SUMMARY
FANCM is a Fanconi anemia nuclear core complex protein required for the functional integrity of the FANC-BRCA pathway of DNA damage response and repair. Here we report the isolation and characterization of two histone-fold-containing FANCM-associated proteins, MHF1 and MHF2. We show that suppression of MHF1 expression results in 1) destabilization of FANCM and MHF2, 2) impairment of DNA damage-induced monoubiquitination and foci formation of FANCD2, 3) defective chromatin localization of FA nuclear core complex proteins, 4) elevated MMC-induced chromosome aberrations, and 5) sensitivity to MMC and camptothecin. We also provide biochemical evidence that MHF1 and MHF2 assemble into a heterodimer that binds DNA and enhances the DNA branch migration activity of FANCM. These findings reveal critical roles of the MHF1-MHF2 dimer in DNA damage repair and genome maintenance through FANCM.
HIGHLIGHTS
MHF1 and MHF2 are histone-fold-containing components of the FA core complex
MHF1 is essential for the stability and chromatin association of FANCM
MHF1 is required for the normal function of the FA pathway
MHF1-MHF2 form a complex that binds DNA and stimulates the branch migration activity of FANCM
INTRODUCTION
Fanconi Anemia (FA) is characterized by developmental defects, bone marrow failure and cancers (de Winter and Joenje, 2009). FA cells exhibit hypersensitivity to DNA interstrand cross-linking (ICL) agents and increased chromosomal breaks, hallmark features that serve as a diagnostic test for the disease. FA is genetically heterogeneous, comprising thirteen complementation groups; the genes mutated in these groups have been identified (de Winter and Joenje, 2009). Eight of the FA proteins (FANCA, -B,-C, -E, -F, -G, -L and -M) and three associated factors (FAAP100, FAAP24 and HES1) form the FA nuclear core complex that is required for the monoubiquitination of the FANCD2-FANCI dimer upon DNA damage to result in the activation of downstream DNA damage repair and tolerance reactions (Ali et al., 2009b; de Winter and Joenje, 2009; Thompson and Hinz, 2009). Until the discovery of FANCM, the mechanism by which the FA pathway protects cells from ICL and other DNA damaging agents remains largely undefined, partly because the majority of the FA core complex proteins lack recognizable functional domains that may provide clues into their biological roles (Ali et al., 2009b). The discovery that FANCM possesses a helicase domain (Meetei et al., 2005) and a DNA translocase activity capable of processing branched DNA intermediates (Gari et al., 2008b) represent a notable advancement in delineating FA core complex functions.
FANCM is a large protein of 250 kDa and related to the archaeal DNA helicase/nuclease HEF (Meetei et al., 2005), S. pombe Fml1 protein (Sun et al, 2008), and S. cerevisiae Mph1 protein (Prakash et al., 2009). FANCM exhibits homology to the superfamily 2 (SF2) DEAH-box of helicases and also to the endonuclease domain of ERCC4 (XPF) (Ali et al., 2009b; Meetei et al., 2005). Several interacting partners of FANCM have been reported. HCLK2 interacts with FANCM in DNA damage checkpoint activation (Collis et al., 2008). The C-terminal nuclease-like domain of FANCM interacts with FAAP24, and the complex of the FANCM C-terminus and FAAP24 binds various types of DNA, including replication fork-like structures. FAAP24 knockdown abolishes FANCD2 monoubiquitination and engenders sensitivity to ICL agents (Ciccia et al., 2007). It has been suggested that FANCM, in complex with FAAP24, serves to recruit the FA core complex to chromatin to activate FANCD2 monoubiquitination (Ciccia et al., 2007; Kim et al., 2008). FANCM translocates on dsDNA and can unwind the D-loop and also process branched DNA structures, such as model DNA replication forks and the Holliday junction, by DNA branch migration (Gari et al., 2008a; Gari et al., 2008b; Prakash et al., 2009; Sun et al., 2008). Importantly, genetic studies in yeast have shown that these orthologues promote replication restart and suppress mitotic crossovers (Prakash et al., 2009; Sun et al., 2008).
Our attempt to better define the function of the FANCM protein has led us to identify two associated proteins, termed MHF1 and MHF2. We provide evidence that MHF1 and MHF2 form a heterodimer that binds DNA, enhances the DNA branch migration activity of FANCM, and is indispensable for the functional integrity of the FA pathway. These results uncover the biological role of the MHF1-MHF2 dimer and reveal additional complexity of the FANC-BRCA pathway of DNA damage response and repair.
RESULTS AND DISCUSSION
MHF1 and MHF2 are histone-fold-containing components of the FA core complex
A two-step affinity purification of N-terminally (His)6-FLAG-tagged FANCM coupled with mass spectrometry (MS) was used to identify FANCM-associated proteins (Singh et al., 2008). Known binding partners of FANCM, including components of the FA core complex, were found. Interestingly, we also identified polypeptides of 16 kDa and 10 kDa, named FAAP16 and FAAP10 (Fanconi Anemia Associated Polypeptide 16 kDa and 10 kDa), respectively (Fig. 1A). We renamed FAAP16 and FAAP10 to MHF1 (FANCM interacting histone-fold protein 1) and MHF2 (FANCM interacting histone-fold protein 2) respectively. Based on the band intensity, it appears that MHF1 and MHF2 are stoichiometric components of the FANCM complex (Fig. 1A). Database search revealed that MHF1 (Ref Seq Accession No. NP_954988.1) is identical to the previously described apoptosis-inducing, TAF9-like domain 1 isoform A (APITD1) protein, a candidate neuroblastoma tumor suppressor (Krona et al., 2004). MHF1 has also been identified as a component of the CENPA-CAD complex and named centromere protein S (CENPS) (Foltz et al., 2006). The chicken ortholog of human MHF2 (UniProt Accession No. A8MT69.1) is recently identified as a binding partner of CENP-S and named as CENP-X (Amano et al., 2009). The CENP-S and CENP-X complex is essential for the stable assembly of outer kinetochore structure in DT40 cells (Amano et al., 2009). BLAST analysis identified likely orthologs of MHF1 and MHF2 in other organisms (Fig. S1A & S1B). Sequence analysis using InterProScan revealed a putative histone-fold domain (IPR009072) in both MHF proteins. The Histone-fold is a conserved protein-dimerization motif present in all histone and several non-histone proteins (Arents and Moudrianakis, 1995), and is characterized by a core that consists of three helices, of which the middle helix is longer than the flanking ones (Arents and Moudrianakis, 1995). The predicted triple helical regions in the putative histone-fold domain of MHF1 and MHF2 are shown in Fig. S1A & S1B.
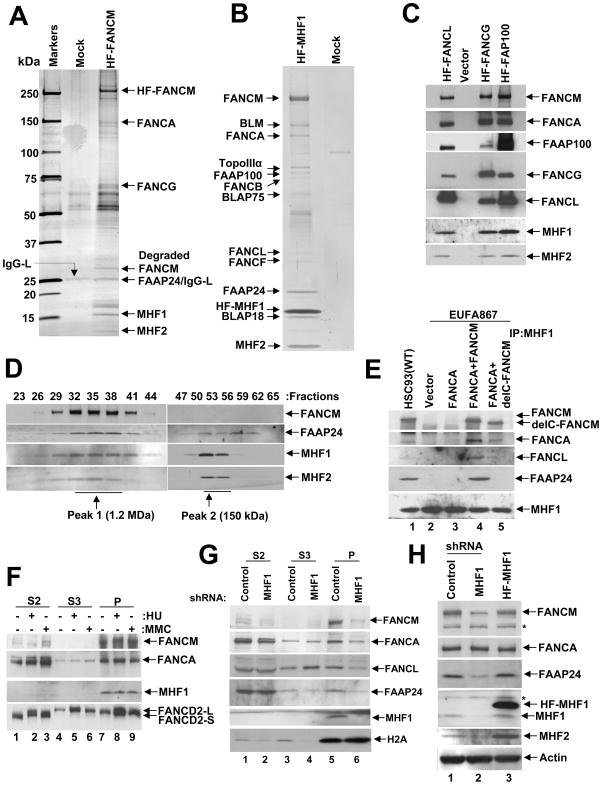
Figure 1. MHF1 and MHF2 are components of the FA core complex.
(A) Silver-stained gel showing the polypeptide bands purified from the nuclear extract of HT1080 cells transduced with vector alone (Mock) and cells expressing (His)6-FLAG-tagged (HF) FANCM. The polypeptides identified by MS analysis are indicated by an arrow. (B) A silver-stained gel showing the BRAFT complex components isolated from HeLa S3 cells expressing HF-MHF1. See also Fig. S1C and S1D. (C) Immunoblot showing the presence of MHF1 and MHF2 in immunoprecipitates of core complex proteins FANCL, FANCG and FAAP100. (D) Immunoblot showing co-fractionation of FAAP24, MHF1 and MHF2 with FANCM by Superose 6 gel filtration. (E) Immunoblot showing co-purification of FA core complex components with MHF1 from EUFA867 cells transduced with vector alone (lane 2), FANCA (lane 3), FANCA and FANCM (lane 4) and FANCA and delta (C-terminus deleted) FANCM (lane 5). (F) HeLa cells were treated with MMC (100 ng/mL for 16 hours) or HU (2 mM for 16 hours) and then fractionated into S2, S3, and P, as described in Material and Methods and immunoblotted with the indicated antibodies. (G) HeLa cells were transduced with control or MHF1 shRNAs, followed by cellular fractionation into S2, S3, and P fractions. The relevant proteins were revealed by immunoblotting. (H) HeLa cells were stably transduced with control-shRNA, MHF1-shRNA or HF-MHF1, and whole-cell lysates were analyzed by immunoblotting with the indicated antibodies. Asterisk (*) denotes a nonspecific cross-reactive band. See also Fig. S1.
When we immunoprecipitated (His)6-FLAG-tagged MHF1 or FLAG-tagged MHF2 using anti-FLAG antibodies, we found the presence of other FA core complex proteins (Fig. S1C & S1D). Likewise, affinity purification of the tagged MHF1 coupled with MS analysis from extracts of HeLa S3 cells revealed its association with MHF2, FANCM and other components of the BRAFT complex that includes the FA proteins (Fig. 1B). In a reciprocal experiment, when we immunoprecipitated several other (His)6-FLAG-tagged FA core complex proteins, FANCL, FANCG or FAAP100 we again found associated MHF1 and MHF2 (Fig. 1C). These results thus identify MHF1 and MHF2 as novel components of the FA core complex.
Next, gel filtration in Superose-6 was used to examine the co-fractionation of FANCM, FAAP24, MHF1 and MHF2. The analysis revealed the co-elution of MHF1 and MHF2 in two separate peaks of average molecular masses of 1.2 MDa (peak 1) and 150 kDa (peak 2), respectively (Fig. 1D). FANCM co-fractionated precisely with MHF1 and MHF2 in peak 1 but was absent in peak 2 (Fig. 1D). As expected, FAAP24 was also found in peak 1 with FANCM and MHF1 and MHF2.
To test the role of FANCM in MHF1’s association with the FA core complex, immunoprecipitation of endogenous MHF1 from EUFA867 (FANCA and FANCM double mutant) and EUFA867-FANCA (deficient in FANCM only) was carried out. MHF1 failed to co-immunoprecipitate other FA core complex proteins from extracts of EUFA867 cells (Fig. 1E, lanes 2, 3), while ectopic expression of either full-length FANCM or C-terminally deleted FANCM restores the association of MHF1 with the FA core complex (Fig. 1E, lanes 4, 5). FAAP24, which binds the C-terminal region of FANCM, was found associated with MHF1 only upon expression of full-length FANCM (Fig. 1E, lane 4). These results suggest that MHF1 is recruited to the FA core complex by FANCM.
Reliance of chromatin association of FANCM and the FA core complex on MHF1
Immunoprecipitation of endogenous MHF1 using anti MHF1 antibody co-depleted FANCM and a significant amount of FAAP24 from the flow-through fraction (Fig. S1E). This association did not change following DNA damage with mitomycin C (MMC) or replication stress with hydroxyurea (HU) (Fig. S1E). Thus, FANCM and MHF1 constitutively associate with each other in vivo. Cellular fractionation was carried out with HeLa cells treated with HU or MMC or left untreated to examine the localization of MHF1 (Kim et al., 2008; Singh et al., 2009). Consistent with previously published results (Kim et al., 2008), FANCM was exclusively detected in the chromatin (P; pellet) fraction under all conditions (Fig. 1F). Monoubiquitinated FANCD2 was found primarily in the P fraction, as previously described (Montes de Oca et al., 2005), while FANCA was distributed among the S2, S3 and P fractions. Interestingly, MHF1 was found only in the P fraction, suggesting that FANCM and MHF1 are localized in the chromatin (Fig. 1F, lanes 7–9).
Since FANCM is required for chromatin association of the FA core complex (Kim et al., 2008), we asked whether MHF1 is also important in this regard. We examined the cellular localization of the FA core complex in HeLa cells transduced with MHF1 shRNA or control shRNA (Fig. 1G). Depletion of MHF1 greatly reduced chromatin-associated FANCM and FAAP24 (Fig. 1G, lanes 5 & 6), and also affected the chromatin localization of FANCA and FANCL (Fig. 1G, lanes 5 & 6). These results thus indicate that MHF1 is needed for optimal chromatin association of the FA core complex.
Stability of FANCM, MHF2 and FAAP24 is affected by MHF1
The absence of a subunit of a multi-component complex often destabilizes the entire complex (Singh et al., 2008). Indeed, FANCM and MHF2 are rendered unstable upon MHF1 depletion, while FAAP24 was only moderately destabilized. No change in the level of FANCA expression was seen (Fig. 1H, lane 2). Over-expression of MHF1 led to an increase in the MHF2 level without affecting the FANCM or FAAP24 level (Fig. 1H, lane 3). Neither the stability nor the interaction of MHF1 and MHF2 is affected by FANCM deficiency (Fig. S1F). These results again indicate an association of MHF1 with FANCM, FAAP24 and MHF2 in a higher order complex.
Dependence of the FA pathway on MHF1
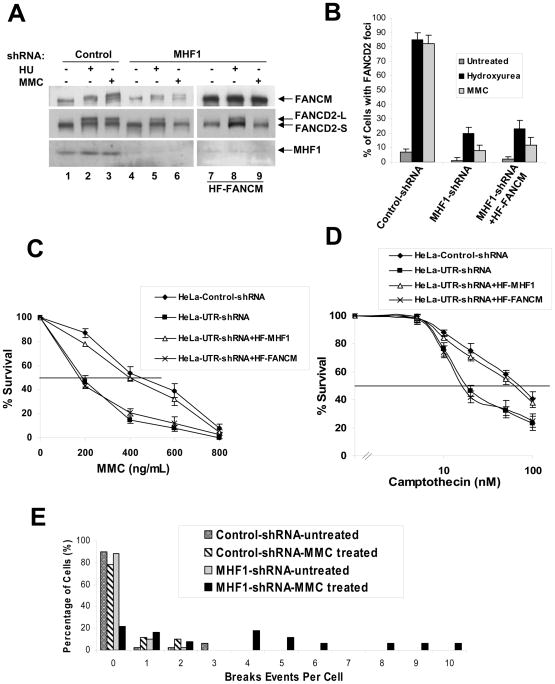
FANCD2 monoubiquitination upon MMC treatment or DNA replication stress requires the FA core complex (de Winter and Joenje, 2009). We wished to determine whether FANCD2 monoubiquitination is similarly dependent on MHF1. As expected, in cells transduced with control shRNA, FANCD2 monoubiquitination occurred normally after MMC or HU treatment (Fig 2A, lanes 2 and 3). In contrast, cells transduced with MHF1 shRNA became impaired for FANCD2 monoubiquitination (Fig. 2A, lanes 5 and 6). Just as previously described for FANCM-deficient cells, MHF1-depleted cells are more severely impaired in FANCD2 monoubiquitination after MMC treatment than after HU treatment (Bakker et al., 2009; Singh et al., 2009). Monoubiquitination of FANCD2 is known to target this protein to repair foci. In control cells, HU or MMC treatment resulted in a dramatic induction in FANCD2 focus formation (Fig. 2B), whereas MHF1 depletion compromised focus formation (Fig. 2B and Fig. S2A). Importantly, cell-cycle analysis indicated that the defect in damage-induced activation of FANCD2 in the absence MHF1 does not result from accumulation of cells at a cell-cycle stage incompatible with FANCD2 monoubiquitination or focus formation (data not shown). Thus, MHF1 is required for efficient damage-induced monoubiquitination and focus formation of FANCD2.
Figure 2. MHF1 is required for the activation of the FA pathway.
(A) Immunoblot showing that ablation of MHF1 expression in HeLa cells by shRNA reduces the level of monoubiquitinated FANCD2 in cells treated with MMC or HU. MHF1 knock down HeLa cells stably overexpressing HF-FANCM were used to show that phenotype observed is not due to the reduced levels of FANCM. (B) Histogram of immunofluorescence analysis of FANCD2 nuclear foci in HeLa cells in which MHF1 expression was knocked down by shRNA. Cells were either left untreated or exposed to MMC or HU, and the percentage of cells with 5 or more FANCD2 foci was determined by examining at least 150 cells. The result shows the average of 3 independent experiments with standard deviations. See also Fig. S2A. (C) & (D) Graph showing MMC and camptothecin survival curve of MHF1 knockdown cells. HeLa cells were transduced with either control or shRNA oligos targeting MHF1 and were subsequently treated with the indicated concentration of MMC and camptothecin. Visible colonies from 200 cells were counted after 10 days. The data represent the percent survival, as compared with untreated cells. Each experiment was performed in triplicate, and mean values are shown with standard deviations. HeLa cells stably expressing shRNA resistant HF-MHF1 were used as a control to show that the phenotype observed is not due to the off-target effect of shRNA. Also, MHF1 knock down HeLa cells stably overexpressing HF-FANCM were used to show that phenotype observed is not due to the reduced levels of FANCM. (E) Chromosomal aberrations following MMC treatment. HeLa cells were transfected with siRNAs and cells were treated with MMC. Metaphase spreads were prepared and scored for chromosomal aberrations. See also Fig. S2E.
Defective FANCD2 monoubiquitination has been previously associated with hypersensitivity to DNA crosslinking agents (de Winter and Joenje, 2009). Specifically, FANCM-deficient cells have been shown to be sensitive to both MMC, an ICL agent, and camptothecin, a topoisomerase I inhibitor (Rosado et al., 2009; Singh et al., 2009). We observed that HeLa cells transduced with MHF1 shRNA are 2 to 3-fold more sensitive to MMC (Fig. 2C) and 3- to 4-fold more sensitive to Camptothecin than the control (Fig. 2D); this drug sensitivity was similar to that engendered by FANCM deficiency (Bakker et al., 2009; Singh et al., 2009). Furthermore, the impaired FANCD2 monoubiquitination (Fig. S2C), MMC (Fig. 2C) and camptothecin sensitivities (Fig. 2D) can be rescued by expressing MHF1 that is resistant to shRNA knockdown suggesting that the observed defects were specific to MHF1 depletion alone and not an off target effect by shRNAs. Also, in order to address the question whether the observed defects viz., impaired FANCD2 monoubiquitination, foci formation and sensitivity to MMC or camptothecin is a direct result of MHF1 knockdown or an indirect effect of the reduced FANCM in MHF1 knockdown cells, we forced overexpressed FANCM in MHF1 knockdown cells to overcome the reduced levels of FANCM in these cells (Fig. S2D). MHF1 knockdown cells overexpressing FANCM still showed impaired FANCD2 monoubiquitination (Fig. 2A, lanes 7–9), reduced foci formation (Fig 2B) and increased sensitivity to MMC (Fig 2C) or camptothecin (Fig 2D) suggesting that the defect were specific to MHF1 and not an indirect effect of FANCM destabilization. Interestingly, overexpression of FANCM in MHF1 knockdown cells also restored the FAAP24 level, suggesting that the instability of the FAAP24 in MHF1 knockdown cells were an indirect effect of FANCM instability (Fig. S2D). Taken together, these data indicate that MHF1 plays a role in ICL repair and in the recovery of replication forks stalled by topoisomerase I-DNA cleavage intermediates induced by camptothecin.
Genomic instability is the hallmark feature of FA cells. FANCM-deleted chicken, mouse and human cells show elevated chromosome aberrations. Accordingly, we tested the MHF1-depleted cells for chromosome aberrations. Indeed MHF1 depletion led to MMC-induced chromosomal instability (Fig. 2E). Even though FANCM-deleted chicken and mouse cells show elevated sister chromatid exchanges (Bakker et al., 2009; Mosedale et al., 2005), FA-M cells or HeLa cells depleted of MHF1 by shRNA do not exhibit this phenotype (Fig. S2E).
MHF1-MHF2 dimer binds DNA
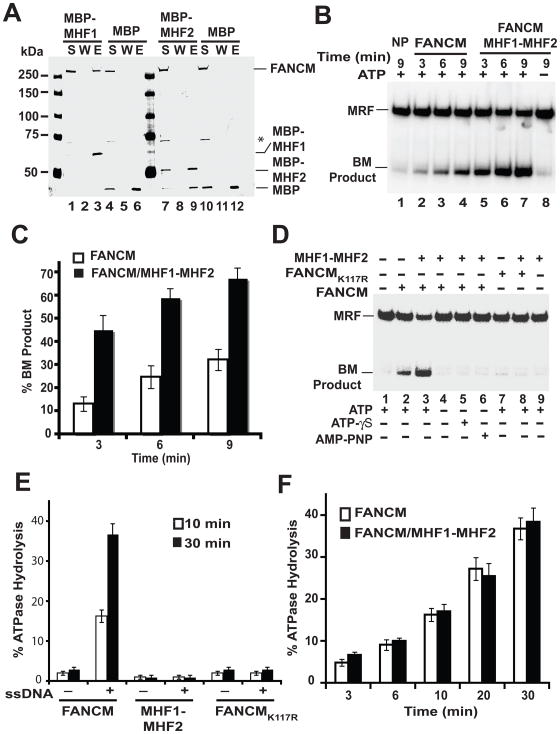
We expressed MHF1 and MHF2 proteins tagged with the maltose binding protein (MBP), and the MHF1-MHF2 complex harboring a GST tag on MHF1 and a (His)6 tag on MHF2 in E. coli and devised procedures for their purification to near homogeneity (Fig. 3A). During MHF1-MHF2 purification, the GST tag was cleaved off the MHF1 subunit. Little or no dissociation of the MHF1-MHF2 complex occurred throughout purification, indicating a high degree of stability of the complex.
Figure 3. DNA binding by the MHF1-MHF2 heterodimer.
(A) Recombinant MHF1-(His)6-MHF2, MBP-MHF1, and MBP-MHF2 were expressed in and purified from E. coli. Purified proteins were analyzed by SDS-PAGE followed by Coomassie blue staining. (B) MHF1-(His)6-MHF2, MBP-MHF1 (1.0 and 2.0 μM), and MBP-MHF2 (1.0 and 2.0 μM) were examined for their DNA binding attribute with a mixture of the four 32P-labeled DNA substrates. (C). The indicated combinations of DNA substrates were incubated with MHF1-(His)6-MHF2 (50, 85, 120, 155, 190, 225, 270 and 305 nM). Quantification of the data is presented in the right panels. Note that treatment of nucleoprotein complexes with SDS and proteinase K (SDS/PK) released the DNA substrates. See also Fig. S3.
We used mobility shift of radiolabeled DNA species - single-stranded (ss), double-stranded (ds), splay arm (SA), and Holliday junction (HJ) in polyacrylamide gels to query whether the purified MHF proteins and complex possess DNA binding activity. We found that neither MBP-MHF1 nor MBP-MHF2 has an appreciable affinity for any of DNA substrates tested (data not shown). In contrast, MHF1-MHF2 heterodimer bound all the DNA substrates (Fig. 3B). We note that the lack of DNA binding in MBP-MHF1 and MBP-MHF2 did not stem from the presence of the MBP tag, as DNA binding activity is seen upon mixing the two MBP-tagged MHF proteins (D.S. and X.F.Z, unpublished data). These data suggest that MHF1 and MHF2 proteins exist and function only as a dimer in DNA binding. Our finding is consistent with the observation that many proteins with histone-fold motifs, such as transcription activating factors (TAFs), function as heterodimers (Arents and Moudrianakis, 1995).
The relative affinities of MHF1-MHF2 toward different DNA substrates were determined by incubating varying amounts of the protein complex with pairs of the substrates (Fig. 3C). These analyses, conducted at 100 mM KCl, revealed the following order of DNA substrate preference: HJ > ds > SA > ss (Fig. 3C). At a higher reaction stringency (150 mM KCl), MHF1-MHF2 showed a clear preference towards HJ over ds (Fig. S3A).
MHF1, but not MHF2, interacts with FANCM
We used affinity pull-down assay with amylose beads (specific for the MBP tag on the MHF proteins) to check for interaction of FANCM with MHF1 and MHF2 proteins. Purified FANCM was retained on the affinity matrix via MHF1 (Fig. 4A, lane 3), but little or no association with MHF2 occurred (Fig. 4A, lane 9). As expected, FANCM did not bind to the amylose beads with MBP present (Fig. 4A, lanes 6 and 12). These results indicate that FANCM interacts with MHF1 but not MHF2.
Figure 4. Enhancement of DNA branch migration by MHF1-MHF2 heterodimer.
(A) Amylose beads pull-down assay with FANCM and MBP-MHF1 or MBP-MHF2. S, W, and E represent supernatant, wash and elution lanes. The asterisk (*) denotes a proteolytic product of FANCM. (B) Stimulation of the branch migration activity of FANCM by the MHF1-MHF2 dimer. ATP was present in all the reactions except the one in lane 8. NP - no protein control; BM, branch migration. (C) Quantification of the results shown in figure 4B. (D) DNA branch migration by FANCM, FANCMK117R, or the combination of FANCM or FANCMK117R and MHF1-MHF2 was examined with or without ATP, ATP-γ-S, or AMP-PNP, as indicated. BM, branch migration. (E) ATP hydrolysis by FANCM, MHF1-MHF2 or FANCMK117R was examined with or without ssDNA, as indicated. (F) Effect of MHF1-MHF2 on the ssDNA-dependent ATPase activity of FANCM. See also Fig. S3 and S4.
Stimulation of FANCM DNA Branch Migration activity by MHF1-MHF2
FANCM has a DNA branch migration activity that can process branched DNA structures such as a movable replication fork (MRF) (Gari et al., 2008b). We tested the effect of MHF1-MHF2 on this activity of FANCM by incubating a radiolabeled MRF with either FANCM alone or FANCM with MHF1-MHF2, and analyzed the reaction mixtures by polyacrylamide gel electrophoresis under non-denaturing conditions and phosphorimaging (Fig. 4B). As expected (Gari et al., 2008b), FANCM alone could process the MRF to yield a branch migration product (Fig. 4C). Importantly, the MHF1-MHF2 dimer, while being devoid of DNA branch migration activity on its own (Fig. 4D, lane 9) had a strong stimulatory effect on this FANCM attribute; a 3.5 fold enhancement was seen at the earliest time point of 3 min. (Fig. 4C). Branch migration by the FANCM-MHF1-MHF2 ensemble is completely dependent on ATP (Fig. 4B), which cannot be substituted by either of the non-hydrolyzable ATP analogues ATP-γ-S and AMP-PNP (Fig. 4D). Consistent with these observations, the ATPase deficient FANCMK117R mutant did not display any DNA branch migration activity even with MHF1-MHF2 present (Fig. 4D & 4E). We note that neither of the purified MHF proteins alone has any DNA branch migration activity (Fig. S3B) nor could they stimulate FANCM’s DNA branch migration activity (data not shown).
MHF1-MHF2 does not affect the FANCM ATPase activity
We asked whether the ATPase activity of FANCM is influenced by the MHF1-MHF2 heterodimer. We first verified that the FANCM ATPase activity is dependent on DNA (Gari et al., 2008b). The results from time course experiments showed that MHF1-MHF2 does not affect the potency of the FANCM ATPase activity, regardless of whether ssDNA or dsDNA was used as cofactor (Fig. 4F and Fig. S3C). We have also examined a possible effect of MHF1-MHF2 on DNA binding by FANCM but have not found a significant effect in this regard (data not shown).
DNA binding by MHF1-MHF2 is required for biological function
Based on sequence alignment ( Supp Fig. 1) of orthologues from higher organisms, the C-terminal region of MHF1 from residue 110 (numbering based on the human protein) onward harbors highly conserved basic residues that could potentially be involved in the interaction with DNA. To test this premise, a truncation mutation, R110X, that deletes the C-terminal portion in question, was generated within the context of the dimeric complex with wild type MHF2 (Fig. S3D). We note that the R110X truncation does not affect the interaction of MHF1 with MHF2 and the mutant MHF1R110X-MHF2 complex can be purified to a nearly homogeneous state using the procedure devised for the wild type counterpart and with a similar yield (Fig. S3D). Importantly, we found that the MHF1R110X-MHF2 complex lacks DNA binding activity (Fig. S4A), thus validating the premise that the C-terminal conserved domain of MHF1 as being crucial for the DNA binding activity of the MHF1-MHF2 complex.
Interestingly, the mutant MHF1R110X- MHF2 complex maintains the ability to interact with FANCM, as verified in an affinity pulldown experiment that made use of the (His)6 tag on the MHF complex (Fig. S3E). However, when tested in the branch migration assay, the MHF1R110X-MHF2 mutant complex failed to stimulate the activity of FANCM in contrast to the wild type MHF1-MHF2 complex (Fig. S4B). Moreover, FLAG-tagged MHF1R110X co-immunoprecipitates with FAAP24, MHF2 and FANCM in cells (Fig. S4C). Expression of MHF1R110X in the MHF1 knockdown HeLa cells, stabilized FANCM and FAAP24 but failed to complement the FA phenotypes of impaired FANCD2 onoubiquitination (Fig. S4D) and sensitivity to MMC and CPT (Fig. S4E &S4F) suggesting that the DNA binding activity of the MHF dimer is critical for biological function.
SYNOPSIS
Our efforts directed at delineating the complexity of FANCM protein complexes have led us to identify two novel associated proteins, which we have named MHF1 and MHF2. Both of these MHF proteins co-exist with FANCM constitutively in the chromatin fraction in cells. Gel filtration analysis of cell extracts showed that a significant fraction of cellular MHF2 and MHF1 proteins co-elute with FANCM in a large complex in excess of 1MDa in mass. Our cell-based studies focusing on the MHF1 protein have provided ample evidence for its involvement in the FA pathway of DNA damage response, such that ablation of MHF1 via shRNA treatment led to a reduction of FANCM and MHF2 protein level and chromatin association, with a companion defect in the monoubiquitination of FANCD2 and increased genomic instability.
Consistent with their possession of the histone-fold, the MHF proteins assemble into a stoichiometric, heterodimeric complex that possesses a DNA binding activity. Our biochemical studies have shown a preference of the MHF1-MHF2 dimer for the Holliday structure over several other DNA substrates. Importantly, our results have revealed that the MHF1-MHF2 dimer interacts directly with FANCM via MHF1, and that the protein dimer strongly enhances the DNA branch migration activity of FANCM in a manner that is dependent on FANCM’s ability to hydrolyze ATP. These properties of the MHF1-MHF2 dimer distinguish it from the FANCM-associated factor FAAP24 (Ciccia et al, 2007), which also binds DNA but, unlike MHF1-MHF2, exerts no stimulatory effect on FANCM’s ability to process branched DNA substrates (Gari et al, 2008b).
In summary, our discovery of the MHF1-MHF2 heterodimeric complex and its biochemical, genetic, and cytological analyses have revealed an additional level of complexity of the FA pathway. More detailed understanding of the architecture and function of the FA core complex that includes MHF1-MHF2 will help elucidate the molecular defects that underlie the pathogenesis of Fanconi anemia.
EXPERIMENTAL PROCEDURES
The following materials and methods were described in the Supplemental Experimental Procedures: cells, cell cultures, chemicals, drug sensitivity assay, cloning, constructs and retroviral production, purification of FANCM and MHF1 complexes, preparation of cellular fractions, FANCD2 immunofluorescence, chromosome breakage and sister chromatid assays, gel filtration analysis, computational analysis, purification of FANCM and FANCMK117R, purification of MHF1-(His)6-MHF2, purification of MBP-MHF1 and MBP-MHF2, DNA substrates, affinity pull-down assays, and ATPase assay.
Protein knockdown by siRNA/shRNA
For stable knockdown of MHF1, pSilencer 5.1-U6 Retro vector system (Ambion, Inc., Austin, TX) was used to express shRNA in mammalian cells. pSilencer 5.1 Retro Scramble supplied with the kit was used as a control. The shRNA sequence targeting 5′UTR of MHF1 is 5′-GATAATGTGTACTGCGTTA -3′. For transient knockdown, ON-TARGETplus SMARTpool siRNAs was used. A nonspecific control, siRNA (Cat. # D-001210-01), was used in all experiments. Cells were transfected as described earlier (Singh et al., 2008).
Antibodies
Rabbit MHF1 and MHF2 polyclonal antibodies were raised against fusion proteins containing full-length MHF1 and MHF2 with maltose binding protein (MBP). These fusion proteins were expressed and purified from E. coli in accordance with the manufacturer’s protocols. Anti-FANCM, -FANCA, -FANCL, -FANCG, -FAAP100, -FANCB and -FANCD2 were described earlier (Ali et al., 2009a; Meetei et al., 2005). Anti-FAAP24 was a kind gift from Dr. Stephen West. Anti-histone H2A antibody was purchased from Millipore (Billerica).
DNA binding assays
DNA binding reactions (10 μL) were carried out for 5 min at 37°C in binding buffer (25mM Tris at pH 7.5, 1mM DTT, 100 μg/mL bovine serum albumin) with the indicated concentration of protein and 30 nM radiolabeled DNA. Where indicated, reactions were treated with 0.5% SDS and 0.5 mg/ml proteinase K at 37°C for 5 min before analysis. After the addition of 5 μL of gel loading buffer (50% glycerol, 20mM Tris-HCl at pH 7.4, 2mM EDTA, 0.05% orange G), the reaction mixtures were resolved in a 4% or 6% native polyacrylamide gel in TBE buffer (45 mM Tris-borate, 1mM EDTA, pH 8.0) at 4°C. Gels were dried onto Whatman DE81 paper (Whatman International Limited) and then analyzed in a Personal Molecular Imager FX PhosphorImager (Bio-Rad).
DNA Branch Migration Assay
The indicated concentration of FANCM (12 nM), FANCM/MHF1/10 (12 nM) or FANCMK117R (20 nM) protein was incubated at 37°C with the radiolabeled DNA substrate (50 nM) in 10 μL of buffer D (25 mM Tris-HCl at pH 7.5, 1 mM DTT, 100 μg/ml BSA) containing 1.5 mM MgCl2, 1 mM ATP, and 50 mM KCl. The reaction was stopped at the indicated times by treatment with SDS (0.5% final) and proteinase K (0.5 mg/ml) for 5 min at 37°C. The reaction mixtures were resolved in an 8% native polyacrylamide gel in TBE buffer at 4°C. Gels were dried and analyzed as above. The branch migration assay with the MHF1R110X-MHF2 mutant was performed as described above for the wild type MHF complex.
Supplementary Material
Acknowledgments
We are grateful to Drs. Weidong Wang and Stephen West for reagents and to Dongqing Liu for the preparation of FANCM baculoviruses. We thank the Cytogenetics Laboratory, Viral Vector Core, Fluorescent Activated Cell Analyzing and Sorting facility of Cincinnati Children’s Research Foundation and the Taplin Biological Mass Spectrometry Facility at Harvard Medical School for their excellent service. This work was supported by National Institutes of Health research grants ES015632, ES07061 (P.S.) and HL084082 (A.R.M), the Fanconi Anemia Research Fund (A.R.M.) and the American Society of Hematology Junior Faculty Award (A.R.M). The authors declare no conflict of interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali AM, Kirby M, Jansen M, Lach FP, Schulte J, Singh TR, Batish SD, Auerbach AD, Williams DA, Meetei AR. Identification and characterization of mutations in FANCL gene: a second case of Fanconi anemia belonging to FA-L complementation group. Hum Mutat. 2009a;30:E761–770. doi: 10.1002/humu.21032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AM, Singh TR, Meetei AR. FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA. Mutat Res. 2009b;668:20–26. doi: 10.1016/j.mrfmmm.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano M, Suzuki A, Hori T, Backer C, Okawa K, Cheeseman IM, Fukagawa T. The CENP-S complex is essential for the stable assembly of outer kinetochore structure. J Cell Biol. 2009;186:173–182. doi: 10.1083/jcb.200903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arents G, Moudrianakis EN. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc Natl Acad Sci U S A. 1995;92:11170–11174. doi: 10.1073/pnas.92.24.11170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ST, van de Vrugt HJ, Rooimans MA, Oostra AB, Steltenpool J, Delzenne-Goette E, van der Wal A, van der Valk M, Joenje H, Te Riele H, de Winter JP. Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum Mol Genet. 2009 doi: 10.1093/hmg/ddp297. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Ling C, Coulthard R, Yan Z, Xue Y, Meetei AR, Laghmani el H, Joenje H, McDonald N, de Winter JP, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol Cell. 2007;25:331–343. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Collis SJ, Ciccia A, Deans AJ, Horejsi Z, Martin JS, Maslen SL, Skehel JM, Elledge SJ, West SC, Boulton SJ. FANCM and FAAP24 function in ATR-mediated checkpoint signaling independently of the Fanconi anemia core complex. Mol Cell. 2008;32:313–324. doi: 10.1016/j.molcel.2008.10.014. [DOI] [PubMed] [Google Scholar]
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668:11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci U S A. 2008a;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol Cell. 2008b;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- Kim JM, Kee Y, Gurtan A, D’Andrea AD. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111:5215–5222. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krona C, Ejeskar K, Caren H, Abel F, Sjoberg RM, Martinsson T. A novel 1p36.2 located gene, APITD1, with tumour-suppressive properties and a putative p53-binding domain, shows low expression in neuroblastoma tumours. Br J Cancer. 2004;91:1119–1130. doi: 10.1038/sj.bjc.6602083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet. 2005;37:958–963. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, Houghtaling S, Grompe M, D’Andrea AD. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- Mosedale G, Niedzwiedz W, Alpi A, Perrina F, Pereira-Leal JB, Johnson M, Langevin F, Pace P, Patel KJ. The vertebrate Hef ortholog is a component of the Fanconi anemia tumor-suppressor pathway. Nat Struct Mol Biol. 2005;12:763–771. doi: 10.1038/nsmb981. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, Ira G. Yeast Mph1 helicase dissociates Rad51-made D-loops: implications for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosado IV, Niedzwiedz W, Alpi AF, Patel KJ. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Ali AM, Busygina V, Raynard S, Fan Q, Du CH, Andreassen PR, Sung P, Meetei AR. BLAP18/RMI2, a novel OB-fold-containing protein, is an essential component of the Bloom helicase-double Holliday junction dissolvasome. Genes Dev. 2008;22:2856–2868. doi: 10.1101/gad.1725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh TR, Bakker ST, Agarwal S, Jansen M, Grassman E, Godthelp BC, Ali AM, Du CH, Rooimans MA, Fan Q, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114:174–180. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson LH, Hinz JM. Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: Mechanistic insights. Mutat Res. 2009;668:54–72. doi: 10.1016/j.mrfmmm.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.