Abstract
Rhodopsin is the prototypical G-protein coupled receptor, coupling light activation with high efficiency to signaling molecules. The dark-state X-ray structures of the protein provide a starting point for consideration of the relaxation from initial light activation to conformational changes that may lead to signaling. In this study we create an energetically unstable retinal in the light activated state and then use molecular dynamics simulations to examine the types of compensation, relaxation, and conformational changes that occur following the cis–trans light activation. The results suggest that changes occur throughout the protein, with changes in the orientation of Helices 5 and 6, a closer interaction between Ala 169 on Helix 4 and retinal, and a shift in the Schiff base counterion that also reflects changes in sidechain interactions with the retinal. Taken together, the simulation is suggestive of the types of changes that lead from local conformational change to light-activated signaling in this prototypical system.
Keywords: membrane protein, simulation, G-protein coupled receptors, rhodopsin, photoisomerization
INTRODUCTION
Rhodopsin is an excellent system for understanding the details of G-protein coupled receptors (GPCR) due to the large amount of experimental information related to both structure and function (for recent reviews, see Refs. 1–6). It is also the first GPCR with a measured tertiary structure7 and is thus an excellent candidate for yielding insight into the molecular details of GPCR function. Explicit, all-atom molecular simulation can provide a view into the choreographic details of the structure–function relationship. A full understanding of these details is difficult, however, due to the large separation in time-scales between the photocycle of rhodopsin and current computational limits in computer simulation of biomolecules. In particular, the full photocycle occurs on the millisecond time scale,8 while the state-of-the-art in computer simulation of large proteins is tens of nanoseconds.
Bovine rhodopsin has served as a model system for the understanding of transduction for many years.8 In particular, studies of bovine rods have led to initial understanding of G-protein coupled systems, to the first GPCR that was sequenced,9 and to understanding of the connections between particular residues and rhodopsin function.3,10 For example, the role of Glu 113 as the counterion,11 the critical role of certain residues in transduction,12,13 and initial suggestions for spectral tuning14,15 all began with rhodopsin. An upcoming frontier is understanding the connections between the photocycle’s underlying conformational changes that lead to activation and signaling, and the structures of the G-protein itself.16,17
A key aspect of understanding rhodopsin and other GPCRs is the dynamic motion of the ligand activation.18,19 In rhodopsin, the cis–trans photoisomerization of retinal is the activation mechanism. The structural and energetic consequences of retinal’s isomerization are of great interest. Molecular dynamics (MD) simulations offer a means of obtaining atomic-scale dynamics of such systems. A main limitation has been the short time scales attainable in all-atom MD simulations. However, we have performed a 150-ns simulation––long enough to examine important dynamic events along the path from the dark-adapted to the light-adapted state. The transition to the LUMI intermediate state takes about 150 ns. Thus, we are within range of an early rhodopsin intermediate and can compare to corresponding experimental data. By constraining the C11–C12 dihedral angle of retinal in the simulation, we force the isomerization. In the subsequent dynamics, the constraint is turned off. We obtain a single trajectory of the consequences of the isomerization on the structure and energetics of rhodopsin. Within this comparison, we recognize that the simulation yields only one pathway to the state at 150 ns, not the statistical ensemble that actually exists. However, some aspects of the dynamics are highly probable and will occur for most trajectories. These aspects can safely be expected in the dynamics of our simulation.
The details of the simulation are given in the Methods section. We then describe the results of the simulation analysis concerning the structural dynamics and the energetics of our model rhodopsin system that are observed after the forced photoisomerization, including changes in retinal’s dihedral angles, a narrowing of the distance between Ala 169 and retinal’s ionone ring, helix kink and tilt angle transitions, a switch in the protonated Schiff base counterion, and changes in retinal’s interaction with its environment, including disengagement from Helix 6. These events are connected to other large-scale transitions in the post-photoisomerized state of rhodopsin and lead towards coupling with transducin.
SIMULATION METHOD
The present work is a continuation of our earlier simulation of the dark-adapted state of rhodopsin.20 In the earlier work, MD simulations were performed using an all-atom representation. The lipid and water environment were explicitly treated. The CHARMM force field was used (version 22 for protein and version 27 for lipids, both released in August of 1999),21,22 which includes parameters defined for retinal.23 All calculations started from the first X-ray structure of rhodopsin (1F88).7 To be consistent with our previous work, we did not use the new rhodopsin dark-adapted structures24–27 as our starting point. Analysis of our earlier dark-adapted rhodopsin simulation has suggested relatively small changes from the original structure. The total system size (41,623 atoms) consisted of protein, 99 DOPC lipids, 100 mM salt concentration (14 sodium, 16 chloride), palmitylated lipids attached to Cys 322 and Cys 323, and 7441 TIP3 waters.
The LAMMPS28 lammps.sandia.gov molecular simulation package was used to produce a 150-ns trajectory. The initial state is the final state of the 40-ns simulation of the dark-adapted rhodopsin.20 Periodic images were used in all directions. The simulation was performed at constant membrane surface area of 55 × 77 Å2. The direction perpendicular to the lipid bilayer was controlled at a constant pressure of 1 atm. The temperature was controlled using the Nose-Hoover thermostat at 307 K. All bonds to H atoms were constrained using the SHAKE algorithm and the equations of motion were integrated using the velocity verlet algorithm with a 2 fs timestep. Electrostatic interactions were treated using the particle–particle particle–mesh (P3M) method.
In the simulation, the cis–trans photoisomerization occurs by constraining the C11–C12 dihedral angle. An MD simulation of 200 fs starts with the C11–C12 dihedral angle constrained in the cis state and ends at the trans state. Thereafter, the 150-ns simulation does not constrain the dihedral angle.
RESULTS
C11–C12 Dihedral Transition
We begin by discussing the postisomerization dynamics of retinal itself. Figure 1(a) shows the dihedral angle of the C11–C12 dihedral in retinal as a function of time, including both the 150 ns after isomerization and the 40 ns before the isomerization (from the earlier simulation of the dark-adapted state).20 The time before isomerization is represented as negative in this plot and subsequent plots. After isomerization, the dihedral angle does not substantially change during the 150-ns simulation. Thus, the C11–C12 dihedral transition is stable. After isomerization, the self-energy rises by about 7 kcal/mol [Figure 1(b)], which is primarily due to the C11–C12 dihedral transition. In the vicinity of t = 20 ns, there are fluctuations that bring the energy below the average cis state energy. By about 30 ns, the self-energy drops sharply and permanently below the average cis state self-energy and subsequently slowly decreases to about 4 kcal/mol below the cis state self-energy. This drop in energy is primarily due to the relaxation of dihedrals other than the C11–C12 dihedral. The C9–C10 dihedral angle transitions from 160° to the lower energy 180° state. Also, before t = 30 ns, the C6–C7 dihedral exhibits oscillatory behavior between 60° and 150°, but settles near 60° after t = 30 ns, thus preventing the β-ionone ring from being coplanar with the retinal chain. In the first 30 ns after isomerization, the dihedral angles C12–C13 and C8–C9 have large fluctuations, although about the average value of the dark adapted state. These large fluctuations end at about t = 30 ns, when the retinal self energy changes, and the average dihedral angle and the fluctuations return to the values of the dark adapted state. The effect of the dihedral motion involving C9 is to move the C19 methyl group to the same side of retinal as the C20 methyl group.
Fig. 1.
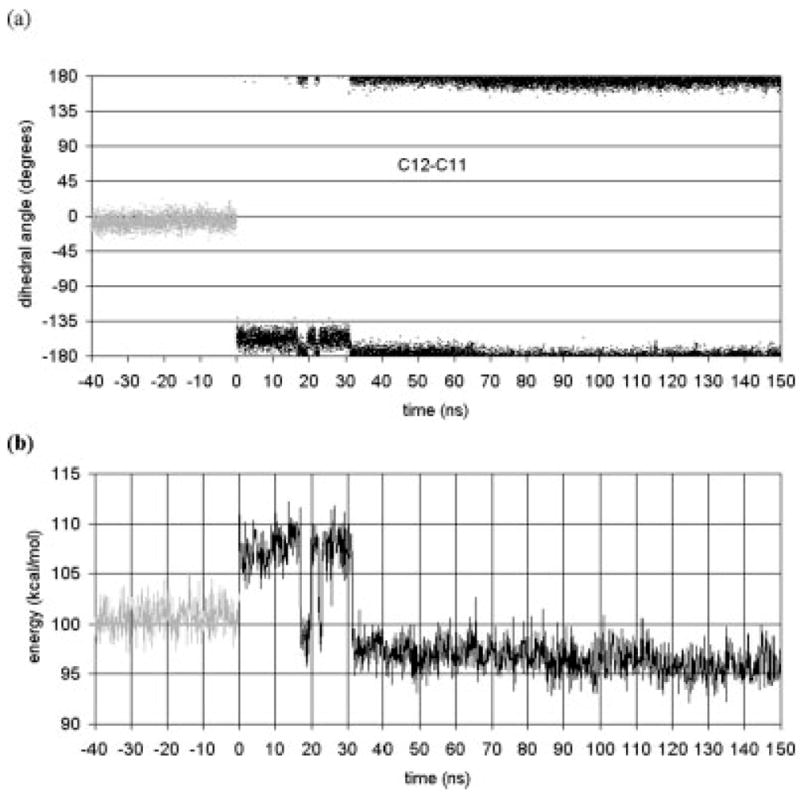
(a) Retinal dihedral angles as a function of time. (b) Retinal’s self energy as a function of time. The gray line represents the dynamics for the 40 ns run of the dark-adapted rhodopsin, i.e., retinal in the cis state and the C11–C12 dihedral has angle 0. The black line represent the dynamics after the cis–trans isomerization, i.e., retinal is in the trans state with the angle at 180 (= −180).
The net structural changes in retinal can be examined by calculating a pseudodihedral angle. The N16 and C7 atoms with their respective H atoms are at opposite ends of the retinal chain. Hence, the H16–N16–C7–H7 dihedral angle is a measure of the linearity of the retinal chain (up to the ionone ring). In the dark-adapted state, the average value is 45°, which indicates the bend and twist of the chain, whereas after photoisomerization, a sudden shift occurs, and the dihedral then fluctuates between 60° and 180° for 30 ns. At t = 30 ns, the dihedral angle changes to 180° and stays there for the remainder of the simulation. In this final conformation, the chain is linear and coplanar. The main relaxation after isomerization is the transition to this final flattened all-trans conformation.
We have also examined retinal’s bond orientations for comparison with available deuterium NMR structural data for retinal in the dark-adapted state of rhodopsin.29 Bond orientation vectors with respect to the bilayer normal were computed for retinal’s methyl groups bonded to C5, C9, and C13 for both the dark-adapted state and post-photoisomerization. Considering the large instantaneous fluctuations in bond orientation and the limited statistical sampling achieved within the 40-ns dark-adapted simulation, results for the dark-adapted state are in good agreement with the NMR measurements (Table I). The C5 and C9 bond vector orientations undergo substantial transitions postphotoisomerization. This observed transition in the simulation could be compared with future NMR measurements of retinal’s methyl bond vectors postphotoisomerization.
TABLE I.
Retinal Bond Orientations, θ (°)
| System | C5 | C9 | C13 |
|---|---|---|---|
| Ground state 2H-NMRa | 70 ± 3 | 128 ± 3 | 68 ± 2 |
| Ground state simulationb,c | 75 ± 11 | 129 ± 11 | 59 ± 14 |
| Postisomerization simulationc,d | 137 ± 15 | 80 ± 35 | 67 ± 15 |
NMR data from Salgado et al.29 Note that the bond orientation θ was not distinguished from its supplement π − θ in the NMR measurements. Errors correspond to inverse curvature matrix of the χ2 hypersurface utilized for the nonlinear regression fits.
40-ns Dark-adapted state simulation data from our previous work in Ref. 20.
Errors correspond to the standard deviation from the average values given.
150-ns Postphotoisomerization simulation data from this work.
Retinal – Ala 169 Gap Narrows
The isomerization of retinal starts a sequence of structural transformations that ultimately results in the light-adapted structure. For comparison to our 150 ns simulation, structural transformations that have occurred in the transition to the LUMI state are the most relevant. The isomerization directly moves the ionone ring. Consequently, the residues neighboring the ionone ring should change after isomerization. We have examined the distance between the ionone ring and Ala 169 as a function of the simulation time. Crosslinking experiments30 find that Ala 169 and the ionone ring can be crosslinked for rhodopsin in the LUMI state. Figure 2 shows that in the simulation, the separation distance decreases after isomerization with the final separation at about 9 Å. The crosslinking experiments imply a shorter separation, which could be achieved via rotation of Helix 4 (since Ala 169 is on the side of Helix 4 opposite to retinal), or by further movement of the ionone ring. One would expect that there is a set of conformations for the LUMI state, and only an (unknown) fraction of them allow the ionone–Ala 169 crosslinking. Our simulation does show that the separation distance is reduced significantly, and that if Helix 4 were to rotate, or if the ionone ring moved further, the crosslinking could occur. In the following sections, we discuss the structural transformations of the helices seen in the simulations. The large-scale motions of helices are naturally slower than the individual motions of residues or of retinal, which limits the helical motion that can be seen in nanosecond-time-scale simulations. However, the 150 ns simulation is sufficiently long to observe transitions in some of the helices.
Fig. 2.
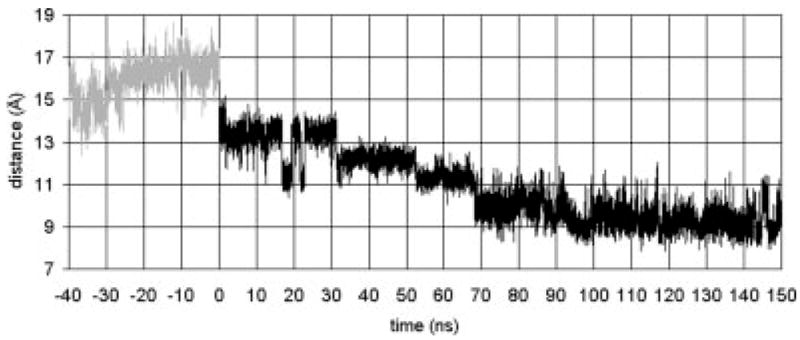
The distance between retinal’s ionone ring and Ala 169. The gray line represents the dynamics for the 40 ns run of the dark adapted rhodopsin, i.e., retinal in the cis state. The black line represents the dynamics after the cis–trans isomerization, i.e., retinal is in the trans state.
Helix Tilt and Kink Angle Transitions
An important characteristic of membrane proteins is the tilt of the transmembrane helices relative to the lipid bilayer. The tilt is an important mechanism by which the helix can match the bilayer thickness and the corresponding hydrophobic/hydrophilic regions. Kinks in the helices play a similar geometric role. Within the membrane protein, there are internal interactions that also influence the structure of the transmembrane helices. The isomerization of retinal alters the interaction between retinal and the helices, which can lead to significant changes in the geometry of the helices. In this manner, the molecular-scale isomerization event can yield subsequent larger-scale transmembrane helix structural changes. Because these are transmembrane helices, the action of the isomerization is propagated across the membrane. Both the cytoplasmic ends of the transmembrane helices and the loops, which interact with the G-protein, are affected. Thus, we have a basic outline of the mechanism by which rhodopsin interacts with the G-protein.
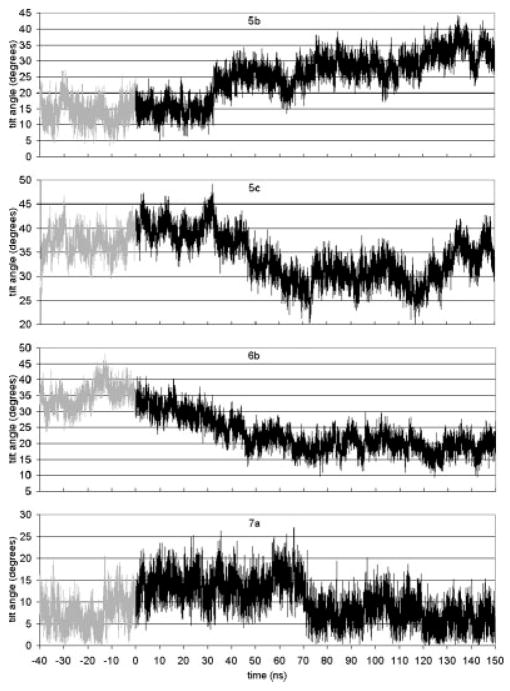
We examined the degree of tilt and kink of each helix as a function of simulation time. Since a kink can split the transmembrane helix into more than one part, with varying tilt angles, we calculate the tilt of each part of the helix. These parts have been defined in terms of the kink centers (Ref. 20 and the figure captions for details of the definitions). Figure 3 shows tilt angles as a function of time for helix segments 5b, 5c, 6b, and 7a. These tilt angles show significant changes during the simulation beyond the short time-scale fluctuations of about ±5°. At about t = 32 ns, the tilt angles for Helices 5b and 5c start to shift considerably. For 5b, the tilt angle rises and continues to slowly rise until the end of the simulation. Segment 5c reaches a steady state by about t = 70 ns and fluctuates by about an average value of 30°. This structural change in tilt angle is also reflected in the change in kink angle for 5b–5c (Fig. 4). There is a sudden drop in the kink angle at t = 32 ns. Ultimately, this kink angle decreases from 25° in the dark-adapted state to about 5° at t = 150 ns. This small kink angle reflects the fact that the tilt angles of 5b and 5c become almost identical after the transition. Thus, the main effect is almost complete removal of the kink in Helix 5.
Fig. 3.
Tilt angles as a function of time for Helices 5b (Phe 203–His 211), 5c (His 211–Leu 226), 6b (Pro 267–Thr 277), and 7a (Ile 286–Pro 291).
Fig. 4.
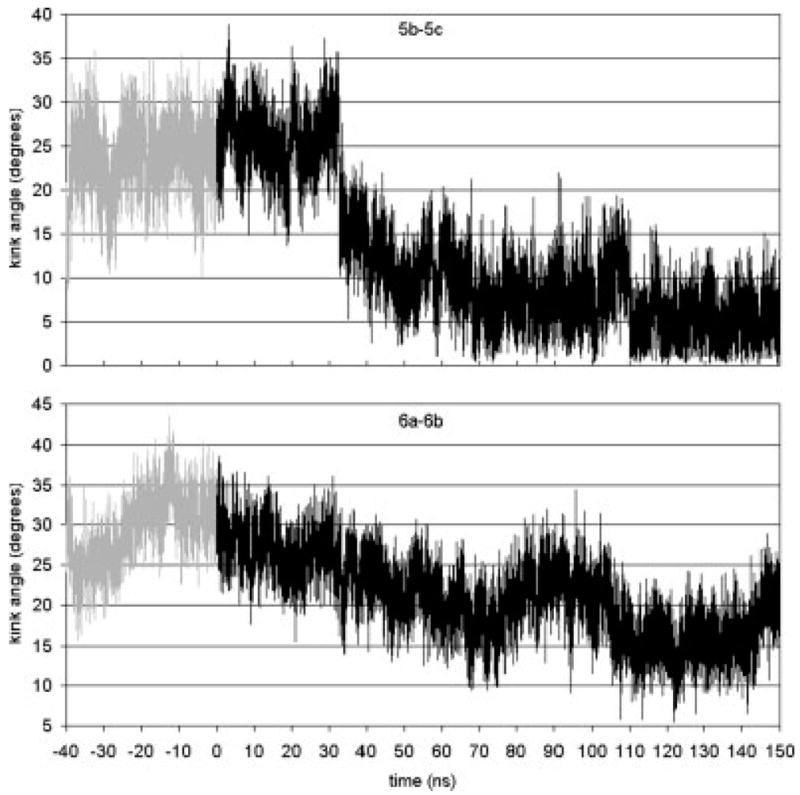
Kink angles as a function of time for Helices 5b–5c (kink at His 211) and 6a–6b (kink at Pro 267).
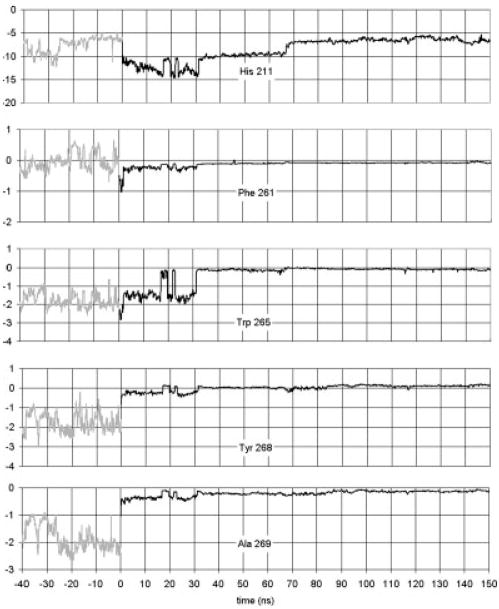
The retinal ionone ring is close to Helix 5. In fact, in the dark state the ionone ring is in contact with Met 207 on Helix 5. The kink in Helix 5 occurs at His 211. In the dark state, Met 207 is between His 211 and retinal. After isomerization, the ionone ring and Met 207 switch sides. His 211 then can and does come in contact with retinal. Simulations of the single Helix 5 in the membrane have the same tilt and kink angles as the helix does in the dark-adapted state.20 This implies that the interactions of Helix 5 with the rest of the rhodopsin protein do not affect the kink angle. The fact that isomerization results in a change in the kink angle implies that the structural changes resulting from isomerization cause the kink angle to almost disappear. For this to happen, some residues or retinal must be influencing the kink. Since His 211 comes into contact with retinal after isomerization, and since His 211 is the hinge point of the kink, the obvious candidate for the kink removal is the interaction of His 211 with the ionone ring. In Figure 5, the interaction energy between His 211 and the ionone ring is given. Before isomerization, the energy switches between two states with energies of −6 and −10 kcal/mol, respectively. After isomerization, the energy drops to −14 kcal/mol. This confirms that the His 211 interaction with the ionone ring increases in strength. In the first 30 ns, this interaction brings His 211 and ionone into position. Near t = 30 ns retinal completes the transition to a straight and mostly planar structure. At about the same time, there is the sharp drop in the Helix 5 kink angle (Fig. 4). The straightening of retinal and the continued strong interaction between His 211 and the ionone ring pulls on the kink’s hinge and reduces the kink angle.
Fig. 5.
Interaction energies (in kcal/mol) between retinal’s β-ionone ring and nearby side chains.
The tilt and kink angles of Helix 6 exhibit a correlated dynamics. Within 70 ns of the forced isomerization, the tilt angle of Helix part 6b decreases from its dark state value of 35° to an average of about 17° (Fig. 3). The kink angle also decreases for the same 70 ns and thereafter oscillates about an average value of 20°. In this case, the tilt angle of section b is the major part of the dynamics; the kink angle change is a result of just 6b’s tilt dynamics. While the ionone ring in the dark state is in contact with Ala 269 of Helix 6, this contact does not appear to be the driver for the change in the tilt of Helix section 6b. Helix section 6b moves away from the ionone ring and there is not a strong interaction between the ionone ring and Ala 269 that would force the whole segment to have such a tilt (cf. Fig. 5). It is thus more likely that the connection to Helix 7 through the short loop E3 exerts the pull on Helix 6 that alters the tilt of segment b.
Following retinal’s cis–trans isomerization, interactions between retinal’s β-ionone ring and the nearby aromatic side chains of Helix 6 diminish. The distance between them grows due to the movement of retinal itself. Figure 5 shows that the interaction energies between retinal’s β-ionone ring and Phe 261, Trp 265, and Tyr 268 on Helix 6 are well below kT after isomerization. This shows the decoupling of Helix 6 from retinal that will allow subsequent large-scale Helix 6 movement,31 which is coupled to C3 motion and transducin coupling.
Since Helix 7 includes Lys 296, it is not surprising that its tilt angle is quickly influenced by the retinal isomerization. Figure 3 shows that after isomerization, the tilt angle of Helix section 7a increases slightly from about 7° to 15°. At about t = 70 ns (where the changes in the Helix 6 tilt angle stop), the tilt angle abruptly returns to the dark state value. Correlation between the dynamics of Helices 6 and 7 likewise is not surprising given the strong interactions between their sidechains.
PSB Counterion Switch
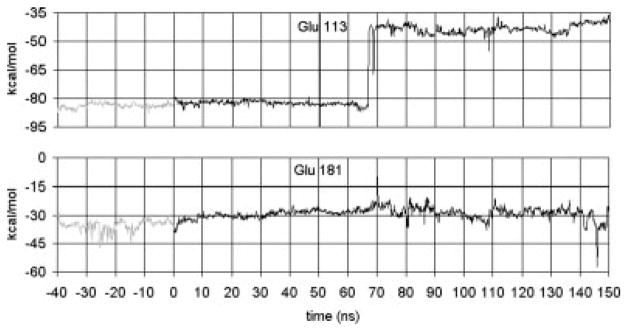
From the helix tilt and kink angle data, it is clear that some event occurs near t = 70 ns that impacts the helix dynamics and structure. The event, a switch in the dominant counterion of the retinal protonated Schiff base (PSB), is itself interesting. In rhodopsin’s dark-adapted state, Glu 113 acts as the counterion to the PSB. Some experimental evidence has suggested that Glu 181 is protonated in the dark-adapted state, but transfers its proton to Glu 113 via Ser 186 and then replaces Glu 113 as the PSB counterion.32 Other work suggests that both Glu 113 and Glu 181 are unprotonated and that both act as the PSB counterion, with Glu 181 dominating in the Meta I state.33 We have simulated both Glu 113 and Glu 181 in their unprotonated states, and we find that structural changes occur that could lead to the PSB counterion switching from Glu 113 to Glu 181. The simulation shows that the salt bridge between the retinal PSB and Glu 113 breaks near t = 70 ns after photoisomerization and does not form again (Figs. 6 and 7). Figure 6 also shows that at t = 146 ns a much stronger (~20 kcal/mol) interaction between Glu 181 and the retinal PSB is briefly established. This occurs because of a dihedral transition in Glu 181 and in Lys 296 simultaneously occurring to shorten the separation between the two residues.
Fig. 6.
Interaction energies (in kcal/mol) between the retinal PSB (including Lys 296) and nearby PSB counterion candidates Glu 113 and Glu 181. Glu 113 acts as the counterion in the dark adapted state and through the first 65 ns after forced photoisomerization. Near t = 70 ns, the salt bridge between Glu 113 and retinal breaks. Later, near t = 146 ns, Glu 181 briefly becomes the PSB counterion, hinting at possible subsequent completion of the transition from Glu 113 to Glu 181 acting as the PSB counterion. The gray lines represent the dynamics for the 40 ns run of the dark adapted rhodopsin, i.e., retinal in the cis state. The black lines represent the dynamics after the cis–trans isomerization, i.e., retinal is in the trans state.
Fig. 7.
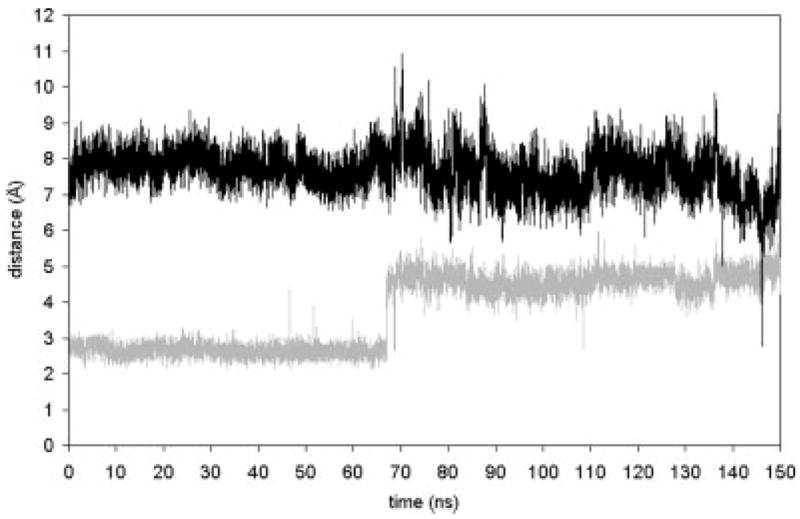
Distance between retinal’s H16 atom (the proton on the PSB), and the Cδ atom of potential counterions Glu 113 (gray) and Glu 181 (black). The minimum value of 2.8 Å in the Glu 181 time series corresponds with the maximum value of 6.3 Å in the Glu 113 time series (both at t = 146 ns).
Important details concerning the breaking of the salt bridge are indicated in the plot of the interaction energies of retinal with its environment (Fig. 8). In these calculations, we take “retinal” to include Lys 296. It does not make sense to separate the PSB, in particular. After isomerization, the energies remain at the same average value as before isomerization until just before t = 70 ns. At this point, the interaction with the solvent gets much stronger, while the interaction with the rest of the protein weakens by about 35 kcal/mol. We have already noted that the salt bridge between retinal and Glu 113 breaks at t = 70 ns. This loss of the strong binding energy results in the weakening of the interaction with the protein. The large strengthening in retinal–solvent interaction energy implies that some water molecules have moved into the protein close to retinal. This indeed is true as images of the region for times near t = 70 ns show (Fig. 9).
Fig. 8.
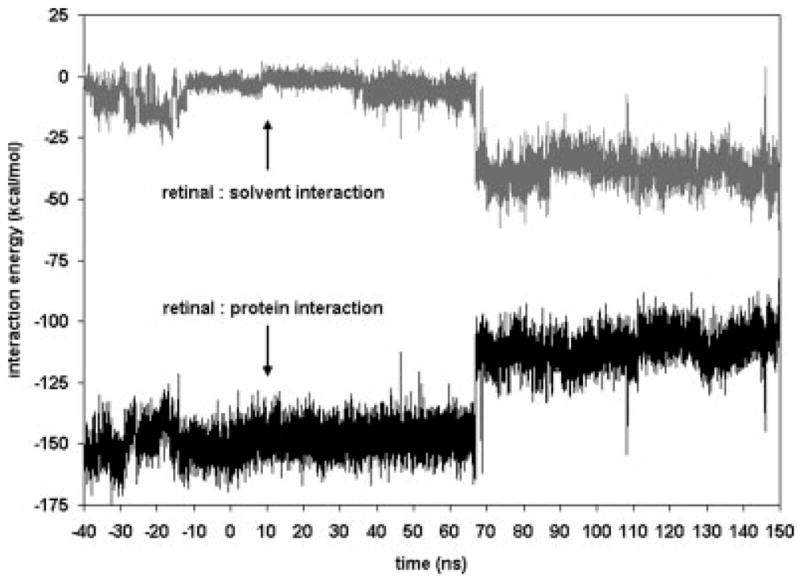
Interaction energy of retinal with the surrounding aqueous solvent, and retinal with the rest of the rhodopsin molecule. Retinal–lipid interaction was negligible. The large shift near 70 ns is due to the breaking of the salt bridge between Glu 113 and the PSB. A water molecule partially compensates for the broken bridge as is evidenced by the lower retinal–solvent interaction energy.
Fig. 9.
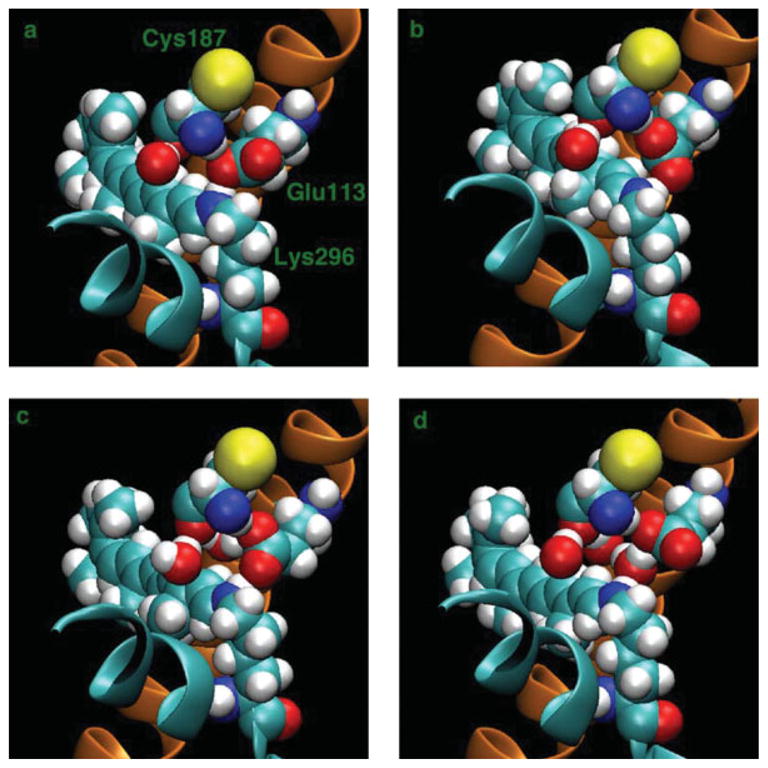
Images34 of the rhodopsin protein, Lys 296 (retinal) with Glu 113 and Cys 187, and nearby waters at different times (a) t = 64 ns, (b) t = 65 ns, (c) t = 66 ns, and (d) t = 67 ns. Helices 3 (orange) and 7 (cyan) are also visible in a cartoon representation. Lys 296 is on Helix 7 and Glu 113 is on Helix 3.
The changing hydrogen bond network involving retinal, including nearby water molecules, is shown in Figure 9, which shows images at t = 64, 65, 66, and 67 ns. At t = 64 ns, the Glu 113 salt bridge with the N of Lys 296 is intact. In addition, there is a hydrogen bond between NH of Cys 187 and the same O of Glu 113 that is part of the salt bridge. At t = 65 ns, the NH of Cys 187 switches to the other O of Glu 113 to form a hydrogen bond. At t = 66 ns, a water molecule is visible behind the Glu 113 and Cys 187. At t = 67 ns, the water breaks the salt bridge by moving between the N of Lys 296 and the O of Glu 113. For the rest of the simulation, a water molecule is between Glu 113 and Lys 296.
C3 Loop
Cytoplasmic loop 3 (C3) is an important loop in the interaction between rhodopsin and its G-protein. Without the presence of the G-protein in the simulation and without being in the active light-adapted state, direct understanding of how isomerization ultimately results in structural changes that impact the G-protein is not possible. Instead, we want to characterize how the structural changes in the helices affect the C3 loop structure. This gives a sense of the dynamic connection between isomerization and the C3 loop configuration.
The C3 loop connects Helices 5 and 6, which undergo substantial structural changes in the 150 ns time period as discussed earlier. These helix structural transformations impact the structure of the connecting loop. Figure 10 shows images of the C3 loop, where the final configuration of our dark-state simulation is compared with the final state of the 150 ns isomerization simulation. Sections of Helices 5 and 6 are also shown. Substantial motion has occurred in the loop, but this is inconclusive since the loop is primarily within solution and therefore has a large configurational space. The more important aspects of the figure are the differences in the helix positions and their effect on the loop. The end of Helix 6, which is attached to the C3 loop, has rotated significantly, bringing the ends of the two helices closer together. The bottom image shows that the C3 loop near Helix 5 is adopting a more helical structure.
Fig. 10.
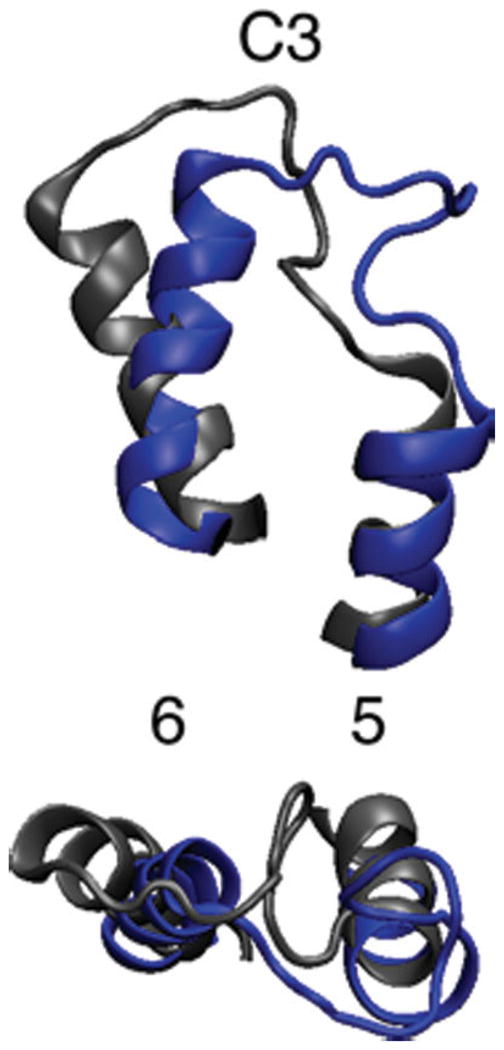
Rhodopsin’s C3 loop in dark state (gray) and after 150 ns of simulation (blue).34 Bottom image is 90° rotation of top image. The label 5 and 6 identify the respective transmembrane helices.
Large-Scale Structural Changes
Figure 11 shows a comparison of the rhodopsin structure in the dark state and at t = 150 ns. To distinguish the two states, the dark state helices are gray in the top image. Otherwise each helix is colored differently to identify it. The structure of transmembrane Helices 1, 2, 3, 4, and 7 do not change much within the 150 ns simulation time. As described earlier, Helices 5 and 6 do undergo significant structural changes. The top image of Figure 11 shows retinal in the cis conformation, while the bottom image shows the trans conformation. The helices that have substantial structural changes are the ones near the ionone ring of retinal, i.e., Helices 5 and 6. However, Helix 4, which is near the ionone ring for trans retinal, does not undergo a structural change. The difference being that Helices 5 and 6 are in contact with retinal at t = 0 and immediately feel the effects of the isomerization of retinal. The changes for these two helices are complete by the time retinal has completed its structural transformations. More specifically, they are complete by t = 70 ns, after the Glu 113–retinal salt bridge has broken. In contrast, Helix 4 is not in contact with retinal at t = 0 and is not strongly affected by the retinal transformations.
Fig. 11.
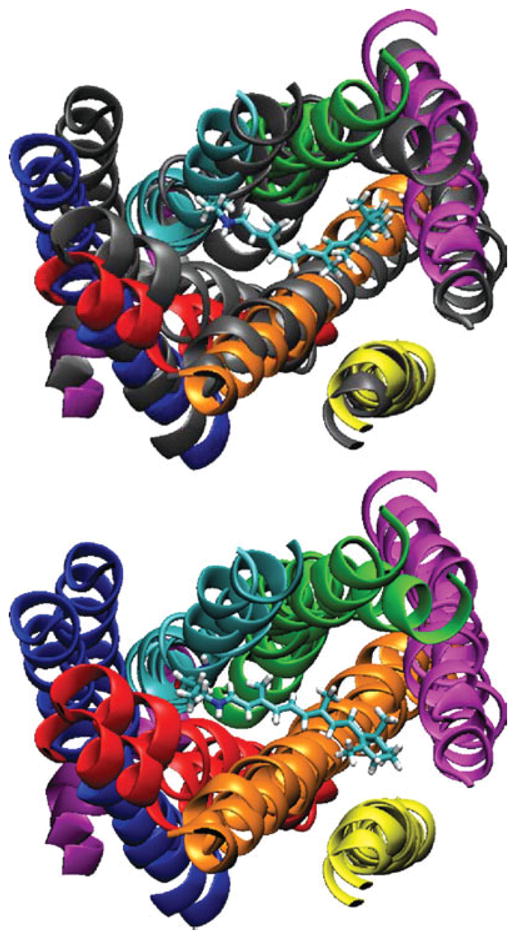
Comparison of helix positions in dark state (gray in top image) and after 150 ns after isomerization (color in top image).34 Top image shows retinal in cis state. The bottom image shows retinal in the trans state with the dark and 150 ns state both colored. The helix number and colors are 1 (blue), 2 (red), 3 (orange), 4 (yellow), 5 (magenta), 6 (cyan), 7 (green), and 8 (purple).
DISCUSSION
The relatively recent X-ray structure of rhodopsin provides an essential starting point for detailed consideration of how tertiary structure is linked to function for this key protein.7 Important issues for understanding this linkage concern how the conformational change in the retinal, induced by light, is coupled into large-scale conformational changes that ultimately lead to G-protein signaling.1 Further and closely related questions concern the effects of mutations, the membrane environment, water molecules, and charges.
Molecular dynamics simulations can begin to address some of these issues, but also have important limitations that should be presented along with the advantages. A particularly important limitation is that the present molecular dynamics capability can explore events near to the starting state that are relatively rapid (up to several tens of nanoseconds). Without a sense of the structure of photo-intermediates, the method can suggest how these intermediates could form, but cannot predict them with confidence. This is related to the same time-scale issue, since the number of possible intermediates formed from the dark-adapted rhodopsin structure is very large. Ideally, it would be the case that several thousand alternative starting conformational changes in the retinal could be performed and the ensemble set of these changes then used to determine a relative free energy surface for the transition. Within that type of framework, the confidence for prediction of intermediate conformational states would be considerably greater. Taking advantage of the considerable computer resources available through Sandia, we are able to explore, with low statistical confidence, a key structural transition within the photocycle. Thus, our simulations are suggestive of the types of changes that might occur after photoisomerization, but should not be considered predictions of intermediate structures.
It might be suggested that the current calculations should be augmented with detailed quantum calculations to understand the initial femtoseconds of excitation and the revised energy surface for the light activated transition. Multiple research groups are performing quantum chemical calculations of the initial events in photoisomerization.35–40 We wanted to address the longer-time scale (nanosecond towards millisecond) relaxation of the system following the initial light activation. While quantum chemical calculations have advanced significantly in the last few decades, they continue to be limited by the number of heavy atoms in the system and to explore very fast (femtosecond to picosecond) time scales. For this reason, we accepted the fact that our characterization of the very early events in light activation are by necessity incomplete. The main point is that the excited, light-activated state will decay quickly, and that how the energy from the isomerized retinal leads to the relaxation and eventual signal activation of G-proteins from the rhodopsin protein are the main targets of our calculation.
Recent work has suggested that rhodopsin may exist in a homodimer state in the native membrane.41,42 The implications of that finding are not directly addressed in these calculations, but we can speculate somewhat, based on the dynamics results, on what might happen within a homodimer model that differ from the results of the monomer. In particular, Helix 4 is believed to be involved in the dimerization. Our results are consistent with experimental work showing that Ala 169 (in Helix 4) moves closer to the β-ionone ring during activation. This may lead to effects on coupling between two monomers.43 While it is less clear how this could impact signaling, it does suggest that activation of one monomer in a dimer pair would be communicated to the other monomer. This could be viewed as a type of allostery, where the activation mechanism of the second monomer might be shifted to a more sensitive state due to the initial activation of the first monomer.
In the remaining discussion, we will start with the early events of excitation and work our way to the longer-time scale events. We will conclude the discussion with more speculative conclusions based on extrapolation of the results.
Initial Events in Photoactivation
A plausible candidate for the initial changes in retinal with light has been suggested based on a minimal motional change induced by light, but still consistent with a cis–trans isomerization.35–40 Our initial model covers that change by forcing the main cis–trans isomerization and then allowing relaxation from that state within the CHARMM potential function. The response to this change is then followed throughout the remainder of the simulation. Similar to ideas examining the effects of the initial velocity distribution on the range of motions examined in a trajectory, we do expect that the starting point will have an impact on the relaxation observed in the calculation. At the same time, there are not sufficient computer resources available to any biophysical research group to create an ensemble set of all candidate transitions and their response properties. Therefore, we took the most straightforwardly possible approach and forced the transition to allow the initial change to be consistent with experimental work. We examine the nature of the coupling between the local conformational change and the larger-scale conformational rearrangements that ultimately lead to G-protein activation. While we fully accept that this candidate starting transition may be inaccurate on the femtosecond time-scale, we want to emphasize that the relaxation from this excited state conformational change can still help us to understand how a local conformational change (in the retinal) can lead to larger-scale conformational change (and ultimately to G-protein activation).
It is interesting to speculate, however, on the types of cis–trans isomerizations that are not allowed––either quantum mechanically or from the molecular dynamics viewpoint. In either formulation, the type of large cis–trans motion that would occur with a rigid retinal moiety would create very large and unfavorable van der Waals clashes within the protein cavity. Thus, it makes biophysical sense that the type of motion adopted by the retinal chain would be as minimal as possible to allow the sensitivity of the response to be magnified and coupled to larger-scale motion. Phrased in another way, the quantum chemical findings suggest that the rapid-relaxation of the retinal molecule is due to the effects of the protein cavity environment in prepriming the conformation of the retinal such that the efficiency of cis–trans change is very high. Furthermore, once the trans state has been reached, there is a very high chance of the energy stored in the initially local conformational change of the retinal being carried over to larger changes in the coupling of G-proteins. Thus, we argue that evolution has created an extremely sensitive molecular instrument for converting the energy of photon activation to protein conformational change and signal activation. That the motional changes in the retinal are small, is significant, because it creates a system where the photon energy can be readily and efficiently absorbed and then the isomerized reaction readily focused on conformational change.
We emphasize that the dark-adapted state of rhodopsin was the starting point for the calculations. If structures of the intermediates are determined, it would be possible to perform calculations exploring the whole range of conformational change that is initiated by the cis–trans photo-conversion. In other words, the prediction of intermediate conformations is not nearly as reliable as the experimental determination of those intermediates. From the experimentally determined intermediates, simulations would be possible that create a thermodynamic picture of the molecular changes that occur during the photocycle. Without X-ray structures for the intermediates, the nature of the changes are inferred from the current calculations and those changes should be seen as suggestive, rather than definitive predictions.
Several other research groups have also performed molecular dynamics calculations of rhodopsin.44–49 Each of these studies has something of value to add to our understanding of the molecular nature of rhodopsin and its activation by light. Each group has made different choices with respect to membrane representation, length of simulation, size of simulation, and treatment of electrostatics. It is too early to tell whether a subset of these models is more right or more wrong than others, due to the problem of sampling that all of the computational groups face. It is possible to read each paper as a possible molecular story for how the system might behave. We suggest that the full set of these papers is very valuable for bringing new insights into the nature of the molecular behavior, in part because of the different characterization of the system (i.e., pK states, environment conditions, etc.).
Nature of Cis–Trans Potential Function
There has been considerable discussion in the literature about the nature of the changes in the retinal during photoactivation. These changes are likely to be minimal in overall conformation, but in the absence of a light-adapted structure, it is difficult to be certain about the scale of the overall change with light activation. In our simulations, the change was forced within 200 fs. We emphasize that the potential energy surface used during the isomerization, as well as for the relaxation, is the ground-state surface defined by the CHARMM potential function. That is, there is no attempt made to define a light-activated surface for the isomerization reaction and then to determine the longer-time relaxation process. Papers that examine the quantum dynamics on a short-time scale and connect to the FTIR spectra should be examined for this type of analysis.38
Time-Scale for Isomerization
Experimental work50 with femtosecond stimulated Raman spectroscopy has shown that there are significant changes in the retinal spectra during the transition from the dark-adapted state to the stable bathorhodopsin intermediate.51 The experimental work suggests that the full transition from dark-adapted to bathorhodopsin state is about 1 ps in length and that the efficiency of the process, as well as the capture of the energy for large-scale conformational change is very high. In particular, the results suggest changes in C=C, C—C, and C—H regions of the spectra that relax very quickly while also showing environmental effects suggestive of a change in interaction between retinal and the protein environment. The bathorhodopsin state is then reached, the effective starting point for our calculations. The conformation of this state is not fully determined experimentally, but may reflect a series of changes along the retinal structure, supporting a minimal change in the retinal overall shape. This is consistent with detailed quantum chemical calculations as well as findings from lipid bilayer simulations that suggest that correlated dihedral changes can be made that prevent very large conformational changes. As noted earlier, while we share an interest in determining the details of this bathorhodopsin starting stage, we elected not to explore the quantum details of the excited surface, nor to optimize all possible starting points. Instead, our starting point (the bathorhodopsin state) is consistent with experimental information and is thus plausible as an energy storage state intermediate to the signaling cascade and thus to M-II.
Initial Relaxation Events
In our simulations, there is a time-delay from the initial rapid isomerization to the larger-scale conformational transitions and the relaxation of the retinal conformation. This makes sense, since the excited state surface for the initial conformational change will act as a trigger to create the larger-scale conformational changes that are seen experimentally in later stages of the photocycle. In order for those longer-term events to occur, there must be a period of relaxation after the isomerization, during which the retinal conformational change is adjusted and adapted to by the protein compartment. In the initial stages of relaxation (roughly 30 ns in our simulation), the protein adjusts to the retinal. This can be seen in the energy changes that occur within the retinal: initially 7 kcal/mol higher in energy than the dark-adapted self-energy and then a relaxation, after about 30 ns to a lower self energy state. At the same time, within that 30 ns window, the coupling of the retinal to other sidechains near to the dark-adapted cavity is shifting and the β-ionone ring is moving from being near one location to being near another. While the nature of these changes is subtle, the overall effect is very large. This magnification is due to the hydrogen bonding nature of the protein, resembling a large allosteric network, with the changes in one location being communicated energetically to other locations. The net effect is that within 30 ns, in this particular simulation, the initial relaxation events from the cis–trans isomerization have already been communicated to many other parts of the protein.
Changes by End of 30 ns
We find it fascinating that large scale changes occur from the small scale changes initiated by the cis–trans isomerization of retinal. The nature of these changes, subject to our sampling limitations imposed by molecular dynamics calculations, suggest a tight coupling within the rhodopsin–retinal system.52,53 Thus, the large tilt changes in Helix 6 are well underway by 30 ns, the changes in the G-protein activation region (Helices 5 and 6 and cytoplasmic loop C3) have also started. But, the largest indicator of the nature of these shifts is the changes in sidechain-to-retinal coupling. The almost complete decoupling of the retinal interaction energy between retinal and Phe 261, Trp 265 and Tyr 268 is already indicative of the types of changes that occur later in the cycle.
Shift of β-Ionone Ring and Ala 169
Experimental work has suggested that at least part of the photocycle involves a shift in Helix 4 with Ala 169 and its interaction with retinal.30 The detailed molecular nature of the distance shift is not resolved in the experimental work, but it is suggested in the molecular dynamics simulations. In particular, the shift is initiated by the decoupling of the retinal from the dark-adapted state side-chain interactions. This loosening of dark-state interactions enables a relatively modest shift in conformational space of the retinal and a much larger shift in the packing of the protein around the trans retinal ring system. The movement of Ala 169 to increase interaction with the retinal ring and the shift in Helix 4 is thus consistent with a shift in the overall energetics of the retinal/rhodopsin system, following the relaxation from the trans state.
Counterion Switch
Experimental work has suggested that part of the light activation could involve a switch of the Schiff base counterion from the Glu 113 to Glu 181.11,32,54–65 This would involve a change in protonation states during the photocycle33 and experimental work has confirmed that Glu 113 is the dark-state counterion and been suggestive, but not conclusive, that Glu 181 is active during the light-activated stages.66,67 The simulation results are intriguing in suggesting that this counterion switch mechanism could be present as part of the relaxation mechanism of the protein to the retinal conformational change. We should emphasize that in our calculations there is no change in the charge state of Glu 181 during or after the forced isomerization.57,68 Thus, the driving force for the change is not wholly from charge transfer during the photocycle, but could also be related to the coupled relaxation of the full system from the initial events of light activation. In this regard, quantum calculations about the effect of the counterion switch are also suggestive of the types of changes occurring with a counterion switch that could underlie function.35,69
Longer (30–150 ns) Changes
Activation of G-protein coupled receptors will depend on changes in Helices 5 and 6 along with the C3 loop regions.31,70–76 These locations have been implicated, by mutagenesis studies, as key players in the coupling of the activated rhodopsin to G-protein signaling.77 For this reason, it is intriguing to note that the simulation results suggest large-scale changes in all three of these locations. Helix 5 shows an increased degree of bending between upper and lower parts of the helices, and that drives further changes in the C3 loop regions. At the same time, Helix 6 shows a decrease in the kink angle and a shift to a more perpendicular orientation of the helix axis. These three sets of coupled changes are all a relaxation response to the forced isomerization change. It is not hard to imagine that the nature of these changes could lead to a shift in binding affinity for a G-protein system and thus to activation.78 At the same time, we emphasize that the simulations do not include the G-protein binding, since the details of this interaction are still not known experimentally, and that the timescale of activation within this system is clearly beyond the timescale of the current calculations. Thus, the nature of these changes should be seen as suggestive of the types of shifts that are likely to occur with activation of the system.
Implications for Rhodopsin Function
Rhodopsin has evolved to be highly tuned to converting light energy to conformational change and signal amplification. The simulations that we have performed are suggestive of some of the types of coupling and change that underlie this tight connection between structure and function. This underlies much experimental work with mutants suggesting that disruption of the tight conformation of rhodopsin will lead to a loss of function.12 An example is the Cys 110–Cys 187 pair.79 In particular, if the retinal cis–trans isomerization were not coupled into a protein cavity setting that adjusted to support the transition, then the signal amplification and coupling would not be so efficient. In other words, the nature of the coupled set of changes induced by light activation is amplified by the design of the retinal cavity to induce a set of protein changes that lead to activation. In this sense, the rhodopsin system is very much like the switch mechanism suggested by experimental work.80–82 The relatively small change introduced by the retinal conformational change is then amplified through the hydrogen bond and sidechain interaction network into a shift in helix orientations, Schiff base partners, and eventual G-protein coupling.
An emerging frontier is then to understand more about the nature of the coupling between the rhodopsin protein and G-protein. Several groups have started to attempt this model, based on the X-ray structures of G-proteins16,17,83–88 and we believe that the simulation results here may lead to speculation about the nature of the interfacial coupling between these domains.89–95 In particular, we suggest that the shift in the surface properties of the rhodopsin protein on activation will be found to create a much less favorable environment for protein–protein interaction. In this regard, the mutation studies and their effects on the surface of dark-adapted and light-activated (inferred) could be interesting96–98 as well as studies of the role of the cytoplasmic loop region.99 Several groups are working to understand more about protein–protein interactions and this could be a model system for understanding how the modulation of protein–protein interactions lead to shifts in function.
Implications for Spectral Tuning
While the current molecular dynamics calculations do not address the changes in the binding cavity that underlie spectral response, it is interesting to suggest several implications for spectral tuning from the results. In particular, the results strongly underlie the importance of the protein cavity in forming a part of the solvation environment dictating the type of conformational change that will be determined by the light-activated retinal.15,100–108 In that sense, the nature of these sidechain and main chain interactions drive a preference for a particular type of excited state and a particular type of relaxation response. Experimental work that shifts the nature of retinal further supports this type of thinking.109–113 It can also be inferred that spectral tuning will lead to a similar type of excited state conformation for retinal and, following that transition, to a similar set of relaxation events that lead to G-protein activation.114 It is thus intriguing to consider the nature of the changes possibly induced by different environments. An example is the set of mutants looking at hydroxyl changes.115 We would suggest that by shifting the energy required for isomerization and forcing a similar pathway for relaxation from the excited state, the retinal/rhodopsin system has been engineered for maximum flexibility by evolution.14
Effect of the Membrane Environment
While the current simulations did not directly compare different models for the membrane with the transition, it is clear from experimental work that rhodopsin is very sensitive to the details of the lipid setting.52,116–124 We can speculate, from our current and previous121 work about why this may be important. We suggest that the nature of the coupling from the cis–trans activated state to the relaxed state will depend on an ability of the helices to reorient, the sidechains to shift, and the cytoplasmic loops to adjust their relative positions. The lipid environment then provides a resistance to this motion that can be either supportive of the change, making the free energy change for the large-scale motion relatively easy, or providing a setting that makes the relative shift in conformation much more difficult, allowing a change, but only with much greater resistance and thus with a decrease in signal to noise outcome. We suggest that the effects of DHA on rhodopsin are to make the transitions between helix orientations (e.g., the kink changes in Helices 5 and 6) much easier and thus to support the nature of the large scale changes underlying function in this system.124–127
Implications for Other GPCR Systems
We believe that study of the rhodopsin system leads to insights important for other GPCR systems as well.3,128–130 This is especially important due to the large number of important pharmacological targets within the large GPCR family.131–142 In our simulation results, the nature of the coupling between a local conformational change and a large-scale helix and loop change may be similar across the GPCR family. This would imply that the effect of ligand binding to a GPCR is tightly regulated by a coupled set of energetic interactions, in a similar manner to that found within the retinal-rhodopsin system.143 The intriguing result is that the nature of the second extracellular loop (containing the β-bulge region) may thus be critical in determining the nature of the coupling between local conformational binding induced changes and large-scale signaling induced changes.144–147 We would suggest that this is similar, in some ways, to the nature of spectral tuning in that adjustments to the binding cavity can support the relative efficiency and recognition of certain ligands relative to others.
CONCLUSIONS
The high quantum efficiency of rhodopsin is coupled to an ability to capture local changes in retinal behavior and induce conformational changes that lead to signaling.1,148–152 How this happens remains a mystery that will require more computational and experimental work.4,153 The molecular details of how the stable bathorhodopsin intermediate (about 1 ps after light absorption and cis–trans isomerization) leads to conformational changes in the Meta II state were the target of our calculations in this study. What we find emphasizes the nature of the coupling throughout the protein that we had found in our previous simulation result, which is consistent with the general behavior of other protein systems. In particular, the cis–trans isomerization leads to a change in the sidechain interactions with retinal, and eventually to large-scale changes in tilt and conformation of the system. While the details of these changes may not be fully sampled in the current simulations, the results that we report are suggestive of the types of compensation that may occur in the system upon light activation.
The preliminary analysis of the long simulation of rhodopsin after isomerization shows that significant structural changes occur in the 150 ns timeframe. The isomerization results in some of the seven transmembrane helices undergoing tilt and kink angle changes that are well beyond their fluctuation range in the equilibrium dark state. One of the key structural changes is the motion of the ionone ring of retinal. The ring progressively gets closer to Ala 169. This is consistent with crosslink experiments30 where a crosslink forms between the ring and Ala 169.
A strong energetic transition is observed to occur at t = 70 ns after isomerization. The majority of the energy of this transition involves breaking of the salt bridge between Glu 113 and the protonated Schiff base. Raman spectroscopy51 has shown that the protonated Schiff base stretching modes are indicative of very different Schiff base environments. This implies that the transition from bathorhodopsin to the LUMI state involves chromophore relaxation and dramatic changes in the Schiff base region. Our results are in agreement with these experimental observations. Furthermore, besides the structural dynamics, we show the connection to the energetics that drives the changes.
Acknowledgments
Sandia is a multiprogram laboratory operated by Sandia Corporation, a Lockheed Martin Company for the United States Department of Energy’s National Nuclear Security Administration under contract DE-AC04-94AL85000.
References
- 1.Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: insights from recent structural studies. Annu Rev Biophys Biomol Struct. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- 2.Shi L, Javitch JA. The binding site of aminergic G protein-coupled receptors: the transmembrane segments and second extracellular loop. Annu Rev Pharmacol Toxicol. 2002;42:437–467. doi: 10.1146/annurev.pharmtox.42.091101.144224. [DOI] [PubMed] [Google Scholar]
- 3.Filipek S, Stenkamp RE, Teller DC, Palczewski K. G protein-coupled receptor rhodopsin: a prospectus. Annu Rev Physiol. 2003;65:851–879. doi: 10.1146/annurev.physiol.65.092101.142611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hubbell WL, Altenbach C, Hubbell CM, Khorana HG. Rhodopsin structure, dynamics, and activation: a perspective from crystallography, site-directed spin labeling, sulfhydryl reactivity, and disulfide cross-linking. Adv Protein Chem. 2003;63:243–290. doi: 10.1016/s0065-3233(03)63010-x. [DOI] [PubMed] [Google Scholar]
- 5.Burns ME, Baylor DA. Activation, deactivation, and adaptation in vertebrate, photoreceptor cells. Annu Rev Neurosci. 2001;24:779–805. doi: 10.1146/annurev.neuro.24.1.779. [DOI] [PubMed] [Google Scholar]
- 6.Lamb TD, Pugh EN. Dark adaptation and the retinoid cycle of vision. Prog Retin Eye Res. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 7.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: a G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 8.Sakmar TP. Rhodopsin: a prototypical G protein-coupled receptor. Prog Nucleic Acid Res Mol Biol. 1998;59:1–34. doi: 10.1016/s0079-6603(08)61027-2. [DOI] [PubMed] [Google Scholar]
- 9.Ferretti L, Karnik SS, Khorana HG, Nassal M, Oprian DD. Total synthesis of a gene for bovine rhodopsin. Proc Natl Acad Sci USA. 1986;83:599–603. doi: 10.1073/pnas.83.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung BKK, Stryer L. Photolyzed rhodopsin catalyzes the exchange of Gtp for bound Gdp in retinal rod outer segments. Proc Natl Acad Sci USA. 1980;77:2500–2504. doi: 10.1073/pnas.77.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakmar TP, Franke RR, Khorana HG. Glutamic acid-113 serves as the retinylidene Schiff-base counterion in bovine rhodopsin. Proc Natl Acad Sci USA. 1989;86:8309–8313. doi: 10.1073/pnas.86.21.8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke RR, Konig B, Sakmar TP, Khorana HG, Hofmann KP. Rhodopsin mutants that bind but fail to activate transducin. Science. 1990;250:123–125. doi: 10.1126/science.2218504. [DOI] [PubMed] [Google Scholar]
- 13.Fahmy K, Sakmar TP, Siebert F. Structural determinants of active state conformation of rhodopsin: molecular biophysics approaches. Methods Enzymol. 2000;315:178–196. doi: 10.1016/s0076-6879(00)15843-4. [DOI] [PubMed] [Google Scholar]
- 14.Yokoyama S. Molecular evolution of color vision in vertebrates. Gene. 2002;300:69–78. doi: 10.1016/s0378-1119(02)00845-4. [DOI] [PubMed] [Google Scholar]
- 15.Kuwayama S, Imai H, Hirano T, Terakita A, Shichida Y. Conserved proline residue at position 189 in cone visual pigments as a determinant of molecular properties different from rhodopsins. Biochemistry. 2002;41:15245–15252. doi: 10.1021/bi026444k. [DOI] [PubMed] [Google Scholar]
- 16.Noel JP, Hamm HE, Sigler PB. The 2.2-Å crystal-structure of transducin-α complexed with gtp-γ-s. Nature. 1993;366:654–663. doi: 10.1038/366654a0. [DOI] [PubMed] [Google Scholar]
- 17.Lambright DG, Noel JP, Hamm HE, Sigler PB. Structural determinants for activation of the α-subunit of a heterotrimeric G-protein. Nature. 1994;369:621–628. doi: 10.1038/369621a0. [DOI] [PubMed] [Google Scholar]
- 18.Mathies R, Freedman TB, Stryer L. Resonance Raman studies of conformation of retinal in rhodopsin and isorhodopsin. J Mol Biol. 1977;109:367–372. doi: 10.1016/s0022-2836(77)80040-5. [DOI] [PubMed] [Google Scholar]
- 19.Hayward G, Carlsen W, Siegman A, Stryer L. Retinal chromophore of rhodopsin photoisomerizes within picoseconds. Science. 1981;211:942–944. doi: 10.1126/science.7466366. [DOI] [PubMed] [Google Scholar]
- 20.Crozier PS, Stevens MJ, Forrest LR, Woolf TB. Molecular dynamics simulation of dark-adapted rhodopsin in an explicit membrane bilayer: coupling between local retinal and larger scale conformational change. J Mol Biol. 2003;333:493–514. doi: 10.1016/j.jmb.2003.08.045. [DOI] [PubMed] [Google Scholar]
- 21.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 22.Schlenkrich M, Brickmann J, MacKerell AD, Jr, Karplus M. An empirical potential energy function for phospholipids: criteria for parameter optimization and applications. In: Merz KMJ, Roux B, editors. Biological membranes: a molecular perspective from computation and experiment. New York: Birkhauser; 1996. pp. 31–81. [Google Scholar]
- 23.Nina M, Smith JC, Roux B. Ab-initio quantum–chemical analysis of schiff-base water interactions in bacteriorhodopsin. Theochem J Mol Struct. 1993;105:231–245. [Google Scholar]
- 24.Edwards PC, Li J, Burghammer M, McDowell JH, Villa C, Hargrave PA, Schertler GFX. Crystals of native and modified bovine rhodopsins and their heavy atom derivatives. J Mol Biol. 2004;343:1439–1450. doi: 10.1016/j.jmb.2004.08.089. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 26.Okada T. X-ray crystallographic studies for ligand–protein interaction changes in rhodopsin. Biochem Soc Trans. 2004;32:738–741. doi: 10.1042/BST0320738. [DOI] [PubMed] [Google Scholar]
- 27.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 Å crystal structure. J Mol Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 28.Plimpton SJ. Fast parallel algorithm for short-range molecular dynamics. J Comp Phys. 1995;117:1–19. [Google Scholar]
- 29.Salgado GFJ, Struts AV, Tanaka K, Fujioka N, Nakanishi K, Brown MF. Deuterium NMR structure of retinal in the ground state of rhodopsin. Biochemistry. 2004;43:12819–12828. doi: 10.1021/bi0491191. [DOI] [PubMed] [Google Scholar]
- 30.Borhan B, Souto ML, Imai H, Shichida Y, Nakanishi K. Movement of retinal along the visual transduction path. Science. 2000;288:2209–2212. doi: 10.1126/science.288.5474.2209. [DOI] [PubMed] [Google Scholar]
- 31.Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG. Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science. 1996;274:768–770. doi: 10.1126/science.274.5288.768. [DOI] [PubMed] [Google Scholar]
- 32.Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan DH, Chang BSW, Sakmar TP, Mathies RA. Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci USA. 2003;100:9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritter E, Zimmermann K, Heck M, Hofmann KP, Bartl FJ. Transition of rhodopsin into the active metarhodopsin II state opens a new light-induced pathway linked to Schiff base isomerization. J Biol Chem. 2004;279:48102–48111. doi: 10.1074/jbc.M406857200. [DOI] [PubMed] [Google Scholar]
- 34.Humphrey W, Dalke A, Schulten K. VMD—visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 35.Cembran A, Bernardi F, Olivucci M, Garavelli M. Counterion controlled photoisomerization of retinal chromophore models: a computational investigation. J Am Chem Soc. 2004;126:16018–16037. doi: 10.1021/ja048782+. [DOI] [PubMed] [Google Scholar]
- 36.Blomgren F, Larsson S. Using 1,3-butadiene and 1,3,5-hexatriene to model the cis–trans isomerization of retinal, the chromophore in the visual pigment rhodopsin. Int J Quantum Chem. 2002;90:1536–1546. [Google Scholar]
- 37.Cembran A, Bernardi F, Olivucci M, Garavelli M. The retinal chromophore/chloride ion pair: structure of the photo isomerization path and interplay of charge transfer and covalent states. Proc Natl Acad Sci USA. 2005;102:6255–6260. doi: 10.1073/pnas.0408723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buss V, Weingart O, Sugihara M. Fast photoisomerization of a rhodopsin model–an ab initio molecular dynamics study. Angew Chemie Int Ed Engl. 2000;39:2784–2786. doi: 10.1002/1521-3773(20000804)39:15<2784::aid-anie2784>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 39.Blomgren F, Larsson S. Initial step of the photoprocess leading to vision only requires minimal atom displacements in the retinal molecule. Chem Phys Lett. 2003;376:704–709. [Google Scholar]
- 40.Blomgren F, Larsson S. Primary photoprocess in vision: minimal motion to reach the photo-and bathorhodopsin intermediates. J Phys Chem B. 2005;109:9104–9110. doi: 10.1021/jp040693w. [DOI] [PubMed] [Google Scholar]
- 41.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 42.Javitch JA. The ants go marching two by two: oligomeric structure of G-protein-coupled receptors. Mol Pharmacol. 2004;66:1077–1082. doi: 10.1124/mol.104.006320. [DOI] [PubMed] [Google Scholar]
- 43.Jensen AA, Spalding TA. Allosteric modulation of G-protein coupled receptors. Eur J Pharm Sci. 2004;21:407–420. doi: 10.1016/j.ejps.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 44.Huber T, Botelho AV, Beyer K, Brown MF. Membrane model for the G-protein-coupled receptor rhodopsin: hydrophobic interface and dynamical structure. Biophys J. 2004;86:2078–2100. doi: 10.1016/S0006-3495(04)74268-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohrig UF, Guidoni L, Laio A, Frank I, Rothlisberger U. A molecular spring for vision. J Am Chem Soc. 2004;126:15328–15329. doi: 10.1021/ja048265r. [DOI] [PubMed] [Google Scholar]
- 46.Lemaitre V, Yeagle P, Watts A. Molecular dynamics simulations of retinal in rhodopsin: from the dark-adapted state towards lumirhodopsin. Biochemistry. 2005;44:12667–12680. doi: 10.1021/bi0506019. [DOI] [PubMed] [Google Scholar]
- 47.Pitman MC, Grossfield A, Suits F, Feller SE. Role of cholesterol and polyunsaturated chains in lipid–protein interactions: molecular dynamics simulation of rhodopsin in a realistic membrane environment. J Am Chem Soc. 2005;127:4576–4577. doi: 10.1021/ja042715y. [DOI] [PubMed] [Google Scholar]
- 48.Rohrig UF, Guidoni L, Rothlisberger U. Solvent and protein effects on the structure and dynamics of the rhodopsin chromophore. Chemphyschem. 2005;6:1836–1847. doi: 10.1002/cphc.200500066. [DOI] [PubMed] [Google Scholar]
- 49.Schlegel B, Sippl W, Holtje HD. Molecular dynamics simulations of bovine rhodopsin: influence of protonation states and different membrane-mimicking environments. J Mol Model. 2005;12:49–64. doi: 10.1007/s00894-005-0004-z. [DOI] [PubMed] [Google Scholar]
- 50.Kukura P, McCamant DW, Yoon S, Wandschneider DB, Mathies RA. Structural observation of the primary isomerization in vision with femtosecond-stimulated Raman. Science. 2005;310:1006–1009. doi: 10.1126/science.1118379. [DOI] [PubMed] [Google Scholar]
- 51.Pan DH, Mathies RA. Chromophore structure in lumirhodopsin and metarhodopsin I by time-resolved resonance Raman micro-chip spectroscopy. Biochemistry. 2001;40:7929–7936. doi: 10.1021/bi010670x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown MF. Modulation of rhodopsin function by properties of the membrane bilayer. Chem Phys Lipids. 1994;73:159–180. doi: 10.1016/0009-3084(94)90180-5. [DOI] [PubMed] [Google Scholar]
- 53.Janz JM, Farrens DL. Role of the retinal hydrogen bond network in rhodopsin Schiff base stability and hydrolysis. J Biol Chem. 2004;279:55886–55894. doi: 10.1074/jbc.M408766200. [DOI] [PubMed] [Google Scholar]
- 54.Zvyaga TA, Min KC, Beck M, Sakmar TP. Movement of the retinylidene Schiff-base counterion in rhodopsin by one helix turn reverses the pH-dependence of the metarhodopsin-I to metarhodopsin-Ii transition. J Biol Chem. 1993;268:4661–4667. [PubMed] [Google Scholar]
- 55.Hufen J, Sugihara M, Buss V. How the counterion affects ground-and excited-state properties of the rhodopsin chromophore. J Phys Chem B. 2004;108:20419–20426. [Google Scholar]
- 56.Sugihara M, Buss V, Entel P, Hafner J. The nature of the complex counterion of the chromophore in rhodopsin. J Phys Chem B. 2004;108:3673–3680. [Google Scholar]
- 57.Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc Natl Acad Sci USA. 2004;101:941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Birge RR, Knox BE. Perspectives on the counterion switch-induced photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci USA. 2003;100:9105–9107. doi: 10.1073/pnas.1733801100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jager S, Lewis JW, Zvyaga TA, Szundi I, Sakmar TP, Kliger DS. Time-resolved spectroscopy of the early photolysis intermediates of rhodopsin Schiff base counterion mutants. Biochemistry. 1997;36:1999–2009. doi: 10.1021/bi962320u. [DOI] [PubMed] [Google Scholar]
- 60.Sakmar TP, Franke RR, Khorana HG. The Role of the retinylidene Schiff-base counterion in rhodopsin in determining wavelength absorbency and Schiff-base pKa. Proc Natl Acad Sci USA. 1991;88:3079–3083. doi: 10.1073/pnas.88.8.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahmy K, Jager F, Beck M, Zvyaga TA, Sakmar TP, Siebert F. Protonation states of membrane-embedded carboxylic-acid groups in rhodopsin and metarhodopsin-Ii–a Fourier-transform infrared-spectroscopy study of site-directed mutants. Proc Natl Acad Sci USA. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Arnis S, Fahmy K, Hofmann KP, Sakmar TP. A conserved carboxylic-acid group mediates light-dependent proton uptake and signaling by rhodopsin. J Biol Chem. 1994;269:23879–23881. [PubMed] [Google Scholar]
- 63.Fahmy K, Siebert F, Sakmar TP. A Mutant rhodopsin photo-product with a protonated Schiff-base displays an active-state conformation–a Fourier-transform infrared-spectroscopy study. Biochemistry. 1994;33:13700–13705. doi: 10.1021/bi00250a021. [DOI] [PubMed] [Google Scholar]
- 64.Jager F, Fahmy K, Sakmar TP, Siebert F. Identification of glutamic-acid-113 as the Schiff-base proton acceptor in the meta-rhodopsin-Ii photointermediate of rhodopsin. Biochemistry. 1994;33:10878–10882. doi: 10.1021/bi00202a005. [DOI] [PubMed] [Google Scholar]
- 65.Kim JM, Altenbach C, Kono M, Oprian DD, Hubbell WL, Khorana HG. Structural origins of constitutive activation in rhodopsin: role of the K296/E113 salt bridge. Proc Natl Acad Sci USA. 2004;101:12508–12513. doi: 10.1073/pnas.0404519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan ECY, Kazmi MA, De S, Chang BSW, Seibert C, Marin EP, Mathies RA, Sakmar TP. Function of extracellular loop 2 in rhodopsin: glutamic acid 181 modulates stability and absorption wavelength of metarhodopsin II. Biochemistry. 2002;41:3620–3627. doi: 10.1021/bi0160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ludeke S, Beck R, Yan ECY, Sakmar TP, Siebert F, Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J Mol Biol. 2005;353:345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 68.Lewis JW, Szundi I, Fu WY, Sakmar TP, Kliger DS. pH Dependence of photolysis intermediates in the photoactivation of rhodopsin mutant E113Q. Biochemistry. 2000;39:599–606. doi: 10.1021/bi991860z. [DOI] [PubMed] [Google Scholar]
- 69.Schreiber M, Buss V, Sugihara M. Exploring the opsin shift with ab initio methods: geometry and counterion effects on the electronic spectrum of retinal. J Chem Phys. 2003;119:12045–12048. [Google Scholar]
- 70.Franke RR, Sakmar TP, Graham RM, Khorana HG. structure and function in rhodopsin—studies of the interaction between the rhodopsin cytoplasmic domain and transducin. J Biol Chem. 1992;267:14767–14774. [PubMed] [Google Scholar]
- 71.Fahmy K, Sakmar TP. Regulation of the rhodopsin transducin interaction by a highly conserved carboxylic-acid group. Biochemistry. 1993;32:7229–7236. doi: 10.1021/bi00079a020. [DOI] [PubMed] [Google Scholar]
- 72.Min KC, Zvyaga TA, Cypess AM, Sakmar TP. characterization of mutant rhodopsins responsible for autosomal-dominant retinitis-pigmentosa—mutations on the cytoplasmic surface affect transducin activation. J Biol Chem. 1993;268:9400–9404. [PubMed] [Google Scholar]
- 73.Han M, Lin SW, Minkova M, Smith SO, Sakmar TP. Functional interaction of transmembrane helices 3 and 6 in rhodopsin—replacement of phenylalanine 261 by alanine causes reversion of phenotype of a glycine 121 replacement mutant. J Biol Chem. 1996;271:32337–32342. doi: 10.1074/jbc.271.50.32337. [DOI] [PubMed] [Google Scholar]
- 74.Han M, Lin SW, Smith SO, Sakmar TP. The effects of amino acid replacements of glycine 121 on transmembrane helix 3 of rhodopsin. J Biol Chem. 1996;271:32330–32336. doi: 10.1074/jbc.271.50.32330. [DOI] [PubMed] [Google Scholar]
- 75.Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR. Rhodopsin activation blocked by metal-ion-binding sites linking transmembrane helices C and F. Nature. 1996;383:347–350. doi: 10.1038/383347a0. [DOI] [PubMed] [Google Scholar]
- 76.Abdulaev NG. Building a stage for interhelical play in rhodopsin. Trends Biochem Sci. 2003;28:399–402. doi: 10.1016/S0968-0004(03)00164-6. [DOI] [PubMed] [Google Scholar]
- 77.Franke RR, Sakmar TP, Oprian DD, Khorana HG. A single amino-acid substitution in rhodopsin (lysine-248-]leucine) prevents activation of transducin. J Biol Chem. 1988;263:2119–2122. [PubMed] [Google Scholar]
- 78.Ernst OP, Meyer CK, Marin EP, Henklein P, Fu WY, Sakmar TP, Hofmann KP. Mutation of the fourth cytoplasmic loop of rhodopsin affects binding of transducin and peptides derived from the carboxyl-terminal sequences of transducin α and γ subunits. J Biol Chem. 2000;275:1937–1943. doi: 10.1074/jbc.275.3.1937. [DOI] [PubMed] [Google Scholar]
- 79.Karnik SS, Sakmar TP, Chen HB, Khorana HG. Cysteine residue-110 and residue-187 are essential for the formation of correct structure in bovine rhodopsin. Proc Natl Acad Sci USA. 1988;85:8459–8463. doi: 10.1073/pnas.85.22.8459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shieh T, Han M, Sakmar TP, Smith SO. The steric trigger in rhodopsin activation. J Mol Biol. 1997;269:373–384. doi: 10.1006/jmbi.1997.1035. [DOI] [PubMed] [Google Scholar]
- 81.Yeagle PL, Albert AD. A conformational trigger for activation of a G protein by a G protein-coupled receptor. Biochemistry. 2003;42:1365–1368. doi: 10.1021/bi0270539. [DOI] [PubMed] [Google Scholar]
- 82.Spooner PJR, Sharples JM, Goodall SC, Bovee-Geurts PHM, Verhoeven MA, Lugtenburg J, Pistorius AMA, DeGrip WJ, Watts A. The ring of the rhodopsin chromophore in a hydrophobic activation switch within the binding pocket. J Mol Biol. 2004;343:719–730. doi: 10.1016/j.jmb.2004.08.049. [DOI] [PubMed] [Google Scholar]
- 83.Sondek J, Lambright DG, Noel JP, Hamm HE, Sigler PB. Gtpase mechanism of Gproteins from the 1.7-Å crystal-structure of transducin α-Center-Dot-Gdp-Center-Dot-Alf4(−) Nature. 1994;372:276–279. doi: 10.1038/372276a0. [DOI] [PubMed] [Google Scholar]
- 84.Lambright DG, Sondek J, Bohm A, Skiba NP, Hamm HE, Sigler PB. The 2.0 angstrom crystal structure of a heterotrimeric G protein. Nature. 1996;379:311–319. doi: 10.1038/379311a0. [DOI] [PubMed] [Google Scholar]
- 85.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein β γ dimer at 2.1 angstrom resolution. Nature. 1996;379:847–847. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 86.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G(A) protein β γ dimer at 2.1 angstrom resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 87.Filipek S, Krzysko KA, Fotiadis D, Liang Y, Saperstein DA, Engel A, Palczewski K. A concept for G protein activation by G protein-coupled receptor dimers: the transducin/rhodopsin interface. Photochem Photobiol Sci. 2004;3:628–638. doi: 10.1039/b315661c. [DOI] [PubMed] [Google Scholar]
- 88.Ling Y, Ascano M, Robinson P, Gregurick SK. Experimental and computational studies of the desensitization process in the bovine rhodopsin-arrestin complex. Biophys J. 2004;86:2445–2454. doi: 10.1016/S0006-3495(04)74301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zvyaga TA, Fahmy K, Sakmar TP. Characterization of rhodopsin-transducin interaction—a mutant rhodopsin photoproduct with a protonated Schiff-base activates transducin. Biochemistry. 1994;33:9753–9761. doi: 10.1021/bi00198a046. [DOI] [PubMed] [Google Scholar]
- 90.Ernst OP, Hofmann KP, Sakmar TP. Characterization of rhodopsin mutants that bind transducin but fail to induce Gtp nucleotide uptake—classification of mutant pigments by fluorescence nucleotide release, and flash-induced light-scattering assays. J Biol Chem. 1995;270:10580–10586. doi: 10.1074/jbc.270.18.10580. [DOI] [PubMed] [Google Scholar]
- 91.Yang K, Farrens DL, Hubbell WL, Khorana HG. Structure and function in rhodopsin. Single cysteine substitution mutants in the cytoplasmic interhelical E–F loop region show position-specific effects in transducin activation. Biochemistry. 1996;35:12464–12469. doi: 10.1021/bi960848t. [DOI] [PubMed] [Google Scholar]
- 92.Beck M, Sakmar TP, Siebert F. Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin. Biochemistry. 1998;37:7630–7639. doi: 10.1021/bi9801560. [DOI] [PubMed] [Google Scholar]
- 93.Han M, Smith SO, Sakmar TP. Constitutive activation of opsin by mutation of methionine 257 on transmembrane helix 6. Biochemistry. 1998;37:8253–8261. doi: 10.1021/bi980147r. [DOI] [PubMed] [Google Scholar]
- 94.Gurevich VV, Gurevich EV. The new face of active receptor bound minireview arrestin attracts new partners. Structure. 2003;11:1037–1042. doi: 10.1016/s0969-2126(03)00184-9. [DOI] [PubMed] [Google Scholar]
- 95.Smith WC, Dinculescu A, Peterson JJ, McDowell JH. The surface of visual arrestin that binds to rhodopsin. Mol Vision. 2004;10:392–398. [PubMed] [Google Scholar]
- 96.Marin EP, Krishna KG, Zvyaga TA, Isele J, Siebert F, Sakmar TP. The amino terminus of the fourth cytoplasmic loop of rhodopsin modulates rhodopsin-transducin interaction. J Biol Chem. 2000;275:1930–1936. doi: 10.1074/jbc.275.3.1930. [DOI] [PubMed] [Google Scholar]
- 97.Marin EP, Krishna AG, Archambault V, Simuni E, Fu WY, Sakmar TP. The function of interdomain interactions in controlling nucleotide exchange rates in transducin. J Biol Chem. 2001;276:23873–23880. doi: 10.1074/jbc.M101197200. [DOI] [PubMed] [Google Scholar]
- 98.Marin EP, Krishna AG, Sakmar TP. Disruption of the alpha 5 helix of transducin impairs rhodopsin-catalyzed nucleotide exchange. Biochemistry. 2002;41:6988–6994. doi: 10.1021/bi025514k. [DOI] [PubMed] [Google Scholar]
- 99.Raman D, Osawa S, Gurevich VV, Weiss ER. The interaction with the cytoplasmic loops of rhodopsin plays a crucial role in arrestin activation and binding. J Neurochem. 2003;84:1040–1050. doi: 10.1046/j.1471-4159.2003.01598.x. [DOI] [PubMed] [Google Scholar]
- 100.Han M, Groesbeek M, Sakmar TP, Smith SO. The C9 methyl group of retinal interacts with glycine-121 in rhodopsin. Proc Natl Acad Sci USA. 1997;94:13442–13447. doi: 10.1073/pnas.94.25.13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jager S, Han M, Lewis JW, Szundi I, Sakmar TP, Kliger DS. Properties of early photolysis intermediates of rhodopsin are affected by glycine 121 and phenylalanine 261. Biochemistry. 1997;36:11804–11810. doi: 10.1021/bi971122f. [DOI] [PubMed] [Google Scholar]
- 102.Lin SW, Kochendoerfer GG, Carroll HS, Wang D, Mathies RA, Sakmar TP. Mechanisms of spectral tuning in blue cone visual pigments—visible and Raman spectroscopy of blue-shifted rhodopsin mutants. J Biol Chem. 1998;273:24583–24591. doi: 10.1074/jbc.273.38.24583. [DOI] [PubMed] [Google Scholar]
- 103.Kochendoerfer GG, Lin SW, Sakmar TP, Mathies RA. How color visual pigments are tuned. Trends Biochem Sci. 1999;24:300–305. doi: 10.1016/s0968-0004(99)01432-2. [DOI] [PubMed] [Google Scholar]
- 104.Cowing JA, Poopalasundaram S, Wilkie SE, Robinson PR, Bowmaker JK, Hunt DM. The molecular mechanism for the spectral shifts between vertebrate ultraviolet-and violet-sensitive cone visual pigments. Biochem J. 2002;367:129–135. doi: 10.1042/BJ20020483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Janz JM, Farrens DL. Assessing structural elements that influence Schiff base stability: mutants E113Q and D190N destabilize rhodopsin through different mechanisms. Vision Res. 2003;43:2991–3002. doi: 10.1016/j.visres.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 106.Lewis JW, Szundi I, Kazmi MA, Sakmar TP, Kliger DS. Time-resolved photointermediate changes in rhodopsin glutamic acid 181 mutants. Biochemistry. 2004;43:12614–12621. doi: 10.1021/bi049581l. [DOI] [PubMed] [Google Scholar]
- 107.Patel AB, Crocker E, Eilers M, Hirshfeld A, Sheves M, Smith SO. Coupling of retinal isomerization to the activation of rhodopsin. Proc Natl Acad Sci USA. 2004;101:10048–10053. doi: 10.1073/pnas.0402848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rader AJ, Anderson G, Isin B, Khorana HG, Bahar I, Klein-Seetharaman J. Identification of core amino acids stabilizing rhodopsin. Proc Natl Acad Sci USA. 2004;101:7246–7251. doi: 10.1073/pnas.0401429101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Han M, Groesbeek M, Smith SO, Sakmar TP. Role of the C-9 methyl group in rhodopsin activation: characterization of mutant opsins with the artificial chromophore 11-cis-9-demethyl-retinal. Biochemistry. 1998;37:538–545. doi: 10.1021/bi972060w. [DOI] [PubMed] [Google Scholar]
- 110.Stecher H, Prezhdo O, Das J, Crouch RK, Palczewski K. Isomerization of all-trans-9-and 13-desmethylretinol by retinal pigment epithelial cells. Biochemistry. 1999;38:13542–13550. doi: 10.1021/bi9913294. [DOI] [PubMed] [Google Scholar]
- 111.Hirano T, Lim IT, Kim DM, Zheng XG, Yoshihara K, Oyama Y, Imai H, Shichida Y, Ishiguro M. Constraints of opsin structure on the ligand-binding site: studies with ring-fused retinals. Photochem Photobiol. 2002;76:606–615. doi: 10.1562/0031-8655(2002)076<0606:coosot>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 112.Das J, Crouch RK, Ma JX, Oprian DD, Kono M. Role of the 9-methyl group of retinal in cone visual pigments. Biochemistry. 2004;43:5532–5538. doi: 10.1021/bi036097u. [DOI] [PubMed] [Google Scholar]
- 113.Vogel R, Siebert F, Ludeke S, Hirshfeld A, Sheves M. Agonists and partial agonists of rhodopsin: retinals with ring modifications. Biochemistry. 2005;44:11684–11699. doi: 10.1021/bi0508587. [DOI] [PubMed] [Google Scholar]
- 114.Hunt DM, Dulai KS, Cowing JA, Julliot C, Mollon JD, Bowmaker JK, Li WH, Hewett-Emmett D. Molecular evolution of trichromacy in primates. Vision Res. 1998;38:3299–3306. doi: 10.1016/s0042-6989(97)00443-4. [DOI] [PubMed] [Google Scholar]
- 115.Chan T, Lee M, Sakmar TP. Introduction of hydroxyl-bearing amino-acids causes bathochromic spectral shifts in rhodopsin—amino-acid substitutions responsible for red-green color pigment spectral tuning. J Biol Chem. 1992;267:9478–9480. [PubMed] [Google Scholar]
- 116.Beck M, Siebert F, Sakmar TP. Evidence for the specific interaction of a lipid molecule with rhodopsin which is altered in the transition to the active state metarhodopsin II. FEBS Lett. 1998;436:304–308. doi: 10.1016/s0014-5793(98)01156-9. [DOI] [PubMed] [Google Scholar]
- 117.Isele J, Sakmar TP, Siebert F. Rhodopsin activation affects the environment of specific neighboring phospholipids: an FTIR spectroscopic study. Biophys J. 2000;79:3063–3071. doi: 10.1016/S0006-3495(00)76541-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Litman BJ, Niu SL, Polozova A, Mitchell DC. The role of docosahexaenoic acid containing phospholipids in modulating G protein-coupled signaling pathways—Visual transduction. J Mol Neurosci. 2001;16:237–242. doi: 10.1385/JMN:16:2-3:237. [DOI] [PubMed] [Google Scholar]
- 119.Mitchell DC, Niu SL, Litman BJ. Optimization of receptor-G protein coupling by bilayer lipid composition I—kinetics of rhodopsin-transducin binding. J Biol Chem. 2001;276:42801–42806. doi: 10.1074/jbc.M105772200. [DOI] [PubMed] [Google Scholar]
- 120.Niu SL, Mitchell DC, Litman BJ. Optimization of receptor-G protein coupling by bilayer lipid composition II—formation of metarhodopsin II-transducin complex. J Biol Chem. 2001;276:42807–42811. doi: 10.1074/jbc.M105778200. [DOI] [PubMed] [Google Scholar]
- 121.Feller SE, Gawrisch K, Woolf TB. Rhodopsin exhibits a preference for solvation by polyunsaturated docosohexaenoic acid. J Am Chem Soc. 2003;125:4434–4435. doi: 10.1021/ja0345874. [DOI] [PubMed] [Google Scholar]
- 122.Mitchell DC, Niu SL, Litman BJ. DHA-rich phospholipids optimize G-protein-coupled signaling. J Pediatr. 2003;143:S80–S86. doi: 10.1067/s0022-3476(03)00405-0. [DOI] [PubMed] [Google Scholar]
- 123.Mitchell DC, Niu SL, Litman BJ. Enhancement of G protein-coupled signaling by DHA phospholipids. Lipids. 2003;38:437–443. doi: 10.1007/s11745-003-1081-1. [DOI] [PubMed] [Google Scholar]
- 124.Grossfield AF, Feller SE, Pitman MC. A role for the direct interactions in the modulation of rhodopsin by ω-3 polyunsaturated lipids. Proc Natl Acad Sci USA. 2006;103:4888–4893. doi: 10.1073/pnas.0508352103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Feller SE, Gawrisch K, MacKerell AD. Polyunsaturated fatty acids in lipid bilayers: intrinsic and environmental contributions to their unique physical properties. J Am Chem Soc. 2002;124:318–326. doi: 10.1021/ja0118340. [DOI] [PubMed] [Google Scholar]
- 126.Gerken U, Jelezko F, Gotze B, Branschadel M, Tietz C, Ghosh R, Wrachtrup J. Membrane environment reduces the accessible conformational space available to an integral membrane protein. J Phys Chem B. 2003;107:338–343. [Google Scholar]
- 127.Carrillo-Tripp M, Feller SE. Evidence for a mechanism by which ω-3 polyunsaturated lipids may affect membrane protein function. Biochemistry. 2005;44:10164–10169. doi: 10.1021/bi050822e. [DOI] [PubMed] [Google Scholar]
- 128.Sakmar TP. Structure of rhodopsin and the superfamily of seven-helical receptors: the same and not the same. Curr Opin Cell Biol. 2002;14:189–195. doi: 10.1016/s0955-0674(02)00306-x. [DOI] [PubMed] [Google Scholar]
- 129.Archer E, Maigret B, Escrieut C, Pradayrol L, Fourmy D. Rhodopsin crystal: new template yielding realistic models of G-protein-coupled receptors. Trends Pharmacol Sci. 2003;24:36–40. doi: 10.1016/s0165-6147(02)00009-3. [DOI] [PubMed] [Google Scholar]
- 130.Mirzadegan T, Benko G, Filipek S, Palczewski K. Sequence analyses of G-protein-coupled receptors: similarities to rhodopsin. Biochemistry. 2003;42:2759–2767. doi: 10.1021/bi027224+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ballesteros JA, Shi L, Javitch JA. Structural mimicry in G protein-coupled receptors: implications of the high-resolution structure of rhodopsin for structure-function analysis of rhodopsin-like receptors. Mol Pharmacol. 2001;60:1–19. [PubMed] [Google Scholar]
- 132.Becker OM, Shacham S, Marantz Y, Noiman S. Modeling the 3D structure of GPCRs: advances and application to drug discovery. Curr Opin Drug Discov Dev. 2003;6:353–361. [PubMed] [Google Scholar]
- 133.Bissantz C, Bernard P, Hibert M, Rognan D. Protein-based virtual screening of chemical databases. II. Are homology models of G-protein coupled receptors suitable targets? Proteins: Struct Funct Genet. 2003;50:5–25. doi: 10.1002/prot.10237. [DOI] [PubMed] [Google Scholar]
- 134.Cavasotto CN, Orry AJW, Abagyan RA. Structure-based identification of binding sites, native ligands and potential inhibitors for G-protein coupled receptors. Proteins: Struct Funct Genet. 2003;51:423–433. doi: 10.1002/prot.10362. [DOI] [PubMed] [Google Scholar]
- 135.Filipek S, Teller DC, Palczewski K, Stenkamp R. The crystallographic model of rhodopsin and its use in studies of other G protein-coupled receptors. Annu Rev Biophys Biomol Struct. 2003;32:375–397. doi: 10.1146/annurev.biophys.32.110601.142520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mosberg HI, Fowler CB. Do GPCR models derived by homology from the rhodopsin X-ray structure correctly predict helix irregularities? Biopolymers. 2003;71:387–387. [Google Scholar]
- 137.Bartfai T, Benovic JL, Bockaert J, Bond RA, Bouvier M, Christopoulos A, Civelli O, Devi LA, George SR, Inui A, Kobilka B, Leurs R, Neubig R, Pin JP, Quirion R, Roques BP, Sakmar TP, Seifert R, Stenkamp RE, Strange PG. The state of GPCR research in 2004. Nat Rev Drug Discov. 2004;3:574–626. [Google Scholar]
- 138.Becker OM, Marantz Y, Shacham S, Inbal B, Heifetz A, Kalid O, Bar-Haim S, Warshaviak D, Fichman M, Noiman S. G protein-coupled receptors: in silico drug discovery in 3D. Proc Natl Acad Sci USA. 2004;101:11304–11309. doi: 10.1073/pnas.0401862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hillisch A, Pineda LF, Hilgenfeld R. Utility of homology models in the drug discovery process. Drug Discov Today. 2004;9:659–669. doi: 10.1016/S1359-6446(04)03196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oliveira L, Hulsen T, Hulsik DL, Paiva ACM, Vriend G. Heavier-than-air flying machines are impossible. FEBS Lett. 2004;564:269–273. doi: 10.1016/S0014-5793(04)00320-5. [DOI] [PubMed] [Google Scholar]
- 141.Yohannan S, Faham S, Yang D, Whitelegge JP, Bowie JU. The evolution of transmembrane helix kinks and the structural diversity of G protein-coupled receptors. Proc Natl Acad Sci USA. 2004;101:959–963. doi: 10.1073/pnas.0306077101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bosch L, Iarriccio L, Garriga P. New prospects for drug discovery from structural studies of rhodopsin. Curr Pharm Des. 2005;11:2243–2256. doi: 10.2174/1381612054367436. [DOI] [PubMed] [Google Scholar]
- 143.Bissantz C. Conformational changes of G protein-coupled receptors during their activation by agonist binding. J Recept Signal Transduct Res. 2003;23:123–153. doi: 10.1081/rrs-120025192. [DOI] [PubMed] [Google Scholar]
- 144.Mehler EL, Periole X, Hassan SA, Weinstein H. Key issues in the computational simulation of GPCR function: representation of loop domains. J Comput Aided Mol Des. 2002;16:841–853. doi: 10.1023/a:1023845015343. [DOI] [PubMed] [Google Scholar]
- 145.Forrest LR, Woolf TB. Discrimination of native loop conformations in membrane proteins: decoy library design and evaluation of effective energy scoring functions. Proteins: Struct Funct Genet. 2003;52:492–509. doi: 10.1002/prot.10404. [DOI] [PubMed] [Google Scholar]
- 146.Lawson Z, Wheatley M. The third extracellular loop of G-protein-coupled receptors: more than just a linker between two important transmembrane helices. Biochem Soc Trans. 2004;32:1048–1050. doi: 10.1042/BST0321048. [DOI] [PubMed] [Google Scholar]
- 147.Nikiforovich GV, Marshall GR. Modeling flexible loops in the dark-adapted and activated states of rhodopsin, a prototypical G-protein-coupled receptor. Biophys J. 2005;89:3780–3789. doi: 10.1529/biophysj.105.070722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 149.Okada T, Palczewski K. Crystal structure of rhodopsin: implications for vision and beyond. Curr Opin Struct Biol. 2001;11:420–426. doi: 10.1016/s0959-440x(00)00227-x. [DOI] [PubMed] [Google Scholar]
- 150.Teller DC, Okada T, Behnke CA, Palczewski K, Stenkamp RE. Advances in determination of a high-resolution three-dimensional structure of rhodopsin, a model of G-protein-coupled receptors (GPCRs) Biochemistry. 2001;40:7761–7772. doi: 10.1021/bi0155091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stenkamp RE, Filipek S, Driessen C, Teller DC, Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim Biophys Acta Biomembr. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 152.Fishkin N, Berova N, Nakanishi K. Primary events in dim light vision: a chemical and spectroscopic approach toward understanding protein/chromophore interactions in rhodopsin. Chem Rec. 2004;4:120–135. doi: 10.1002/tcr.20000. [DOI] [PubMed] [Google Scholar]
- 153.Liu RSH, Colmenares LU. The molecular basis for the high photosensitivity of rhodopsin. Proc Natl Acad Sci USA. 2003;100:14639–14644. doi: 10.1073/pnas.2536769100. [DOI] [PMC free article] [PubMed] [Google Scholar]