Abstract
Multiple clinical trials are investigating the use of the DNA methylation inhibitors azacitidine and decitabine for the treatment of solid tumors. Clinical trials in hematological malignancies have shown that optimal activity does not occur at their maximum tolerated doses but selection of an optimal biological dose and schedule for use in solid tumor patients is hampered by the difficulty of obtaining tumor tissue to measure their activity. Here we investigate the feasibility of using plasma DNA to measure the demethylating activity of the DNA methylation inhibitors in patients with solid tumors. We compared four methods to measure LINE-1 and MAGE-A1 promoter methylation in T24 and HCT116 cancer cells treated with decitabine treatment and selected Pyrosequencing for its greater reproducibility and higher signal to noise ratio. We then obtained DNA from plasma, peripheral blood mononuclear cells, buccal mucosa cells and saliva from ten patients with metastatic solid tumors at two different time points, without any intervening treatment. DNA methylation measurements were not significantly different between time point 1 and time point 2 in patient samples. We conclude that measurement of LINE-1 methylation in DNA extracted from the plasma of patients with advanced solid tumors, using Pyrosequencing, is feasible and has low within patient variability. Ongoing studies will determine whether changes in LINE-1 methylation in plasma DNA occur as a result of treatment with DNA methylation inhibitors and parallel changes in tumor tissue DNA.
Keywords: DNA methylation, plasma DNA, biomarker, cancer, repetitive elements, DNA methylation inhibitors, solid tumors, repetitive DNA elements, LINE, MAGE-A1
Introduction
Azacitidine and decitabine, two DNA methylation inhibitors, recently obtained FDA approval for the treatment of myelodysplastic syndromes. The activity of these and other novel “demethylating” agents is being investigated in different disease settings in multiple clinical trials. In an effort to correlate the inhibition of DNA methylation with the clinical activity of these drugs, many investigators have sought to measure changes in DNA methylation in tumor and non-tumor tissues. However, a variety of target sequences in DNA obtained from different tissues as well as diverse methods of measurement have been used in the different studies, leading to frequently conflicting results. For example, a decrease in p15 promoter methylation measured in bone marrow mononuclear cells by methylation-specific PCR (MSP),1 bisulfite sequencing,1 and MsSNuPE2 appeared to be associated with a clinical response in patients with hematological diseases, but when measured in peripheral blood mononuclear cells (PBMC) by COBRA,3 and Pyrosequencing4, 5 this association was not found. In solid tumor trials, the MAGE-A1 promoter methylation has been measured in peripheral blood by methylation-sensitive restriction enzyme digestion coupled with PCR,6 and more recently, by MSP and Pyrosequencing.7 In this case, all methods revealed a decrease in MAGE-A1 promoter methylation in peripheral blood, but this decrease ranged from a maximum of 6.8% in the latter study to a maximum of approximately 90% in the former. In these studies, consistent decreases in 5-methylcytosine content of PBMC DNA were observed using high-performance liquid chromatography (HPLC) but did not correlate with response to treatment.6 7 It is clear that the choice of assay as well as the tissue in which methylation is measured bear heavily on the results that are obtained.
Different assays measure different aspects of DNA methylation.8 For example, MSP and MethyLight generally measure the relative amounts of fully methylated (or fully unmethylated) sequences in a pool of DNA, although they can also be designed to measure the relative amounts of a specific partially methylated sequence. In contrast, COBRA, MsSNuPE and Pyrosequencing measure the methylation level at several single CpG sites, across multiple DNA molecules within a given pool of DNA. If DNA methylation inhibitors are expected to convert one methylation pattern into another (for example, convert all fully methylated MAGE-A1 promoters into all fully unmethylated MAGE-A1 promoters) then methods that quantitate the amount of a given methylation pattern will be useful to measure their effect. However, if DNA methylation inhibitors are expected to “demethylate” some, but not all the CpG sites of a given sequence, then methods that quantitate the methylation level of several specific CpG sites in a given sample will be better suited to detect their effect.
Obtaining samples from solid tumors for correlative studies is difficult and cannot be done repeatedly. Plasma DNA is enriched with tumor DNA in patients with advanced solid tumors and numerous genetic and epigenetic alterations associated with the tumor of origin have been described in the circulating DNA of patients with cancer.9, 10 Our hypothesis is that changes in the methylation of plasma DNA will parallel changes in methylation induced by the DNA methylation inhibitors in the tumor DNA. Abnormal gene promoter methylation occurs in tumor-type specific patterns such that a panel of three to four gene promoters defines an abnormality in 70–90% of each cancer type.11 However, there is no single gene promoter that is hypermethylated in all tumor types or even in all samples of one single tumor type. Therefore, to measure changes in methylation after treatment with DNA methylation inhibitors, a panel of markers would have to be defined for each tumor type, and still, 10–30% of the time, they would not be informative. In addition, substantial amounts of DNA would be needed to be able to amplify all these single-copy sequences from clinical samples. Although measurement of total 5-methylcytosine content by HPLC has proven an economical and reliable method to investigate the effects of DNA methylation inhibitors, this method requires microgram quantities of DNA which cannot be consistently obtained from plasma samples.6 7 Yang et al.12 showed that changes in the methylation levels of the LINE-1 repetitive elements could be used as a surrogate marker of genome-wide methylation changes. Given the abundance of LINE-1 elements in the genome, minimum amounts of DNA are required for their amplification and analysis. The objectives of the current study were to determine the optimal method to measure changes in repetitive element methylation and to describe the inter- and intra-patient variability of the assay in DNA obtained from the plasma of patients with advanced solid tumors. We also measured LINE-1 methylation in DNA extracted from peripheral blood mononuclear cells (PBMC), buccal mucosa cells (Bucc) and saliva and compared it to the methylation of the MAGE-A1 promoter, a single-copy DNA sequence that is normally methylated in non-tumor tissues.
Results
Pyrosequencing Measurements of LINE-1 Methylation Show Lower Variability and Higher Signal to Noise Ratio than Measurements by COBRA, MsSNuPE or MethyLight
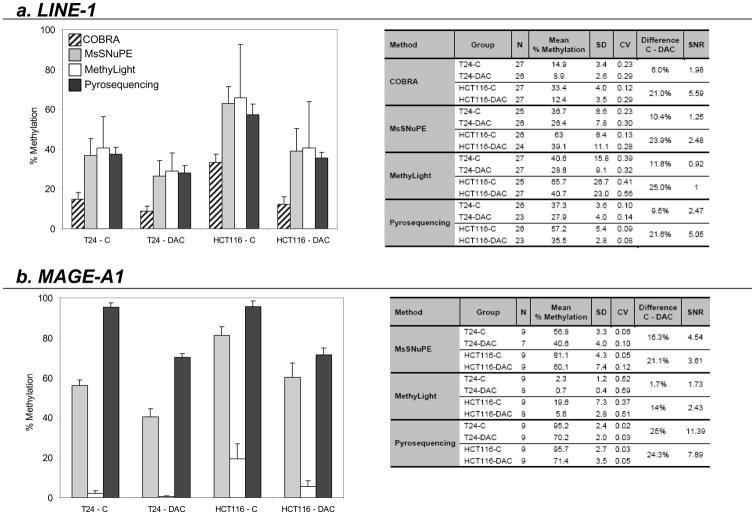
After a 24-hour treatment with 1μM decitabine, average LINE-1 methylation in T24 bladder cancer cell DNA decreased by 6% as measured by COBRA (from 14.9±3.4% to 8.9±2.6%), by 10.4% as measured by MsSNuPE (from 36.7±8.6% to 26.4±7.8%), by 11.8% as measured by MethyLight (from 40.6±15.8% to 28.8±9.1%) and by 9.5% as measured by Pyrosequencing (from 37.3±3.6% to 27.9±4.0%) (Fig. 2a).
Figure 2. LINE-1 and MAGE-A1 promoter methylation changes in T24 and HCT116 cell lines after treatment with decitabine.
Bar graphs represent methylation levels of untreated cells (T24-C and HCT116-C) and cells treated for 24 hours with 1μM of decitabine (T24-DAC and HCT116-DAC). The tables present the summary statistics of the results obtained (N: number of measurements; SD: standard deviation; CV: coefficient of variability; Difference C-DAC: difference in methylation between treated and untreated cells; SNR: signal to noise ratio).
In HCT116 colon cancer cells, average LINE-1 methylation was higher than in T24 bladder cancer cells at baseline, as measured by all methods, but the decrease after treatment was also larger: 21% by COBRA (from 33.4±4.0% to 12.4±3.5%), 23.9% by MsSNuPE (from 63.0±8.4% to 39.1±11.1%), 25% by MethyLight (from 65.7±26.7% to 40.7±23.0%) and 21.6% by Pyrosequencing (from 57.2±5.4% to 35.5±2.8%). (Note that data from MethyLight are expressed relative to a fully methylated reference whilst data from the other assays are expressed in absolute terms.)
Coefficients of variation were lowest with Pyrosequencing (between 0.08 and 0.14) and highest with MethyLight (between 0.32 and 0.56) while COBRA and MsSNuPE displayed similar coefficients of variation (between 0.12 – 0.29 and 0.13 – 0.30 respectively). Signal to noise ratio was highest for COBRA and Pyrosequencing in the HCT116 experiment (5.59 and 5.05 respectively), but in the T24 experiment, signal to noise ratio was greater for Pyrosequencing than for COBRA (2.47 and 1.98 respectively).
Pyrosequencing Measurements of MAGE-A1 Promoter Methylation Show Lower Variability and Higher Signal to Noise Ratio than Measurements by MsSNuPE or MethyLight
After treatment of T24 bladder cancer cells with decitabine, the average MAGE-A1 promoter methylation decreased by 16.3% (from 56.2±2.9% to 40.6±4%) as measured by MsSNuPE, by 1.7% (from 2.3±1.2% to 0.7±0.4%) as measured by MethyLight and by 25% (from 95.2±2.4% to 70.2±2.0%) as measured by Pyrosequencing (Fig. 2b). In HCT116 cells, MAGE-A1 promoter methylation after treatment with decitabine decreased by 21.1% (from 81.1±4.3% to 60.1±7.4%) as measured by MsSNuPE, by 14% (from 19.6±7.3% to 5.6±2.8%) as measured by MethyLight and by 24.3% (from 95.7±2.7% to 71.4±3.5%) as measured by Pyrosequencing. Coefficients of variation ranged from 5.1 to 12.3 with MsSNuPE, 37.1 to 58.5 with MethyLight and 2.5 to 4.9 with Pyrosequencing. Signal to noise ratios in the T24 and HCT116 experiments respectively were 1.73 and 2.43 with MethyLight, 4.54 and 3.61 with MsSNuPE and 11.39 and 7.89 with Pyrosequencing.
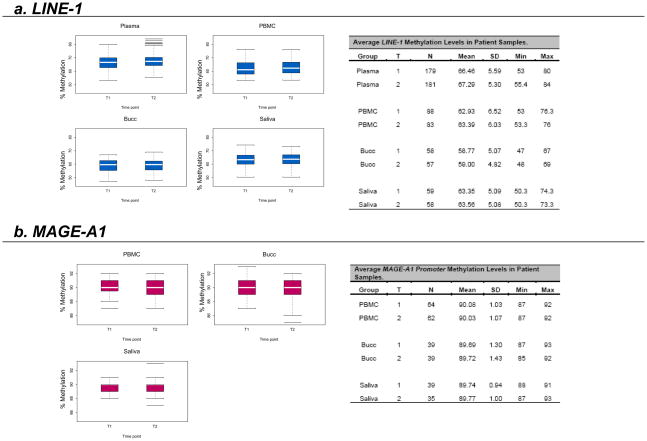
LINE-1 Methylation Measurements Remain Stable Across Time in Plasma, PBMC, Bucc and Saliva DNA of Patients with Metastatic Solid Tumors
Plasma, PBMC, Bucc and saliva were collected at two different time points (without any intervening treatment) from seven men and three women with metastatic solid tumors (9 renal, 1 prostate). The median age at time of enrollment was 57 years (range 41 to 75 years) and the median number of days between the two collection time points was 5 (range 1 to 91 days). The amount of DNA obtained ranged between 0.4–5.6ng/μL (median 1.0ng/μL) from plasma, 1.9–71.9μg (median 23μg) from PBMC, 0.5–21.7μg (median 3.1μg) from Bucc and 8.7–72.6μg (median 23.5μg) from saliva.
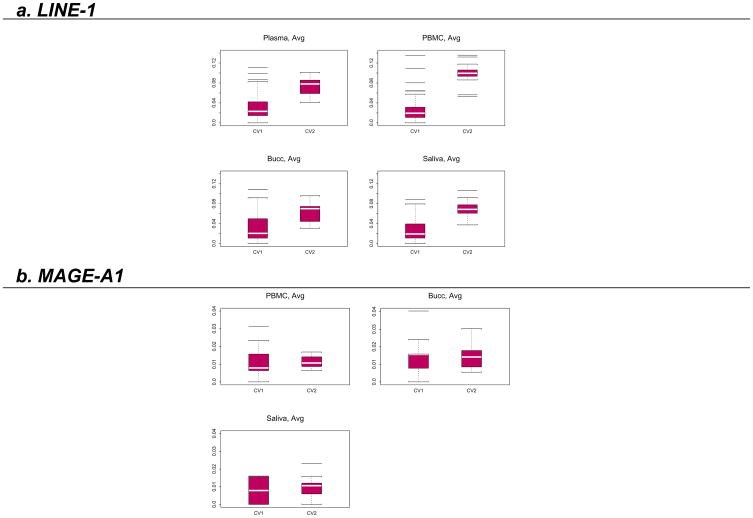
Each DNA sample was analyzed on at least two separate runs by Pyrosequencing, and each run contained at least two replicates of each sample. Results are presented as the average of the methylation levels at P1, P2 and P3 (shown in Fig. 1) and summarized in Fig. 3. The box plots in Fig. 3 show that there was no time effect in the measured LINE-1 methylation levels. To confirm this finding, a linear mixed effect model was fit for the data as described in the statistical section. The fitted model showed there was no difference between the two time points in terms of LINE-1 methylation (p=0.49). However, LINE-1 methylation was found to be higher in plasma DNA compared to PBMC DNA (p<0.0001), higher in PBMC DNA compared to Bucc DNA (p<0.0001) and higher in saliva DNA compared to Bucc DNA (p<0.0001). There was no difference between LINE-1 methylation levels in PBMC compared to saliva DNA (p=0.66). The within-run variation, as measured by the coefficient of variation, was much smaller than the between-run variation in LINE-1 methylation for all types of samples (Fig. 4).
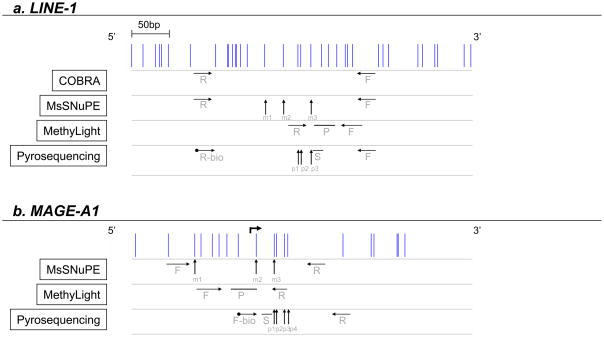
Figure 1. Primer maps.
1a. CpG island promoter of the full length LINE-1 (GenBank accession no.X58075, nucleotide position 108–520, complementary strand). 1b. Promoter region of the MAGE-A1 gene (GenBank accession no.GN013581, nucleotide position 136–276). Vertical lines represent single CpG sites. Horizontal arrows indicate the location of primers (F: forward primer; R: reverse primer; P: probe; S: sequencing primer; -bio: biotinylated primer). Vertical arrows point to the CpG sites analyzed by MsSNuPE (m1–m3) and by Pyrosequencing (p1–p3 for LINE-1; p1–p4 for MAGE-A1).
Figure 3. LINE-1 and MAGE-A1 promoter methylation measurements in patient samples at two separate time points (T1 & T2).
Box plots of the LINE-1 and MAGE-A1 promoter methylation levels observed in patient samples (Plasma; PBMC: peripheral blood mononuclear cells; Bucc: buccal mucosa cells; Saliva) at two different time points (T1 and T2). White horizontal bars represent the medians. The tables present summary statistics of the results obtained (T: time point; N: number of measurements; SD: standard deviation; Min: lowest value obtained; Max: highest value obtained).
Figure 4. Within- (CV1) and between-run (CV2) variability of LINE-1 and MAGE-A1 promoter methylation measurements in patient samples.
Box plots of the within-run (CV1) and the between-run (CV2) coefficients of variation of the LINE-1 and MAGE-A1 promoter methylation measurements in patient samples (Plasma; PBMC: peripheral blood mononuclear cells; Bucc: buccal mucosa cells; Saliva). White horizontal bars represent the averages.
MAGE-A1 Promoter Methylation Measurements Did Not Change Across Runs, Across Time and Across Different Samples
MAGE-A1 promoter methylation was measured in the DNA obtained from the PBMC, Bucc and saliva collected from the same 10 patients as above. Results are presented as the average of the methylation levels at P1, P2, P3 and P4 (shown in Fig. 1) and summarized in Fig. 3. The box plots in Fig. 3 again show no time effect in the measured levels. This finding was confirmed using the same linear mixed effect model as above (p=0.91), but in this case, there were no detectable differences in MAGE-A1 promoter methylation between PBMC, Bucc and saliva DNA. In contrast to the findings with LINE-1 methylation, the within- and between-run variations were not different for the MAGE-A1 promoter methylation in any of the samples (Fig. 4).
Discussion
The possibility of reversing the aberrant silencing of tumor suppressor genes in cancer cells with DNA methylation inhibitors is exciting. However, finding a dose and schedule that optimally demethylates cancer cells in solid tumors has been difficult. We hypothesized that changes in methylation in plasma DNA, which is enriched with tumor DNA in patients with advanced solid tumors, should provide a window into the demethylating dynamics of DNA methylation inhibitors in patients with these malignancies. We selected the repetitive element of the genome LINE-1 because of its abundance and ease of amplification. This study was performed to select an optimal method of measurement to detect changes in repetitive element methylation and to show the feasibility and the operating characteristics of repetitive element methylation measurements in plasma DNA. We also analyzed MAGE-A1 promoter methylation as a comparison.
Our in vitro studies showed that, of the four methods, Pyrosequencing showed the greatest reproducibility in measuring changes in LINE-1 and MAGE-A1 promoter methylation. Pyrosequencing also produced the highest signal to noise ratios, even at low baseline levels of methylation as was the case for LINE-1 methylation in T24 bladder cancer cells. The difference in methylation detected at the MAGE-A1 promoter with the different methods was remarkable and highlights once again how the technique and assay design can influence the result obtained. As discussed in the Introduction, MethyLight provides a different perspective on DNA methylation determination from COBRA, MsSNuPE and Pyrosequencing. The latter methods will quantify the percentage of individual CpGs that are methylated across a pool of DNA molecules. In contrast, the MethyLight assays used in this study measure the percentage of molecules in which all the CpGs of the amplified sequence are methylated. If a single one is unmethylated, that molecule will be counted as unmethylated. In assessing the effects of DNA methylation inhibitors, MethyLight will inform of the percentage of “fully methylated” molecules that become “not fully methylated” which is not necessarily equivalent to “unmethylated”. The advantage of MethyLight is its higher sensitivity, which allows for the detection of single copy genes in clinical samples with small quantities of DNA, but its high variability might make changes difficult to detect and quantify.13, 14
Previous studies have measured LINE-1 methylation levels in whole blood,15 serum,16 PBMCs16 and buccal mucosa cells17 in an effort to develop novel markers for early detection of malignant diseases. In contrast, our intent is to develop a marker of activity of DNA methylation inhibitors that can guide the clinical development of these drugs in patients with solid tumors and thus our focus was to determine the variability of the assay in patient samples. We show that LINE-1 and MAGE-A1 promoter methylation levels measured by Pyrosequencing do not vary significantly with time within an individual, and therefore changes in their levels could potentially be attributed to the effect of a drug. It is noteworthy that the variability of LINE-1 methylation measurements, in both cells in culture and in the patient samples, is larger than that of the MAGE-A1 promoter methylation. This is not surprising if one considers that the assay used in this study measures LINE-1 methylation at a consensus sequence located in the 5′UTR, but there are up to 600,000 copies of LINE-1 present in the human genome,18 and it has been shown that methylation levels of unique LINE-1 elements are different at different loci.17 The relative amounts of each unique LINE-1 element are likely to vary from sample to sample and probably account for the observed variability. Taking this variability into account, this assay should reliably detect a change greater than 5–10% in LINE-1 methylation in plasma DNA.
It is well established that patients with cancer have increased levels of circulating DNA compared to healthy volunteers and that this DNA is largely of tumor origin.19, 20 One source of controversy is whether plasma or serum is the optimal source of the nucleic acids for the purpose of developing clinically useful diagnostic or prognostic tests. While it has been shown repeatedly that larger quantities of DNA can be extracted from serum than from plasma,19, 21 the concern is that a large part of that additional DNA may come from the lysis of circulating blood cells, which could impact the results of the assays.22 Although this has not been definitively proven,21 the fact that more prolonged processing times of both plasma and serum result in the isolation of not only larger quantities but also larger fragments of DNA would support this view.19, 23 In this study, we processed the samples within one hour of collection and plasma was centrifuged a second time immediately after the first to clean it of cellular debris, such that if lysis of leukocytes is a significant source of “normal DNA contamination”, it was avoided. In this setting we observed that LINE-1 methylation levels were different in PBMC versus plasma, suggesting that if substantial lysis of leukocytes were allowed to occur, the results obtained might be significantly impacted. Therefore, we recommend that close attention be paid to the sample collection procedure, ensuring that it is consistent across the samples and that processing occurs as soon after collection as possible, in order to eliminate potential sources of variability.
An unexpected observation was that in our study LINE-1 methylation levels in plasma were in fact higher than those observed in the PBMC, Bucc and saliva samples. Global hypomethylation in cancer occurs predominantly at the expense of decreased methylation of repetitive elements.24 However, it has been shown that LINE-1 methylation in cancer is in fact highly variable 25 and renal cancers in particular appear to lack LINE-1 hypomethylation.16, 26 Therefore, this observation may be explained by the fact that nine out of the ten patients in our study had metastatic renal cell carcinoma.
We conclude that Pyrosequencing is preferable to COBRA, MsSNuPE or MethyLight for the purpose of measuring changes in repetitive element methylation and that measurement of LINE-1 methylation in plasma DNA from patients with advanced solid tumors using Pyrosequencing is feasible and has low within patient variability. Therefore, decreases in LINE-1 methylation in plasma DNA after treatment with a DNA methylation inhibitor could be attributable to the effect of the drug. Given the high between-run variability of the assay compared to the within-run variability, in assessing changes due to methylation inhibitors, all samples for a given patient should be run at the same time and in triplicate or more replicates, rather than in duplicate in multiple runs. Collection of blood samples from patients with solid tumors in clinical trials using DNA methylation inhibitors is ongoing.
Methods
Tissue Culture
T24 (bladder transitional cell carcinoma cells) and HCT116 (colon carcinoma cells) were obtained from the American Type Culture Collection (Manassas, VA). They were seeded at 1×105 cells per 100mm dish 24 hours prior to treatment in McCoy’s 5A medium supplemented with 10% heat-inactivated FCS, penicillin 100U/mL and streptomycin 100μg/mL and grown in a humidified 37°C incubator containing 5% CO2. Cells were treated with 1μmol/L 5-aza-CdR (Sigma-Aldrich Chemical Company, St.Louis, MO) for 24 hours. At 72 h, the cells were trypsinized and harvested. This treatment was repeated in three independent cell culture experiments, each consisting of three separate dishes per group.
Patient Sample Collection, Processing and Storage
Patients with advanced solid tumors presenting to the Genitourinary Cancer Center at MDACC were asked to participate in the IRB approved LAB 06-848 protocol, if they had not received prior treatment or had progressed on a prior treatment and were undergoing evaluation for subsequent therapy. After informed consent was obtained, the patients were asked to provide peripheral blood, saliva and buccal mucosa samples on two separate days without intervening treatment, as described below.
Venous blood was collected in two 10mL Vacutainer® K3 EDTA tubes (BD Medical 366457) and transferred into two 50mL polypropelene (PP) tubes (BD Medical 352098). After centrifugation at 1,450rpm for 10min at 25°C (Centrifuge, ALC PK 130R; Biomedical Solutions), the plasma was transferred to one 15mL PP tube (Corning 430052) and centrifuged again at 2,050rpm for 10min at 4°C to remove any contaminating debris. Equal aliquots were then transferred to capped cryovials (Sarstedt 72.694.006) and frozen at −80°C. HBSS (Sigma, H9394) was added to the remaining cellular pellet in the 50mL PP tubes, to a total volume of 35mL. After gentle mixing, 10mL of Histopaque-1077 (Sigma 1077-1) were underlain in each centrifuge tube, which was then spun at 1,450rpm for 30min at 25°C. The cloudy monolayer of PBMC was collected and washed with an equal volume of DPBS (Invitrogen 14190-250). The vials were centrifuged at 1,150rpm for 10min at 25°C, the supernatant was discarded and the pellet resuspended in 2mL of DPBS at room temperature (RT). Equal aliquots were frozen initially at −80°C and then transferred to a liquid nitrogen tank.
After abstaining from food and drink for 1 hour and swishing their mouth with an alcohol-free mouthwash, patients were instructed to swish with 15mL of normal saline for 10–15 seconds and then to spit the saline solution, which in turn was frozen at −80°C. Following this procedure, two cytology brushes (Fisher 14-959-103) were rubbed across the inside of each cheek (one for each side) and each agitated in 15mL of DPBS to rinse cells from the brush. The brushes were discarded and the tubes frozen at −80°C.
DNA Extraction and Bisulfite Conversion
Genomic DNA from T24 and HCT116 cells was obtained by standard proteinase K and phenol-chloroform extraction and subject to bisulfite conversion, as previously described.12, 27
Plasma samples were divided into 300μL aliquots in 1.5mL tubes, to which a 300μL of a sodium iodide solution containing 7.1M sodium iodide crystalline (Fisher Chemical cat#S324-100), 40mM Tris, 20mM EDTA and 200μg/mL glycogen was added. After gentle mixing, the solution was incubated at 60°C for 15min. After cooling to RT, the DNA was precipitated by adding 500μL isopropranol and incubating at RT for 15min. It was then washed, first with 40% isopropranol then with 70% ethanol, and finally dissolved in 15μL of TE. The quality and quantity of DNA were measured on a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Inc. Wilmington, DE). This was followed by bisulfite conversion of 500ng of DNA using the EpiTect Bisulfite Kit (Qiagen cat#59104).
Primer Design
Primers for MsSNuPE, COBRA and MethyLight were designed using Methyl Primer Express version 1.0 (Applied Biosystems, Foster City, USA). Primers for Pyrosequencing were designed using the PSQ assay design software version 1.0.6 (Biotage, Uppsala, Sweden). Primers are listed in Table 1. Figure 1 depicts the location of the primers.
Table 1.
Primer Sequences.
| Sequence | Assay | Primer Sequence (5′–3′) | |
|---|---|---|---|
| LINE-1 | MsSNuPE | F1 | 5′-TTTTTTGAGTTAGGTGTGGG-3′ |
| R1 | 5′-CATCTCACTAAAAAATACCAAACAA-3′ | ||
| F2 | 5′-GGGTGGGAGTGATT-3′ | ||
| R2 | 5′-GAAAGGGAATTTTTTGATTTTTTG-3′ | ||
| R2′ | 5′-TTTTTTAGGTGAGGTAATGTTT-3′ | ||
| COBRA | F | TTTTTTGA GTTAGGTGTGGG | |
| R | CATCTCACTAAAAAATACCAAACAA | ||
| Pyrosequencing | F1 | 5′-TTTTTTGA GTTAGGTGTGGG -3′ | |
| R1 | 5′-biotin-TCTCACTAAAAAATACCAAACAA-3′ | ||
| S | 5′-GGGTGGGAGTGAT-3′ | ||
| MethyLight | F1 | 5′-GTGGGATATAGTTTCGTGGTGCGCGTTT-3′ | |
| R1 | 5′-AAAAAAAAATAACGAACGCACCTAAA-3′ | ||
| P | 5′-6FAM- TTGAAAAGCGTAATATTCGGGTGGGAGTGATT- BHQ-1-3′ | ||
| MAGE-A1 | MsSNuPE | F1 | GTTTATTTTTATTTTTATTTAGGTAGGATT |
| R1 | TTACCTCCTCACAAAACCTAAA | ||
| F2 | TTTTATTTTTATTTAGGTAGGATT | ||
| R2 | TGGGGTAGAGAGAAG | ||
| R2′ | AGGTTTTTATTTTGAGGGA | ||
| Pyrosequencing | F1 | biotin-TATTGTGGGGTAGAGAGAAG | |
| R1 | AAATCCTCAATCCTCCCTCAA | ||
| S | AACCTAAATCAAATTCCTT | ||
| MethyLight | F1 | TCGGTCGAATTTTACGTCGTT | |
| R1 | AAATCCGATTCCCGCCA | ||
| P | 6FAM-CATCCGAATACCCGAATATAACG CCACTAACT-BHQ-1 | ||
| COL2A1 | MethyLight | F1 | TCTAACAATTATAAACTCCAACCACCAA |
| R1 | GGGAAGATGGGATAGAAGGGAATAT | ||
| P | 6FAM-CCTTCATTCTAACCCAATACCTATCCC ACCTCTAAA-BHQ-1 | ||
MsSNuPE
For both Ms-SNuPE and the COBRA assays, the LINE-1 element was first amplified in a 25μL PCR containing 2μL of bisulfite-treated DNA, 0.25μM of each primer (Table 1), 100μM of each dNTP, 1U Taq DNA polymerase (Promega, cat.#M166B) and 2.5μL 10x PCR buffer. PCR conditions were 95°C for 3min followed by 40 cycles of denaturation at 95°C for 1min, annealing at 52°C for 1min, and extension at 72°C for 1min, followed by a final extension at 72°C for 10min. The PCR product was run in a 2% agarose gel at 100V with 1×TAE buffer for 30min and then isolated using the QIAquick Gel Extraction Kit (Qiagen, cat.#28706). The purified PCR product was resuspended in 40μL of water.
A second 10μL PCR was prepared in duplicate, containing 4μL of the recovered PCR product, 1x PCR buffer, 0.1μL of each oligonucleotide (10μM final concentration, Table 1), 0.1μL (1μCi) of either [32P]dCTP or [32P]dTTP and 0.5U of Taq polymerase. The MsSNuPE was run at 95°C for 2min, 50°C for 1min and 72°C for 1min. The product was denatured for 5min at 95°C and electrophoresed on a 15% acrylamide gel, using a vertical gel electrophoresis apparatus, at 100watts for 1 hour with 1×TBE buffer. The gel was dried at 80°C for 30min and then exposed to a phosphorimaging cassette for 1 hour. The ratio of [32P]dCTP versus [32P]dTTP incorporation was measured with ImageQuant software to produce a percent methylation value for each CpG site within the sample.28
The MAGE-A1 promoter sequence was amplified using the following PCR conditions: 94°C for 4min followed by 40 cycles of denaturation at 94°C for 1min, annealing at 53°C for 1min, and extension at 72°C for 1min, with a final extension at 72°C for 1min. The MsSNuPE conditions were: 95°C for 2min, 50°C for 2min and 72°C for 1min.
COBRA
LINE-1 elements were amplified in the same PCR reaction used as a first step for MsSNuPE, described above. 40μl of this PCR product was dehydrated to ~20μl, and then incubated with 2 μl of the restriction enzyme Hinf I (New England BioLabs, cat#R0155S) for 2 hours at 37°C. The resultant product was electrophoresed on a 2.5% agarose gel and band intensity was measured by densitometry (KODAK 1D v. 3.6.1). The sum of the intensities of the cut bands was divided by the total intensity, or sum of cut and uncut bands, to yield the percent methylation value of the sample.29
Pyrosequencing
For Pyrosequencing, LINE-1 elements were amplified in a 25μL PCR containing 2μL of bisulfite-treated DNA, 60mmol/L Tris HCl (pH 8.8), 15mmol/L ammonium sulfate, 0.5mmol/L MgCl2, 1mmol/L deoxynucleotide triphosphate (dNTP) mix, 1unit of Taq polymerase, 1pmol forward and 1 pmol biotin labeled reverse primers (Table 1). PCR conditions were 95°C, 54°C and 72°C for 30sec for a total of 50 cycles. The PCR product was purified using Streptavidin Sepharose High Performance (Amersham Biosciences, Uppsala, Sweden) and the Sepharose beads containing the immobilized PCR products were purified, washed and denatured using 0.2mol/L of NaOH solution, and then washed again using the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Inc., Westborough, MA) as per the manufacturer’s recommendations. Next, 0.3μmol/L of the sequencing primer (see Table 1) was annealed to the purified single-stranded PCR product and the Pyrosequencing reaction was performed using the PSQ HS 96 Pyrosequencing System (Pyrosequencing, Inc., Westborough, MA). The level of methylation was expressed for each DNA locus as the percentage of methylated cytosines over the sum of methylated and unmethylated cytosines. Non-CpG cytosine residues were used as built in controls to verify bisulfite conversion.30 Results are presented as the average of the methylation at P1, P2 and P3 (Fig. 1).
For the MAGE-A1 promoter sequence, a 25μL PCR was carried out as above, using 10pmol biotinylated forward primer and 10pmol reverse primer. PCR conditions were 95°C, 59°C and 72°C for 30sec for a total of 50 cycles. Results are presented as the average of the methylation at P1, P2 and P3 (Fig. 1).
MethyLight
A real-time PCR was performed using bisulfite- and CpG methylation-dependent primers, so that only fully methylated fragments of LINE-1 and the MAGE-A1 promoter would be amplified. The nested probe was designed to be independent of these factors. A 30μL PCR was run with 0.3μM primers and 0.1μM probe (Table 1), 3.5mM MgCl2, 0.625U AmpliTaq Gold Polymerase (Applied Biosystems, cat.# N808-0248), 0.20 mM dNTPs, and 1X PCR buffer. The PCR conditions consisted of an initial denaturation at 95°C for 10 minutes followed by 50 cycles of denaturation at 95°C for 15sec and annealing/extension at 60°C for 1min. Reactions were also performed for non-CpG-containing regions of the control gene collagen IIA (COL2A1) Male genomic DNA (Promega, cat#G1471) treated with M.SssI DNA methylase (New England Biolab, cat#M0226) was used as a fully methylated reference. The percentage of fully methylated DNA was expressed as the percentage of fully methylated reference (PMR) and calculated as the ratio of the methylation of the gene of interest normalized to each control gene relative to that of the SssI-treated sample. (i.e. PMR=[(GENE/CONTROL)sample/(GENE/CONTROL)M.SssI-Reference] × 100)
Statistical Analysis
Descriptive statistics, such as mean, standard deviation, minimum and maximum values, were calculated for each group and the coefficient of variation (CV) was used as an index for the variability. The signal to noise ratio was calculated as (% methylation of control - % methylation of treated)/pooled standard deviation. Box plots were prepared to demonstrate the distribution of the methylation levels or CVs, where the white bar in each box plot represents the median. To determine the effect of time and sample type on the methylation levels, a linear mixed effect model was fit for the data, using the average methylation as the end point. All available data from multiple runs were used when fitting the model and the association among the repeated measures within the same patient was taken into account by specifying the appropriate variance-covariance matrix. All statistical analyses were carried out in Splus 8.0 (Insightful Corporation, Seattle, Washington. 2007).
Acknowledgments
Funding:
Robert E. and May R. Wright Foundation, 7/2004-6/2005.
American Society of Clinical Oncology - Career Development Award, 7/2006-6/2009.
Abbreviations
- DNA
Deoxyribonucleic acid
- LINE
Long interspersed nuclear elements
- MAGE-A
Melanoma antigen family A gene
- MsSNuPE
Methylation-sensitive single-nucleotide primer extension
- COBRA
Combined bisulfite restriction analysis
- PCR
Polymerase chain reaction
- Bucc
Buccal mucosa cells
Footnotes
Peter Jones
Conflicts of interest: Speakers Bureau: Merck & Co; Stock Options or Bond Holding: TherEpi Inc. and Epigenomics AG; Ownership or Partnership: TherEpi Inc.; Consulting Fees: Epigenomics AG; Officer, Board Member, Trustee: Epigenomics AG
Contributor Information
Ana Aparicio, Email: aaparicio@mdanderson.org, Department of Genitourinary Medical Oncology, University of Texas, MD Anderson Cancer Center, Houston, TX
Brittany North, Email: bnorth@mdanderson.org, Department of Genitourinary Medical Oncology, University of Texas, MD Anderson Cancer Center, Houston, TX.
Lindsey Barske, Email: lindsey.barske@duke.edu, Developmental Biology Program, Dept of Cell Biology, Duke University Medical Center, Durham, NC.
Xuemei Wang, Email: xuewang@mdanderson.org, Department of Biostatistics, University of Texas, MD Anderson Cancer Center, Houston, TX.
Valentina Bollati, Email: valentina.bollati@unimi.it, Department of Environmental and Occupational Health, University of Milan, Milan, Italy.
Daniel Weisenberger, Email: weisenbe@usc.edu, Department of Surgery, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA.
Christine Yoo, Email: cyoo@usc.edu, Department of Biochemistry and Molecular Biology, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA.
Nizar Tannir, Email: ntannir@mdanderson.org, Department of Genitourinary Medical Oncology, University of Texas, MD Anderson Cancer Center, Houston, TX.
Erin Horne, Email: ehorne@mdanderson.org, Department of Genitourinary Medical Oncology, University of Texas, MD Anderson Cancer Center, Houston, TX.
Susan Groshen, Email: groshen_s@ccnt.norccc.usc.edu, Department of Preventive Medicine, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA.
Peter Jones, Email: jones_p@ccnt.norccc.usc.edu, Department of Urology, Biochemistry and Molecular Biology, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA.
Allen Yang, Email: yang_a@ccnt.norccc.usc.edu, Division of Hematology, University of Southern California, Norris Comprehensive Cancer Center, Los Angeles, CA.
Jean-Pierre Issa, Email: jissa@mdanderson.org, Department of Leukemia, University of Texas, MD Anderson Cancer Center, Houston, TX.
References
- 1.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9. doi: 10.1158/0008-5472.CAN-06-0080. [DOI] [PubMed] [Google Scholar]
- 2.Daskalakis M, Nguyen TT, Nguyen C, Guldberg P, Kohler G, Wijermans P, et al. Demethylation of a hypermethylated P15/INK4B gene in patients with myelodysplastic syndrome by 5-Aza-2′-deoxycytidine (decitabine) treatment. 2002;100:2957–64. doi: 10.1182/blood.V100.8.2957. [DOI] [PubMed] [Google Scholar]
- 3.Issa JP, Garcia-Manero G, Giles FJ, Mannari R, Thomas D, Faderl S, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–40. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 4.Yang AS, Doshi KD, Choi SW, Mason JB, Mannari RK, Gharybian V, et al. DNA methylation changes after 5-aza-2′-deoxycytidine therapy in patients with leukemia. Cancer Res. 2006;66:5495–503. doi: 10.1158/0008-5472.CAN-05-2385. [DOI] [PubMed] [Google Scholar]
- 5.Kantarjian H, Oki Y, Garcia-Manero G, Huang X, O’Brien S, Cortes J, et al. Results of a randomized study of 3 schedules of low-dose decitabine in higher-risk myelodysplastic syndrome and chronic myelomonocytic leukemia. Blood. 2007;109:52–7. doi: 10.1182/blood-2006-05-021162. [DOI] [PubMed] [Google Scholar]
- 6.Samlowski WE, Leachman SA, Wade M, Cassidy P, Porter-Gill P, Busby L, et al. Evaluation of a 7-day continuous intravenous infusion of decitabine: inhibition of promoter-specific and global genomic DNA methylation. J Clin Oncol. 2005;23:3897–905. doi: 10.1200/JCO.2005.06.118. [DOI] [PubMed] [Google Scholar]
- 7.Appleton K, Mackay HJ, Judson I, Plumb JA, McCormick C, Strathdee G, et al. Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol. 2007;25:4603–9. doi: 10.1200/JCO.2007.10.8688. [DOI] [PubMed] [Google Scholar]
- 8.Siegmund KD, Laird PW. Analysis of complex methylation data. Methods. 2002;27:170–8. doi: 10.1016/s1046-2023(02)00071-3. [DOI] [PubMed] [Google Scholar]
- 9.Esteller M, Sanchez-Cespedes M, Rosell R, Sidransky D, Baylin SB, Herman JG. Detection of aberrant promoter hypermethylation of tumor suppressor genes in serum DNA from non-small cell lung cancer patients. 1999;59:67–70. [PubMed] [Google Scholar]
- 10.Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, et al. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–9. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 11.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 12.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aparicio A, Weber JS. Review of the clinical experience with 5-azacytidine and 5-aza-2′-deoxycytidine in solid tumors. 2002;3:627–33. [PubMed] [Google Scholar]
- 14.Widschwendter A, Muller HM, Fiegl H, Ivarsson L, Wiedemair A, Muller-Holzner E, et al. DNA methylation in serum and tumors of cervical cancer patients. Clin Cancer Res. 2004;10:565–71. doi: 10.1158/1078-0432.ccr-0825-03. [DOI] [PubMed] [Google Scholar]
- 15.Hsiung DT, Marsit CJ, Houseman EA, Eddy K, Furniss CS, McClean MD, et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev. 2007;16:108–14. doi: 10.1158/1055-9965.EPI-06-0636. [DOI] [PubMed] [Google Scholar]
- 16.Chalitchagorn K, Shuangshoti S, Hourpai N, Kongruttanachok N, Tangkijvanich P, Thong-ngam D, et al. Distinctive pattern of LINE-1 methylation level in normal tissues and the association with carcinogenesis. Oncogene. 2004;23:8841–6. doi: 10.1038/sj.onc.1208137. [DOI] [PubMed] [Google Scholar]
- 17.Phokaew C, Kowudtitham S, Subbalekha K, Shuangshoti S, Mutirangura A. LINE-1 methylation patterns of different loci in normal and cancerous cells. Nucleic Acids Res. 2008;36:5704–12. doi: 10.1093/nar/gkn571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Taback B, O’Day SJ, Hoon DS. Quantification of circulating DNA in the plasma and serum of cancer patients. Ann N Y Acad Sci. 2004;1022:17–24. doi: 10.1196/annals.1318.004. [DOI] [PubMed] [Google Scholar]
- 20.Gahan PB, Swaminathan R. Circulating nucleic acids in plasma and serum. Recent developments. Ann N Y Acad Sci. 2008;1137:1–6. doi: 10.1196/annals.1448.050. [DOI] [PubMed] [Google Scholar]
- 21.Umetani N, Hiramatsu S, Hoon DS. Higher amount of free circulating DNA in serum than in plasma is not mainly caused by contaminated extraneous DNA during separation. Ann N Y Acad Sci. 2006;1075:299–307. doi: 10.1196/annals.1368.040. [DOI] [PubMed] [Google Scholar]
- 22.Jen J, Wu L, Sidransky D. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann N Y Acad Sci. 2000;906:8–12. doi: 10.1111/j.1749-6632.2000.tb06581.x. [DOI] [PubMed] [Google Scholar]
- 23.Page K, Powles T, Slade MJ, MTDEB, Walker RA, Coombes RC, et al. The importance of careful blood processing in isolation of cell-free DNA. Ann N Y Acad Sci. 2006;1075:313–7. doi: 10.1196/annals.1368.042. [DOI] [PubMed] [Google Scholar]
- 24.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 25.Estecio MR, Yan PS, Ibrahim AE, Tellez CS, Shen L, Huang TH, et al. High-throughput methylation profiling by MCA coupled to CpG island microarray. Genome Res. 2007;17:1529–36. doi: 10.1101/gr.6417007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Florl AR, Lower R, Schmitz-Drager BJ, Schulz WA. DNA methylation and expression of LINE-1 and HERV-K provirus sequences in urothelial and renal cell carcinomas. Br J Cancer. 1999;80:1312–21. doi: 10.1038/sj.bjc.6690524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) 1997;25:2529–31. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang AS, Estecio MR, Garcia-Manero G, Kantarjian HM, Issa JP. Comment on “Chromosomal instability and tumors promoted by DNA hypomethylation” and “Induction of tumors in nice by genomic hypomethylation”. 2003;302:1153. doi: 10.1126/science.302.5648.1153a. [DOI] [PubMed] [Google Scholar]
- 30.Barske LBV, Yang AS, Aparicio AM. A comparison of bisulfite-based DNA methylation assays for the measurement of DNA methylation changes induced by decitabine. AACR-NCI-EORTC International Conference Molecular Targets and Cancer Therapeutics: Discovery, Biology and Clinical Applications; Philadelphia. 2005. [Google Scholar]