Abstract
Background
Alterations of endothelial nitric oxide synthase (eNOS) enzyme activity via eNOS gene polymorphisms have been associated with significant cardiovascular morbidity and mortality. Both the thymidine to cytosine transition mutation (T−786→C) in the promoter region and the missense mutation in the exon 7 coding region of the eNOS gene (G894→T) have been associated with several cardiovascular disease states. We hypothesized that heart transplant recipients who carried at least one allele of either of the polymorphisms would have reduced myocardial tissue expression of eNOS measured in the explanted heart.
Methods/Results
Genomic DNA was isolated from myocardial tissue samples obtained from 43 explanted human hearts using standard methods. Regions of the eNOS gene were amplified from genomic DNA with a polymerase chain reaction using specific primers. Protein expression of eNOS was measured by Western blot analysis. There was a statistically significant decrease in mean eNOS expression in samples containing at least one allele for the T−786→C promoter polymorphism (p = 0.04) compared to patients homozygous for the T allele. There was no change in eNOS expression associated with the G894→T exonic polymorphisms. Conclusions: Our data show in failing human myocardium that the T−786→C promoter polymorphism is associated with reduced eNOS expression whereas the G894→T polymorphism of exon 7 is not associated with change in either eNOS mRNA or protein expression. Reduced eNOS expression associated with the promoter polymorphism may contribute to the vascular, contractile, and autonomic responses to ventricular failure.
Keywords: Nitric Oxide, Genetics, Cardiomyopathy, Pharmacogenetics
INTRODUCTION
The role of nitric oxide (NO) in the regulation of vascular homeostasis is well known. In addition, NO has been found to have significant influence in maintaining cardiac autonomic balance as well as playing an important role in myocardial contractility. [1–5] Given the effects of NO on the peripheral vasculature and autonomic tone, an increase in NO production is thought to have a cardioprotective effect. [3, 4]
Endothelial nitric oxide synthase, encoded by a 26-exon gene located on chromosome 7, is one of a triad of enzymes that facilitate production of NO through an L-arginine mediated pathway. [1] Impaired eNOS mediated NO synthesis is associated with significant cardiovascular risk factors (glucose intolerance, arterial hypertension and hyperlipidemia) in animal models. [3,4] Additionally, it has been shown that reductions in eNOS activity have an adverse impact on event-free survival in patients with congestive heart failure.[6] In humans, several polymorphisms of the eNOS gene have been described and are thought to lead to altered NO bioavailability. As a result, these polymorphisms may lead to increased disease risk in parallel with that demonstrated in animal models. In particular, a thymidine to cytosine (T to C) transition mutation (T−786 →C) in the promoter region of this gene has been described. This polymorphism has been associated with reduced eNOS expression in cell reporter systems and has been linked to spontaneous coronary artery vasospasm, atherosclerosis, and autonomic imbalance in patients with heart failure [6, 7]. A second polymorphism involving a missense mutation in the exon 7 coding region of the eNOS gene (G894→T) has been associated with a higher prevalence of hypertension, coronary artery disease and myocardial infarction [8–10].
It has been presumed that the deleterious effects of these polymorphisms are the result of reduced eNOS mRNA expression, yet this has not been confirmed by analysis of adult human tissue. In vivo demonstration of reduced eNOS expression associated with single nucleotide polymorphisms (SNPs) would provide important mechanistic evidence for their impact on a variety of cardiovascular disease states. To test whether these polymorphisms determine eNOS expression in vivo, we performed quantitative polymerase chain reaction (QPCR) for eNOS mRNA expression and Western analysis for eNOS protein in samples of myocardial tissue obtained from the explanted hearts of patients undergoing heart transplantation.
MATERIALS AND METHODS
Subjects
Approval for use of human subjects was obtained from the Institutional Review Board of The Ohio State University. Left Ventricular tissue was obtained from the explanted hearts of forty-three consecutive patients undergoing orthotopic heart transplantation through The Cooperative Human Tissue Network: Midwestern Division at the Ohio State University. Tissue was stored at −80°C until further processing as described below.
Genotyping
Extraction and amplication of DNA
A standard salting-out method was used to extract Genomic DNA from tissue. The T−786→C polymorphism genotypes of eNOS were determined by polymerase chain reaction (PCR) amplification using the primers 5′-TGG AGA GTG CTG GTG TAC CCC A-3′ (sense) and 5′-GCC TCC ACC CCC ACC CTG TC-3′ (antisense). PCR reactions were performed in 50 μl volumes that included approximately 300ng of genomic DNA template, 18μM of primers, 2.5mM of each dNTP, 5μl 1.5mM MgCl2 ammonium buffer (Gene Choice, Frederick, MD), and 2.5U of DNA Taq Polymerase(Promega, Madison, WI). The PCR mixtures were heated to 94°C for 4min and underwent 35 cycles at 94°C for 30s for denaturation, 65°C for 30s for annealing, and 72°C for 1minute for extension. Following the 35 cycles, the final product was obtained from a ten minute extension cycle at 72 °C.
The eNOS genotypes for the G-T substitution in exon 7 (G894→T) were determined by PCR amplification using the primers 5′-GAA ACG GTC GCT TCG ACG T-3′ (sense) and 5′ – ATC CCA CCC AGT CAA TCC CT-3′ (anti-sense). Polymerase chain reaction was performed in a 50 μL reaction volume that included approximately 300 ng of template genomic DNA, 18 μM of each primer, 2.5 mM of each dNTP, 21 mmol/mL MgCl2, 5 μL of 10x PCR buffer, and 2.5 U of Taq DNA polymerase. Samples were amplified for 35 cycles as follows: 94°C for 45 seconds for denaturation, 47°C for 2 minutes for annealing, and 72°C for 2 minutes for extension. Extension was conducted at 72°C for 10 minutes.
Determination of promoter and exonic genotypes
DNA amplified for the T−786→C promoter polymorphism was digested with Msp I (New England Bio-labs, Ipswich, MA) for at least 3 hours at 37 °C. Amplified DNA targeting the G894→T polymormphism was digested with MboI for 3 hours at 37°C. Digested fragments for both polymorphisms were separated by electrophoresis in 4% agarose and visualized using ethidium bromide staining. Allele base pair measurement was determined by using the ϕX174 RF DNA/Hae III Fragments standard ladder (Promega, Madison, WI). For the promoter polymorphism, fragments of 140 and 40bp were determined to be the wild-type allele (allele “T”),and fragments of 90, 50, and 40bp indicated presence of the “C” allele. [11] For the G894→T exonic polymorphism, fragments of 206 bp indicate the wild-type allele (allele “G”), and fragments of 119 and 87 bp indicated the polymorphic variant (allele “T”).[11]
Quantitative PCR (Real Time PCR)
Extraction of mRNA
Total RNA was isolated with the use of RNA-BEE(Teltest, Friendswood, TX) and quantified by using a spectrophotometer with 260–280nm ratios. Reverse transcription was performed using 2μg of total RNA and random hexamers (PerkinElmer, Norwalk, CT) as primers for first-strand synthesis of cDNA and the following conditions: 70 °C for 2 minutes, 42 °C for 60 minutes, and 94 °C for 5 minutes.[12]
Quantification of mRNA
Quantitative PCR was used to measure transcript levels of eNOS genes in left ventricular heart tissue. For all assays, commercially available pre-designed primer/probe sets were utilized per the manufacturer’s recommendations (Assays On Demand; Applied Biosystems, Foster City, CA) and 2X TaqMan Universal PCR Master Mix (Applied Biosystems). Pre-designed primer/probe sets were used (Applied Biosystems) with human β-actin serving as an internal control in a separate reaction well. Data were analyzed using Sequence Detector software (version 1.6; Applied Biosystems).
Expression levels were quantified as the difference between number of cycles required to achieve threshold for the target eNOS mRNA and that required for the beta actin house keeping gene (delta threshold).[12] Messenger RNA expression quantified by this method is inversely proportional to the value of the expression measure. Therefore, to allow a more intuitive representation, values are expressed as the reciprocal of the delta threshold (mRNA expression is directly related to the magnitude of the measure).
Protein Quantification
Western blot analysis was performed to determine whether eNOS protein expression corresponded to differences in mRNA expression. Western blots were performed on homogenates of human heart tissue with antibodies to eNOS (Santa Cruz, Calif) and GAPDH (Cell Signaling, MA). Proteins were separated on 7.5% SDS-PAGE gels and transferred to nitrocellulose membranes. The membranes were probed with the primary antibodies: eNOS (1:2000, overnight, 4°C) and GAPDH (1:5000). The eNOS protein expression was normalized to GAPDH.
Statistics
Statistical analysis was performed using the two-sample Wilcoxon rank-sum (Mann-Whitney) test to evaluate differences in mean eNOS expression. Kruskal Wallis nonparametric analysis was used to evaluate eNOS expression among multiple genotypic combinations. Non-parametric analyses were used given the relatively small sample numbers precluding confidence in the assumption of any parametric distribution of the data.
RESULTS
Demographic characteristics, ejection fractions, New York Heart Association Functional Class (NYHA FC), UNOS status, and use of positive inotropic agents in the different polymorphic groups are shown in Table 1. There were no signficiant differences in NYHA FC, UNOS status, ejection fractions, proportion of Caucasian subjects, proportion of patients with ischemic etiology of cardiomyopathy, or patients treated with positive inotropic agents among the different polymorphic groups. Patients homozygous for the wild type exonic gene were significantly (p = 0.05) younger than those with at least one copy of the polymorphic variant. There was a significantly greater (p = 0.03) proportion of women in those subjects homozygous for the wild type promoter gene as compared to those with at least one copy of the polymorphic variant. However, neither of these variables was significant in multivariable statistical models or showed significant interactions with the gene polymorphisms in terms of mRNA or protein expression.
Table 1.
Demographic and Functional Characteristics of the Genotpyic Groups
| Promoter Polymorhism | Exon 7 | |||
|---|---|---|---|---|
| Genotypes | TC/CC | TT | GT/TT | GG |
| Age | 46±4 yrs | 52±2 yrs | 55±2 yrs | 47±3 yrs* |
| Caucasian | 89% | 73% | 89% | 81% |
| Female | 21% | 53%** | 26% | 35% |
| Ischemic | 57% | 47% | 58% | 54% |
| VAD | ||||
| Ejection Fraction | 19±1% | 25±6% | 22±4% | 21±3% |
| Inotropic Support | 57% | 60% | 53% | 62% |
| UNOS CLASS | ||||
| 1+ | 25% | 25% | 27% | 22% |
| 1A | 17% | 17% | 27% | 9% |
| 1B | 33% | 25% | 20% | 39% |
| 2 | 25% | 33% | 26% | 30% |
| NYHA FC | ||||
| II | 56% | 53% | 65% | 52% |
| III | 44% | 47% | 35% | 48% |
p= 0.05 difference between Exon 7 Genotypes
= p = 0.03 between Promoter Genotypes
= Original UNOS Class 1
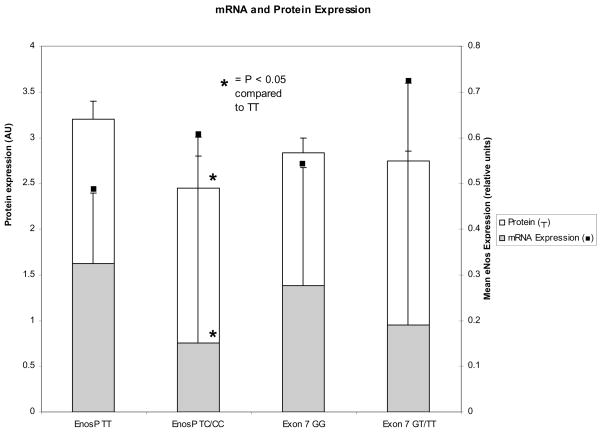
Twenty five samples were found to be heterozygous for the T−786→C eNOS promoter polymorphism (TC). Three samples were found to be homozygous for the polymorphism (Table 2). This distribution of genotypes was not consistent with Hardy-Weinberg proportions (Chi Square p < 0.05). Compared to data reported in multinational populations (www.ncbi.nlm.nih.gov/SNP), the sample in this investigation was characterized by a higher proportion of patients carrying at least one copy of the polymorphism. Messenger RNA expression in the three samples homozygous for the promoter polymorphism did not differ from those measured in heterozygotes. Therefore, the three homozygous samples were grouped with the heterozygous samples for comparison with the homozygous wild type samples. Using the two-sample Wilcoxon rank-sum test there was a statistically significant difference (p = 0.04) in mean eNOS expression in the combined (TC/CC groups) compared to wild type (TT) genotype. Samples containing the eNOS promoter polymorphism were found to have a significant (p= 0.04) >50% decrease in mRNA expression compared to samples that were homozygous for the wild type gene (0.23 ± 0.32 versus 0.42 ± 0.29 reciprocal expression units relative to beta actin house keeping gene; Figure 1).
Table 2.
Proportion of Alleles and Genotypes of a Promoter and Exonic Polymorphism of the eNos Gene
| Promoter |
Exon 7 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| T(%) | C(%) | TT(%) | TC(%) | CC(%) | G(%) | T(%) | GG(%) | GT(%) | GG(%) | |
| Cardiomyopathy |
||||||||||
| Observed | 61 | 39 | 32 | 57 | 11 | 76 | 24 | 56 | 39 | 5 |
| Expected |
37 | 48 | 15 | 58 | 36 | 6 | ||||
| Multinational* Population | 71 | 29 | 56 | 30 | 14 | 81 | 19 | 67 | 28 | 5 |
Promoter = T−786→C polymorphism; Exon 7 = G894→T polymorphism; Cardiomyopathy = transplant recipients
Figure 1.
Mean and standard deviation endothelial nitric oxide mRNA expression (solid bars) in reciprocal delta thresholds relative to beta-actin and protein expression (arbitrary units relative to GAPDH) in myocardial tissue obtained from patients with end-stage heart failure. Patients heterozygous or homozygous for the promoter polymorphism (EnosP) were found to have a significant (p<0.05) reduction in mRNA and protein expression compared to patients homozygous for the T allele. No significant changes in mRNA or protein expression were observed with the polymorphism in exon 7.
Eighteen samples were heterozygous for the G to T eNOS exon 7 polymorphism (Table 2). One sample was homozygous for the polymorphism (TT) and was combined with the heterozygous samples for comparison with the homozygous wild type samples. The distribution of these alleles was consistent with Hardy-Weinberg proportions (p = 0.5). There was no statistically significant difference in mean eNOS expression in the combined (GT/TT) groups compared to the wild type (GG) genotype (p > 0.5; Figure 1).
Seven samples were either heterozygous (TC) or homozygous (CC) for the eNOS promoter polymorphism and were homozygous for the wild type exon 7 gene (GG). Three samples were heterozygous (GT) for the eNOS exon 7 polymorphism and were also homozygous for the wild type promoter (TT). Kruskal Wallis nonparametric analysis revealed a statistically significant decrease (p < 0.05) in mean eNOS expression in samples which had at least one allele of the T−786→C eNOS promoter polymorphism and which were homozygous for the wild type exon 7 gene vs. samples having one allele for the G894→T eNOS exon7 polymorphism but homozygous for the wild type promoter.
Sixteen samples had at least one copy of both the T−786→C and G894→T eNOS polymorphisms. There was a trend (p = 0.1) toward a reduction of mean eNOS expression in those samples having at least one copy of both polymorphisms vs. all others including wild-type homozygous samples.
The results of Western blot analysis are shown in Figure 1. In parallel with the mRNA expression, there was a significant (p<0.05) reduction in eNOS protein normalized to GAPDH in patients who were heterozygous or homozygous for the promoter polymorphism compared to patients homozygous for the T allele. In contrast, there were no differences in eNOS protein expression with the different genotypes for the exonic polymorphism.
Etiology of Cardiomyopathy
Ischemic or non-ischemic etiology of cardiomyopathy was not found to be a determinant of differences in mRNA or protein expression in any of the above analyses for either of the gene polymorphisms and was not found to have a significant interaction with genotypes in the statistical models.
DISCUSSION
Our data provide the first demonstration in human myocardial tissue that the T−786→C promoter polymorphism is associated with a significant reduction of eNOS mRNA expression and a corresponding reduction in eNOS protein. In contrast, the G894→T polymorphism in exon 7 was not associated with altered eNOS expression. This finding is critical in extending the link between gene polymorphisms and clinical outcomes beyond purely associative studies by identifying the impact of a polymorphism on gene transcription and protein expression.
We show direct evidence for reduced mRNA expression in myocardial tissue obtained from humans with dilated cardiomyopathy and therefore the observations reflect the in vivo tissue environment and do not rely on artificial cell reporter systems or animal models. [13] The observations are, however, consistent with the reported influence of the promoter polymorphism on transcriptional activity in these model systems. In addition, our data are consistent with the reported reduction of eNOS mRNA expression in human platelets exhibiting the T−786→C promoter polymorphism.[14]
The expression data we report provides a molecular basis for prior studies associating eNOS polymorphisms with physiologic and clinical outcomes. Several studies have described a link between the T−786→C promoter polymorphism and the prevalence of coronary artery vasospasm, peripheral vascular disease and myocardial infarction.[13, 15, 16] In accordance with our mRNA and expression data, Rossi and colleagues demonstrated significant changes in vascular reactivity in association with the eNOS promoter polymorphism. [16] They did not find an independent association of the exon 7 polymorphism and vascular reactivity, which is in agreement with the mRNA and protein expression data in our study. Cattaruzza and coworkers found that human umbilical vein endothelial cells demonstrated decreased eNOS mRNA and protein expression in response to shear stress in the presence of the promoter polymorphism. [17] We have reported that patients with congestive heart failure who are homozygous for the T−786→C promoter polymorphism of the eNOS gene have significantly advanced autonomic imbalance.[7] This autonomic imbalance is consistent with that which would be predicted for a reduction in NO availability resulting from a loss of function eNOS polymorphism. It is noted that in the current study, we combined the TC and CC groups for statistical purposes owing to the relatively small number of subjects homozygous for the polymorphism. The hypothesis of the study of autonomic imbalance was that two copies of the promoter polymorphism would be required to produce a measureable change in autonomic balance considering the multiple inputs to autonomic regulation including other enzyme sources of nitric oxide. Therefore, the expression data reported in the current study in adult human myocardial tissue are consistent with the role of the eNOS promoter polymorphism in modifying cardiovascular function proposed in the above studies.
Although we did not identify significant differences in mRNA or protein expression associated with the G894→T polymorphism in exon 7, some studies have shown reduced event free survival associated with this gene variant in the heart failure population [4]. However, there is also considerable conflicting data in regards to cellular and clinical associations with this polymorphism.[18–20] The G894→T polymorphism results in the substitution of a glutamate residue for an aspartate at position 298 within the protein and is not likely to alter protein function to a considerable extent.[21] Initially it was suggested that this polymorphism resulted in eNOS intracellular cleavage by an unknown protease, hypothesizing a potential mechanism for enzyme impairment. [22] Subsequent studies have not reproduced these findings and have provided evidence for extracellular eNOS cleavage. This suggests that the reported relationship of the G894→T eNOS polymorphism to cardiovascular outcomes cannot be explained by cleavage or impaired enzyme function.[18] In the current study, there was no reduction in eNOS mRNA or protein expression in human myocardial tissue associated with the G894→T polymorphism corroborating studies which have excluded altered mRNA expression as a cause for reported associated clinical outcomes.
Recent studies have suggested there is linkage disequilibrium between both the T−786 →C and G894→T eNOS polymorphisms, therefore making it difficult to discern which plays the greater role in altering eNOS activity.[20, 23] Based on our statistical analysis it is evident that both polymorphisms are prevalent in the heart failure population. However, we did not find complete linkage disequilibrium based on our sample with a D′ of 0.65 (analysis with Helix Tree software; Golden Helix, Bozeman, MT). However, this does exceed the calculated D′ of 0.26 using the CEPH samples available in the HapMap Project. The fact that the promoter polymorphism did not show Hardy-Weinberg proportions but the exonic polymorphism did further suggests the absence of linkage disequilibrium between these polymorphisms. Nevertheless, our statistical analysis was able to separate samples with the two polymorphic variants and demonstrate a reduction in mRNA and protein expression restricted to the promoter polymorphism. Based on our experience, it is conceivable that some studies reporting an association of clinical outcome with the exon 7 polymorphism may have had significant coexistence of the promoter polymorphism that may truly govern the response. In addition, the exon 7 polymorphism may serve as an effect modifier similar to the interaction described by Cattaruzza.[17]
Experimental Limitations
This study examined human failing myocardial tissue, and therefore is subject to the technical limitations attendant to sampling of and measurement in human tissue. It is possible that some variation in measures of mRNA and protein are ascribed to sampling areas of fibrosis, especially in those patients with ischemic cardiomyopathy. However, the same tissue samples were used to measure expression for both the promoter and the exonic polymorphism. If expression differences were purely due to sampling, then it would be more likely that the same changes in expression would be noted in both polymorphic groups.
Measurement of eNOS enzyme activity as well as protein expression would further solidify the relation between the polymorphisms and their ultimate impact on NO bioavailability. The volume of tissue available through the tissue procurement process was not adequate to perform this analysis in addition to mRNA and protein expression.
A strength of this investigation is that it provides expression data in human myocardial tissue rather than examining expression in model systems. It is possible that the impact of either polymorphism may be different in normal tissue. The variety of environmental stresses in disease states may place greater demands on enzyme synthesis and activity than normal conditions. The difficulty in obtaining normal human myocardial tissue obviously limits the capacity to compare our findings to the normal setting.
The promoter polymorphism was not found to achieve Hardy-Weinberg proportions in contrast to the exonic polymorphism. This may be due to the small sample size; however, the object of this investigation was to test eNOS message expression in the presence of different genotypes rather than to extrapolate findings to general populations. It is intriguing that the promoter polymorphism was more prevalent in this small sample of patients with end-stage heart failure than the reported prevalence in normal populations. This raises the possibility that the promoter polymorphism may be related to the heart failure phenotype, but this is a speculation that must be examined in larger population studies.
CONCLUSIONS
This investigation is the first to show in adult human tissue that there is reduced mRNA and protein expression associated with the T−786→C polymorphism of the eNOS gene. Conversely, there is not a significant change in expression levels with the G894→T polymorphism in exon 7. These findings provide an essential mechanistic basis for reported changes in physiologic cardiovascular responses and outcomes associated with the promoter polymorphism. Presence of the promoter polymorphism may identify patients with heart failure who will benefit from treatment with NO donors or related therapeutic interventions. As such, this study provides a foundation for future gene based outcome studies and pharmacogenetic investigations targeting therapeutic interventions that may compensate for deficient eNOS expression.
Acknowledgments
Dennis Mathias for his assistance in preparing graphics for this manuscript. Dr. Wolfgang Sadee and his staff at The Ohio State University School of Biomedical Sciences/Pharmacogenomics Core Lab for their assistance with genotyping methods.
Funding Sources: Dr. Philip F. Binkley is supported by National Heart, Lung and Blood Institute/National Institutes of Health Grant K24-HL04208 and the James H. and Ruth J. Wilson Professorship. Dr. Ziolo is supported by National Institutes of Health Grant HL079283 and HLO94692 Dr. Wang is supported by The American Heart Association Post-Doctoral Fellowship 0725560B
Footnotes
Disclosures: The authors have no disclosures to report
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Amit A Doshi, Division of Cardiovascular Medicine and the Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH, USA.
Mark T. Ziolo, Department of Physiology and Cell Biology and the Davis Heart and Lung Research Institute, Columbus, OH, USA.
Honglan Wang, Department of Physiology and Cell Biology and the Davis Heart and Lung Research Institute, Columbus, OH, USA.
Emily Burke, Division of Cardiovascular Medicine, The Ohio State University, Columbus, OH, USA.
Amanda Lesinski, Division of Cardiovascular Medicine and Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH, USA.
Philip F. Binkley, Division of Cardiovascular Medicine and the Davis Heart and Lung Research Institute, The Ohio State University, Columbus, OH, USA.
References
- 1.Cook S. Coronary artery disease, nitric oxide and oxidative stress: the “Yin-Yang” effect--a Chinese concept for a worldwide pandemic. Swiss Med Wkly. 2006;136:103–13. doi: 10.4414/smw.2006.11068. [DOI] [PubMed] [Google Scholar]
- 2.Cooke GE, Doshi A, Binkley PF. Endothelial nitric oxide synthase gene: prospects for treatment of heart disease. Pharmacogenomics. 2007;8:1723–34. doi: 10.2217/14622416.8.12.1723. [DOI] [PubMed] [Google Scholar]
- 3.Cooke JP, Losordo DW. Nitric oxide and angiogenesis. Circulation. 2002;7;105(18):2133–5. doi: 10.1161/01.cir.0000014928.45119.73. [DOI] [PubMed] [Google Scholar]
- 4.McNamara DM, Holubkov R, Postava L, Ramani R, Janosko K, Mathier M, MacGowan GA, Murali S, Feldman AM, London B. Effect of the Asp298 variant of endothelial nitric oxide synthase on survival for patients with congestive heart failure. Circulation. 2003;107:1598–602. doi: 10.1161/01.CIR.0000060540.93836.AA. [DOI] [PubMed] [Google Scholar]
- 5.Ziolo MT, Kohr MJ, Wang H. Nitric oxide signaling and the regulation of myocardial function. J Mol Cell Cardiol. 2008;45:625–32. doi: 10.1016/j.yjmcc.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stein B, Frank P, Schmitz W, Scholz H, Thoenes M. Endotoxin and cytokines induce direct cardiodepressive effects in mammalian cardiomyocytes via induction of nitric oxide synthase. J Mol Cell Cardiol. 1996;28:1631–9. doi: 10.1006/jmcc.1996.0153. [DOI] [PubMed] [Google Scholar]
- 7.Binkley PF, Nunziatta E, Liu-Stratton Y, Cooke G. A polymorphism of the endothelial nitric oxide synthase promoter is associated with an increase in autonomic imbalance in patients with congestive heart failure. Am Heart J. 2005;149:342–8. doi: 10.1016/j.ahj.2004.05.059. [DOI] [PubMed] [Google Scholar]
- 8.Cai H, Wilcken DE, Wang XL. The Glu-298-->Asp (894G-->T) mutation at exon 7 of the endothelial nitric oxide synthase gene and coronary artery disease. J Mol Med. 1999;77:511–4. doi: 10.1007/s001099900020. [DOI] [PubMed] [Google Scholar]
- 9.Hingorani AD, Liang CF, Fatibene J, Lyon A, Monteith S, Parsons A, Haydock S, Hopper RV, Stephens NG, O’Shaughnessy KM, Brown MJ. A common variant of the endothelial nitric oxide synthase (Glu298-->Asp) is a major risk factor for coronary artery disease in the UK. Circulation. 1999;100:1515–20. doi: 10.1161/01.cir.100.14.1515. [DOI] [PubMed] [Google Scholar]
- 10.Shimasaki Y, Yasue H, Yoshimura M, Nakayama M, Kugiyama K, Ogawa H, Harada E, Masuda T, Koyama W, Saito Y, Miyamoto Y, Ogawa Y, Nakao K. Association of the missense Glu298Asp variant of the endothelial nitric oxide synthase gene with myocardial infarction. J Am Coll Cardiol. 1998;31:1506–10. doi: 10.1016/s0735-1097(98)00167-3. [DOI] [PubMed] [Google Scholar]
- 11.Marroni AS, Metzger IF, Souza-Costa DC, Nagassaki S, Sandrim VC, Correa RX, Rios-Santos F, Tanus-Santos JE. Consistent interethnic differences in the distribution of clinically relevant endothelial nitric oxide synthase genetic polymorphisms. Nitric Oxide. 2005;12:177–82. doi: 10.1016/j.niox.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Pfaffl MW. Quantification strategies in real-time PCR. In: Bustin S, editor. A-Z of quantitative PCR. La Jolla: International University Line; 2004. [Google Scholar]
- 13.Nakayama M, Yasue H, Yoshimura M, Shimasaki Y, Kugiyama K, Ogawa H, Motoyama T, Saito Y, Ogawa Y, Miyamoto Y, Nakao K. T-786-->C mutation in the 5′-flanking region of the endothelial nitric oxide synthase gene is associated with coronary spasm. Circulation. 1999;99:2864–70. doi: 10.1161/01.cir.99.22.2864. [DOI] [PubMed] [Google Scholar]
- 14.Dosenko VE, Zagoriy VY, Haytovich NV, Gordok OA, Moibenko AA. Allelic polymorphism of endothelial NO-synthase gene and its functional manifestations. Acta biochimica Polonica. 2006;53:299–302. [PubMed] [Google Scholar]
- 15.Gomma AH, Elrayess MA, Knight CJ, Hawe E, Fox KM, Humphries SE. The endothelial nitric oxide synthase (Glu298Asp and −786T>C) gene polymorphisms are associated with coronary in-stent restenosis. Eur Heart J. 2002;23:1955–62. doi: 10.1053/euhj.2002.3400. [DOI] [PubMed] [Google Scholar]
- 16.Rossi GP, Cesari M, Zanchetta M, Colonna S, Maiolino G, Pedon L, Cavallin M, Maiolino P, Pessina AC. The T-786C endothelial nitric oxide synthase genotype is a novel risk factor for coronary artery disease in Caucasian patients of the GENICA study. J Am Coll Cardiol. 2003;41:930–7. doi: 10.1016/s0735-1097(02)03012-7. [DOI] [PubMed] [Google Scholar]
- 17.Cattaruzza M, Guzik TJ, Slodowski W, Pelvan A, Becker J, Halle M, Buchwald AB, Channon KM, Hecker M. Shear stress insensitivity of endothelial nitric oxide synthase expression as a genetic risk factor for coronary heart disease. Circ Res. 2004;15;95:841–7. doi: 10.1161/01.RES.0000145359.47708.2f. [DOI] [PubMed] [Google Scholar]
- 18.Fairchild TA, Fulton D, Fontana JT, Gratton JP, McCabe TJ, Sessa WC. Acidic hydrolysis as a mechanism for the cleavage of the Glu(298)-->Asp variant of human endothelial nitric-oxide synthase. J Biol Chem. 2001;276:26674–9. doi: 10.1074/jbc.M103647200. [DOI] [PubMed] [Google Scholar]
- 19.Kato N, Sugiyama T, Morita H, Nabika T, Kurihara H, Yamori Y, Yazaki Y. Lack of evidence for association between the endothelial nitric oxide synthase gene and hypertension. Hypertension. 1999;33:933–6. doi: 10.1161/01.hyp.33.4.933. [DOI] [PubMed] [Google Scholar]
- 20.Rossi GP, Maiolino G, Zanchetta M, Sticchi D, Pedon L, Cesari M, Montemurro D, De Toni R, Zavattiero S, Pessina AC. The T(−786)C endothelial nitric oxide synthase genotype predicts cardiovascular mortality in high-risk patients. J Am Coll Cardiol. 2006;48:1166–74. doi: 10.1016/j.jacc.2006.05.046. [DOI] [PubMed] [Google Scholar]
- 21.Raman CS, Li H, Martasek P, Kral V, Masters BS, Poulos TL. Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center. Cell. 1998;95:939–50. doi: 10.1016/s0092-8674(00)81718-3. [DOI] [PubMed] [Google Scholar]
- 22.Tesauro M, Thompson WC, Rogliani P, Qi L, Chaudhary PP, Moss J. Intracellular processing of endothelial nitric oxide synthase isoforms associated with differences in severity of cardiopulmonary diseases: cleavage of proteins with aspartate vs. glutamate at position 298. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:2832–5. doi: 10.1073/pnas.97.6.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rossi GP, Cesari M. Reply. J Am Coll Cardiol. 2007;49:1226–7. [Google Scholar]