Abstract
A key question in eukaryotic differentiation is whether there are common regulators or biochemical events that are required for diverse types of differentiation or whether there is a core mechanism for differentiation. The unicellular model organism Saccharomyces cerevisiae undergoes filamentous differentiation in response to environmental cues. Because conserved cell cycle regulators, the mitotic cyclin-dependent kinase Clb2/Cdc28, and its inhibitor Swe1 were found to be involved in both nitrogen starvation- and short chain alcohol-induced filamentous differentiation, they were identified as components of the core mechanism for filamentous differentiation. We report here that slowed DNA synthesis also induces yeast filamentous differentiation through conserved checkpoint proteins Mec1 and Rad53. Swe1 and Clb2 are also involved in this form of differentiation, and the core status of Swe1/Clb2/Cdc28 in the mechanism of filamentous differentiation has therefore been confirmed. Because the cAMP and filamentous growth mitogen-activated protein kinase pathways that mediate nitrogen starvation-induced filamentous differentiation are not required for slowed DNA synthesis-induced filamentous growth, they can therefore be excluded from the core mechanism. More significantly, slowed DNA synthesis also induces differentiation in mammalian cancer cells, and such stimulus conservation may indicate that the core mechanism for yeast filamentous differentiation is conserved in mammalian differentiation.
INTRODUCTION
Unicellular Saccharomyces cerevisiae undergoes developmental switches between two differentiation states in response to environmental cues. For example, under nitrogen starvation, diploid S. cerevisiae cells switch from the yeast form (growth as single oval cells) to the filamentous or pseudohyphae form (growth as elongated cell chains that retain physical attachment between the mother and daughter cells) (Gimeno et al., 1992). Besides nitrogen starvation, other environmental stimuli (such as short chain alcohols and mating pheromones) have also been found to induce filamentous growth (Dickinson, 1996; Lorenz et al., 2000; Erdman and Snyder, 2001). Filamentous growth of S. cerevisiae has been considered as a potential model system for eukaryotic differentiation. This idea was initially supported by the findings that two conserved signaling pathways (the mitogen-activated protein kinase [MAPK] and cAMP pathways) that mediate mammalian cell differentiation also mediate nitrogen starvation-induced filamentous differentiation of S. cerevisiae (Liu et al., 1993; Roberts and Fink, 1994; Ward et al., 1995; Lorenz and Heitman, 1997). The idea was better supported by the fact that cell cycle regulation that is highly conserved in eukaryotic organisms plays a key role in filamentous growth (Rua et al., 2001). Conserved cell cycle regulators, the mitotic cyclin-dependent kinase Clb2/Cdc28, and its inhibitor Swe1 were found to be involved in both nitrogen starvation- and short chain alcohol-induced filamentous differentiation, and they were identified as components of the core mechanism for filamentous differentiation. We report here that slowed DNA synthesis induces filamentous differentiation in yeast, and it involves the conserved cell cycle regulators (Swe1 and Clb2) but not the MAPK or cAMP pathway.
DNA integrity checkpoints are conserved signaling pathways that are activated by DNA damage or replication blocks to delay cell cycle progression until DNA repair or replication is finished. At the heart of the checkpoints are highly conserved proteins that are required for cell cycle arrest: the mammalian ATM and ATR protein kinases and their homolog Mec1 in S. cerevisiae (Paulovich and Hartwell, 1995); the downstream mammalian CHK2/CDS1 protein kinase is homologous to yeast Rad53 (Matsuoka et al., 1998). In S. cerevisiae, Rad53 is activated by two distinct pathways: DNA replication stress activates Rad53 in the DNA replication checkpoint that involves Sgs1 and Mrc1, whereas DNA damage activates Rad53 in a distinct Rad9-dependent checkpoint that causes mitotic arrest in G1 and G2 phases (Frei and Gasser, 2000; Alcasabas et al., 2001; Myung et al., 2001). The checkpoint proteins have more widespread roles than originally thought with some functions such as the regulation of dNTP synthesis that are independent of cell cycle per se (Michelson and Weinert, 1999). The checkpoint mechanisms play a role in cancer suppression. Mutations in the human p53, ATM, and BRCA1 genes have been tied to cancer as well as checkpoint defects. We report here that the checkpoint proteins Mec1 and Rad53 have one additional function: to initiate filamentous differentiation in S. cerevisiae in response to slowed DNA synthesis.
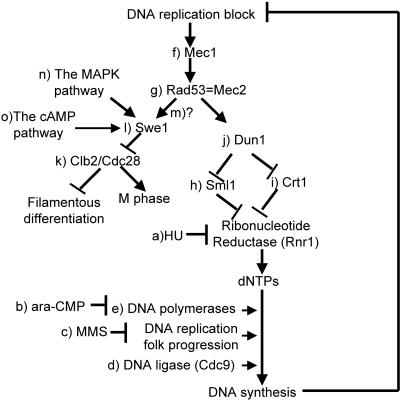
DNA synthesis involves multiple steps, and inhibitors or inhibitory conditions have been used to block or slow DNA synthesis at distinct steps. Hydroxyurea (HU) can block DNA synthesis by inhibiting ribonucleotide reductase that catalyzes dNTP synthesis (Figure 1a). ara-CTP is an analog of dCTP (a natural substrate of DNA polymerases), and it can block DNA fragment synthesis by inhibiting DNA polymerases through competition with dCTP (Hatse et al., 1999) (Figure 1b). DNA-alkylating agent methyl methansulfonate (MMS) can block DNA synthesis by stalling DNA replication forks (Tercero and Diffley, 2001) (Figure 1c). DNA ligase III ligates newly synthesized DNA fragments to complete DNA synthesis. Shifting the temperature of cells with a cdc9-1 allele that encodes a temperature-sensitive DNA ligase III blocks the completion of DNA synthesis (Game et al., 1979) (Figure 1d). DNA synthesis can also be blocked in cells with a temperature-sensitive DNA polymerase when they are grown at restrictive temperatures (Figure 1e). We will demonstrate that moderate application of these conditions slows DNA synthesis and induces filamentous differentiation in yeast.
Figure 1.
Signaling pathways involved in filamentous differentiation of S. cerevisiae.
MATERIALS AND METHODS
YEPD and SC media preparation, cloning, yeast transformation, yeast mating, yeast sporulation, and tetrad dissection were performed as described previously (Jiang, 2002). S. cerevisiae deletion strains were purchased from Research Genetics (Huntsville, AL).
DNA Oligomer Primers for Polymerase Chain Reaction (PCR) Reactions
DNA oligomer primers for PCR reactions were as follows: D1083 (5′-GCT GGA CAA CAA GAA CGA CAT ACA CCG CGT AAA GGC CCA CAA GAC TGC tcg aat tcc tgc agc cc-3′, D1084 (5′-GTT AGA TCA AGA GGA AGT TCG TCT GTT GCC GAA AAT GGT GGA AAG TCG tac gac tca cta tag gg-3′, D1903 (5′-ACACTTTTTTTTCCCCGCCGATGAGAAAGTG-3′, and D1904 (5′-GTTATTGGATTATTTATACAATGCGGCCCATAAGCAC-3′.
Plasmid Construction
D1100 (dun1::kanMX4 in Bluescript) was created as follows. The genomic DNA of S. cerevisiae deletion strain 33576 (dun1::kanMX4) was digested with EcoRV + KpnI and ligated into Bluescript, and the DNA was used to transform DH5α for ampicillin and kanamycin-resistant transformants from which D1100 was isolated. pBS1479 contains TRP1KL, the TRP1 gene from Kluvermyces lactis (Rigaut et al., 1999). D1069 (YEp[LEU2] sml1::kanMX4) was created as follows. The genomic DNA of S. cerevisiae deletion strain 30512 was digested with XbaI + SacII and ligated into a YEp[LEU2] plasmid D102 digested with XbaI + SacII, and the DNA was used to transform DH5α for ampicillin and kanamycin-resistant transformants from which D1069 was isolated.
Strain Construction
Yeast strains used in this report are listed in Table 1. A364a strains DLY264 (originally from T. Weinert, University of Arizona, Tuscon, AZ), Y2032, and Y821 were kindly provided by Tim Formosa (University of Utah, Salt Lake City, UT). Y256 and Y71 are MATa and MATa/α versions of Y33, respectively (Jiang, 2002). Y256 was transformed with D1100 (containing dun1::kanMX4) digested with EcoRV and KpnI for G418 resistance to create Y990. Y71 was transformed with mec1::TRP1KL as a PCR product of D1083, D1084 (as the primers) and pBS1479 (containing TRP1KL as the template) for Trp+ transformants to create Y933. Y933 was transformed with D1069 (containing sml1::kanMX4) digested with XbaI and SacII for G418 resistance to create Y951. Y951 was sporulated to create Y962 and Y958. Σ1278b diploid strains MLY61a/a, MLY183a/a, XPY95a/α were kindly provided by Joe Heitman (Duke University Medical Center, Durham, NC), and they were sporulated to create Y1724, Y1730, Y1726, and Y1727. Y1730 was mated with Y1727 to produce a diploid, and the diploid was sporulated to produce Y1745. The swe1ΔkanMX4 gene disruptor was PCR-amplified from yeast deletion strain 1238 with primers D1903 and D1904 and introduced into Y1893 to create Y2204.
Table 1.
Yeast strains
| Strain | Genotype | Background | Source |
|---|---|---|---|
| Y821 | MATa his3 leu2 trp1 ura3 | A364a | T. Formosa |
| Y2032 | MATa cdc9-1 his3 leu2 trp1 ura3 | A364a | T. Formosa |
| DLY264 | MATa mec2-1::URA3 his3 leu2 trp1 ura3 | A364a | T. Weinert |
| W303A | MATα ade2 can1 his3 leu2 trp1 ura3 | W303 | R. Rothstein |
| W303B | MATa ade2 can1 his3 leu2 trp1 ura3 | W303 | R. Rothstein |
| Y33 | MATα Ty1-URA3-33 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | Y.W. Jiang |
| Y256 | MATa Ty1-URA3-33 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | This study |
| Y71 | MATa/α Ty1-URA3-33 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | This study |
| Y990 | MATa Ty1-URA3-33 dun1::kanMX4 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | This study |
| Y958 | MATa Ty1-URA3-33 sml1:: :kanMX4 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | This study |
| Y962 | MATα Ty1-URA3-33 mec1::TRP1KL sml1:: :kanMX4 ade2 can1 his3 leu2 trp1 ura3-1 | W303 | This study |
| MLY61a/α | MATa/α ura3-52 | Σ1278b | J. Heitman |
| MLY183a/α | MATa/α ura3-52 tec1::kan/tec1::kan | Σ1278b | J. Heitman |
| XPY95a/α | MATa/α ura3-52 flo8::HgyB/flo8::HgyB | Σ1278b | J. Heitman |
| Y1724 | MATa ura3-52 | Σ1278b | This study |
| Y1726 | MATa ura3-52 flo8::HgyB | Σ1278b | This study |
| Y1727 | MATα ura3-52 flo8::HgyB | Σ1278b | This study |
| Y1730 | MATa ura3-52 tec1::kan | Σ1278b | This study |
| Y1745 | MATa ura3-52 flo8::HgyB tec1::kan | Σ1278b | This study |
| ATCC 208922 | MATa/α his3::hisG/+ leu2Δ/+ trp1::hisG/+ ura3Δ/+ | Σ1278b | ATCC |
| Y1893 | MATa leu2Δ | Σ1278b | This study |
| Y2195 | MATa URA3::pGAL:CLB2 | Σ1278b | This study |
| Y2204 | MATa leu2Δ swe1ΔkanMX4 | Σ1278b | This study |
| YPH499 | MATa ade2 gal3 his3 leu2 lys2 trp1Δ1 ura3-52 | S288C | P. Heiter |
| YHA300 | MATa ade5-1 leu2-3,112 trp1-289 ura3-52 pol2-3::LEU2 γCp[TRP1] POL2 | YHA | T. Formosa |
| YHA302 | MATa ade5-1 leu2-3,112 trp1-289 ura3-52 pol2-3::LEU2 γCp[TRP1] pol2-18 | YHA | T. Formosa |
RESULTS
Hydroxyurea-induced Filamentous Growth
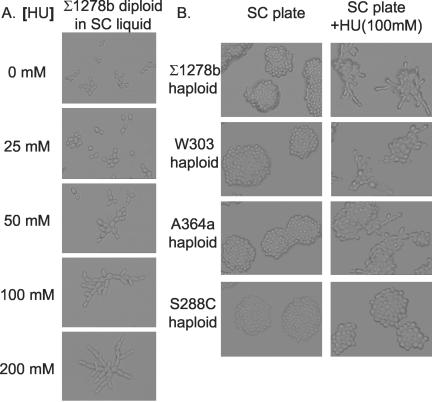
An effective way to inhibit DNA synthesis is to block the synthesis of dNTPs that are the substrates for DNA synthesis. Ribonucleotide reductase is the key enzyme in dNTP synthesis. Hydroxyurea (an inhibitor of ribonucleotide reductase) is known to inhibit DNA synthesis and arrest cells in S phase at concentrations above 300 mM. Cells exposed to sublethal concentrations of hydroxyurea (50, 100, and 150 mM) are slow to complete DNA synthesis and display an extended S phase (Clarke et al., 2001). We examined the effects of hydroxyurea at different concentrations on yeast growth and morphology. Yeast Σ1278b diploid cells grown in liquid SC medium with sublethal concentrations (50-200 mM) of hydroxyurea underwent apparent filamentous growth to form chains of elongated cells (Figure 2A). Filamentation efficiencies were 100% in the cultures with 100 or 200 mM hydroxyurea. We were able to observe similar filamentous growth of a Σ1278b haploid strain on solid SC medium with 100 mM hydroxyurea (Figure 2B), indicating that this type of filamentous growth was not specific to diploids or the liquid culture condition. Three observations convinced us that the elongated cells were cells undergoing bona fide filamentous growth rather than dead or dying cells with unusual morphology. First, the elongated cells were alive and able to divide. Increases in optical density were recorded in the liquid cultures that contained pure filamentous cells, and macrocolonies containing elongated cell chains were formed on solid SC + HU medium (our unpublished data). Second, filamentous differentiation was reversed when hydroxyurea was removed. When the elongated cells from a liquid culture or a colony were plated onto a SC plate, they all gave rise to nonfilamentous yeast-form progenies (our unpublished data). Third, hydroxyurea-induced filamentous growth showed dependence on Swe1 that is essential for nitrogen starvation- and short chain alcohol-induced filamentous growth (see below). These results are consistent with the idea that slowed DNA synthesis caused by lowered dNTP levels induces differentiation in S. cerevisiae.
Figure 2.
Concentration-dependent induction of yeast filamentous growth by hydroxyurea. (A) Diploid Σ1278b cells (ATCC 208922) were grown at 30°C to mid-log phase in liquid SC (Jiang, 2002) containing hydroxyurea at indicated concentrations. The cells were studied under a light microscope and photographed. In this and subsequent experiments of this report, at least six fields were randomly chosen and examined for each culture. In each case, cells in all fields displayed highly similar cell morphology and the photograph of a typical field is shown here. (B) Haploid Σ1278b (Y1724), W303B, A364a (Y821), and S228C (YPH499) cells were grown on SC and SC + HU (100 mM) plates at 30°C overnight. The growth patterns of microcolonies were examined under a light microscope and photographed.
Because filamentous growth cannot be induced in certain laboratory strains such as S288C with nitrogen starvation or short chain alcohols (Liu et al., 1996; Lorenz et al., 2000), we wondered whether hydroxyurea could induce filamentous growth in laboratory strains other than Σ1278b. Hydroxyurea at 100 mM induced moderate filamentous growth in W303 and A364a (Y821) cells but not in S228C (YPH499) cells at all (Figure 2B). The S228C cells remained unresponsive even when they were treated with higher sublethal concentrations of hydroxyurea (our unpublished data).
Slowed DNA Synthesis-induced Filamentous Growth
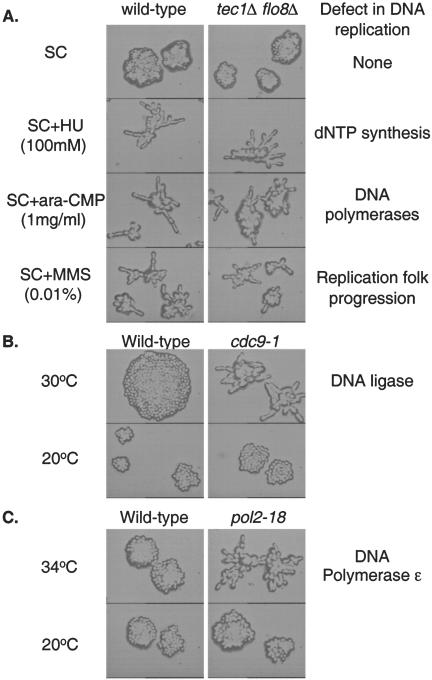
To confirm that slowed DNA synthesis is indeed responsible for hydroxyurea-induced filamentous differentiation, we tested the other conditions that slow DNA synthesis to see whether they could also induce filamentous differentiation. DNA fragment synthesis is catalyzed by DNA polymerases. ara-CTP, the triphosphorylated metabolite of the deoxycytidine analog ara-C, is a potent inhibitor of mammalian DNA polymerases through competition with the natural substrate dCTP. When incorporated into the nascent DNA chain, ara-CMP residues strongly obstruct further elongation by DNA polymerase, causing accumulation of short strands of DNA. ara-CMP is a potent inhibitor of yeast DNA polymerases (McIntosh et al., 1986), it inhibits yeast growth at 2 mg/ml. We treated yeast cells with different concentrations of ara-CMP and discovered that ara-CMP induced uniform filamentous differentiation at a sublethal concentration of 1 mg/ml (Figure 3A).
Figure 3.
Induction of filamentous growth by slowed DNA synthesis. (A) Isogenic Σ1278b haploid wildtype (Y1724) and tec1Δ flo8Δ double mutant (Y1745) cells were grown at 30°C on SC plates with HU (100 mM), ara-CMP (1 mg/ml), and MMS (0.01%). The growth patterns of microcolonies were examined under a light microscope and photographed. (B) Isogenic haploid A364a strains Y821 (wild-type) and temperature sensitive Y2032 (cdc9-1) were grown on SC plates at 30 and 20°C overnight. 30°C is semirestrictive for Y2032. The growth patterns of microcolonies were examined under a light microscope and photographed. (C) Isogenic haploid strains YHA300 (wild-type) and temperature-sensitive YHA302 (pol2-18) were grown on SC plates at 34 and 20°C overnight; 34°C is semirestrictive for YHA302. The growth patterns of microcolonies were examined under a light microscope and photographed.
MMS is a DNA-alkylating agent that can slow DNA replication fork progression (Tercero and Diffley, 2001). MMS inhibits cell growth at 0.03%. We discovered that MMS induced uniform filamentous growth at 0.01% (Figure 3A).
The last step of DNA synthesis is ligation of newly synthesized DNA fragments (especially Okazaki fragments of the lagging strand). The reaction is catalyzed by DNA ligase III that is encoded by CDC9. Cells carrying a temperature-sensitive cdc9-1 allele can grow at 25°C but not 37°C. The completion of DNA synthesis is slowed in cdc9-1 cells at semirestrictive temperatures (Game et al., 1979). We predicted that such delayed completion of DNA synthesis would also induce filamentous growth. As shown in Figure 3B, cdc9-1 mutant cells underwent apparent filamentous growth at semirestrictive 30°C. These results confirmed that slowed DNA synthesis induces filamentous growth in S. cerevisiae.
We also tested whether DNA polymerase temperature-sensitive mutants undergo filamentous growth at a semipermissive temperature. As shown in Figure 3C, pol2-18 mutant cells (with a restrictive temperature of 37°C) underwent uniform filamentous growth at semirestrictive 34°C. Similar results were obtained with other DNA polymerase temperature-sensitive mutants (i.e., pol2-9, pol1-17, pol1/cdc17-1, pol3/cdc2-1, and pol3/cdc2-2 mutants; our unpublished data).
Again, the filamentous cells induced by the these conditions were alive and able to divide to form macrocolonies, and all filamentous cells gave rise to nonfilamentous cells when the conditions that slowed DNA synthesis were removed (our unpublished data). These results indicate that slowed DNA synthesis induces filamentous differentiation in S. cerevisiae.
Because the MAPK and cAMP signaling pathways mediate nitrogen starvation-induced filamentous growth, we investigated their possible involvement in slowed DNA synthesis-induced filamentous growth. TEC1 encodes a DNA-binding transcriptional regulator that serves as the effector for the MAPK pathway, whereas FLO8 encodes a DNA-binding transcriptional regulator that serves as the effector for the cAMP pathway. tec1Δ flo8Δ mutant cells are defective in both signaling pathways, and they responded to slowed DNA synthesis with normal filamentous growth (Figure 3A, right). These results indicate that neither the MAPK nor the cAMP pathway is involved in slowed DNA synthesis-induced filamentous growth. The MAPK and cAMP pathways can therefore be excluded from the core mechanism of filamentous differentiation.
Involvement of the Mec1/Rad53 Checkpoint Proteins in Slowed DNA Synthesis-induced Filamentous Growth
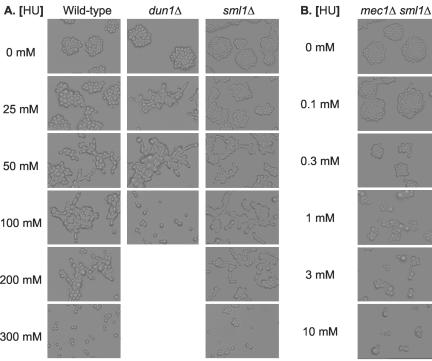
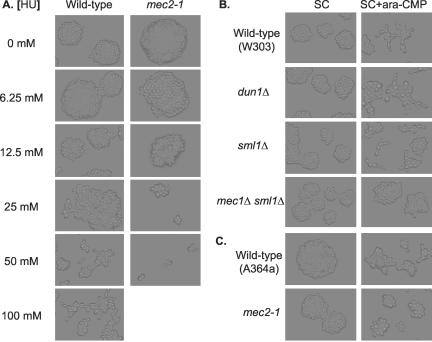
When DNA replication is blocked or DNA is damaged, the Mec1/Rad53 checkpoint proteins (Figure 1, f and g) are activated to halt cell cycle progression and enhance DNA synthesis (Weinert et al., 1994). We wondered whether slowed DNA synthesis per se was sufficient to induce filamentous growth or whether the Mec1/Rad53 checkpoint proteins were involved. To distinguish these two possibilities, we tested whether slowed DNA synthesis could induce filamentous growth in the absence of functional Mec1 or Rad53. MEC1 is an essential gene, and mec1Δ cells are not viable. However, the requirement of Mec1 for viability can be bypassed by overexpression of RNR1 that encodes the rate-limiting large subunit of ribonucleotide reductase (Desany et al., 1998), by deletion of SML1 (Figure 1h) that encodes a protein inhibitor of Rnr1 (Zhao et al., 1998), or by removal of CRT1 (Figure 1i) that encodes the transcriptional repressor of RNR1 (Huang et al., 1998). We tested the possible Mec1 dependence by comparing responses of mec1Δ sml1Δ and sml1Δ cells to slowed DNA synthesis. The response profile of sml1Δ cells to hydroxyurea was indistinguishable from that of isogenic wild-type cells: 300 mM hydroxyurea arrested growth of both wild-type and sml1Δ cells, and they responded to sublethal concentrations of hydroxyurea by undergoing filamentous growth (Figure 4A). When mec1Δ sml1Δ cells were treated with hydroxyurea, we observed the following two phenotypes. First, growth of the mec1Δ sml1Δ cells was arrested by as little as 10 mM hydroxyurea (Figure 4B). Such hypersensitivity to hydroxyurea in the mec1Δ sml1Δ cells was expected, because the cells are defective in transcriptional activation of RNR1 in response to a DNA replication block. Second, we were unable to find a sublethal concentrations of hydroxyurea that induced filamentous growth in the mec1Δ sml1Δ cells. RAD53 is also known as MEC2. We tested a strain with a mec2-1 mutation and observed similar hypersensitivity to hydroxyurea and absence of filamentous differentiation induced by sublethal concentrations of hydroxyurea (Figure 5A).
Figure 4.
Haploid W303 strains Y256 (wildtype), Y990 (dun1Δ) Y958 (sml1Δ), and Y962 (mec1Δ sml1Δ) were grown overnight at 30°C on SC plates with indicated concentrations of hydroxyurea. The growth patterns of microcolonies were examined under a light microscope and photographed.
Figure 5.
A key role of the Mec1/Rad53 checkpoint pathway in slowed DNA synthesis-induce filamentous growth. (A) Cells of isogenic haploid A364a strains Y821 (wildtype) and DLY264 (mec2-1) were grown on SC plates with indicated concentrations of hydroxyurea overnight and photographed under a light microscope. (B) Cells of isogenic haploid W303 strains Y256 (wild-type), Y990 (dun1Δ), Y958 (sml1Δ), and Y962 (mec1Δ sml1Δ) were grown on SC and SC + ara-CMP (1 mg/ml) plates at 30°C overnight and then photographed under a light microscope. (C) Cells of isogenic haploid A364a strains Y821 (wild-type) and DLY264 (mec2-1) were grown on SC and SC + ara-CMP (1 mg/ml) plates at 30°C overnight and photographed under a light microscope.
The failure to induce filamentous growth in the mec1Δ sml1Δ and mec2-1 cells with any concentrations of hydroxyurea agrees well with a critical role of the Mec1/Rad53 checkpoint proteins in slowed DNA synthesis-induced differentiation. However, one might argue that the hydroxyurea hypersensitivity in the mec1Δ sml1Δ and mec2-1 mutants interfered with induction of the filamentous differentiation. We addressed this concern by examining the cells' responses to ara-CMP (1 mg/ml) to which neither mec1Δ sml1Δ nor mec2-1 cells showed hypersensitivity (Figure 5, B and C). ara-CMP at 1 mg/ml induced filamentous growth in the sml1Δ and wild-type cells, but not in the mec1Δ sml1Δ or mec2-1 cells. These results clearly established an essential role of the Mec1/Rad53 checkpoint proteins in the induction of filamentous growth by slowed DNA synthesis.
Moreover, we discovered that hypersensitivity to hydroxyurea per se does not necessarily interfere with differentiation. DUN1 (Figure 1j) encodes a downstream kinase that enhances RNR1 gene expression (and the ribonucleotide reductase activity) at both transcriptional and posttranslational levels (by opposing Crt1 and Sml1, respectively) in Mec1/Rad53-mediated responses to DNA replication block or DNA damage (Zhou and Elledge, 1993; Zhao and Rothstein, 2002). It takes longer for dun1Δ cells to complete DNA replication due to lowered dNTP levels. dun1Δ cells also showed hypersensitivity to hydroxyurea. Hydroxyurea (100 mM) arrested the growth of the dun1Δ cells (Figure 4A). However, 25 mM hydroxyurea (that could not induce filamentous growth in wildtype cells) induced filamentous growth in the dun1Δ cells, despite the hydroxyurea hypersensitivity. These results indicate that hydroxyurea hypersensitivity and the inability to undergo hydroxyurea-induced filamentous growth are distinct biological phenomena. These results also indicate that the Dun1 branch of the Mec/Rad53 checkpoint pathway is not directly involved in slowed DNA synthesis-induced filamentous growth (Figure 1).
DNA Damage Response and Filamentous Growth
The above-mentioned results indicate that activation of the Mec1/Rad53 checkpoint proteins is necessary for filamentous growth, and one may wonder whether activation of the checkpoint proteins per se is sufficient for filamentous growth. Because the Mec1/Rad53 DNA integrity checkpoint proteins can also be activated by DNA damage via the Rad9/Chk1-dpendent DNA damage response checkpoint, we decided to test whether activation of the Mec1/Rad53 proteins by sublethal levels of DNA-damaging agents could also induce filamentous growth. We tested several DNA-damaging agents (bleomycin, etoposide, and hydrogen peroxide) and failed to find any sublethal concentrations that induced filamentous growth (our unpublished data). The results indicate that activation of the checkpoint proteins per se is not sufficient to induce filamentous growth. Because these DNA-damaging agents activate the Mec1/Rad53 checkpoint proteins to delay cell cycle progression in a cell cycle stage-nonspecific manner, their inability to induce filamentous growth may suggest that an S phase delay be key to the Mec1/Rad53-mediated filamentous growth.
The inability of DNA-damaging agents to induce filamentous differentiation also suggested an absent role of the Rad9/Chk1-dependent DNA damage response checkpoint in Mec1/Rad53-mediated filamentous differentiation. We created and tested Σ1278b strains with rad9Δ and chk1Δ mutations. The strains responded to sublethal levels of hydroxyurea with normal filamentous growth (our unpublished data).
Mitotic CDK Clb2/Cdc28 and Slowed DNA Synthesis-induced Filamentous Growth
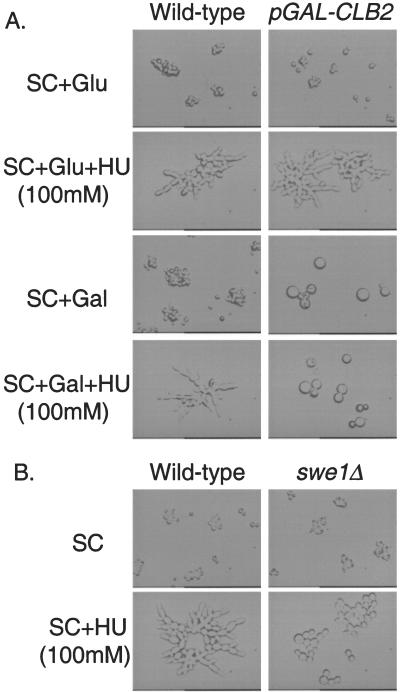
The S phase delay requirement in the Mec1/Rad53-mediated filamentous growth underscores that importance of cell cycle regulation. The evolutionarily conserved cyclin-dependent kinase Cdc28 that regulates cell cycle progression plays a key role in yeast differentiation. Inhibition of the mitotic cyclin-dependent kinase Clb2/Cdc28 activity is believed to be the pivotal event leading to filamentous differentiation (Figure 1k). Constitutive filamentous growth can be obtained by slowing G2/M transition, either with a defective Cdc28 kinase or deletion of the CLB2 gene (Ahn et al., 1999). SWE1 encodes a kinase that phosphorylates Cdc28 to inhibit the Clb2/Cdc28 activity (Figure 1l). Overexpression of Clb2 or deletion of the SWE1 gene inhibits nitrogen starvation- and short chain alcohol-induced filamentous growth (La Valle and Wittenberg, 2001). These observations led to a proposition that there is a core mechanism of filamentous differentiation that consists of Swe1, Clb2/Cdc28 (Rua et al., 2001). We tested the effects of Clb2 overexpression on hydroxyurea-induced filamentous growth. As shown in Figure 6A, cells overexpressing Clb2 (from pGAL-CLB2) grew as enlarged yeast-form cells and were not able to undergo hydroxyurea-induced filamentous growth. To rule out the possibility that the Clb2 overexpression-caused cell enlargement played a role in abolishing hydroxyurea-induced filamentous growth, we tested the effect of swe1Δ. As shown in Figure 6B, swe1Δ abolished hydroxyurea-induced filamentous growth without significant cell enlargement. Because overexpression of MIH1 (which encodes the Swe1-antagonizing Cdc28 phosphotase) did not inhibit HU-induced filamentous growth (our unpublished data), it is unlikely that Mih1 is involved in HU-induced filamentous growth. These results confirmed the core status of Swe1/Clb2/Cdc28 in the mechanism of filamentous growth. We propose that in response to slowed DNA synthesis the Mec1/Rad53 checkpoint proteins activate Swe1 to inhibit Clb2, cause an S phase delay and effect filamentous growth (Figure 1m). Cds1 (the Schizosaccharomyces pombe homolog of Rad53) seems to directly phosphorylate Wee1 (the S. pombe homologs of Swe1) in response to HU treatment (Boddy et al., 1998). It will be interesting to determine whether Rad53 directly phosphorylates Swe1 in slowed DNA synthesis-induced filamentous growth of S. cerevisiae.
Figure 6.
Involvement of Clb2 and Swe1 in HU-induced filamentous differentiation. (A) Isogenic wild-type (Y1724) and pGAL-CLB2 (Y2195) cells were grown at 30°C to mid-log phase in indicated media. The cells were examined under a light microscope and photographed. The expression of pGAL-CLB2 was repressed by glucose (Glu) and induced by galactose (Gal). (B) Wild-type (Y1724) and swe1Δ (Y2204) cells were grown at 30°C to mid-log phase in indicated media. The cells were examined under a light microscope and photographed.
DISCUSSION
Slowed DNA Synthesis-induced Filamentous Differentiation in S. cerevisiae
In this study, we used five conditions (HU, MMS, ara-CMP, cdc9-1, and DNA polymerase temperature-sensitive alleles) to slow yeast DNA synthesis and all five conditions induced filamentous growth. This form of filamentous growth showed dependence on the conserved Mec1, Rad53, and Swe1 checkpoint proteins as well as the Clb2 cell cycle regulator but not the MAPK or cAMP pathway. The requirement of Swe1 and Clb2 for slowed DNA synthesis-induced filamentous growth has confirmed the core status of Swe1/ Clb2/Cdc28 in the mechanism of filamentous differentiation. Because the cAMP and filamentous MAPK pathways that mediate nitrogen starvation-induced filamentous differentiation are not required for slowed DNA synthesis-induced filamentous growth, they can therefore be excluded from the core mechanism. We also found that an S phase delay seemed to be important, and the Rad9/Chk1-dependent DNA damage checkpoint pathway is not required for the Mec1/Rad53-mediated filamentous growth.
As mentioned above, we have determined that Mec1/Rad53-mediated filamentous growth does not involve the MAPK and cAMP pathways, and we have also determined that the MAPK and cAMP pathways mediate filamentous growth in a MEC1-independent manner. Butanol-induced filamentous growth involves the MAPK pathway but not the cAMP pathway (Lorenz et al., 2000; La Valle and Wittenberg, 2001). We have learned that haploid mec1Δ sml1Δ cells undergo normal butanol-induced filamentous growth (our unpublished data). This result indicates that the Mec1/Rad53 pathway is not required for butanol-induced filamentous growth; therefore, the MAPK pathway functions in a Mec1/Rad53-independent manner (Figure 1n). The MAPK and cAMP pathways function in parallel in nitrogen starvation-induced filamentous growth. Diploid mec1Δ sml1Δ cells undergo normal nitrogen starvation-induced filamentous growth (our unpublished data). This result indicates that the Mec1/Rad53 pathway is not involved in nitrogen starvation-induced filamentous growth and therefore the MAPK and cAMP pathways function in a Mec1/Rad53-independent manner (Figure 1o).
The Relationship with the Replication Checkpoint
Because DNA replication stress activates Rad53 in the DNA replication checkpoint that involves Sgs1 and Mrc1 (Frei and Gasser, 2000; Alcasabas et al., 2001), we examined possible roles of Sgs1 and Mrc1 in HU-induced filamentous growth. We created and tested sgs1Δ and mrc1Δ mutations. The strains responded to sublethal levels of hydroxyurea with normal filamentous growth (our unpublished data). There are two possible explanations for these results. First, the signaling pathway for slowed DNA synthesis-induced filamentous differentiation is distinct from the replication checkpoint despite the fact that they share Mec1 and Rad53. Second, there are signaling components that are functionally redundant to Sgs1 and Mrc1 in the replication checkpoint. This idea is supported by the fact that RAD53 is essential for cell viability, whereas neither SGS1 nor MRC1 is essential. More studies are needed to better define and understand the similarities and differences between the pathway for slowed DNA synthesis-induced filamentous differentiation and the DNA replication checkpoint.
Differentiation Therapies for Cancers
Defective control of cell differentiation plays an important role in cancerous growth of mammalian cells. For example, several forms of leukemia may arise from a disruption of the normal program of differentiation, such that a committed progenitor of a particular type of blood cell continues to divide indefinitely, instead of differentiating terminally and dying after a strictly limited number of division cycles. It has been documented that certain leukemia and other types of cancer cells can return to their former differentiation programs when they are treated with DNA synthesis inhibitors (such as ara-C, deoxyadenosine analogs, and hydroxyurea) at sublethal concentrations (Griffin et al., 1982; Kaplinsky et al., 1987; Arbiser et al., 1993; Hatse et al., 1999; Niitsu et al., 2001). Although the underlying mechanism for restoring differentiation commitment in the cancer cells is not known, certain types of cancer have been successfully treated with DNA synthesis inhibitors at sublethal levels in “differentiation therapies” (Leszczyniecka et al., 2001). The similarities between slowed DNA induced differentiation in yeast and mammals deserve further studies.
A Conserved Core Mechanism for Eukaryotic Cell Differentiation?
Filamentous growth of S. cerevisiae has been viewed as a potential model for mammalian cell differentiation. Such hope is based on the assumption that the mechanism(s) of cell differentiation is conserved in eukaryotic systems. Because the mechanism(s) of eukaryotic cell differentiation is not well understood, the only programmatic way to test the idea is to see whether there are common stimuli that use conserved signaling pathways to induce differentiation in both mammalian and yeast cells. We have showed here that slowed DNA synthesis induces yeast filamentous growth in yeast through conserved cell cycle regulators of Swe1/Clb2/Cdc28. Induction of yeast differentiation by the mammalian differentiation stimuli through the conserved cell cycle regulator proteins strongly suggests that the core mechanism (Swe1/Clb2/Cdc28) for cell differentiation may be evolutionarily conserved in mammals and yeast.
Acknowledgments
We thank Tim Formosa and Joe Heitman for supplying yeast strains. We thank Tim Formosa for suggesting the idea of using MMS to induce filamentous growth. We deeply appreciate insightful comments and helpful suggestions from Sandy Johnson, David J. Stillman, Jeff Kapler, and Michael Polymenis. We thank David J. Stillman for helping edit the manuscript. This work was supported by a Damon Runyon Scholarship awarded to Y.W.J.
References
- Ahn, S.H., Acurio, A., and Kron, S.J. (1999). Regulation of G2/M progression by the STE mitogen-activated protein kinase pathway in budding yeast filamentous growth. Mol. Biol. Cell 10, 3301-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alcasabas, A.A., Osborn, A.J., Bachant, J., Hu, F., Werler, P.J., Bousset, K., Furuya, K., Diffley, J.F., Carr, A.M., and Elledge, S.J. (2001). Mrc1 transduces signals of DNA replication stress to activate Rad53. Nat. Cell Biol. 3, 958-965. [DOI] [PubMed] [Google Scholar]
- Arbiser, J.L., Arbiser, Z.K., and Majzoub, J.A. (1993). Differential regulation of choriocarcinoma gene expression by DNA synthesis inhibitors. Endocr. J. 40, 263-268. [DOI] [PubMed] [Google Scholar]
- Boddy, M.N., Furnari, B., Mondesert, O., and Russell, P. (1998). Replication checkpoint enforced by kinases Cds1 and Chk1. Science 280, 909-912. [DOI] [PubMed] [Google Scholar]
- Clarke, D.J., Segal, M., Jensen, S., and Reed, S.I. (2001). Mec1p regulates Pds1p levels in S phase: complex coordination of DNA replication and mitosis. Nat. Cell Biol. 3, 619-627. [DOI] [PubMed] [Google Scholar]
- Desany, B.A., Alcasabas, A.A., Bachant, J.B., and Elledge, S.J. (1998). Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 12, 2956-2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson, J.R. (1996). 'Fusel' alcohols induce hyphal-like extensions and pseudohyphal formation in yeasts. Microbiology 142, 1391-1397. [DOI] [PubMed] [Google Scholar]
- Erdman, S., and Snyder, M. (2001). A filamentous growth response mediated by the yeast mating pathway. Genetics 159, 919-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei, C., and Gasser, S.M. (2000). The yeast Sgs1p helicase acts upstream of Rad53p in the DNA replication checkpoint and colocalizes with Rad53p in S-phase-specific foci. Genes Dev. 14, 81-96. [PMC free article] [PubMed] [Google Scholar]
- Game, J.C., Johnston, L.H., and von Borstel, R.C. (1979). Enhanced mitotic recombination in a ligase-defective mutant of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 76, 4589-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno, C.J., Ljungdahl, P.O., Styles, C.A., and Fink, G.R. (1992). Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68, 1077-1090. [DOI] [PubMed] [Google Scholar]
- Griffin, J., Munroe, D., Major, P., and Kufe, D. (1982). Induction of differentiation of human myeloid leukemia cells by inhibitors of DNA synthesis. Exp. Hematol. 10, 774-781. [PubMed] [Google Scholar]
- Hatse, S., De Clercq, E., and Balzarini, J. (1999). Role of antimetabolites of purine and pyrimidine nucleotide metabolism in tumor cell differentiation. Biochem. Pharmacol. 58, 539-555. [DOI] [PubMed] [Google Scholar]
- Huang, M., Zhou, Z., and Elledge, S.J. (1998). The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 94, 595-605. [DOI] [PubMed] [Google Scholar]
- Jiang, Y.W. (2002). Transcriptional cosuppression of yeast Ty1 retrotransposons. Genes Dev. 16, 467-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplinsky, C., Estrov, Z., Freedman, M.H., Gelfand, E.W., and Cohen, A. (1987). Effect of deferoxamine on DNA synthesis, DNA repair, cell proliferation, and differentiation of HL-60 cells. Leukemia 1, 437-441. [PubMed] [Google Scholar]
- La Valle, R., and Wittenberg, C. (2001). A role for the Swe1 checkpoint kinase during filamentous growth of Saccharomyces cerevisiae. Genetics 158, 549-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leszczyniecka, M., Roberts, T., Dent, P., Grant, S., and Fisher, P.B. (2001). Differentiation therapy of human cancer: basic science and clinical applications. Pharmacol. Ther. 90, 105-156. [DOI] [PubMed] [Google Scholar]
- Liu, H., Styles, C.A., and Fink, G.R. (1993). Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262, 1741-1744. [DOI] [PubMed] [Google Scholar]
- Liu, H., Styles, C.A., and Fink, G.R. (1996). Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144, 967-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M.C., Cutler, N.S., and Heitman, J. (2000). Characterization of alcohol-induced filamentous growth in Saccharomyces cerevisiae. Mol. Biol. Cell 11, 183-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz, M.C., and Heitman, J. (1997). Yeast pseudohyphal growth is regulated by GPA2, a G protein alpha homolog. EMBO J. 16, 7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, S., Huang, M., and Elledge, S.J. (1998). Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science 282, 1893-1897. [DOI] [PubMed] [Google Scholar]
- McIntosh, E.M., Kunz, B.A., and Haynes, R.H. (1986). Inhibition of DNA replication in Saccharomyces cerevisiae by araCMP. Curr. Genet. 10, 579-585. [DOI] [PubMed] [Google Scholar]
- Michelson, R., and Weinert, T. (1999). Sensor-less checkpoint activation? Nat. Cell Biol. 1, E177-E179. [DOI] [PubMed] [Google Scholar]
- Myung, K., Datta, A., Chen, C., and Kolodner, R.D. (2001). SGS1, the Saccharomyces cerevisiae homologue of BLM and WRN, suppresses genome instability and homeologous recombination. Nat. Genet. 27, 113-116. [DOI] [PubMed] [Google Scholar]
- Niitsu, N., Ishii, Y., Matsuda, A., and Honma, Y. (2001). Induction of differentiation of acute promyelocytic leukemia cells by a cytidine deaminase-resistant analogue of 1-beta-D-arabinofuranosylcytosine, 1-(2-deoxy-2-methylene-beta-D-erythro-pentofuranosyl)cytidine. Cancer Res. 61, 178-185. [PubMed] [Google Scholar]
- Paulovich, A.G., and Hartwell, L.H. (1995). A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell 82, 841-847. [DOI] [PubMed] [Google Scholar]
- Rigaut, G., Shevchenko, A., Rutz, B., Wilm, M., Mann, M., and Seraphin, B. (1999). A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030-1032. [DOI] [PubMed] [Google Scholar]
- Roberts, R.L., and Fink, G.R. (1994). Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8, 2974-2985. [DOI] [PubMed] [Google Scholar]
- Rua, D., Tobe, B.T., and Kron, S.J. (2001). Cell cycle control of yeast filamentous growth. Curr. Opin. Microbiol. 4, 720-727. [DOI] [PubMed] [Google Scholar]
- Tercero, J.A., and Diffley, J.F. (2001). Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412, 553-557. [DOI] [PubMed] [Google Scholar]
- Ward, M.P., Gimeno, C.J., Fink, G.R., and Garrett, S. (1995). SOK2 may regulate cyclic AMP-dependent protein kinase-stimulated growth and pseudohyphal development by repressing transcription. Mol. Cell. Biol. 15, 6854-6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert, T.A., Kiser, G.L., and Hartwell, L.H. (1994). Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 8, 652-665. [DOI] [PubMed] [Google Scholar]
- Zhao, X., Muller, E.G., and Rothstein, R. (1998). A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell. 2, 329-340. [DOI] [PubMed] [Google Scholar]
- Zhao, X., and Rothstein, R. (2002). The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc. Natl. Acad. Sci. USA 99, 3746-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Z., and Elledge, S.J. (1993). DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell 75, 1119-1127. [DOI] [PubMed] [Google Scholar]