Abstract
The quantitative assessment of the anatomic consequences of cerebral infarction is critical in the study of the etiology and therapeutic response in patients with stroke. We present here an overview of the operation of “WebParc,” a computational system that provides measures of stroke lesion volume and location with respect to canonical forebrain neural systems nomenclature. Using a web-based interface, clinical imaging data can be registered to a template brain that contains a comprehensive set of anatomic structures. Upon delineation of the lesion, we can express the size and localization of the lesion in terms of the regions that are intersected within the template. We demonstrate the application of the system using MRI-based diffusion-weighted imaging and document measures of the validity and reliability of its uses. Intra- and inter-rater reliability is demonstrated, and characterized relative to the various classes of anatomic regions that can be assessed. The WebParc system has been developed to meet criteria of both efficiency and intuitive operator use in the real time analysis of stroke anatomy, so as to be useful in support of clinical care and clinical research studies. This article is an overview of its base-line operation with quantitative anatomic characterization of lesion size and location in terms of stroke distribution within the separate gray and white matter compartments of the brain.
Keywords: Stroke, Embolism, Image processing, Computer assisted, Magnetic resonance imaging, Cerebral anatomy
1 Introduction
Quantitative lesion analysis is increasingly important for the characterization of stroke in clinical neuroimaging studies [1–4]. It is potentially important to prediction of stroke outcome and the effectiveness of therapeutic interventions in stroke [1–3, 5, 6]. Following image acquisition, quantitative analysis of the specific structures involved in stroke requires lesion segmentation. In this article, we demonstrate a freely available, modular system for lesion analysis.
The rationale for the use of such analytic techniques comes from the principle that the neurological deficit and the quantitative characterization of the stroke that causes it should provide similar predictions of long-term functional outcome and act as a guide for therapeutic monitoring. The neurological deficit according to the NIH Stroke Scale (NIHSS) in the acute phase of stroke is predictive of long-term disability [7, 8]. This scale integrates judgments following the standard neurological examination of the relative degrees of impairment levels of consciousness, attention, language compression and production, visual fields and gaze, motor paresis and coordination paresis and somatic sensory function. Furthermore, the volume of tissue destroyed by stroke as estimated in the acute phase from diffusion-weighted MR imaging is closely predictive of the stable, long-term volume of tissue loss from stroke (>1 month) [5]. However, this volumetric loss has not proved to be as reliably predictive of long-term disability as the specific neurological deficits as characterized by clinical measures, in particular, the NIHSS [9, 10].
Acutely administered thrombolytic and anticoagulant agents may restore the sufficiency of perfusion and thus conserve brain tissue thereby lessening the degree of neurological deficit [11–18]. However, pharmacologic agents protective against excitotoxic tissue dissolution, shown unequivocally to have brain tissue sparing effects in experimental animals [12, 19–21], given acutely to patients with stroke have had no identifiable brain sparing effect in so far as this could be detected by measures of stroke volume or long-term disability [3, 7, 12, 19–29]. Among the reasons that clinical trials have failed to confirm the predictions from the experimental studies may be limitations of the sensitivity and anatomic specificity of the methods and tools that have been used to determine the tissue damage effects in human studies. Such limitations relate in particular to estimates of the components and degree of the neurological deficit as the surrogate estimates of the volume and distribution. There may be tissue salvage due to neuroprotective agents that are not detected by current methods of analysis if the topographic details and volumes of the stroke are not accounted for.
The weight of evidence coming from perfusion–diffusion mismatch in acute human stroke unequivocally indicates that brain tissue remains at risk and is potentially salvageable for many hours and possibly days after the acute event [3, 24–29]. The neurological deficit, as characterized by the clinical examination is variably predictive of the location and size of the stroke. It is most strongly predictive of relatively small infarctions limited to the primary motor, somatosensory or visual cortices (M1, S1, or V1, respectively), but less well to the degree that the lesion involves non-primary or association areas [30–41]. Overall, however, taking into consideration the location and size of the lesion improves the reliability of the prediction of the severity of the neurological deficit [39]. The progression of ischemia and infarction may vary among cerebral regions with quite different implications for clinical outcome [42].
We present here an overview of the operation of “WebParc,” a freely available computational system that provides measures of stroke volume and lesion location with respect to canonical forebrain neural systems nomenclature. Similar in concept to the methods of Nowinski et al. [43–45], it is implemented in the template registration style of localization analysis [46, 47]. In the absence of direct segmentation of structures in the patient images, template registration provides one of the only ways to determine topographic location properties of the lesion. Differences with these prior methods include use of alternate topographically defined anatomic templates, use of a freely distributed modular architecture that is suitable for testing and documenting relative performance and improvements of each module with respect to human expert analysis, and provision of a baseline set of validation and reliability measures of this implementation. It is complementary to the methods of Nowinski et al. [43–45] in that many of the advances in automation of segmentation and speed of registration can be quantified using this system. Indeed any modular improvement over the baseline implementation can be readily accomplished. It is envisioned to be user friendly and efficient in application, making it potentially useful in assessment of the topography and progression of stroke over the interval that a patient is acutely hospitalized and through the period of follow up.
2 Methods
2.1 Overview
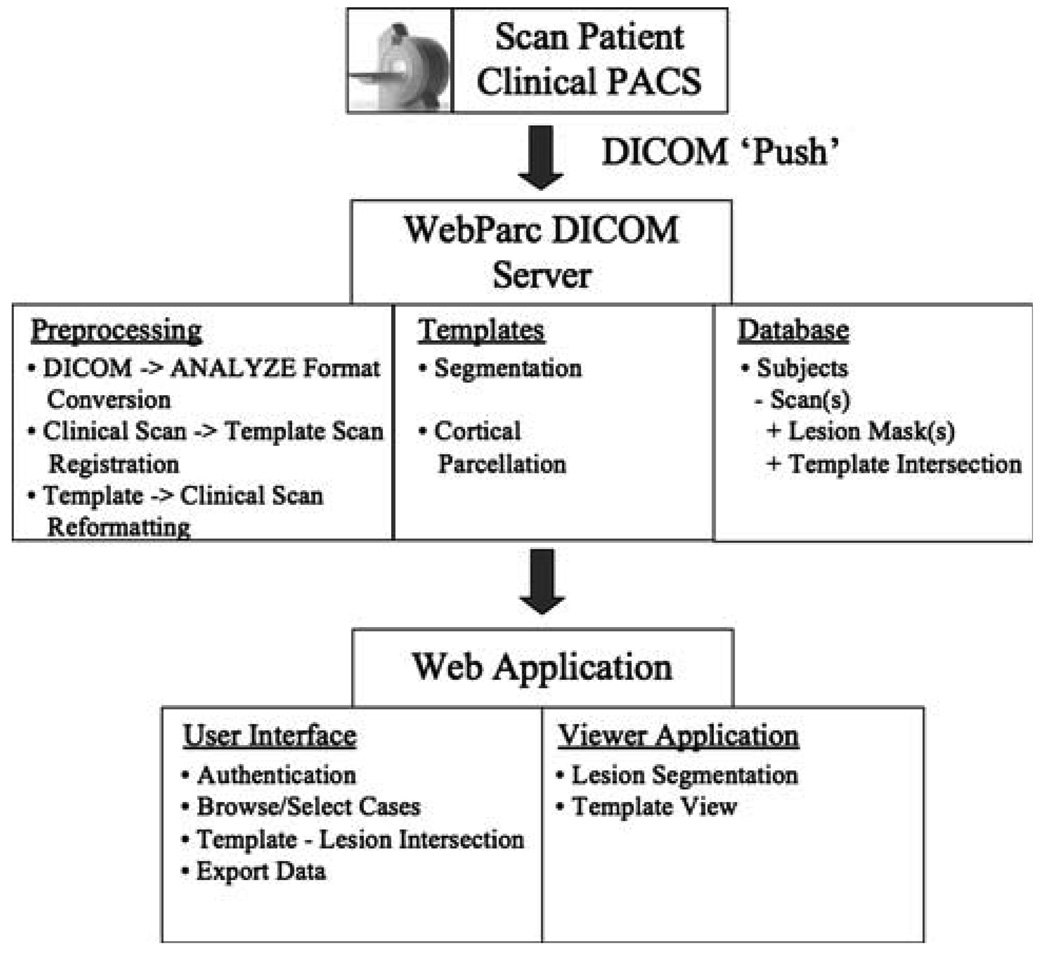
The application is simple in concept and designed for the quick estimation of size and location of lesions observed in clinical images. In brief, WebParc is a data management system that securely receives a clinical imaging data set, receives lesion annotation from the clinician, and expresses this lesion information with respect to a set of co-registered anatomic templates of detailed anatomic structures. The output of the system includes an estimate of lesion volume with respect to the anatomic structures in the templates. The system is modular, and includes a DICOM server to receive the clinical imaging data, a set of anatomic templates, an automated preprocessing and registration stream, a lesion annotation web application, a database to support the data management, and a web-based user interface for execution of the system functions (see Fig. 1). We present here the system invoked using very basic functionality for each of these modules to provide a baseline measure of system performance. It is acknowledged that virtually every module can be improved, and the quantification of these improvements over baseline will be the important objectives of future studies.
Fig. 1.
Overview figure showing the data flow and modular design of the WebParc system
2.2 WebParc modules
2.2.1 DICOM server
The clinical image set is uploaded from the clinical PACS system to the WebParc DICOM server using the DICOM shareware package from Central Test Node (CTN) at http://erl.wustl.edu/research/dicom/ctn.html [48]. The program simple_storage from this package receives the DI-COM images from the push and stores them to disk.
2.2.2 Templates
In this implementation of the system, we utilize two anatomic templates. The first is a “general segmentation” template that consists of the following structures in their entirety: cerebral cortex, cerebral white matter, thalamus, caudate, putamen, pallidum, acumbens, hippocampus, amygdala, lateral, third and fourth ventricles, cerebellum cortex, cerebellum white matter, and brainstem. These regions are defined using the methods of Filipek et al. [49, 50]. The template is built from a coronal, 1.5-mm slice-thickness volumetric T1-weighted magnetic resonance imaging (MRI) scan of an individual, 32-year-old, right-handed male subject. The second template is a “cortical parcellation” template, where the cerebral cortex is divided into the canonical forebrain gyral pattern [51, 52]. Table 1 includes a listing of these regions and their abbreviations. This template was generated from the same subject and images as the general segmentation template above. Each template has an “n-ary” region label mask (each unique region has a unique fill value), which we refer to as the “template,” and the corresponding, registered T1-weighted anatomic grayscale image that underlies the template, referred to as the “template image”. The template image is identical for the general segmentation and cortical parcellation templates.
Table 1.
Abbreviations
| Abbreviations | Structure: full name | Abbreviations | Structure: full name |
|---|---|---|---|
| AG | Angular gyrus | PHa | Parahippocampal gyrus, anterior division |
| CALC | Intracalcarine cortex | PHp | Parahippocampal gyrus, posterior division |
| CGa | Cingulate gyrus, anterior division | PO | Parietal operculum cortex |
| CGp | Cingulate gyrus, posterior division | POG | Postcentral gyrus |
| CN | Cuneal cortex | PP | Planum polare |
| CO | Central opercular cortex | PRG | Precentral gyrus |
| F1 | Superior frontal gyrus | PT | Planum temporale |
| F2 | Middle frontal gyrus | SC | Subcallosal cortex |
| F3o | Inferior frontal gyrus, pars opercularis | SCLC | Supracalcarine cortex |
| F3t | Inferior frontal gyrus, pars triangularis | SGa | Supramarginal gyrus, anterior division |
| FMC | Frontal medial cortex | SGp | Supramarginal gyrus, posterior division |
| FO | Frontal operculum cortex | SPL | Superior parietal lobule |
| FOC | Frontal orbital cortex | T1a | Superior temporal gyrus, anterior division |
| FP | Frontal pole | T1p | Superior temporal gyrus, posterior division |
| H | Heschl’s gyrus (includes H1 and H2) | T2a | Middle temporal gyrus, anterior division |
| INS | Insular cortex | T2p | Middle temporal gyrus, posterior division |
| JPL | Juxtapositional lobule cortex | T3a | Inferior temporal gyrus, anterior division |
| LG | Lingual gyrus | T3p | Inferior temporal gyrus, posterior division |
| OF | Occipital fusiform gyrus | TFa | Temporal fusiform cortex, anterior division |
| OLi | Lateral occipital cortex, inferior division | TFp | Temporal fusiform cortex, posterior division |
| OLs | Lateral occipital cortex, superior division | TO2 | Middle temporal gyrus, temporooccipital part |
| OP | Occipital pole | TO3 | Inferior temporal gyrus, temporooccipital part |
| PAC | Paracingulate gyrus | TOF | Temporal occipital fusiform cortex |
| PCN | Precuneous cortex | TP | Temporal pole |
2.2.3 Preprocessing steps
Upon image receipt at the WebParc server, a number of preprocessing steps are automatically initiated. This includes conversion of the DICOM input data into the Analyze image format, using the program mri_convert from the FreeSurfer software package [53] (http://www.nitrc.org/projects/freesurfer/). No additional parameters are provided, as we desire only simple output format conversion. Next, we perform brain extraction in the clinical image using the Brain Extraction Tool (BET) from the FSL software toolkit [54, 55] (http://www.nitrc.org/projects/fsl/) with no additional parameters. This is followed by linear affine registration of the clinical scan to the template image using FMRIB’s Linear Image Registration Tool (FLIRT), also from the FSL software toolkit. An initialization matrix is provided in order to direct the registration from the clinical acquisition orientation (i.e., axial) into the coronal orientation of the template image, and the -nosearch option is selected. Once completed, the saved transformation matrix is inverted (using convert_xfm –inverse from the FSL package) and applied to the templates to cast them in registration with the clinical scan in its native acquisition space. This template reformatting is accomplished using the FLIRT program with the -applyxfm –init {inverse_transformation_matrix} options. This complete set of pre-processing currently requires approximately 10 min of elapsed time. The database, see below, is updated to include this study and scan as available to the user interface.
2.2.4 Lesion annotation
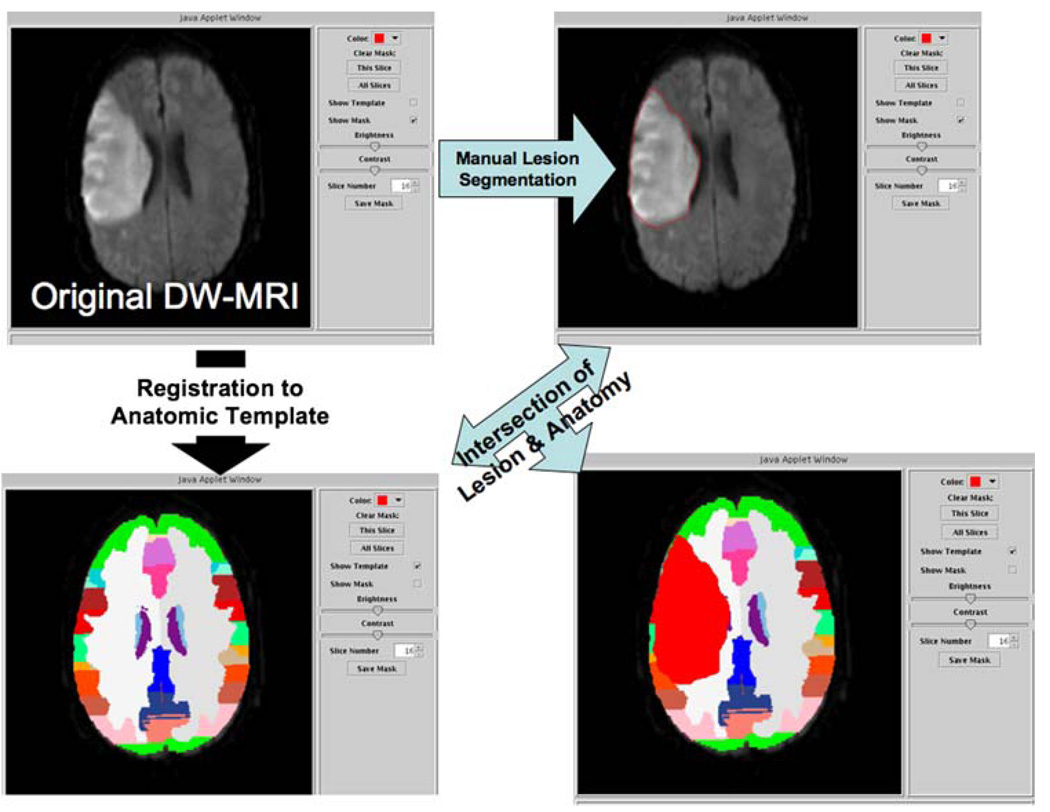
The lesion is segmented in the clinical imaging set by hand with a Java applet that provides web-based visualization of the clinical scans (Fig. 2). In this simple interface, the user can step through the clinical scan slices, manipulate the window and level settings, invoke the lesion drawing function, and toggle on/off the template and lesion masks. Registration quality is manually verified prior to lesion segmentation. If the visual alignment of the template brain exterior border were to be visually appreciated to be greater than 5 mm from the brain exterior border as seen in the clinical image, then the remaining lesion annotation process is not performed. “Off-line” improvements in registration can be performed, and the processing re-instated upon successful registration. This off-line correction was not needed in either the validation or reliability studies reported herein. In the lesion-mask draw mode, the user right-clicks points around the periphery of the lesion. Upon leaving the draw mode, the first and last click points are connected. The lesion mask can be deleted for the current slice, or all slices, if desired. The lesion mask can be saved at any point in the process. The lesion segmentation mask can be traced from a series of 10–20 images in approximately 10 min, depending upon lesion extent. Each saved mask updates the database (see below).
Fig. 2.
WebParc visualization interface. The user can visualize the clinical images (upper left), visually validate the registration between clinical case and template (lower left), and then proceed to make the lesion segmentation using mouse-driven cursor (upper right). Upon completion of the segmentation, the user initiates the intersection of the lesion mask with the template-based regions of anatomic interest, yielding a “lesion volume” for each anatomic region that the lesion intersects with (lower right)
2.2.5 Database
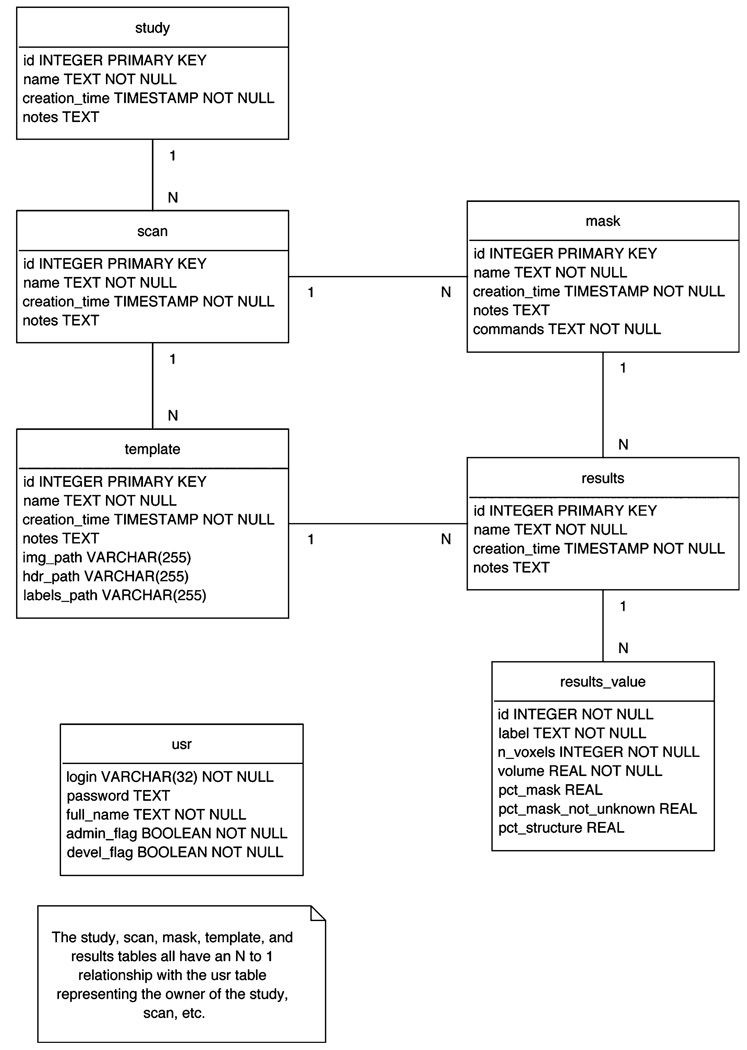
Data management is controlled by a back-end database built in PostgreSQL. The study is the principal element of the database structure. Each study can have multiple scans and each scan, in turn, can have multiple masks created from it and multiple templates registered to it. Any available mask can be intersected with any available template for a scan, in order to generate a new result. There is a separate table for registered users of the system. See Fig. 3 for the detailed database schema.
Fig. 3.
WebParc database schema. Detailed depictions of the study, scan, template, mask, result and usr database constructs are displayed
2.2.6 User interface
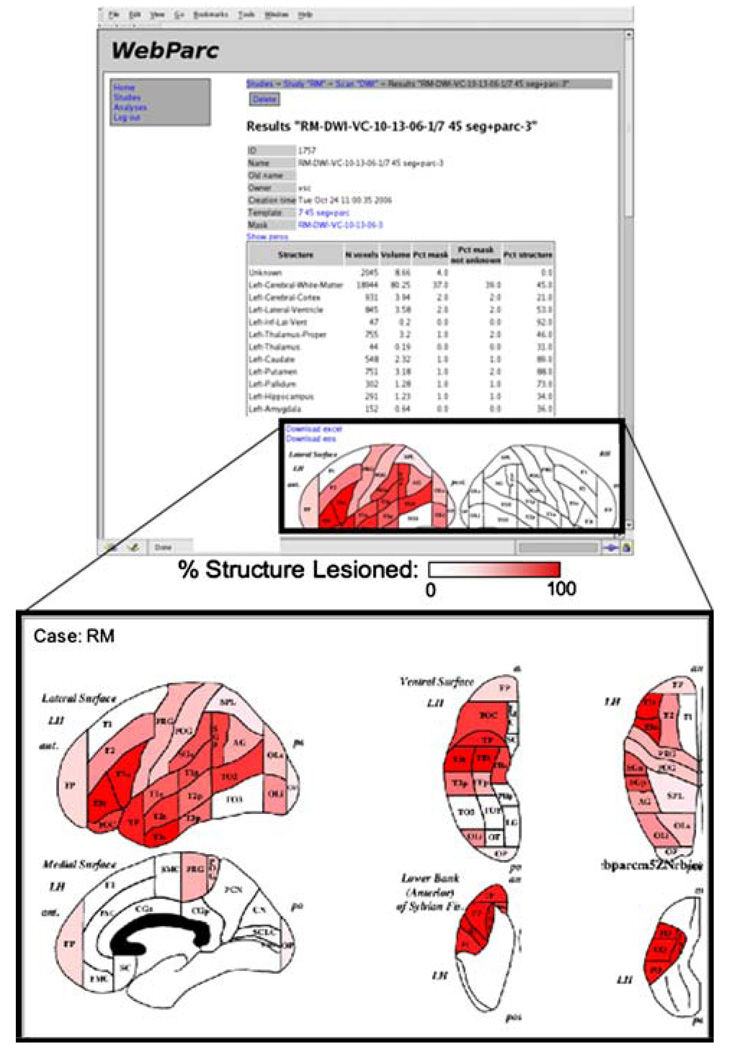
The user interface controls all access to the images, lesion annotation, result generation, and data export. Owing to the potential sensitive nature of the data contained, all users must be registered and authenticate their session. Patient privacy is ensured using secure connections, registered users with passwords, and a system of user permissions. Once logged in, the user can navigate the existing studies and scans, identifying cases for which lesion masks need to be generated, and execute the lesion mask–template intersections to generate the desired quantitative representations of the lesion. As the template anatomy is registered with the clinical images, the intersection of the lesion with the template provides a measure of volume with respect to specific structures and the percentage of that structure that is involved, and is executed in near real time. The system includes a schematic graphic display for percent involvement of the neocortical parcellation units (Fig. 4). The resulting data can be exported for further statistical treatment outside of the WebParc system. Intersections between multiple lesion masks can also be performed to support inter- and intra-rater comparisons. The interface is coded in HTML+CSS. The webserver is implemented in Python on top of Apache (using mod_python).
Fig. 4.
Web-based interface to WebParc results. Top panel shows the quantitative results of the lesion intersection with the specified template. The lower panel shows the cortical parcellation intersection results on an idealized template for simplified visual interpretation. Regional abbreviations are provided in Table 1
2.3 Validation study
The series of images selected for validation of this approach includes MRI-based three-dimensional (1 × 1 × 3 mm resolution) T1-weighted coronal SPGR imaging sequences in 21 subjects ranging in age from 34 to 75 years and presumed from the clinical analysis to have had MCA embolic strokes. These patients have been the basis of prior studies [30, 31]. The topographic and volumetric analyses in each were performed approximately 6 months after the acute event. The semi-automated cardviews software package, taken as the standard for expert clinical analysis for this study, was used to perform general anatomic segmentation and cortical parcellation of the stroke lesion and all spared tissue in each case [56]. The segmentation operation for lesion analysis using cardviews is detailed in Caviness et al. [31]. In short, the perimeter of the principal infarction was traced using the full dynamic range of signal intensity allowed by the imaging parameters. Frank cavitation and a surrounding rim of altered signal intensity were readily distinguished from adjacent brain tissue. General segmentation of the spared brain tissue was performed as detailed in Filipek et al. [49]. The procedure involved user-guided local intensity contouring and manual refinement based upon a comprehensive set of standardized and validated anatomic “rules” for each of the anatomic structures. Cortical parcellation of the spared tissue followed the procedure of Rademacher et al. [52], and involved tracing a canonical set of limiting sulci and a rule-based system for creating gyral-based cortical regions throughout the hemisphere. The complete set of analyses in these patients, including complete anatomic segmentation, parcellation, and lesion analysis is knowledge and labor intensive, and requires up to 2 days per brain to accomplish.
Our comparisons here between the cardviews and WebParc estimates include overall lesion volume, as well as the lesioned volume percent of the general segmentation and cortical parcellation regions as obtained using these two different systems. We generate the Pearson product moment correlation coefficient for correspondence between the two measures.
2.4 Reliability study
For the assessment of methodological reliability, diffusion-weighted imaging (DWI) data was obtained from 10 patients imaged in the 24–48 h interval following acute infarction in the centro-sylvian region of the cerebrum as a result of embolus to the middle cerebral artery. Images were performed on a 1.5T Siemens Sonata system and included axial 5-mm skip 1-mm diffusion-weighted acquisitions with TR = 5,000 ms, TE = 80 ms and b-value = 1,000. Windowing with respect to image, signal intensity and contrast were operator controlled by a slide bar where these parameters were optimized visually with respect to the operator-perceived definition of the boundary of thalamus and posterior limb of the internal capsule of the hemisphere opposite the acute lesion. The infarctions selected for analysis had been initially established to be at least 15 cc in volume. For each case, there was only the single salient infarction although brains with minimal amounts of ipsilateral or contralateral leucomalacia were not excluded. The complete sequence of image planes presenting the region of stroke-induced elevation of DWI signal is segmented. For estimates of intra-rater reliability, one rater (VSC) performed the analysis three times. The mean and standard deviation of the coefficients of variation for all measures of the lesioned volume of each structure were calculated. For inter-rater reliability, a second rater (NM) analyzed each of the cases. Mean value of lesion volume by structure was calculated. The mean inter-rater difference in absolute value and percent of structure was calculated per structure, and the Pearson product moment correlation coefficient between the two raters generated.
3 Results
A prototype of this application has been created in a secure web-based environment. This prototype application was used to generate the results of the validation and reliability studies to be presented.
3.1 Validation
The cardviews estimate provides segmentation and parcellation of the entire brain and stroke volume [31]. The fully segmented and parcellated cardviews-defined lesion mask is registered and intersected with the WebParc templates (which use the same segmentation and parcellation scheme) to give the estimates of stroke tissue volume and localization. Thus, this method compares estimates of volume of specific anatomic structures normalized to a template to volumetric measures of these same structures which were precisely and accurately determined by a segmentation method applied directly to the individual brains. In some of these, the lesions the topography were mildly distorted by leptomeningal scarring of the gyration pattern and shrinkage of the hemisphere. The measures of variation reflect these distortions. In none was there swelling, hernation, or a secondary hemorrhagic mass. Such distortions are of course regularly encountered in stroke and may be expected to affect the accuracy of estimates of volume of specific neurological structures as estimated by normalization to a template when this occurs.
3.1.1 General segmentation
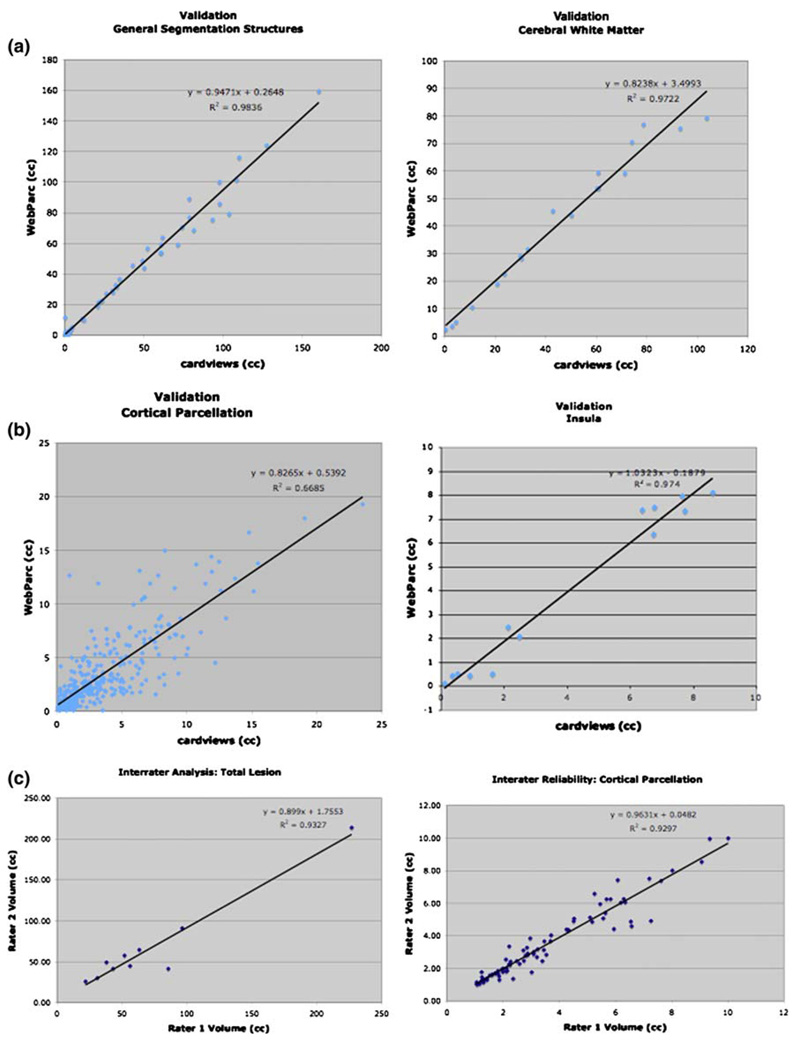
The agreement across all measures between cardviews- and WebParc-based (CV–WP) estimates of volumes of cerebral structures destroyed by stroke is high with r2 > 0.98 (Fig. 5a; Table 2). The structures with the largest lesion mass, the cerebral cortex and the central cerebral white matter, demonstrate r2 > 0.95. Estimates for pallidum and putamen, in which estimated infarction was only of the order of 1–3 cc, still provided r2 > 0.6.
Fig. 5.
cardviews–WebParc regression plots. a “General segmentation”: regression for all (N = 67) general segmentation measures across all subjects (left) and regression for the volume estimate of cerebral white matter involvement across the 21 subjects (right). b Cortical parcellation: regression for all (N = 336) parcellation unit measures across all subjects (left) and regression for the volume estimate of insula involvement across the 16 of the 21 subjects that had involvement of this region (right). c Intrarater correlation plots for total lesion (left) and all parcellation units (right). Note that the r2 value for the intrarater correlation plots for total lesion plot goes to 0.356 when the large lesion is excluded
Table 2.
cardviews (CV)–WebParc (WP) validation results
| Anatomic structurea | N | CV volume Average (cc) |
WP volume Average (cc) |
r2 |
|---|---|---|---|---|
| Cerebral cortex | 19 | 61.8 | 61.3 | 0.986 |
| Cerebral white matter | 19 | 44.9 | 40.5 | 0.972 |
| Putamen | 13 | 2.4 | 1.9 | 0.628 |
| Pallidum | 8 | 0.7 | 0.6 | 0.689 |
| INS | 16 | 4.2 | 4.0 | 0.974 |
| CO | 14 | 3.0 | 3.2 | 0.629 |
| F3o | 14 | 3.7 | 2.9 | 0.544 |
| PRG | 14 | 6.4 | 6.8 | 0.808 |
| PT | 14 | 1.7 | 1.9 | 0.447 |
| SGp | 14 | 5.0 | 6.5 | 0.565 |
| F2 | 13 | 4.6 | 4.8 | 0.743 |
| FO | 13 | 1.6 | 1.4 | 0.387 |
| POG | 13 | 5.8 | 9.4 | 0.728 |
| FOC | 12 | 2.9 | 3.9 | 0.740 |
| H1 | 12 | 1.1 | 1.0 | 0.734 |
| PO | 12 | 2.3 | 1.8 | 0.466 |
| PP | 12 | 1.7 | 1.2 | 0.534 |
| TO2 | 12 | 4.9 | 2.2 | 0.568 |
| F3t | 11 | 2.4 | 2.1 | 0.620 |
| OLi | 11 | 2.7 | 2.6 | 0.933 |
| OLs | 11 | 4.9 | 5.9 | 0.860 |
| T1p | 11 | 3.4 | 2.5 | 0.428 |
| AG | 10 | 2.5 | 1.3 | 0.452 |
| SGa | 10 | 4.6 | 1.6 | 0.581 |
| T2p | 10 | 3.5 | 3.1 | 0.433 |
| FP | 9 | 8.1 | 7.6 | 0.944 |
| T1a | 8 | 0.9 | 1.1 | 0.148 |
| SPL | 7 | 3.6 | 2.8 | 0.759 |
| TO3 | 6 | 2.5 | 1.7 | 0.974 |
| TP | 6 | 4.5 | 3.7 | 0.938 |
Data are shown for anatomic structures that were lesioned in six or more of the 21 subjects in this cohort. The anatomic regional names are provided in Table 1
3.1.2 Parcellation units
The agreement between cardviews and WebParc cortical parcellation values, although less strong than that for general segmentation, is still good but with much greater variance (r2 approximately 0.7; Table 2; Fig. 5b). For PU found to be involved in at least 6 of the brains, the r2 generally ranged from ~0.5 to >0.9 with the average falling at ~0.65. This is illustrated by an example plot for the insula (INS) (Fig. 5b), a PU at the center of the Sylvian area and involved in most of the strokes. It is pertinent here to note that that INS was frequently totally involved in the lesion.
3.2 Reliability
3.2.1 Overview of lesion distribution
The reliability study was based on 10 brains with MCA embolic strokes that were not included in previously published studies. All the 10 involved cerebral cortex and white matter while 4 patients also involved subcortical nuclei. Most of the strokes were centered in the centro-Sylvian territory, although with substantial stroke-to-stroke variation with respect to specific topography. Volumes of strokes varied from 25 to 213 cc (mean = 66.0 cc). Such variation, we have earlier emphasized, is consistent with embolic occlusion of the MCA [30].
3.2.2 Intra-rater reliability
A single operator (vsc) analyzed each of the 10 brains three times. The average coefficient of variation of the total lesion volume was 5% (range 1–12%) and that for total cerebral cortex and white matter involvement was similar (Table 3). The intrarater variability increases in the sub-cortical regions, where total volume of lesion involvement is small, and is only affected in a small subset of the cases. The average intrarater coefficient of variation (CV) across all cortical parcellation units involved in at least four cases is 6%, and ranges from a minimum of 3%, for the insula (INS) and the anterior portion of the supramarginal gyrus (SGa) to a maximum of 13% for the posterior supramarginal gyrus (SGp) (Table 3).
Table 3.
Intrarater reliability
| Anatomic structurea | N | Volume |
Intrarater coefficient of variation |
||
|---|---|---|---|---|---|
| Mean (cc) | Range (cc) | Mean (%) | SD (%) | ||
| Total lesion | 10 | 66.0 | 25.9–214.0 | 5 | 3 |
| Cerebral cortex | 10 | 32.7 | 11.8–106.0 | 5 | 3 |
| Cerebral white matter | 10 | 24.9 | 9.5–79.9 | 6 | 4 |
| Caudate | 4 | 1.9 | 1.8–2.2 | 9 | 8 |
| Putamen | 4 | 3.1 | 2.8–3.3 | 4 | 2 |
| Pallidum | 4 | 1.2 | 1.1–1.2 | 9 | 6 |
| Thalamus | 4 | 2.5 | 1.5–3.2 | 10 | 3 |
| PRG | 4 | 5.4 | 1.7–10.0 | 7 | 9 |
| SGa | 4 | 1.8 | 1.2–2.5 | 3 | 2 |
| SGp | 4 | 3.7 | 1.3–7.4 | 13 | 9 |
| TP | 4 | 3.6 | 1.3–10.0 | 4 | 2 |
| PO | 5 | 1.5 | 1.1–2.5 | 6 | 5 |
| CO | 6 | 3.4 | 1.1–5.1 | 4 | 5 |
| INS | 7 | 5.7 | 2.8–8.0 | 3 | 4 |
| POG | 7 | 4.1 | 1.1–6.6 | 8 | 5 |
Data are shown for anatomic structures that were lesioned in four or more of the 10 subjects in this cohort. The anatomic regional names are provided in Table 1
3.2.3 Inter-rater reliability
A second investigator (nm) executed a single mask in each of the 10 image sets from the DWI series. It was done without reference to the masks executed on the comparisons images sets by the first operator. In general, there was excellent concordance between the average of three values by rater 1 and the single values by rater 2 (Table 4; r2 = 0.933; Fig. 5c). The two raters averaged a volume difference of approximately 5 cc, representing approximately 6% of the total lesion estimate. As with the intra-rater data, the inter-rater concordance is reduced in the subcortical gray matter structures, again due principally to their small size and infrequent occurrence. With respect to overall cortical parcellation units, the correlation between the two raters is again very high (r2 = 0.930; Fig. 5c; Table 4). Volumetric agreement is more variable, ranging from 3 to 22% in these structures.
Table 4.
Interrater reliability
| Structurea | N | Rater 1 Mean (cc) |
Rater 2 Mean (cc) |
Absolute difference |
r2 | |
|---|---|---|---|---|---|---|
| Value (cc) | % | |||||
| Total lesion | 10 | 66.0 | 71.5 | 5.5 | 8 | 0.933 |
| Cerebral cortex | 10 | 32.7 | 37.4 | 4.7 | 13 | 0.849 |
| Cerebral white matter | 10 | 24.9 | 26.1 | 1.2 | 5 | 0.935 |
| Caudate | 4 | 1.9 | 1.4 | 0.6 | 35 | 0.426 |
| Putamen | 4 | 3.1 | 2.6 | 0.5 | 18 | 0.063 |
| Pallidum | 4 | 1.2 | 1.3 | 0.1 | 8 | 0.375 |
| Thalamus | 4 | 2.5 | 2.4 | 0.1 | 4 | 0.990 |
| PRG | 4 | 5.4 | 5.1 | 0.3 | 7 | 0.997 |
| SGa | 4 | 1.8 | 1.4 | 0.4 | 22 | 0.911 |
| SGp | 4 | 3.7 | 4.6 | 0.9 | 21 | 0.957 |
| TP | 4 | 3.6 | 4.5 | 0.9 | 21 | 0.974 |
| PO | 5 | 1.5 | 1.6 | 0.05 | 3 | 0.984 |
| CO | 6 | 3.4 | 3.3 | 0.1 | 3 | 0.992 |
| INS | 7 | 5.6 | 5.3 | 0.3 | 6 | 0.947 |
| POG | 7 | 4.1 | 3.9 | 0.2 | 6 | 0.883 |
Data are shown for anatomic structures that were lesioned in four or more of the 10 subjects in this cohort. The anatomic regional names are provided in Table 1
4 Discussion
This initial version of WebParc meets satisfactorily its first-order objectives. Being web-based, the system may be used at any computer with a web browser and is well suited to a clinical setting. It validly approximates the volumes of strokes where these have been estimated by the time-intensive cardviews approach [30, 31] where measures are made on the individual brain rather than from normalization to a template. Thus, despite what must be topographic distortion of the abnormal brain with reference to its topography prior to stroke and other errors inherent in the template-registration method, the WebParc approximation for total stroke volume and for that of the major forebrain anatomic compartments is still highly correlated with direct observation and demonstrates inter-rater reliability in the range of 5% or better. The reliability of volume estimates for the separate gray matter forebrain structures and cortical parcellation units were more variable. Coefficients of variation were generally less than 20% and for structures greater than 1–2 cc in volume, it approached the 5–10% level achieved with the general segmentation.
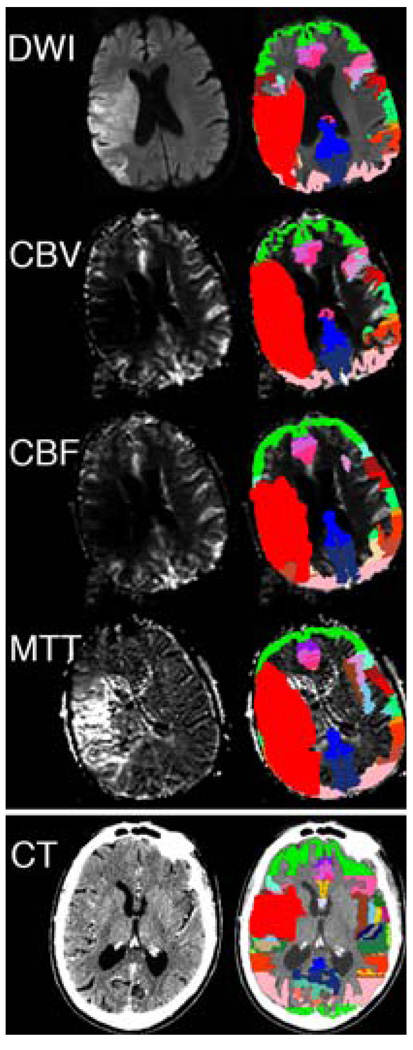
The observed level of reliability using this method is only modestly less favorable than that obtained by Luby et al. [57], using a system where data were obtained by extensively trained operators working with workstations with state of art display properties generally available only in the radiology departments of advanced hospitals. In the present case, the lesion segmentations were done on standard issue desktop computers where the resolution of viewing screens would be comparable to that on general desktop models available on the clinical floors of our hospital for use in support of patient care. The investigators were familiar with reading brain MRIs in general for clinical usage and also with the general operation of the software. Training for the application of WebParc only comprised several images of sets that had been traced in part multiple times during software development to set criteria for windowing and to familiarize the operators with its specific operations. The data from WebParc analysis is readily displayed for visualization. Other potential operational advantages are that image data can originate from any imaging modality, e.g., all MRI, CT or PET based, provided that the basis for a sufficient registration to the standard templates can be achieved. Figure 6 demonstrates the general versatility of the platform in application to diffusion, as well as numerous MR-based perfusions measures in a single patient, as well as CT in a separate patient. While detailed discussion of the reliability or validation of these quantitative measures is beyond the scope of this presentation, these topics will need to be addressed within the context of the specific details of these additional imaging modalities in terms of issues pertaining to registration, lesion segmentation, etc.
Fig. 6.
Multimodality examples of WebParc use. The top four panels represent different MR imaging modes in a single patient, and include diffusion-weighted imaging (DWI), cerebral blood volume (CBV). Cerebral blood flow (CBF), and mean transit time (MTT). Spatial details of the user-perceived lesion in each of these modalities can be different, and these differences were quantified using this application. The bottom panel demonstrates the WebParc functionality on CT imaging
The technical design of the WebParc system intentionally employs a modular architecture that permits future enhancements. The templates are modular, and a variety of anatomic analysis systems derived from individual and probabilistic representations from any demographic population can be used. Estimates of vascular territories, as in Nowinski et al. [43–45] are also highly informative. The segmentation module can be augmented with more automated methods for lesion extraction. The linear registration module can be replaced with more complex registration schemes that may enhance overall accuracy. The current study provides an assessment of the simplest instantiation of this approach that should serve as a validation for the general approach, and provide a baseline upon which future technical advanced can be measured.
While automated segmentation of infarct has been achieved in some cases [43–45], there still is a broad class of clinically appreciable infarct presentation for which there is not yet a routinely available automated approach. Despite the inherently larger potential variation that may come from manual techniques, these techniques can still provide clinically viable infarct quantification. They also provide a baseline expert-observer set of observations upon which the benefits of enhanced technology (in automation, registration, etc.) can be quantified. Our demonstration of this software instance in relatively high-contrast datasets that are potentially amenable to automated approaches is necessary to minimize clinical decision variance and expose the variances that are imparted by the processing scheme. As such, these measures provide a “best-case” estimate of these errors. Whatever the quantification system, we must provide a set of baseline reliability measures to determine those clinical questions to which the solution has potential applicability.
This version of WebParc, as noted, requires only a matter of 10 min for data transfer and preprocessing, and an additional 10 min for segmentation of a mask outlining a volume according to DWI signal. Thus, depending upon when the user initiates the process, quantitative results can be obtained in as little as 20 min from the end of the scanning session. The present version is replete with routines that generate all derivative data sets included in this manuscript. This type of system can be used for calculation of progression of lesion volumes across a series of scans in the same patient as a function of elapsed time following the estimated time of the stroke event.
The operational features make the program suitable for monitoring the apparent volume of stroke according to any template structure or compartment. It is logistically not possible to do serial imaging and to process these according to WebParc in support of minute-to-minute assessments during the immediate acute interval after hospital admission. However, this type of analysis could well be useful for monitoring effects of interventions or changes in the medical or neurological state over days during the acute hospital admission. It is well suited, moreover, for monitoring the longer-term evolution of the lesion as by the FLAIR or T1-weighted modalities in relation to neurological recovery.
4.1 Limitations
We have not as yet undertaken a systematic analysis of the reasons for the inter-rater variation estimated in this analysis. We suspect that it relates to variations in operator judgment at the margins of DWI abnormalities where signal intensity typically fades in a graded fashion. Thus, the apparent margins may vary depending upon “windowing” levels chosen by different investigators. It may vary depending upon the resolution of the workstation available for the analysis. Such variability could in principle be lessened by an automated system that segmented the stroke computationally [43–45, 58, 59] although such an approach would demand informed investigator supervision to edit out the abundant high signal artifact characteristic of DWI, for example, the one typically observed near the base of the temporal lobes.
In the clinical setting, the imaging plane thickness can be typically 5 mm, and there may be gaps between the series of image planes. WebParc treats the subject data as a continuous block at the effective slice separation (slice thickness plus gap). Such conditions obviously limit the sensitivity of the method, and its utility will rest with the judgment of the clinician or investigator as slice separation becomes large and higher-order interpolation and resampling schemes could be envisioned. However, this factor has not been demonstrated to be overly problematic in data with 6 mm (5 mm skip 1 mm) effective separation as used here.
Higher-order spatial registration schemes can also be implemented. It is noted, however, that there will typically be a throughput time “cost” and additional potential modes of failure as registration becomes more complex. However, the ultimate resolution of this type of system is predicated upon the quality of the underlying anatomic registration. Significant spatial variability is known to exist under linear or affine transformation techniques and is reduced using various non-linear applications [60]. We note that using linear registration techniques, approximately 65% of large structures such as cerebral cortex and white matter will overlap, whereas for smaller subcortical regions, this overlap may be reduced to approximately 55%. Anatomic overlap is a useful basis for evaluation of the underlying registration technique for any implementation of this type of solution.
WebParc obviously addresses only a limited, but potentially critical, subset of the various limitations in linking the animal and human study. For example, the topography of MCA stroke is profoundly different from that of perfusion compromise in animal studies as usually designed. The stroke volumes in a series of dysphasic subjects studied here, all with MCA embolic strokes varied over two orders of magnitude with the spatial distribution of infarction ranging over the entire territory of the MCA [30, 31]. The strokes variably involved superficial and deep cerebral structures. Only an approach that provides morphometric and topographic representations of the lesion, will allow a study design to control for the variables of volume with respect to specific structure, tissue type or location within the territories of a given vascular perfusion domain. WebParc is an example of a system that brings the advantages of simplicity of application, efficiency, and acceptable levels of reliability to these objectives in stroke research.
Whereas the present analysis treats only MCA strokes, the method is equally applicable to other forebrain vascular territories. We have used it on our clinical service in analyses of strokes in both the PCA and ACA territories (data not illustrated). We have not as yet applied it to analyses of stroke in the cerebellum but do not foresee difficulty in doing so in that it can be based upon a comprehensive parcellation system previously worked out in our laboratory [61]. Also we have not applied it to strokes in the brain stem where the method may be less reliably matched to a normalized template because of small size of the brain stem and its variable angulation with respect to the forebrain coordinates. We also add that the method is viewed in principle as having general applicability to any image sets where there is a topographically definable abnormality imposed upon a brain that is suitable to normalization to a template of the normal brain, for example, tumor, hemorrhage, or multiple sclerosis (MS) lesions. It would not be suitable to analysis of gross developmental malformations such as holoprosencephaly or lisencephaly where the topology of structure is not comparable to that of the normal brain.
4.2 Prospects for future use
We envision the method as having application primarily in the study of stroke and stroke treatment; but the potential exists for the study of any condition where the topography of abnormality is defined by an imaging method. This would of course include non-embolic vascular disorders such as leucoaraiosis which may be expected significantly to affect the evaluation of stroke as well as the degree of recovery from disability [62–65] or hemorrhage. With respect to stroke, we propose that the approach can be a component of future trials of neuroprotective agents where effectiveness can be judged both in terms of tissue salvage with respect to specific structures as well as neurological recovery [66, 67].
Acknowledgments
This study was supported in part by NIH grants PO1 NS27950, EB005149, and DA09467, by Human Brain Project Grant NS34189, and by grants from the Fairway Trust and the Giovanni Armenise Harvard Foundation for Advanced Scientific Research.
Contributor Information
David N. Kennedy, Department of Psychiatry, University of Massachusetts Medical School, Worcester, MA, USA David.Kennedy@umassmed.edu Center for Morphometric Analysis, Massachusetts General Hospital, 149 13th Street, Charlestown, Boston, MA, USA; Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA.
Christian Haselgrove, Department of Psychiatry, University of Massachusetts Medical School, Worcester, MA, USA; Center for Morphometric Analysis, Massachusetts General Hospital, 149 13th Street, Charlestown, Boston, MA, USA; Department of Neurology, Massachusetts General Hospital, Boston, MA, USA.
Nikos Makris, Center for Morphometric Analysis, Massachusetts General Hospital, 149 13th Street, Charlestown, Boston, MA, USA; Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA.
Donald M. Goldin, Center for Morphometric Analysis, Massachusetts General Hospital, 149 13th Street, Charlestown, Boston, MA, USA
Michael H. Lev, Harvard Medical School, Boston, MA, USA Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA; Department of Radiology, Massachusetts General Hospital, Boston, MA, USA.
David Caplan, Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA.
Verne S. Caviness, Center for Morphometric Analysis, Massachusetts General Hospital, 149 13th Street, Charlestown, Boston, MA, USA Department of Neurology, Massachusetts General Hospital, Boston, MA, USA; Harvard Medical School, Boston, MA, USA; Athinoula A. Martinos Center for Biomedical Imaging, Massachusetts General Hospital, Boston, MA, USA.
References
- 1.Sorensen AG, Wu O, Copen WA, Davis TL, Gonzalez RG, Koroshetz WJ, Reese TG, Rosen BR, Wedeen VJ, Weisskoff RM. Human acute cerebral ischemia: detection of changes in water diffusion anisotropy by using MR imaging. Radiology. 1999;212:785–792. doi: 10.1148/radiology.212.3.r99se24785. [DOI] [PubMed] [Google Scholar]
- 2.Sorensen AG, Copen WA, Ostergaard L, Buonanno FS, Gonzalez RG, Rordorf G, Rosen BR, Schwamm LH, Weisskoff RM, Koroshetz WJ. Hyperacute stroke: simultaneous measurement of relative cerebral blood volume, relative cerebral blood flow, and mean tissue transit time. Radiology. 1999;210:519–527. doi: 10.1148/radiology.210.2.r99fe06519. [DOI] [PubMed] [Google Scholar]
- 3.Schwamm LH, Koroshetz WJ, Sorensen AG, Wang B, Copen WA, Budzik R, Rordorf G, Buonanno FS, Schaefer PW, Gonzalez RG. Time course of lesion development in patients with acute stroke: serial diffusion- and hemodynamic-weighted magnetic resonance imaging. Stroke. 1998;29:2268–2276. doi: 10.1161/01.str.29.11.2268. [DOI] [PubMed] [Google Scholar]
- 4.Schaefer PW, Hunter GJ, He J, Hamberg LM, Sorensen AG, Schwamm LH, Koroshetz WJ, Gonzalez RG. Predicting cerebral ischemic infarct volume with diffusion and perfusion MR imaging. AJNR Am J Neuroradiol. 2002;23:1785–1794. [PMC free article] [PubMed] [Google Scholar]
- 5.Rivers CS, Wardlaw JM, Armitage PA, Bastin ME, Carpenter TK, Cvoro V, Hand PJ, Dennis MS. Do acute diffusion-and perfusion-weighted MRI lesions identify final infarct volume in ischemic stroke? Stroke. 2006;37:98–104. doi: 10.1161/01.STR.0000195197.66606.bb. [DOI] [PubMed] [Google Scholar]
- 6.Lovblad KO, Baird AE, Schlaug G, Benfield A, Siewert B, Voetsch B, Connor A, Burzynski C, Edelman RR, Warach S. Ischemic lesion volumes in acute stroke by diffusion-weighted magnetic resonance imaging correlate with clinical outcome. Ann Neurol. 1997;42:164–170. doi: 10.1002/ana.410420206. [DOI] [PubMed] [Google Scholar]
- 7.Sulter G, Steen C, De Keyser J. Use of the Barthel index and modified Rankin scale in acute stroke trials. Stroke. 1999;30:1538–1541. doi: 10.1161/01.str.30.8.1538. [DOI] [PubMed] [Google Scholar]
- 8.Baird AE, Dashe J, Connor A, Burzynski C, Schlaug G, Warach S. Comparison of retrospective and prospective measurements of the national institutes of health stroke scale. Cerebrovasc Dis. 2000;10:80–81. doi: 10.1159/000016031. [DOI] [PubMed] [Google Scholar]
- 9.Thijs VN, Lansberg MG, Beaulieu C, Marks MP, Moseley ME, Albers GW. Is early ischemic lesion volume on diffusion-weighted imaging an independent predictor of stroke outcome? A multivariable analysis. Stroke. 2000;31:2597–2602. doi: 10.1161/01.str.31.11.2597. [DOI] [PubMed] [Google Scholar]
- 10.Hand PJ, Wardlaw JM, Rivers CS, Armitage PA, Bastin ME, Lindley RI, Dennis MS. MR diffusion-weighted imaging and outcome prediction after ischemic stroke. Neurology. 2006;66:1159–1163. doi: 10.1212/01.wnl.0000202524.43850.81. [DOI] [PubMed] [Google Scholar]
- 11.Group NIoNDaSRTPSS. Tissue plasminogen activator for acute ischemic stroke. N Engl J Med. 1995;333:1581–1587. doi: 10.1056/NEJM199512143332401. [DOI] [PubMed] [Google Scholar]
- 12.Higashida RT, Furlan AJ, Roberts H, Tomsick T, Connors B, Barr J, Dillon W, Warach S, Broderick J, Tilley B, Sacks D. Trial design and reporting standards for intra-arterial cerebral thrombolysis for acute ischemic stroke. Stroke. 2003;34:e109–e137. doi: 10.1161/01.STR.0000082721.62796.09. [DOI] [PubMed] [Google Scholar]
- 13.del Zoppo GJ, Higashida RT, Furlan AJ, Pessin MS, Rowley HA, Gent M. PROACT: a phase II randomized trial of recombinant pro-urokinase by direct arterial delivery in acute middle cerebral artery stroke. PROACT investigators. Prolyse in acute cerebral thromboembolism. Stroke. 1998;29:4–11. doi: 10.1161/01.str.29.1.4. [DOI] [PubMed] [Google Scholar]
- 14.Furlan A, Higashida R, Wechsler L, Gent M, Rowley H, Kase C, Pessin M, Ahuja A, Callahan F, Clark WM, Silver F, Rivera F. Intra-arterial prourokinase for acute ischemic stroke. The PROACT II study: a randomized controlled trial. Prolyse in acute cerebral thromboembolism. JAMA. 1999;282:2003–2011. doi: 10.1001/jama.282.21.2003. [DOI] [PubMed] [Google Scholar]
- 15.Mani RL, Eisenberg RL, McDonald EJ, Jr, Pollock JA, Mani JR. Complications of catheter cerebral arteriography: analysis of 5,000 procedures. I. Criteria and incidence. AJR Am J Roentgenol. 1978;131:861–865. doi: 10.2214/ajr.131.5.861. [DOI] [PubMed] [Google Scholar]
- 16.Skalpe IO. Complications in cerebral angiography with iohexol (omnipaque) and meglumine metrizoate (isopaque cerebral) Neuroradiology. 1988;30:69–72. doi: 10.1007/BF00341947. [DOI] [PubMed] [Google Scholar]
- 17.Heiserman JE, Dean BL, Hodak JA, Flom RA, Bird CR, Drayer BP, Fram EK. Neurologic complications of cerebral angiography. AJNR Am J Neuroradiol. 1994;15:1401–1407. discussion 1408–1411. [PMC free article] [PubMed] [Google Scholar]
- 18.Waugh JR, Sacharias N. Arteriographic complications in the DSA era. Radiology. 1992;182:243–246. doi: 10.1148/radiology.182.1.1727290. [DOI] [PubMed] [Google Scholar]
- 19.Dyker AG, Lees KR. Duration of neuroprotective treatment for ischemic stroke. Stroke. 1998;29:535–542. doi: 10.1161/01.str.29.2.535. [DOI] [PubMed] [Google Scholar]
- 20.McCulloch J, Dewar D. A radical approach to stroke therapy. Proc Natl Acad Sci USA. 2001;98:10989–10991. doi: 10.1073/pnas.211430898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCulloch J. Excitatory amino acid antagonists and their potential for the treatment of ischaemic brain damage in man. Br J Clin Pharmacol. 1992;34:106–114. doi: 10.1111/j.1365-2125.1992.tb04118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonas S, Aiyagari V, Vieira D, Figueroa M. The failure of neuronal protective agents versus the success of thrombolysis in the treatment of ischemic stroke. The predictive value of animal models. Ann N Y Acad Sci. 2001;939:257–267. doi: 10.1111/j.1749-6632.2001.tb03633.x. [DOI] [PubMed] [Google Scholar]
- 23.Singhal AB, Lo EH, Dalkara T, Moskowitz MA. Advances in stroke neuroprotection: hyperoxia and beyond. Neuroimaging Clin N Am. 2005;15:697–720. xii–xiii. doi: 10.1016/j.nic.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Rivers CS, Wardlaw JM, Armitage PA, Bastin ME, Carpenter TK, Cvoro V, Hand PJ, Dennis MS. Persistent infarct hyperintensity on diffusion-weighted imaging late after stroke indicates heterogeneous, delayed, infarct evolution. Stroke. 2006;37:1418–1423. doi: 10.1161/01.STR.0000221294.90068.c4. [DOI] [PubMed] [Google Scholar]
- 25.Lansberg MG, O’Brien MW, Tong DC, Moseley ME, Albers GW. Evolution of cerebral infarct volume assessed by diffusion-weighted magnetic resonance imaging. Arch Neurol. 2001;58:613–617. doi: 10.1001/archneur.58.4.613. [DOI] [PubMed] [Google Scholar]
- 26.Eastwood JD, Lev MH, Provenzale JM. Perfusion CT with iodinated contrast material. AJR Am J Roentgenol. 2003;180:3–12. doi: 10.2214/ajr.180.1.1800003. [DOI] [PubMed] [Google Scholar]
- 27.Baird AE, Benfield A, Schlaug G, Siewert B, Lovblad KO, Edelman RR, Warach S. Enlargement of human cerebral ischemic lesion volumes measured by diffusion-weighted magnetic resonance imaging. Ann Neurol. 1997;41:581–589. doi: 10.1002/ana.410410506. [DOI] [PubMed] [Google Scholar]
- 28.Barber P, Darby D, Desmond P, Yang Q, Gerraty R, Jolley D, Donnan G, Tress B, Davis S. Prediction of stroke outcome with echoplanar perfusion- and diffusion-weighted MRI. Neurology. 1998;51:418–426. doi: 10.1212/wnl.51.2.418. [DOI] [PubMed] [Google Scholar]
- 29.Neumann-Haefelin T, Wittsack HJ, Wenserski F, Siebler M, Seitz RJ, Modder U, Freund HJ. Diffusion- and perfusion-weighted MRI. The DWI/PWI mismatch region in acute stroke. Stroke. 1999;30:1591–1597. doi: 10.1161/01.str.30.8.1591. [DOI] [PubMed] [Google Scholar]
- 30.Caviness VS, Makris N, Montinaro E, Sahin NT, Bates JF, Schwamm L, Caplan D, Kennedy DN. Anatomy of stroke, part II: volumetric characteristics with implications for the local architecture of the cerebral perfusion system. Stroke. 2002;33:2557–2564. doi: 10.1161/01.str.0000036084.82955.c7. [DOI] [PubMed] [Google Scholar]
- 31.Caviness VS, Makris N, Montinaro E, Sahin NT, Bates JF, Schwamm L, Caplan D, Kennedy DN. Anatomy of stroke, part I: an MRI-based topographic and volumetric system of analysis. Stroke. 2002;33:2549–2556. doi: 10.1161/01.str.0000036083.90045.08. [DOI] [PubMed] [Google Scholar]
- 32.Bogousslavsky J, Van Melle G, Regli F. The Lausanne Stroke Registry: analysis of 1,000 consecutive patients with first stroke. Stroke. 1988;19:1083–1092. doi: 10.1161/01.str.19.9.1083. [DOI] [PubMed] [Google Scholar]
- 33.Caplan L, Babikian V, Helgason C, Hier DB, DeWitt D, Patel D, Stein R. Occlusive disease of the middle cerebral artery. Neurology. 1985;35:975–982. doi: 10.1212/wnl.35.7.975. [DOI] [PubMed] [Google Scholar]
- 34.Catani M, ffytche DH. The rises and falls of disconnection syndromes. Brain. 2005;128:2224–2239. doi: 10.1093/brain/awh622. [DOI] [PubMed] [Google Scholar]
- 35.Geschwind N. Disconnexion syndromes in animals and man. I. Brain. 1965;88:237–294. doi: 10.1093/brain/88.2.237. [DOI] [PubMed] [Google Scholar]
- 36.Geschwind N. Disconnexion syndromes in animals and man. II. Brain. 1965;88:585–644. doi: 10.1093/brain/88.3.585. [DOI] [PubMed] [Google Scholar]
- 37.Mesulam MM. From sensation to cognition. Brain. 1998;121(Pt 6):1013–1052. doi: 10.1093/brain/121.6.1013. [DOI] [PubMed] [Google Scholar]
- 38.Mesulam MM. Large-scale neurocognitive networks and distributed processing for attention, language, and memory. Ann Neurol. 1990;28:597–613. doi: 10.1002/ana.410280502. [DOI] [PubMed] [Google Scholar]
- 39.Menezes NM, Ay H, Wang Zhu M, Lopez CJ, Singhal AB, Karonen JO, Aronen HJ, Liu Y, Nuutinen J, Koroshetz WJ, Sorensen AG. The real estate factor: quantifying the impact of infarct location on stroke severity. Stroke. 2007;38:194–197. doi: 10.1161/01.STR.0000251792.76080.45. [DOI] [PubMed] [Google Scholar]
- 40.Bilello M, Lao Z, Krejza J, Hillis AE, Herskovits EH. Statistical atlas of acute stroke from magnetic resonance diffusion-weighted-images of the brain. Neuroinformatics. 2006;4:235–242. doi: 10.1385/NI:4:3:235. [DOI] [PubMed] [Google Scholar]
- 41.Letovsky SI, Whitehead SH, Paik CH, Miller GA, Gerber J, Herskovits EH, Fulton TK, Bryan RN. A brain image database for structure/function analysis. AJNR Am J Neuroradiol. 1998;19:1869–1877. [PMC free article] [PubMed] [Google Scholar]
- 42.Ay H, Arsava EM, Koroshetz WJ, Sorensen AG. Middle cerebral artery infarcts encompassing the insula are more prone to growth. Stroke. 2008;39:373–378. doi: 10.1161/STROKEAHA.107.499095. [DOI] [PubMed] [Google Scholar]
- 43.Nowinski WL, Qian G, Kirgaval Nagaraja BP, Thirunavuukarasuu A, Hu Q, Ivanov N, Parimal AS, Runge VM, Beauchamp NJ. Analysis of ischemic stroke MR images by means of brain atlases of anatomy and blood supply territories. Acad Radiol. 2006;13:1025–1034. doi: 10.1016/j.acra.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Nowinski W, Qian G, Bhanu Prakash K, Volkau I, Hu Q, Thirunavuukarasuu A, Ivanov N, Parimal A, Qiao Y, Ananthasubramaniam A, Huang S, Runge V, Beauchamp N. Design and development of a computer aided diagnosis system for processing of acute ischemic stroke MR images. WSEAS Trans Biol Biomed. 2006;3:401–407. [Google Scholar]
- 45.Nowinski W, Qian G, Bhanu Prakash K, Volkau I, Thirunavuukarasuu A, Hu Q, Ananthasubramaniam A, Liu J, Gupta V, Ng T, Leong W, Beauchamp N. A CAD system for acute ischemic stroke image processing. Int J Comput Assist Radiol Surg. 2007;2:220–222. [Google Scholar]
- 46.Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B. A probabilistic atlas and reference system for the human brain: international consortium for brain mapping (ICBM) Philos Trans R Soc Lond B Biol Sci. 2001;356:1293–1322. doi: 10.1098/rstb.2001.0915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins DL, Holmes CJ, Peters TM, Evans AC. Automatic 3D model-based neuroanatomical segmentation. Hum Brain Mapp. 1995;3:190–208. [Google Scholar]
- 48.Moore SM, Beecher DE, Hoffman SA. DICOM shareware: a public implementation of the DICOM standard. Proc SPIE. 2003;2165:772. [Google Scholar]
- 49.Filipek PA, Richelme C, Kennedy DN, Caviness VS., Jr The young adult human brain: an MRI-based morphometric analysis. Cereb Cortex. 1994;4:344–360. doi: 10.1093/cercor/4.4.344. [DOI] [PubMed] [Google Scholar]
- 50.Filipek PA, Kennedy DN, Caviness VS, Jr, Rossnick SL, Spraggins TA, Starewicz PM. Magnetic resonance imaging-based brain morphometry: development and application to normal subjects. Ann Neurol. 1989;25:61–67. doi: 10.1002/ana.410250110. [DOI] [PubMed] [Google Scholar]
- 51.Caviness VS, Makris N, Meyer J, Kennedy D. MRI-based parcellation of human neocortex: an anatomically specified method with estimate of reliability. J Cogn Neurosci. 1996;8:566–588. doi: 10.1162/jocn.1996.8.6.566. [DOI] [PubMed] [Google Scholar]
- 52.Rademacher J, Galaburda AM, Kennedy DN, Filipek PA, Caviness V. Human cerebral cortex: localization, parcellation, and morphometry with magnetic resonance imaging. J Cogn Neurosci. 1992;4:352–374. doi: 10.1162/jocn.1992.4.4.352. [DOI] [PubMed] [Google Scholar]
- 53.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 54.Smith SM. Fast robust automated brain extraction. Hum Brain Mapp. 2002;17:143–155. doi: 10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, Matthews PM.Advances in functional and structural MR image analysis and implementation as FSL Neuroimage 2004. 23 Suppl 1S208–S219. [DOI] [PubMed] [Google Scholar]
- 56.Worth AJ, Makris N, Caviness VS, Jr, Kennedy DN. Neuroanatomical segmentation in MRI: technological objectives. Int J Pattern Recognit Artif Intell. 1997;11:1161–1187. [Google Scholar]
- 57.Luby M, Bykowski JL, Schellinger PD, Merino JG, Warach S. Intra- and interrater reliability of ischemic lesion volume measurements on diffusion-weighted, mean transit time and fluid-attenuated inversion recovery MRI. Stroke. 2006;37:2951–2956. doi: 10.1161/01.STR.0000249416.77132.1a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hevia-Montiel N, Jimenez-Alaniz JR, Medina-Banuelos V, Yanez-Suarez O, Rosso C, Samson Y, Baillet S. Robust nonparametric segmentation of infarct lesion from diffusion-weighted MR images; Conf Proc IEEE Eng Med Biol Soc; 2007. pp. 2102–2105. [DOI] [PubMed] [Google Scholar]
- 59.Li W, Tian J, Li E, Dai J. Robust unsupervised segmentation of infarct lesion from diffusion tensor MR images using multiscale statistical classification and partial volume voxel reclassification. Neuroimage. 2004;23:1507–1518. doi: 10.1016/j.neuroimage.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 60.Hellier P, Barillot C, Corouge I, Gibaud B, Le Goualher G, Collins DL, Evans A, Malandain G, Ayache N, Christensen GE, Johnson HJ. Retrospective evaluation of intersubject brain registration. IEEE Trans Med Imaging. 2003;22:1120–1130. doi: 10.1109/TMI.2003.816961. [DOI] [PubMed] [Google Scholar]
- 61.Makris N, Hodge SM, Haselgrove C, Kennedy DN, Dale A, Fischl B, Rosen BR, Harris G, Caviness VS, Jr, Schmahmann JD. Human cerebellum: surface-assisted cortical parcellation and volumetry with magnetic resonance imaging. J Cogn Neurosci. 2003;15:584–599. doi: 10.1162/089892903321662967. [DOI] [PubMed] [Google Scholar]
- 62.Kuller LH, Arnold AM, Longstreth WT, Jr, Manolio TA, O’Leary DH, Burke GL, Fried LP, Newman AB. White matter grade and ventricular volume on brain MRI as markers of longevity in the cardiovascular health study. Neurobiol Aging. 2007;28:1307–1315. doi: 10.1016/j.neurobiolaging.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 63.Au R, Massaro JM, Wolf PA, Young ME, Beiser A, Seshadri S, D’Agostino RB, DeCarli C. Association of white matter hyperintensity volume with decreased cognitive functioning: the Framingham Heart Study. Arch Neurol. 2006;63:246–250. doi: 10.1001/archneur.63.2.246. [DOI] [PubMed] [Google Scholar]
- 64.Fisher CM. Binswanger’s encephalopathy: a review. J Neurol. 1989;236:65–79. doi: 10.1007/BF00314400. [DOI] [PubMed] [Google Scholar]
- 65.Ay H, Arsava EM, Rosand J, Furie KL, Singhal AB, Schaefer PW, Wu O, Gonzalez RG, Koroshetz WJ, Sorensen AG. Severity of leukoaraiosis and susceptibility to infarct growth in acute stroke. Stroke. 2008;39:1409–1413. doi: 10.1161/STROKEAHA.107.501932. [DOI] [PubMed] [Google Scholar]
- 66.Gladstone DJ, Black SE, Hakim AM. Toward wisdom from failure: lessons from neuroprotective stroke trials and new therapeutic directions. Stroke. 2002;33:2123–2136. doi: 10.1161/01.str.0000025518.34157.51. [DOI] [PubMed] [Google Scholar]
- 67.Ginsberg MD. Adventures in the pathophysiology of brain ischemia: penumbra, gene expression, neuroprotection: the 2002 Thomas Willis lecture. Stroke. 2003;34:214–223. doi: 10.1161/01.str.0000048846.09677.62. [DOI] [PubMed] [Google Scholar]