C-terminus; thus, we selected this region as ‘bait’. Epitope (HA)-tagged Dvl3 C-terminus (residues 497-716) was expressed in human embryonic kidney (HEK293) cells, affinity purified with anti-HA antibodies, and the trapped Dvl3-based complexes were analyzed using ion-trap mass spectrometry. We identified the K-homology splicing regulator protein (KSRP) as a novel Dvl3-interacting protein. We demonstrate here that the Dvl3-KSRP interaction regulates canonical Wnt/β-catenin signaling by stabilization of CTNNB1 (β-catenin) mRNA, revealing a new dimension in the control of the canonical Wnt pathway.
Results
Identification of Dvl-interacting proteins
To isolate the protein complexes associated with the C-terminus of Dvl3, lysates of human embryonic kidney 293 cells (HEK293) expressing either empty vector (pCMV-HA) or Dvl3-CTM [pCMV-HA-Dvl3-CTM(497-716)] were affinity purified using anti-HA affinity matrix. The proteins isolated by such affinity pull-downs were identified by trypsin digestion followed by liquid chromatography-mass spectrometry (LC-MS/MS) using an LTQ ion-trap mass spectrometer equipped with a nano LC electrospray ionization source. Peptides that occurred only in the Dvl3-CTM lysate affinity pull-downs but not in the pCMV-HA control lysate pull-downs were considered as ‘positive’.
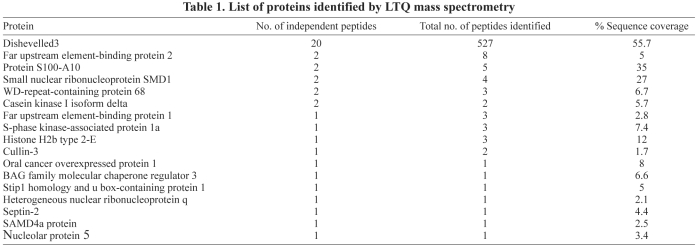
Affinity pull-down reactions with Dvl3-CTM resulted in the identification of the K-homology splicing regulator protein (KSRP) [also known as Far upstream element binding protein 2 (FUBP2)], among the group of interacting proteins by numerous ‘hits’ (Table 1). KSRP is a member of the family of AU-rich-element-binding proteins (ARE-BPs), and possesses four K-homology (KH) motifs. The KH motifs recognize ARE-containing mRNAs and promote mRNA decay by recruiting the targeted mRNA to the degradation machinery (Gherzi et al., 2004; Gherzi et al., 2006).
Table 1.
List of proteins identified by LTQ mass spectrometry
KSRP associates with the C-terminus of Dvl3
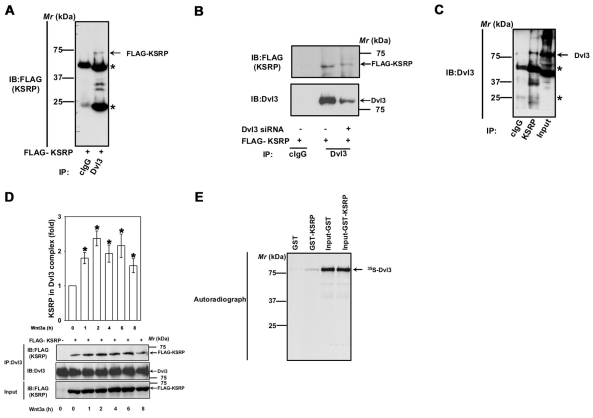
To test the endogenous interaction of Dvl3 and KSRP, lysates of F9 cells were immunoprecipitated with monoclonal anti-Dvl3 or rabbit polyclonal anti-KSRP antibodies. KSRP was detected in the Dvl3 immunoprecipitates, but not in the control IgG immunoprecipitations (Fig. 1A). Depletion of Dvl3 (by treatment with siRNA) from F9 cells before the pull-downs reduced the amount of the FLAG-KSRP found in the complex (Fig. 1B). These data argue in favor of the specificity of the Dvl3-KSRP interaction. Similarly, Dvl3 was also detected in the KSRP immunoprecipitates, but not in the control IgG immunoprecipitations (Fig. 1C), confirming the interaction of Dvl3 and KSRP. Interestingly, stimulation of the cells with Wnt3a increased association of KSRP with Dvl3 (Fig. 1D). We next tested whether the Dvl3-KSRP interaction was direct or indirect by using 35S-labeled Dvl3, which was produced in vitro using rabbit reticulocyte lysates, versus GST-KSRP that was isolated from bacteria in an in vitro assay. Again, Dvl3 bound specifically to GST-KSRP, but not to GST alone (Fig. 1E), strongly suggesting a direct interaction.
Fig. 1.
KSRP interacts with Dvl3. (A) F9 cells were transiently transfected with FLAG-KSRP for 24 hours followed by cell lysis and affinity pull-downs with either mouse control IgG or anti-Dvl3 mouse monoclonal antibody. Interaction of KSRP with Dvl3 was visualized by probing the blots with anti-FLAG antibody. Asterisks indicate the bands of immunoglobulin heavy and light chains. (B) F9 cells were treated with 100 nM of Dvl3 siRNA for 24 hours followed by transient expression of FLAG-KSRP for 24 hours followed by cell lysis and affinity pull-downs with either mouse control IgG or anti-Dvl3 mouse monoclonal antibody. Interaction of KSRP with Dvl3 was visualized by probing the blots with anti-FLAG antibody. (C) F9 cell lysates were immunoprecipitated with either rabbit control IgG or rabbit anti-KSRP polyclonal antibody and the interaction of KSRP with Dvl3 was visualized by probing the blots with anti-Dvl3 mouse monoclonal antibody. (D) F9 cells were transiently transfected with empty vector or FLAG-KSRP for 24 hours. The cells were then treated with Wnt3a (10 ng/ml) for indicated period of time followed by cell lysis and affinity pull-downs with anti-Dvl3 specific antibodies followed by immunoblotting with anti-FLAG antibodies. (E) To test the direct interaction of Dvl3 with KSRP, in vitro synthesized 35S-labeled Dvl3 was used in pull-down experiments with either GST- or GST-KSRP-Sepharose beads in the presence of 0.8% BSA. The interaction was visualized by SDS-PAGE and autoradiography. Representative blots of three independent experiments that proved highly reproducible are shown. *P<0.05 versus control (−Wnt3a).
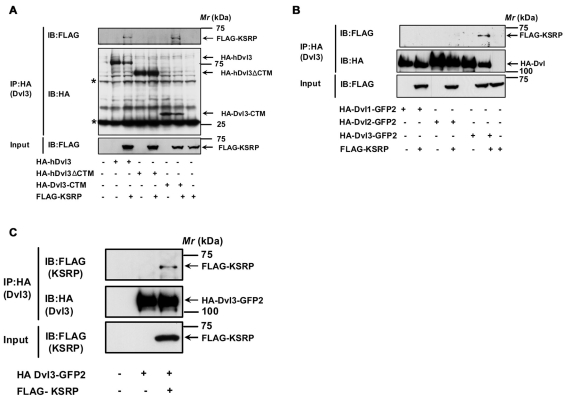
We sought to identify the region of Dvl3 that mediates interaction with KSRP. For these experiments, the endogenous Dvl3 from F9 cells was depleted by siRNA-mediated knockdown. Expression of either HA-Dvl3, HA-Dvl3-ΔCTM(1-496) or HA-Dvl3-CTM(497-716) alone or together with FLAG-KSRP was then used to address this question. The cell extracts were subjected to pull-down with anti-HA antibodies and the Dvl3-based complexes were identified by SDS-PAGE followed by immunoblotting with anti-FLAG antibodies. The mRNA binding protein KSRP was observed in the HA antibody pull-downs of the cell lysates co-expressing Dvl3-KSRP or Dvl3-CTM-KSRP but not Dvl3-ΔCTM-KSRP (Fig. 2A), indicating that KSRP interacts with Dvl3 and that the interaction is mediated through the C-terminus of Dvl3. We also tested whether or not KSRP interacts with different Dvl isoforms (Dvl1, Dvl2 and Dvl3). Interestingly, KSRP displayed the greatest association with the Dvl3 isoform, but not to Dvl1 or Dvl2 isoforms (Fig. 2B). Consistent with the F9 cells, KSRP-Dvl3 interaction also was observed in HEK293 cells (Fig. 2C). This KSRP-Dvl3 interaction in HEK293 cells, similarly to the interaction in F9 cells, was enhanced upon Wnt3a stimulation of the cells (data not shown).
Fig. 2.
KSRP-Dvl3 interaction is mediated through the C-terminus of Dvl3. (A) F9 cells were treated with 100 nM Dvl3 siRNA for 24 hours followed by transient expression of either HA-hDvl3 or HA-hDvl3-ΔCTM (1-496) or HA-Dvl3-CTM (497-716) alone or together with FLAG-KSRP for 24 ho urs followed by cell lysis and affinity pull-downs with anti-HA-affinity matrix. Interaction of KSRP with Dvl3 or its mutants was visualized by probing the blots with anti-FLAG antibody. Asterisks indicate the bands of immunoglobulin hea vy and light chains. (B) F9 cells were transiently transfected with either HA-Dvl1-GFP2 or HA-Dvl2-GFP2 or HA-Dvl3-GFP2 alone or with FLAG-KSRP for 24 hours followed by cell lysis and affinity pull-downs with anti-HA-affinity matrix. Interaction of KSRP with Dvl isoforms was visualized by probing the blots with anti-FLAG antibody. (C) HEK293 cells were transiently transfected with HA-Dvl3-GFP2 alone or with FLAG-KSRP for 24 hours followed by cell lysis and affinity pull-downs with anti-HA-affinity matrix. Interaction of KSRP with Dvl3 was visualized by probing the blots with anti-FLAG antibody. Representative blots of three independent experiments that proved highly reproducible are displayed.
Dvl3 interacts with KSRP via N-terminal KH domains
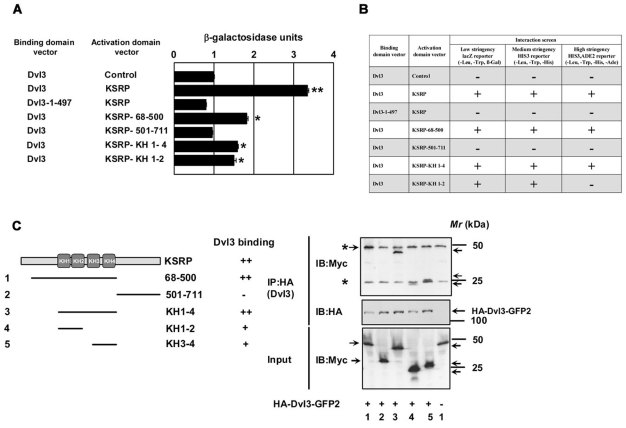
We sought to identify the region(s) of KSRP that interacts with Dvl3 by using limited deletion mutagenesis of KSRP. The binding of the KSRP deletion mutants to full-length Dvl3 was subjected to yeast-two hybrid analysis. The ability of KSRP deletion mutants to interact with Dvl3 was assessed through measurement of reconstituted lacZ reporter activity, using a quantitative β-galactosidase assay (Fig. 3A), histidine selection and adenine selection (Fig. 3B). Consistent with the in vivo and in vitro affinity pull-down data, yeast-two hybrid analysis revealed a positive interaction of KSRP with Dvl3 (Fig. 3A,B). Dvl3-ΔCTM, by contrast, displayed no such activity (Fig. 3A,B). Among the various deletion mutants tested, those mutants harboring the N-terminus of KSRP with KH motifs [residues 68-500, KH1-4 (146-500) and KH1-2 (146-300)] displayed positive interaction with full-length Dvl3 (Fig. 3A,B). Binding of full-length Dvl3 to KSRP and the constructs harboring the N-terminal KH motifs [residues 68-500 and KH1-4 (146-500)] was robust. These transformants alone could survive high-stringency selection (i.e. synthetic medium lacking leucine, tryptophan, histidine and adenine, Fig. 3B). These observations demonstrate that Dvl3 docks to KSRP (at the KH1-4 motif region, residues 146-500). The C-terminus of KSRP (residues 501-711) without KH motifs failed to interact with Dvl3, providing a negative control (Fig. 3A,B). Yeast transformants with the capacity to express Dvl3 and the KSRP-KH3-4 (325-500) uniquely were not viable (data not shown).
Fig. 3.
Dvl3 interacts with KSRP via N-terminal KH domains. Yeast-two hybrid assays were performed using full-length Dvl3 and deletion mutant of Dvl3 (1-497) in the DNA-binding domain vector and the full-length KSRP and the deletion mutants of KSRP in the activation domain vector. (A) To test for positive protein-protein interaction, quantitative β-galactosidase assays were performed. The results from liquid ONPG assays (displayed in β-galactosidase units) are represented in the graph. Dvl3-pGAD-AD (control) was used as a control for self-activation. The data represent mean ± s.e.m. obtained from two independent experiments performed in triplicate. *P<0.05; **P<0.01 versus control (Dvl3-pGAD-AD control). (B) Growth of yeast strains expressing fusion proteins on selective medium were also tested for studying protein-protein interactions. All the yeast cultures used for liquid β-galactosidase assays were normalized to similar OD600 densities and spotted onto selection plates [synthetic minimal medium lacking leucine, tryptophan and histidine (medium stringency)] or [synthetic minimal medium lacking leucine, tryptophan, histidine and adenine (high stringency)]. Yeast transformants that survive on the selection plates (medium and high stringency) were considered positive for an interaction, as shown in B. (C) To test the interaction of Dvl3 and the deletion mutants of KSRP in vivo, F9 cells were transiently transfected with the indicated Myc-tagged KSRP deletion constructs (left panel) along with HA-Dvl3-GFP2 for 24 hours followed by cell lysis and affinity pull-downs with anti-HA affinity matrix. Interaction of Dvl3 with deletion mutants of KSRP was visualized by probing the blots with anti-Myc antibody. The immunoreactive band for the N-terminus of KSRP (68-500) protein was observed to migrate along with the immunoglobulin heavy chain protein (as revealed by the darker intensity in lane 1 when compared with others). Asterisks indicate the bands of immunoglobulin heavy and light chains. Representative blots of two independent experiments that proved highly reproducible are displayed.
To further test the results of yeast two-hybrid analysis, the KSRP deletion mutants were subcloned into a mammalian expression vector with a Myc tag and expressed in F9 cells together with HA-Dvl3. The lysates were assessed for protein-protein interaction by affinity pull-downs. All of the KSRP deletion fragments studied in the yeast-two hybrid screening were successfully expressed in F9 cells (Fig. 3C). A positive interaction of Dvl3 with either the N-terminus of KSRP displaying the KH motifs (residues 68-500) or with the KH motifs of KSRP [KH1-4 (146-500), KH1-2 (146-300) and KH 3-4 (325-500)] was obtained (Fig. 3C). The C-terminus of KSRP (residues 501-711), by contrast, failed to bind Dvl3 under these conditions (Fig. 3C). In total, the results from several distinct and powerful analyses reveal a consistent story: Dvl3 docks to KSRP, especially through N-terminal KH motifs in KSRP.
KSRP negatively regulates canonical Wnt/β-catenin signaling
Since Wnt3a regulates the Dvl3-KSRP association, we examined next whether KSRP can modulate canonical Wnt/β-catenin signaling. When compared with control siRNA-treated cells, suppression of KSRP expression with siRNAs specific to mouse Khsrp provoked a twofold increase in basal Ctnnb1 mRNA levels (Fig. 4A). Interestingly, there also was a modest increase in basal Ctnnb1 mRNA levels upon Dvl3 depletion, which was not seen when Dvl2 was depleted (Fig. 4A). Consistent with the effects of KSRP depletion on basal Ctnnb1 mRNA levels, suppression of KSRP expression also provoked a fivefold increase in β-catenin protein levels, an increase that was similar in magnitude to that provoked by Wnt3a itself (Fig. 4B). Would KSRP knockdown impact the ability of Wnt3a to act at the level of Lef/Tcf-sensitive transcription? Knockdown of KSRP alone increased Lef/Tcf-sensitive transcription twofold (Fig. 4C). In cells treated with siRNAs specific to Khsrp, Wnt3a increased Lef/Tcf-sensitive transcription further (Fig. 4C). We next probed whether or not KSRP exerts an effect on the ability of Wnt3a to stimulate primitive endoderm (PE) formation. Knockdown of KSRP alone induced PE formation in either the absence or presence of Wnt3a (Fig. 4D). Conversely, overexpression of KSRP was found to attenuate Wnt3a-induced β-catenin accumulation and Lef/Tcf-sensitive transcription (Fig. 4E-G). Expression of KSRP likewise attenuated the normal increase in Lef/Tcf-sensitive transcription stimulated by the overexpression of Dvl3 (Fig. 4H). However, expression of an unrelated protein, the mouse S-phase kinase-associated protein 1 (SKP1A), had no apparent effect either on the basal or Wnt3a-stimulated Lef/Tcf-sensitive transcription. Thus, the effects observed with KSRP overexpression are specific (Fig. 4I).
Fig. 4.
KSRP negatively regulates Wnt/β-catenin signaling. (A) F9 cells were treated with either control siRNAs (100 nM) or siRNAs specific to Khsrp, Dvl2 or Dvl3 (100 nM) for 48 hours and the Ctnnb1 and Ppia (cyclophilin A) mRNA levels were quantified using quantitative PCR. The data represent Ctnnb1 mRNA normalized to the Ppia (cyclophilin A) mRNA levels (mean values ± s.e.m.) from three independent experiments whose results were in high agreement. F9 cells were treated with either control siRNAs (100 nM) or siRNAs specific for mouse Khsrp (100 nM) for 48 hours and the lysates were assayed either for cytosolic β-catenin levels (B) or Lef/Tcf-sensitive transcription (C). Upper panel displays mean values ± s.e.m. from three independent experiments; the lower panel displays representative blots. (D) F9 cells were treated with control siRNAs (100 nM) or siRNAs specific for mouse Khsrp (100 nM) for 48 hours before stimulation with Wnt3a for 4 days. Subsequently, formation of primitive endoderm was assayed by immunostaining the cells with anti-cytokeratin endo A antibody (TROMA-1), a hallmark of PE formation. Typical phase-contrast images (PC) and the indirect immunofluorescence images (IIF) are shown from a single experiment, representative of two independent experiments. F9 cells (E) or HEK293 cells (F) were transfected with FLAG-KSRP for 24 hours and the lysates were assayed for cytosolic β-catenin stabilization. (G) F9 cells were transfected with indicated amounts of FLAG-KSRP for 24 hours and the lysates were assayed for Lef/Tcf-sensitive transcription after stimulation with Wnt3a for 7 hours. (H) F9 cells were transfected with HA-Dvl3-GFP2 alone or together with increasing amounts of FLAG-KSRP for 24 hours and the lysates were assayed for Lef/Tcf-sensitive transcription. (I) F9 cells were transfected with indicated amounts of an unrelated protein, mouse S-Phase kinase associated protein 1A (mSKP1A) for 24 hours and the lysates were assayed for Lef/Tcf-sensitive transcription. The data represents mean values ± s.e.m. from three independent experiments *P<0.05; **P<0.01 versus control (−Wnt3a). #P<0.05; ##P<0.01 versus control (+Wnt3a).
Dvl3-KSRP association is RNA dependent
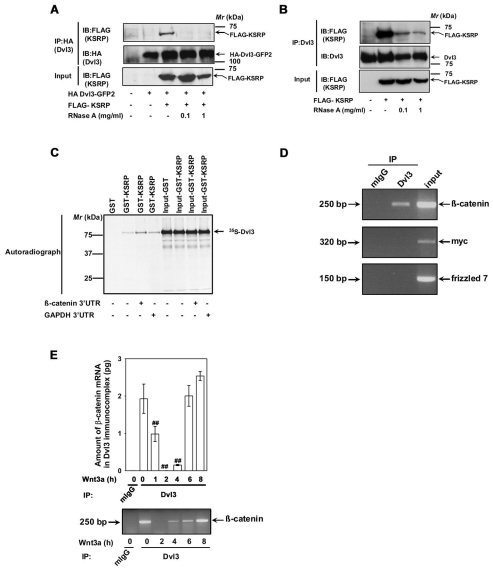
KSRP is known to be an RNA-binding protein and we show that KSRP interacts with Dvl3. We next assessed whether or not the Dvl3-KSRP interaction is dependent on RNA. Lysates prepared from cells co-expressing Dvl3 and KSRP were treated without or with RNaseA before pull-down experiments were carried out. Previous treatment with RNaseA dramatically affected the Dvl3-KSRP interaction. The sensitivity of this interaction was observed in the pull-downs of either exogenous (Fig. 5A) or endogenous (Fig. 5B) Dvl3. To further validate our findings, we tested the interaction of 35S-labeled Dvl3 produced in vitro using rabbit reticulocyte lysates and GST-KSRP in the absence or presence of 3′-untranslated regions (UTR) of either Ctnnb1 or Gapdh mRNA (Fig. 5C). The interaction between Dvl3 and KSRP was observed to be increased in the presence of 3′-UTR of Ctnnb1 mRNA, but not with Gapdh 3′-UTR (Fig. 5C). Taken together, these experiments strongly suggest that the interaction between KSRP and Dvl3 is direct and the presence of Ctnnb1 3′-UTR mRNA appears to enhance the stability of the Dvl3-KSRP complex.
Fig. 5.
Dvl3-KSRP interaction is RNA dependent and Dvl3 complex harbors Ctnnb1 mRNA. (A) F9 cells were co-transfected with HA-Dvl3-GFP2 and FLAG-KSRP for 24 hours followed by cell lysis. The lysates were then incubated without or with indicated amounts of RNaseA at room temperature for 10 minutes followed by affinity pull-downs with anti-HA affinity matrix. Interaction of KSRP with exogenous Dvl3 was probed by immunoblotting with anti-FLAG antibodies. (B) F9 cells were transfected with FLAG-KSRP for 24 hours followed by cell lysis. The lysates were then incubated without or with indicated amounts of RNaseA at room temperature for 10 minutes followed by affinity pull-downs with anti-Dvl3 antibodies. Interaction of KSRP with endogenous Dvl3 was probed by immunoblotting with anti-FLAG antibodies. (C) To test whether the in vitro interaction of KSRP and Dvl3 is also RNA dependent, in vitro synthesized 35S-labeled Dvl3 was used in pull-down experiments with either GST or GST-KSRP-Sepharose beads in the absence or presence (5 μg) of either 3′-UTR of Ctnnb1 or Gapdh. The interaction was visualized by SDS-PAGE and autoradiography. (D) RNA immunoprecipitation assay was performed on F9 cell lysates with either control mouse IgG or anti-Dvl3 antibodies. The RNA isolated from the immunoprecipitates was analyzed by RT-PCR with primers specific for Ctnnb1, Myc or Fzd7. Representative gel of two independent experiments that proved highly reproducible is displayed. (E) F9 cells were treated with Wnt3a (10 ng/ml) for indicated periods of time and RNA immunoprecipitation assay was performed with either control mouse IgG or anti-Dvl3 antibodies. The RNA isolated from the immunoprecipitates was analyzed by quantitative real-time PCR with β-catenin specific primers. Representative blots of two independent experiments that proved highly reproducible were displayed. The data represent mean values ± s.e.m. from two independent experiments whose results were in high agreement. In the lower panel, a representative gel is displayed. ##P<0.01 versus the control (−Wnt3a).
KSRP has been shown to interact with the 3′-UTR of Ctnnb1 mRNA (Gherzi et al., 2006). Hence, we probed for Ctnnb1 mRNA in Dvl3-based complexes that include KSRP (Figs 1, 2 and 3). RNA from the Dvl3-based complexes was extracted and then amplified by RT-PCR. Ctnnb1 mRNA was detected in the Dvl3-KSRP complexes (Fig. 5D). Control IgG pull-downs, by contrast, displayed no Ctnnb1 mRNA (Fig. 5D). Attempts to amplify either Myc or Fzd7 (Frizzled-7) mRNA transcripts (Fig. 5D) in the Dvl3-KSRP complexes isolated in pull-downs served as negative controls. Quantitative real-time PCR established the relative amounts of Ctnnb1 mRNA in the Dvl3-KSRP complexes before and after Wnt3a treatment. Wnt3a treatment provoked a loss of Ctnnb1 mRNA from the Dvl3-KSRP-based complexes. After an initial decline, the Ctnnb1 mRNA displayed a striking recovery during the time-course of Wnt3a stimulation (Fig. 5E).
Wnt3a stimulation stabilizes Ctnnb1 mRNA
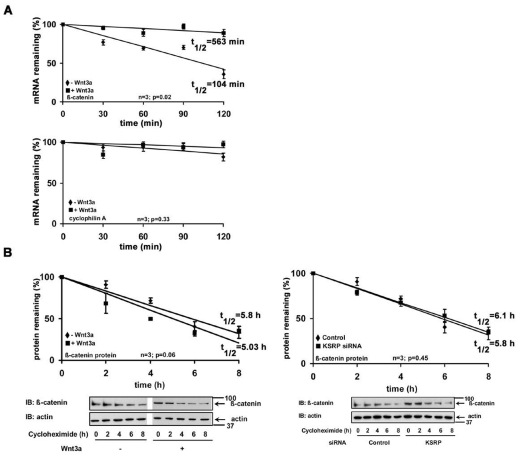
KSRP promotes rapid mRNA decay, so we investigated the stability of Ctnnb1 mRNA in F9 cells. To probe mRNA half-life, F9 cells were treated without or with Wnt3a followed by addition of actinomycin D (a transcription inhibitor). Cells were harvested at various times (0, 30, 60, 90 and 120 minutes) after the addition of actinomycin D. The half-life of Ctnnb1 mRNA was assessed by quantitative real-time PCR. Ctnnb1 mRNA decayed rapidly, displaying a t1/2 of ~100 minutes (Fig. 6A). There were no similar changes in Ppia (cyclophilin A) mRNA levels (Fig. 6A). Ctnnb1 mRNA appeared to undergo rapid decay in unstimulated cells. However, stimulation with Wnt3a stabilized Ctnnb1 mRNA, providing a fivefold post-transcriptional increase in the half-life of the transcript (>560 minutes).
Fig. 6.
Ctnnb1 mRNA has a short half-life. (A) Serum-starved F9 cells were treated without or with Wnt3a (10 ng/ml) for 1 hour, followed by treatment with actinomycin D (5 μg/ml) for indicated periods of time. Ctnnb1 and Ppia (cyclophilin A) mRNA levels were quantified using quantitative PCR. The data represents mean values ± s.e.m. from three independent experiments whose results were in high agreement. (B) β-catenin half-life was estimated either in F9 cells or F9 cells previously treated with Khsrp siRNAs, by treating the cells with cycloheximide (100 μg/ml) for indicated periods of time, followed by immunoblotting and densitometric scanning. β-catenin levels were normalized by the actin levels. The data represents mean values ± s.e.m. from three independent experiments whose results were in high agreement.
The half-life of the β-catenin at the protein level was also of interest. F9 cells were treated with Wnt3a and then harvested at various times (0, 2, 4, 6 and 8 hours) after treatment with the protein synthesis inhibitor, cycloheximide. β-catenin levels were established by immunoblotting from total lysates using β-catenin-specific antibodies. The β-catenin levels were normalized against actin (by making use of anti-actin antibodies, Fig. 6B). In the unstimulated cells, β-catenin decayed with a half-life of ~5.8 hours (Fig. 6B). Neither Wnt3a treatment (t1/2 ~5 hours) nor KSRP knockdown (t1/2 ~6 hours) significantly changed the half-lives of β-catenin (Fig. 6B). The data are in strong agreement with earlier studies performed in murine αT3-1 pituitary cells (Gherzi et al., 2006).
Wnt3a-stimulated β-catenin accumulation is the result of post-transcriptional stabilization of Ctnnb1 mRNA
We next probed whether the Wnt-stimulated increase in β-catenin levels results from either new protein synthesis or inhibition of ubiquitin mediated β-catenin protein degradation, or both. To establish the relative contribution of each, cells were treated with either actinomycin D (a transcription inhibitor) or cycloheximide (a protein synthesis inhibitor), and were then monitored for Lef/Tcf-sensitive transcription. Treatment of cells with either actinomycin D or cycloheximide completely blockedWnt3a-induced Lef/Tcf gene transcription, as expected (Fig. 7A). For cells treated with cycloheximide, but not with actinomycin D, Wnt3a-induced β-catenin accumulation was severely attenuated (Fig. 7B). Thus, the increase in β-catenin in response to Wnt3a stimulation appears to best reflect new protein synthesis. Increased synthesis of β-catenin, in response to Wnt3a stimulation, appears to be a derivative of increased stability of Ctnnb1 mRNA rather than new mRNA synthesis, because actinomycin D failed to impact Wnt3a-induced β-catenin stabilization (Fig. 7B).
Fig. 7.
Wnt3a-induced β-catenin accumulation is the result of post-transcriptional stabilization of Ctnnb1 mRNA. (A) Confluent F9 cells were treated with vehicle (DMSO), actinomycin D (Act D, 5 μg/ml) or cycloheximide (CHX, 100 μg/ml) for 1 hour followed by stimulation with Wnt3a for 7 hours. Lef/Tcf-sensitive transcription was determined. The data represent mean values ± s.e.m. from three separate experiments performed in triplicates. (B) Confluent F9 cells were treated with vehicle (DMSO), actinomycin D (5 μg/ml) or cycloheximide (100 μg/ml) for 1 hour before addition of Wnt3a for indicated periods of time. After stimulation, the lysates were collected and cytosolic β-catenin levels were assayed. Upper panel displays mean values ± s.e.m. obtained from three independent experiments; the lower panel displays representative blots. **P<0.01 versus control (−Wnt3a); #P<0.05; ##P<0.01 versus the control (+Wnt3a).
Overexpression of KSRP effects β-catenin signaling in colon carcinoma cells
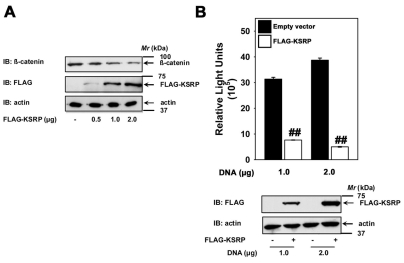
Finally, we probed whether or not KSRP expression might alter β-catenin signaling in colon carcinoma cells. SW480 cells were established from primary adenocarcinoma of the colon and these cells display high levels of endogenous β-catenin- and Tcf-dependent transcription (Fig. 8) (Sinner et al., 2007). Consistent with our observations using F9 and HEK293 cells, overexpression of KSRP in SW480 cells also attenuated cytosolic β-catenin levels (Fig. 8A). The KSRP-overexpression-induced suppression was also observed to be dose-dependent with respect to KSRP expression (Fig. 8A). We also tested the effects of KSRP overexpression on Lef/Tcf-sensitive transcription in SW480 cells. Transfection of SW480 cells with TOPFLASH reporter alone revealed high levels of luciferase activity, suggesting that their elevated levels of β-catenin promote elevated β-catenin/Tcf transcription (Fig. 8B). Overexpression of KSRP was able to strongly attenuate the Lef/Tcf-sensitive transcription in these colon carcinoma cells (Fig. 8B).
Fig. 8.
KSRP overexpression effects β-catenin signaling in colon carcinoma cells. SW480 cells (2.5×105 cells/well in a 12-well cell culture dish) were transfected with indicated amounts of either empty vector or FLAG-KSRP plasmid for 24 hours using a Fugene transfection reagent (Roche applied life sciences) to DNA ratio of 5:2. The lysates were then assayed either for cytosolic β-catenin levels (A) or Lef/Tcf-sensitive transcription (B). Upper panel in B displays mean values ± s.e.m. obtained from three independent experiments; the lower panel displays representative blots. For the β-catenin assay representative blots of two independent experiments that proved highly reproducible are displayed. ##P<0.01 versus the control (empty vector).
Discussion
Dishevelled is a crucial molecule in the regulation of canonical Wnt/β-catenin signaling. The multi-functional nature of Dishevelled proteins continues to be discovered. In the current study, KSRP was identified as a novel Dvl3-interacting protein. KSRP was shown to interact with the C-terminus of Dvl3. Mammalian cells express three Dishevelled isoforms (Dvl1, Dvl2 and Dvl3) (Bikkavilli et al., 2008; Lee et al., 2008). The distinct function(s) of the three isoforms in Wnt signaling are not known. KSRP specifically interacts with Dvl3, but not with Dvl1 or Dvl2. Although the mammalian isoforms share the common domains such as DIX, PDZ and DEP domains, the greatest degree of variation is observed in their C-terminal regions. The C-terminal region of Dvl3 is a largely natively unordered region. Dvl3 displays a short stretch of histidine residues (621-674) that are unique to Dvl3. We speculate that the docking of KSRP to Dvl3 might involve this region. However, in the absence of defined mutations (e.g. point mutations or small internal deletions in the interacting domains of Dvl3 and KSRP) and parallel expression experiments in vivo, functionally relevant versus fortuitous bases for all binding interactions cannot be readily distinguished.
KSRP is an RNA-binding protein that recruits RNA-decay machinery and promotes rapid decay of the selected mRNAs. KSRP has been shown to be capable of influencing CTNNB1 mRNA levels (Gherzi et al., 2006). KSRP is known also to regulate the biogenesis of a subset of miRNAs (Trabucchi et al., 2009). In the present study, we observed that KSRP docks to the C-terminus of Dvl3 and acts as a negative regulator of canonical Wnt/β-catenin signaling. In the absence of Wnt, the Dvl3-KSRP complex probably regulates the stability of Ctnnb1 mRNA to a limited degree. Ctnnb1 mRNA is detected in the Dvl3-KSRP complex. Upon treatment with Wnt3a, a rapid loss of Ctnnb1 mRNA from the Dvl3-KSRP complex was recorded. We speculate that escape from the Dvl3-KSRP complex promotes the stabilization of Ctnnb1 transcripts. This Wnt-stimulated mRNA stabilization probably contributes to the rapid accumulation of β-catenin, triggering downstream events in the canonical Wnt/β-catenin pathway. Exit of Wnt-stimulated Ctnnb1 mRNA from the Dvl3-KSRP complex correlates with β-catenin accumulation. As early as 1 hour following Wnt3a stimulation, a sharp decline was observed for Ctnnb1 mRNA content in the Dvl3-KSRP complex (i.e. mRNA stabilization); a sharp increase in the β-catenin accumulation follows (i.e. protein translation). The increase in the Ctnnb1 mRNA content of the Dvl3-KSRP complex later in the time course (6-8 hours) might well represent a switching ‘off’ of the canonical Wnt pathway signal.
We, and others, thus show that Wnt3a stimulation increases the stability of Ctnnb1 mRNA. How Wnt3a stimulation impairs the mRNA-decay-promoting activities via KSRP remains obscure at the level of fine detail. Both Wnt stimulation and Dishevelled overexpression have been shown to induce AKT activity (Fukumoto et al., 2001). KSRP is phosphorylated by AKT at Ser193; this phosphorylation event induces binding of 14-3-3 to KSRP. The docking of 14-3-3 to KSRP promotes nuclear transport of the complex and impairs the general mRNA-decay-promoting ability of KSRP (Diaz-Moreno et al., 2009; Gherzi et al., 2006). In the present study, we show that Dvl3 associates with KSRP. We observed also a Wnt-stimulated increase in the amount of Dvl3-KSRP complex (current study) and the amount of AKT associated with the KSRP (our unpublished data). Although only a speculation, Dvl3 might regulate the mRNA-decay-promoting role of KSRP via recruitment of AKT. In this regard, Dishevelled proteins are known to form functional polymers or oligomers (Schwarz-Romond et al., 2007). We speculate further that the basal interaction of Dvl3-KSRP that we observed serves as a ‘tag’ for the complex formation aimed at quick recognition and rapid communication with the Dvl3-KSRP complex.
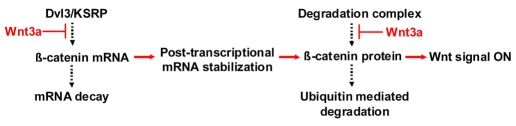
The observations from the current study suggest that β-catenin is regulated in two ways. At the mRNA level, Dvl3-KSRP complex promotes de-stabilization of Ctnnb1 mRNA. At the protein level, GSK3β-mediated phosphorylation regulates degradation of β-catenin. Wnt acts to oppose the ability of the Dvl3-KSRP complex to promote de-stabilization of Ctnnb1 mRNA and to oppose the degradation of β-catenin mediated by GSK3β phosphorylation. Post-transcriptional stabilization and translation of Ctnnb1 mRNA coupled with reduced phosphorylation-mediated degradation of the β-catenin protein in response to Wnt contributes to rapid accumulation of β-catenin. Hence, novel post-transcriptional as well as known post-translational responses are essential for Wnt regulation of canonical gene expression (Fig. 9).
Fig. 9.
Regulation of β-catenin levels in canonical Wnt/β-catenin signaling pathway. Basal levels of β-catenin are regulated at the mRNA level by Dvl3-KSRP complex that promotes de-stabilization of CTNNB1 mRNA, and at the protein level by GSK3β-mediated phosphorylation that promotes degradation of β-catenin. Wnt stimulation contributes to rapid accumulation of β-catenin by opposing the ability of the Dvl3-KSRP complex to promote destabilization of CTNNB1 mRNA and the degradation of β-catenin mediated by GSK3β phosphorylation.
Materials and Methods
Constructs
Mouse Dishevelled isoforms and Dvl3-CTM (497-716) were engineered in-house with GFP2 and HA tags. cDNA fragments encoding mouse Ctnnb1 3′-UTR (2517-3536), and mouse Gapdh 3′-UTR (1011-1230) were subcloned into KpnI and EcoRI sites of pcDNA3.1 vector. FLAG-KSRP expression construct was provided by Roberto Gherzi (Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy). Human KSRP deletion mutants with a Myc tag were engineered in-house into the EcoRI and XhoI sites of pCMV-Myc vector (Clontech). HA-h-Dvl3 and HA-h-Dvl3-ΔCTM were provided by Hsien-yu Wang (Department of Physiology and Biophysics, State University of New York, Stony Brook, NY).
Cell culture
Mouse F9 teratocarcinoma, HEK293 and SW480 cell stocks were obtained from ATCC (Manassas, VA) and were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with either 15% (F9 cells) or 10% (HEK293 cells and SW480) heat-inactivated fetal bovine serum (Hyclone, South Logan, UT) at 37°C in a 5% CO2 incubator. F9 cells stably expressing Rfz1 and pTOPFLASH (M50) luciferase reporter were generated as described earlier (Bikkavilli et al., 2008).
Identification of Dvl3-associated proteins
HEK293 cells were transfected with 12 μg of either pCMV-HA empty vector or pCMV-HA-Dvl3 CTM (497-716) in 150 mm tissue culture dishes using lipofectamine and plus reagents (Invitrogen, Carlsbad) as per the manufacturer's recommendations. After 48 hours of transfection, the cells were lysed in a lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotonin and 1 μg/ml phenlymethylsulphonyl fluoride) followed by incubation on ice for 5 minutes. After clearing the lysates by centrifugation at 14,000 g on a bench-top centrifuge, the supernatants were transfered into fresh Eppendorf tubes and stored at −70°C until further use.
For immunoprecipitations, 10 mg lysate was incubated at 4°C with 40 μl anti-HA high-affinity matrix (Roche) under gentle rotation. After overnight incubation, the resin beads were washed three times with RIPA buffer [20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 5 mM EDTA].
After the third wash, the buffer from the resin beads was completely aspirated and resin beads were resuspended in 0.1M ammonium bicarbonate. The proteins were first reduced by addition of dithiothreitol to a final concentration of 2 mM and incubation at room temperature for 30 minutes, followed by the alkylation by addition of iodoacetamide to a final concentration of 4 mM and incubation at room temperature for 30 minutes in the dark. Tryptic digests were then performed by addition of 0.5 μg trypsin to the resin beads followed by incubation at 37°C. After overnight incubation with trypsin, the samples were acidified with formic acid (final concentration 5%). The tryptic peptides in the solution were separated from the resin beads by spinning in a centrifuge and concentrated in a speed-vac concentrator. The dried peptides were then resuspended in 10 μl buffer A (5% acetonitrile and 0.1% formic acid).
The peptide mixture was then analyzed by automated microcapillary liquid chromatography-tandem mass spectrometry. Fused-silica capillaries (100 m.i.d.) were pulled using a P-2000 CO2 laser puller (Sutter Instruments, Novato, CA) to a 5 m.i.d. tip and packed with 10 cm of 5 m Magic C18 material (Agilent, Santa Clara, CA) using a pressure bomb. This column was then placed in-line with a Dionex Ultimate 3000 equipped with an auto-sampler. The column was equilibrated in buffer A, and the peptide mixture was loaded onto the column using the auto-sampler. The HPLC pump flowed at 100 μl/minute, and the flow rate to the electrospray tip was reduced to approximately 200-300 nl/minute by a split. The HPLC separation was provided by a gradient between Buffer A and Buffer B (90% acetonitrile, 0.1% formic acid).
The HPLC gradient was held constant at 100% buffer A for 5 minutes after peptide loading followed by a 30 minute gradient from 5% buffer B to 40% buffer B. Then, the gradient was switched from 40% to 80% buffer B over 5 minutes and held constant for 3 minutes. Finally, the gradient was changed from 80% buffer B to 100% buffer A over 1 minute, and then held constant at 100% buffer A for 15 minutes. The application of a 1.8 kV distal voltage electrosprayed the eluted peptides directly into the LTQ ion trap mass spectrometer equipped with a nanoLC electrospray ionization source (ThermoFinningan, San Jose, CA). Full masses (MS/MS spectra) were recorded on the peptides over a 400-2000 m/z range, followed by five tandem mass (MS/MS) events sequentially generated in a data-dependent manner on the first, second, third, fourth and fifth most intense ions selected from the full MS spectrum (at 35% collision energy). Mass spectrometer scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (ThermoFinnigan, San Jose, CA).
MS/MS spectra were extracted from the RAW file with Readw.exe (http://www.sourceforge.net/projects/sashimi). The resulting mz XML file contains all the data for all MS/MS spectra and can be read by the subsequent analysis software. The MS/MS data was then searched with Inspect (Tanner et al., 2005) against a human IPI database with optional modifications: +14 on methionine, +57 on cysteine, +80 on threonine, serine, tyrosine. Only the peptides with a P value of at least 0.01 were analyzed further.
Co-immunoprecipitation and immunoblotting
For co-immunoprecipitation experiments, F9 clones stably expressing rat Frizzled 1 or HEK293 cells were transiently transfected for 24 hours with 3 μg each of either HA-tagged Dvl3, Dvl3-ΔCTM or Dvl3 CTM (497-716), Dvl1, Dvl2 either alone or together with FLAG-KSRP or Myc-KSRP deletion constructs in 100 mm culture dishes. After 24 hours, the cells were lysed in 1 ml lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM Na4P2O7, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotonin and 1 μg/ml phenlymethylsulphonyl fluoride). The lysates were cleared by centrifugation at 20,000 g for 15 minutes, twice. The supernatants were transferred into new tubes and protein concentrations were determined by the Lowry method (Lowry et al., 1951). Immunoprecipitations were performed using either rat anti-HA high affinity (Roche), mouse-monoclonal anti-Dvl3 (sc 8027, Santa Cruz) antibodies or rabbit polyclonal anti-KSRP antibodies and protein-G-Sepharose (P3296, Sigma). For immunoblotting, total lysates (30-60 μg protein/lane) were subjected to electrophoresis in 10-12% SDS-PAGE gels. The resolved proteins were transferred electrophoretically to nitrocellulose membrane blots. The blots were incubated with primary antibodies overnight at 4°C and the immunocomplexes were made visible by use of a secondary antibody coupled to horseradish peroxidase and developed using the enhanced chemiluminescence method. The antibodies were purchased from the following sources: anti-HA high affinity antibody (Roche Applied Science, Indianapolis, IN), anti-β-catenin, anti-Myc, anti-FLAG M2 and anti-β-actin antibodies were from Sigma, anti-fibrillarin and anti-GAPDH were obtained from Abcam (Cambridge, MA).
In vitro transcription and translation
3′-UTRs of Ctnnb1 and Gapdh were synthesized by in vitro transcription with T7 RNA polymerase (Roche applied sciences) using pcDNA3.1vector encoding Ctnnb1 and Gapdh UTRs as templates as per the manufacturer's recommendations. The transcription reaction proceeded for 2 hours, after which the reactions were pooled and the RNA was purified with columns used for RNA extraction (Purelink™ RNA mini kit, Invitrogen) and quantified by UV spectrometry. Fusion polypeptides of GST and GST-KSRP were induced in Eschericia coli with 0.1 mM isopropyl-β-galactopyranoside at 30°C for 4 hours and affinity purified using 50% slurry of glutathione-Sepharose 4B (GE Life Sciences) according to the batch purification method suggested by the manufacturer. 35S-labeled Dvl3 was produced by using TNT® T7 Quick Coupled Transcription/Translation System (Promega) as per the vendor's recommendation in the presence of [35S]methionine (43.5 TBq/mmol, NEN radiochemicals, PerkinElmer). Pull-down experiments of in vitro translated Dvl3 and GST-KSRP were performed in RIPA buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA and 1% Triton X-100) in the presence of 0.8% BSA for 2 hours at 4°C. The complexes were washed three times with RIPA buffer and the samples were boiled with 2× SDS-PAGE sample buffer and separated on a SDS-PAGE gel. The gel was then fixed (45% methanol, 10% acetic acid in water, 30 minutes), amplified (Autofluor, National Diagnostics, 30 minutes), dried, and autoradiography was then performed.
Cytosolic β-catenin accumulation assay
To separate the cytosolic β-catenin from membrane-associated β-catenin, lysates were treated with concanavalin-A-Sepharose (Amersham Biosciences, Uppsala, Sweden), as described earlier (Aghib and McCrea, 1995). Briefly, confluent F9 cultures were treated with Wnt3a for 4 hours (Fig. 3B,E,F) and lysed in lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 50 mM NaF, 40 mM Na4P2O7, 50 mM K2HPO4, 10 mM Na2MoO4, 2 mM Na3VO4, 1%, Triton X-100, 0.5% NP40, 1 μg/ml leupeptin, 1 μg/ml aprotonin and 1 μg/ml phenlymethylsulphonyl fluoride). The lysates were transferred to 1.5 ml Eppendorf tubes and rotated at 4°C for 15 minutes followed by centrifugation at 20,000 g for 15 minutes. The supernatants were transferred to new tubes, their protein concentrations were determined and the concentration was adjusted to 2.5 mg/ml with lysis buffer. 60 μl ConA-Sepharose was added to each tube and rotated at 4°C for 1 hour. After a brief centrifugation, the supernatants were transferred to new tubes and 30 μl ConA-Sepharose was added to each tube and rotated at 4°C for a further hour. Finally, after a brief centrifugation, the supernatants were transferred to new tubes and their protein concentration was determined. Immunoblotting was performed with the samples and β-catenin accumulation was analyzed by probing the blots with anti-β-catenin antibodies and normalized by probing with anti-actin antibodies.
Luciferase assays
F9 cells stably expressing Rfz1 and super 8×TOPFLASH (M50) luciferase reporter were seeded into 12-well plates. Following incubation for 24 hours at 37°C, the cells were treated with or without recombinant Wnt3a for 7 hours (R&D systems, Minneapolis, MA). Cells were then directly lysed on the plates by addition of 1× cell culture lysis reagent (Promega, Madison, WI). Lysates were collected into chilled microfuge tubes on ice and centrifuged at 20,000 g for 5 minutes. The supernatant was transferred into a new tube and directly assayed as described below. 20 μl lysate was mixed with 100 μl luciferase assay buffer (20 mM Tricine, pH 7.8, 1.1 mM MgCO3, 4 mM MgSO4, 0.1 mM EDTA, 0.27 mM coenzyme A, 0.67 mM luciferin, 33 mM DTT and 0.6 mM ATP) and the luciferase activities were measured with a luminometer (Berthold Lumat LB 9507). The samples were assayed in triplicate and the luciferase activities were normalized by protein content of the samples and are represented in the figures.
Knockdown protocol
Double-stranded RNAs (siRNAs) targeting mouse Khsrp (Briata et al., 2005) (5′-GGACAGUUUCACGACAACG-3′), mouse Dvl2 (5′-GGAAGAGAUCUCCGAUGAC-3′), mouse Dvl3 (5′-GGAAGAGAUCUCGGACGAC-3′) and control siRNAs (5′-UCUGUGAUUUGAAAGACUAGCCAAG-3′) were synthesized by Invitrogen (Invitrogen, Carlsbad, CA). F9 cells expressing Rfz1 were treated with 100 nM siRNAs by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's protocol. Briefly, siRNAs were incubated with 5 μl Lipofectamine 2000 for 20 minutes in 200 μl Optimem medium (Invitrogen, Carlsbad, CA), and then the mixture was added into 1 ml growth medium in a 12-well plate in which F9 cells expressing Rfz1 were cultured to 80% confluency. After siRNA treatment for 48 hours, the cells were assayed either for β-catenin stabilization, luciferase assays or PE formation, as described earlier.
Indirect immunofluorescence studies
F9 cells stably expressing Rfz1 were transfected with either control siRNAs or Khsrp siRNAs, as described earlier. After 24 hours of transfection, the cells were re-seeded into 24-well plates and incubated at 37°C for 4 hours. After 4 hours, the cells were stimulated with Wnt3a (day 1). On day 2, the cells were given a second siRNA treatment followed by Wnt3a treatment for another day. Finally, after the treatment of the cells with Wnt3a on days 3 and 4, on day 5 the cells were fixed with 3% paraformaldehyde at room temperature for 5 minutes, followed by three washes with MSM-PIPES buffer (18 mM MgSO4, 5 mM CaCl2, 40 mM KCl, 24 mM NaCl, 5 mM PIPES, 0.5% Triton X-100, 0.5% NP40). The cells were then incubated with the TROMA-1 antibody (Developmental Studies Hybridoma Bank, University of Iowa, IA) at 37°C for 30 minutes. After three washes with MSM-PIPES buffer, the cells were incubated with an anti-mouse antibody coupled to Alexa Fluor 488 (Invitrogen) at 37°C for 30 minutes. Finally, the cells were washed in blotting buffer (560 mM NaCl, 10 mM KH2PO4, 0.1% Triton X-100, 0.02% SDS) and images were captured using a Zeiss LSM510 inverted fluorescence microscope.
Immunoprecipitation of ribonucleoprotein complexes
Ribonucleoprotein complexes were immunoprecipitated from F9 cells stably expressing Rfz1 using either control mouse immunoglobulins (IgG) or with mouse monoclonal anti-Dvl3 antibodies (sc-8027, Santa Cruz Biotechnology, CA). Briefly, 6 mg lysate was incubated with 1 μg of either mouse IgG or anti-Dvl3 antibody at 4°C with gentle rotation. After 16 hours, 20 μl protein-G-Sepharose (P3296, Sigma) was added and rotated at 4°C for 4 hours. After 4 hours, the resin beads were washed three times with RIPA buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl and 5 mM EDTA). Total RNA from the immunocomplexes were then isolated using RNA STAT 60 reagent (Tel-test Inc. Friendswood, TX) according to the manufacturer's instructions. After determining the RNA concentrations using a spectrophotometer, first-strand cDNA synthesis was performed using 1 μg total RNA and Superscript II reverse transcriptase (Invitrogen) with oligo dT(18) primer. Real-time quantitative PCR amplification was performed using the DNA engine Opticon continuous fluorescence detection system (MJ Research Inc, Boston, MA). For a 20 μl PCR, 8 μl cDNA template (previously diluted 1:15 with water) was mixed with 6.25 pmol of forward and reverse primers and 2× SYBR PCR master mix (Qiagen, Valencia, CA).
The Light Cycler was programmed such that it included an initial activation step of 95°C for 15 minutes followed by 40 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds and extension at 72°C for 1 minute. Each cDNA sample was analysed in triplicate and the absolute amounts of β-catenin template in the immunocomplexes were determined using an external standard.
Ctnnb1 mRNA stability
The half-life of Ctnnb1 mRNA was determined as described (Gherzi et al., 2006). Briefly, F9 cells stably expressing Rfz1 cells were serum starved for 16 hours followed by without or with Wnt3a (10 ng/ml) treatment for 1 hour. Cells were then harvested from 0 to 120 minutes after addition of actinomycin D (5 μg/ml). Total cellular RNA was isolated using RNA STAT 60 reagent from each of the time points and Ctnnb1 mRNA levels were quantified by quantitative PCR. For quantitative RT-PCR, 250 ng total RNA was reverse transcribed using random primers and PCRs were performed using the Quantitect Sybr Green mix (Qiagen Inc, Valencia, CA) and the Opticon Real-time PCR detection system as described above. The primers used were as follows: Ctnnb1, sense, 5′-gtgcaattcctgagctgaca-3′ and antisense, 5′-cttaaagatggccagcaagc-3′; Dvl2, sense, 5′-acgacgatgctgtacgagtg-3′ and antisense, 5′-atttcgagggagggtgaagt-3′; Myc, sense, 5′-ctccactcaccagcacaactac-3′ and antisense, 5′-ctgcttgaatggacaggatgta-3′; Fzd7, sense, 5′-ccacacgaaccaagaggac-3′ and antisense, 5′-gaacggcacggaggaatg-3′; Ppia (cyclophilin A), sense, 5′-agcatacaggtcctggcatc-3′ and antisense, 5′-ttcaccttcccaaagaccac-3′.
Yeast two-hybrid analysis
The matchmaker yeast two-hybrid kit was used as per the manufacturer's instructions (Clontech, Palo-Alto, CA). Briefly, the yeast strain AH109 was co-transformed with various combinations of truncation mutants of Dvl3 and KSRP cloned into the DNA-binding domain and activating domain vectors respectively and plated on synthetic minimal medium lacking leucine and tryptophan that allows for selection of transformants. To test for positive protein-protein interactions, transformants were cultured in liquid minimal medium lacking leucine and tryptophan for 4 days. All the cultures were adjusted to similar OD600 values and were plated on minimal media lacking leucine, tryptophan, histidine or leucine, tryptophan, histidine and adenine. To quantify the protein-protein interactions, liquid β-galactosidase assays were performed using O-nitrophenol-β-D-galactopyranoside (ONPG, Sigma) as a substrate as mentioned in the yeast protocols handbook (Clontech). Briefly, 5 ml of yeast were grown in selection medium lacking leucine and tryptophan for 4 days. Then, 2 ml of the culture was transferred to 8 ml of YPDA (yeast extract, peptone, dextrose supplemented with adenine) in a 50 ml centrifuge tubes (Corning) and continued to culture with shaking at 30°C for 3 hours. After 3 hours, the exact OD600 values of the cultures were measured. Three aliquots of 1.5 ml of each sample were dispensed into eppendorf tubes. The cells were harvested by centrifugation at 10,000 g for 30 seconds. The cells were washed with 1 ml Z buffer (16.1 g/l Na2HPO4, 5.50 g/l NaH2PO4, 0.75 g/l KCl, 0.246 g/l MgSO4) and finally resuspended in 100 μl Z buffer. The cells were then lysed by three cycles of freezing and thawing. To each tube, 700 μl Z buffer with β-mercaptoethanol and 160 μl of 4 mg/ml ONPG were added and the tubes were incubated at 30°C until a yellow color developed. The reaction was stopped by addition of 400 μl of 1M Na2CO3 followed by high-speed centrifugation at 14,000 g for 10 minutes. The OD420 values of the supernatants were measured and the β-galactosidase units were calculated.
Data analysis
For all of the experiments, data were compiled from at least three independent, replicate experiments performed on separate cultures on separate occasions. In all cases, fold change over the untreated control (set to one) was calculated and displayed. Comparisons of data among groups were performed using one-way analysis of variance (ANOVA) or Student's t-test. The indirect immunofluorescence and phase-contrast images are of representative fields of interest.
Acknowledgments
We thank Antonius Koller (Technical Director, Proteomics Center, SUNY, Stony Brook, NY) for his help with sample preparation and generation and analysis of MS/MS spectra. We thank Roberto Gherzi (Istituto Nazionale per la Ricerca sul Cancro, Genova, Italy) for the generous gift of FLAG-KSRP expression construct and anti-KSRP antibodies and Randall T. Moon (University of Washington, Seattle, WA) for super8xTOPFLASH and pcDNA-FLAG-hDvl3 constructs. We thank Feng-Qian Li and Ken-Itchi Takemaru (Department of Pharmacology, SUNY, Stony Brook, NY) for their technical advice and we also thank Hsien-Yu Wang (Department of Physiology & Biophysics, SUNY, Stony Brook, NY) for critical comments on the manuscript. We thank members of the Malbon and Wang laboratories for their technical advice. This work was supported by USPHS Grant from the NIDDK, National Institutes of Health (to C.C.M.). Deposited in PMC for release after 12 months. This article is freely accessible online from the date of publication.
References
- Aghib D. F., McCrea P. D. (1995). The E-cadherin complex contains the src substrate p120. Exp. Cell Res. 218, 359-369 [DOI] [PubMed] [Google Scholar]
- Axelrod J. D., Miller J. R., Shulman J. M., Moon R. T., Perrimon N. (1998). Differential recruitment of Dishevelled provides signaling specificity in the planar cell polarity and Wingless signaling pathways. Genes Dev. 12, 2610-2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens J., von Kries J. P., Kuhl M., Bruhn L., Wedlich D., Grosschedl R., Birchmeier W. (1996). Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382, 638-642 [DOI] [PubMed] [Google Scholar]
- Bikkavilli R. K., Feigin M. E., Malbon C. C. (2008). G alpha o mediates WNT-JNK signaling through dishevelled 1 and 3, RhoA family members, and MEKK 1 and 4 in mammalian cells. J. Cell Sci. 121, 234-245 [DOI] [PubMed] [Google Scholar]
- Boutros M., Paricio N., Strutt D. I., Mlodzik M. (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94, 109-118 [DOI] [PubMed] [Google Scholar]
- Briata P., Forcales S. V., Ponassi M., Corte G., Chen C. Y., Karin M., Puri P. L., Gherzi R. (2005). p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell 20, 891-903 [DOI] [PubMed] [Google Scholar]
- Diaz-Moreno I., Hollingworth D., Frenkiel T. A., Kelly G., Martin S., Howell S., Garcia-Mayoral M., Gherzi R., Briata P., Ramos A. (2009). Phosphorylation-mediated unfolding of a KH domain regulates KSRP localization via 14-3-3 binding. Nat. Struct. Mol. Biol. 16, 238-246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etheridge S. L., Ray S., Li S., Hamblet N. S., Lijam N., Tsang M., Greer J., Kardos N., Wang J., Sussman D. J., et al. (2008). Murine dishevelled 3 functions in redundant pathways with dishevelled 1 and 2 in normal cardiac outflow tract, cochlea, and neural tube development. PLoS Genet. 4, e1000259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S., Hsieh C. M., Maemura K., Layne M. D., Yet S. F., Lee K. H., Matsui T., Rosenzweig A., Taylor W. G., Rubin J. S., et al. (2001). Akt participation in the Wnt signaling pathway through Dishevelled. J. Biol. Chem. 276, 17479-17483 [DOI] [PubMed] [Google Scholar]
- Gherzi R., Lee K. Y., Briata P., Wegmuller D., Moroni C., Karin M., Chen C. Y. (2004). A KH domain RNA binding protein, KSRP, promotes ARE-directed mRNA turnover by recruiting the degradation machinery. Mol. Cell 14, 571-583 [DOI] [PubMed] [Google Scholar]
- Gherzi R., Trabucchi M., Ponassi M., Ruggiero T., Corte G., Moroni C., Chen C. Y., Khabar K. S., Andersen J. S., Briata P. (2006). The RNA-binding protein KSRP promotes decay of beta-catenin mRNA and is inactivated by PI3K-AKT signaling. PLoS Biol. 5, e5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Heisenberg C. P., Tada M., Rauch G. J., Saude L., Concha M. L., Geisler R., Stemple D. L., Smith J. C., Wilson S. W. (2000). Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature 405, 76-81 [DOI] [PubMed] [Google Scholar]
- Lee Y. N., Gao Y., Wang H. Y. (2008). Differential mediation of the Wnt canonical pathway by mammalian Dishevelleds-1, -2, and -3. Cell Signal. 20, 443-452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan C. Y., Nusse R. (2004). The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 20, 781-810 [DOI] [PubMed] [Google Scholar]
- Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951). Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265-275 [PubMed] [Google Scholar]
- Malbon C. C. (2005). Beta-catenin, cancer, and G proteins: not just for frizzleds anymore. Sci STKE 2005, pe35 [DOI] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering M., Oosterwegel M., Peterson-Maduro J., Godsave S., Korinek V., Roose J., Destree O., Clevers H. (1996). XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86, 391-399 [DOI] [PubMed] [Google Scholar]
- Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004). WNT and beta-catenin signalling: diseases and therapies. Nat. Rev. Genet. 5, 691-701 [DOI] [PubMed] [Google Scholar]
- Polakis P. (2007). The many ways of Wnt in cancer. Curr. Opin. Genet. Dev. 17, 45-51 [DOI] [PubMed] [Google Scholar]
- Reya T., Clevers H. (2005). Wnt signalling in stem cells and cancer. Nature 434, 843-850 [DOI] [PubMed] [Google Scholar]
- Schwarz-Romond T., Fiedler M., Shibata N., Butler P. J., Kikuchi A., Higuchi Y., Bienz M. (2007). The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 14, 484-492 [DOI] [PubMed] [Google Scholar]
- Sinner D., Kordich J. J., Spence J. R., Opoka R., Rankin S., Lin S. C., Jonatan D., Zorn A. M., Wells J. M. (2007). Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Mol. Cell. Biol. 27, 7802-7815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S. Y. (1996). Analysis of Dishevelled signalling pathways during Xenopus development. Curr. Biol. 6, 1456-1467 [DOI] [PubMed] [Google Scholar]
- Tanner S., Shu H., Frank A., Wang L. C., Zandi E., Mumby M., Pevzner P. A., Bafna V. (2005). InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626-4639 [DOI] [PubMed] [Google Scholar]
- Trabucchi M., Briata P., Garcia-Mayoral M., Haase A. D., Filipowicz W., Ramos A., Gherzi R., Rosenfeld M. G. (2009). The RNA-binding protein KSRP promotes the biogenesis of a subset of microRNAs. Nature 459, 1010-1014 [DOI] [PMC free article] [PubMed] [Google Scholar]