Abstract
Polyunsaturated fatty acids are essential to brain functions such as membrane fluidity, signal transduction, and cell survival. It is also thought that low levels of unsaturated lipid in the brain may contribute to Alzheimer’s disease (AD) risk or severity. However, it is not known how accumulation of unsaturated lipids is affected in different regions of the hippocampus, which is a central target of AD plaque pathology, during aging. In this study, we used Fourier Transform Infrared Imaging (FTIRI) to visualize the unsaturated lipid content in specific regions of the hippocampus in the PSAPP mouse model of AD as a function of plaque formation. Specifically, the unsaturated lipid content was imaged using the olefinic =CH stretching mode at 3012 cm−1. The axonal, dendritic, and somatic layers of the hippocampus were examined in the mice at 13 weeks, 24 weeks, 40 weeks and 56 weeks old. Results showed that lipid unsaturation in the axonal layer is significantly increased with normal aging in control (CNT) mice (p < 0.01), but remained low and relatively constant in PSAPP mice. Thus, these findings indicate that unsaturated lipid content is reduced in hippocampal white matter during amyloid pathogenesis and that maintaining unsaturated lipid content early in the disease may be critical in avoiding progression of the disease.
Keywords: Fourier transform infrared imaging, polyunsaturated fatty acids, lipids, Alzheimer’s disease, hippocampus, PSAPP
Introduction
The brain contains the second highest lipid content of any other organ in the body, after adipose tissue. However, adipose tissue utilizes lipids for energy storage, while nearly all lipids in the brain have a role in modifying the structure, fluidity, and function of cellular and subcellular membranes and in myelin1. These membrane lipids are comprised of gangliosides, phospholipids, and cholesterol, which contain various amounts of saturated, monounsaturated, and polyunsaturated fatty acids1.
Polyunsaturated fatty acids (PUFAs) synthesized from ω-3 and ω-6 essential fatty acids are key structural components of phospholipid membranes and are critical for brain function including membrane fluidity and signal transduction2. Normal brain function depends on maintaining homeostatic concentrations of PUFAs both during development and throughout life2. Indeed, impairments in PUFA metabolism have been implicated in many neurological diseases, including Alzheimer’s disease (AD). For example, the ω-3 PUFA docosahexaenoic acid (DHA) is the principle component of neuron membrane phospholipids with the highest concentrations found in the synapses and retinal photoreceptors3. It is also abundant in capillary endothelium3. Serum levels of DHA were found to be lower in AD patients compared to control subjects without cognitive impairment4. Furthermore, lower levels of DHA were associated with the loss of post-synaptic proteins in dendrites in a transgenic mouse model of AD5. The loss of DHA could be due to a number of factors, including decreased dietary intake of essential fatty acids or reduced desaturase enzyme activity4. DHA also was shown to reduce amyloid burden in cell culture6 and in transgenic mouse models of AD7,8, possibly by inhibiting the amyloidogenic pathway or by upregulating amyloid beta (Aβ) clearance mechanisms.
Unsaturated lipids are highly vulnerable to oxidative attack because of their double bond content and the increased degradation of PUFAs in AD may be evidence of lipid peroxidation9. Lipid peroxidation is marked by elevated levels of thiobarbituric acid reactive substances, isoprostanes, and chemically reactive, diffusible aldehydes, such as 4-hydroxy-2-neonenal, malondialdehyde, and acrolein10. Under normal conditions, the brain is generally protected from oxidative damage through a careful balance of physiological amounts of reactive oxygen species and antioxidant defenses11. However, these mechanisms may be impaired in AD and increasing evidence shows elevated lipid peroxidation products and protein carbonyl formation in human AD brain12,13 and in transgenic AD mice14,15, particularly in regions of high amyloid plaque density such as the hippocampus and inferior parietal lobule16.
The hippocampus is of pathological relevance to AD and previous studies have suggested neuronal loss17 and loss of unsaturated lipids in this region18,19. However, previous investigations of unsaturated lipid content in the brain measure serum or blood plasma levels or employ biochemical techniques that require homogenization, eliminating the ability to study and visualize specific brain regions. Fourier Transform Infrared Imaging (FTIRI) is an analytical technique that is used to examine the chemical composition of multiple biological components simultaneously in unfixed, unstained, thin sections of biological tissue. Its usefulness in monitoring unsaturated lipid content by utilizing the olefinic =CH stretching mode (3012 cm−1) has been demonstrated previously20–22.
In this study, we used FTIRI to visualize unsaturated lipid content in specific areas of the hippocampus in a mouse model of AD as a function of animal age. The mouse model, known as the PSAPP mouse, expresses a mutant human presenilin 1 (PS1) gene and a chimeric mouse/human amyloid precursor protein gene (APP), developing amyloid plaques in a similar pattern to human AD by around 6 months of age23,24. The evaluation of unsaturated lipid content and distribution in individual regions of the hippocampus will help understand how different anatomical areas are affected in this critical brain region and how accumulation of unsaturated lipid may be impaired in AD.
EXPERIMENTAL SECTION
Tissue Preparation
Twenty female B6C3-Tg(APPswe, PSEN1dE9)85 Dbo/J (PSAPP) mice and twenty age- and gender-matched B6C3F1/J control (CNT) mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). The mice were cared for and euthanized in accordance with the guidelines set by the Brookhaven National Laboratory (BNL) Institutional Animal Care and Use Committee and were housed at the Brookhaven Laboratory Animal Facility under standard conditions. Animals were fed the standard Mouse Diet 5015 (PMI Nutrition International, St. Louis, MO, USA). At time points representing pre-AD (13 weeks; PSAPP=5, CNT=5), early-AD (24 weeks; PSAPP=5, CNT=5), intermediate-AD (40 weeks; PSAPP=5, CNT=5), and late-AD (56 weeks; PSAPP=5, CNT=5) the mice were weighed and then deeply anesthetized with 100 mg/kg 1:10 ketamine:xylazine administered by intraperitoneal injection. Afterwards, they were perfused transcardially with phosphate buffered saline, resulting in exsanguination and death. The brains were removed, frozen on dry ice, and stored at −80°C until further processing. For each sample, 10 µm thick coronal, whole brain cryosections, which included the hippocampus, were mounted onto Superfrost slides and stained with hematoxylin and eosin (H & E). Adjacent serial sections (10 µm thick) were mounted on IR transparent calcium fluoride (CaF2) slides (Korth Kristalle GmbH, Altenholz, Germany) and dried at room temperature. The CaF2 slides were kept in a dry and dark environment until the FTIRI experiments were carried out.
Fourier Transform Infrared Imaging (FTIRI)
Lipids have characteristic C–H stretching vibrations between 2700 – 3020 cm−1 25. Specifically, the asymmetric and symmetric C–H stretching vibrations of the aliphatic −CH2 functional group fall at ~2920 and 2850 cm−1, respectively. For −CH3 functional groups, the asymmetric and symmetric C–H stretching vibrations are found at ~2957 and 2872 cm−1, respectively. In this study, we examined the unsaturated olefinic C=C–H stretching vibration, which has a unique vibrational frequency of 3012 cm−1 and is well-separated and distinguishable from the saturated aliphatic peaks20.
The FTIRI data were collected using a Bruker imaging system (Bruker Optics, Billerica, MA, USA) consisting of a Vertex 80v Rapid Scan FTIR spectrometer, coupled to a Hyperion 3000 IR microscope, equipped with a 128 × 128 mercury cadmium telluride focal plane array detector (Santa Barbara Focalplane, Goleta, CA, USA). For each sample, an area approximately 2.5 mm × 2 mm encompassing the hippocampus was imaged. At each pixel, absorbance spectra in the mid-infrared spectral range (3800–900 cm−1) were collected using a 15× objective in transmission mode with a spectral resolution of 8 cm−1, 64 scans co-added, and 8 × 8 pixel binning, resulting in a final pixel resolution of 21.6 µm (original pixel resolution was 2.7 µm). A background spectrum was collected from clean CaF2 and ratioed to each of the sample spectra. All spectra then underwent quality tests using Cytospec v1.4.02 to ensure that spectra collected from regions without tissue were not included in the analysis. The remaining spectra were subsequently vector-normalized and converted into first derivative spectra using a 9-point Savitzky-Golay algorithm. These two steps ensured that tissue density or peak intensity differences did not contribute to subsequent analyses.
Separating Histological Structures of the Hippocampus
In order to partition the spectra into different histological structures, i.e. regions of interest (ROIs), unsupervised reduced hierarchical cluster analysis (HCA) was performed on the vector-normalized, first derivative spectra in the 1620 – 1680 cm−1 (i.e. protein) and 2750 – 3020 cm−1 (i.e. lipid) spectral regions using Cytospec v.1.4.02. D-values based on Pearson’s correlation coefficient were used to calculate the distance matrix, which determines the similarity of the spectra. Hierarchical clustering was subsequently performed using Ward’s algorithm, which searches the distance matrix for clusters of similar spectra. On average, five clusters were required to separate the images into histological ROIs, which also corresponded to distinct anatomical regions of the hippocampus based on H & E staining. For each ROI, an average spectrum was calculated. In addition, binary masks were created for each ROI. This was done by converting each ROI pixel in the matrix to ones and the remainder of the pixels in the matrix to zeroes, then repeating the procedure for each ROI separately.
Principal Components Analysis (PCA)
PCA is a technique used to reduce the dimensionality of a large dataset without decreasing the variance. In this case, it was used to determine if one sample set is different from another and which spectral frequencies contribute most to the difference. In order to identify spectral regions as possible sources of maximum variance associated with AD mice and CNT mice, the average spectra from each ROI for each sample were subjected to PCA using The Unscrambler 9.8 (CAMO, Norway). All average spectra were converted into second derivative spectra using a 7-point Savitzky-Golay smoothing algorithm. PSAPP mice were analyzed with their corresponding age-matched controls at each time point in each ROI using a Norris Gap algorithm and a fully cross-validated model. Scores plots were created to identify any clustering of the sample groups, which would indicate variability between the sample groups. If group clusters were identified, a loadings plot of the most influential PC was created to assess the contribution of each wavenumber to the total variance.
Integration Profiles
The degree of unsaturation was investigated by examining the olefinic =CH stretching vibration at 3012 cm−1. The peak intensity at 3012 cm−1 is proportional to the amount of unsaturated lipid in the tissue20, 22. The olefinic content was calculated as a ratio of the integrated area from 3000 – 3020 cm−1 normalized to the total lipid content, i.e. the integrated area of the total C–H stretching region (2750 – 3020 cm−1). A linear baseline in the same spectral region was applied.
After the chemical images for olefinic content were created, the masks for each of the three ROIs described above were applied to each image separately and the resulting images contained only the data points (approximately 1000–4000 pixels) from each ROI. For each ROI, the median olefinic content was determined. The median values were then obtained for PSAPP and CNT mice at each time point. A Kruskal-Wallis test was performed using SPSS v.14.0 to test for significant differences between PSAPP and CNT at each time point, and differences between time points. Post-hoc analyses on significant Kruskal-Wallis tests were performed using Mann-Whitney U tests. A significance level of 0.01 was used for all analysis.
RESULTS AND DISCUSSION
The hippocampus is a critical structure in the brain responsible for certain aspects of learning and declarative memory and is one of the first and most severely affected regions of the brain by amyloid pathology in AD17. The goal of this study was to characterize the unsaturated lipid content and distribution in the intact hippocampus of an aging mouse and to assess changes in unsaturated lipid content in the age-matched PSAPP mouse. We found that regions of the hippocampus containing higher amounts of lipid are affected differently in the PSAPP mice than in normal aging.
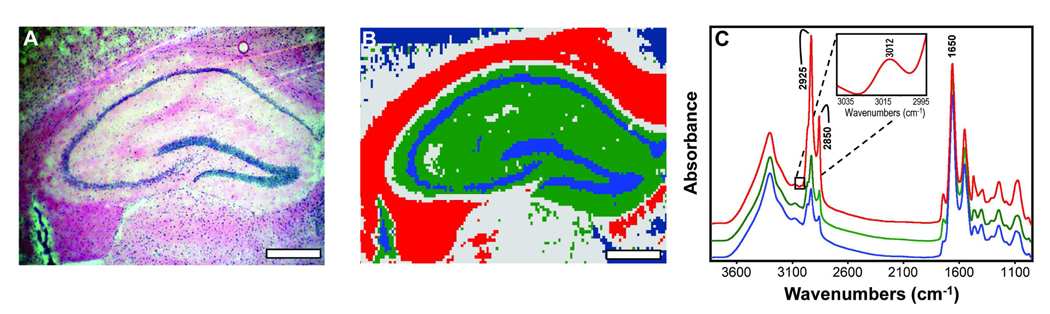
The hippocampal formation consists of the subiculum, CA, and the DG. An H & E stained hippocampal section can be seen in Figure 1A. Using HCA, three hippocampal anatomical ROIs were consistently identified based on their lipid and protein content and are shown in Figure 1B. The red area corresponds to the alveus and fimbria, which are rich in axons (axonal layer), green corresponds to the dendrites of the hippocampus (dendritic layer), and blue corresponds to the cell somata of the granule cells of the DG and pyramidal cells of the CA (somatic layer). The subiculum was not included in the area imaged and was therefore not part of the analysis. Also, the fimbria and alveus are generally not considered a main part of the “hippocampal formation”, but they carry many subcortical afferent and efferent fibers through the hippocampus and are also highly enriched in lipids. Additionally, previous research shows that despite primarily being considered a disease of the grey matter, white matter alterations in other brain regions have also been observed in AD26, 27. Therefore, we included this region in the analyses. Also shown in Figure 1C are representative spectra from each ROI, illustrating the differences in lipid content. The axonal layer spectrum (red) has a strong intensity in the area of the spectrum mainly attributed to lipids (2750 – 3020 cm−1) and corresponds to the high myelin content in this region. The dendritic layer spectrum (green) is intermediate in lipid content, but contains a higher proportion of protein (1620 – 1680 cm−1) relative to the axonal layer spectrum. The somatic layer spectrum (blue) contains the least amount of lipid but a similar amount of protein as the axonal layer spectrum.
Figure 1.
(A) H & E stained hippocampal brain section from a PSAPP mouse. Scale bar = 500 µm. (B) Hierarchical cluster analysis defining three distinct regions of the hippocampus based on lipid content where the axonal layer is red, the dendritic layer is green, and the somatic layer is blue. Scale bar = 500 µm. (C) Average spectrum from each of the three regions illustrating differences in lipid content when normalized to protein content. Important peak frequencies are indicated. Inset shows the olefinic =CH stretching region (3012 cm−1) from the axonal layer spectrum.
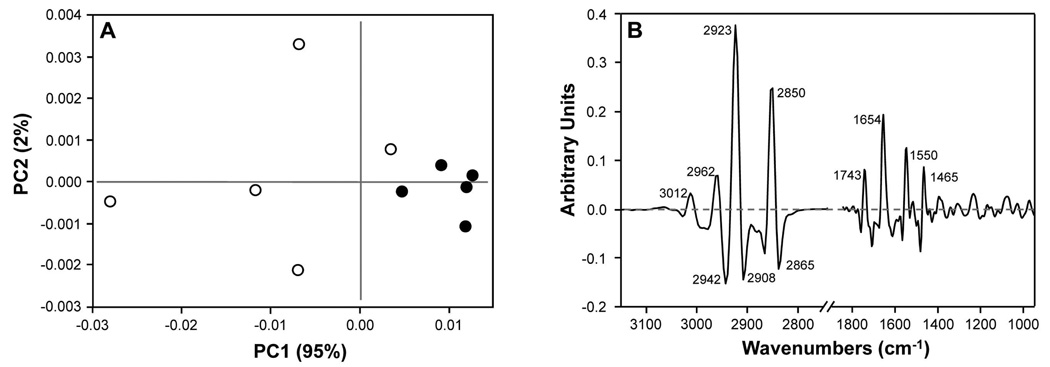
Our first goal was to use PCA to identify the specific regions of the FTIR spectra that contributed to the observed variance in the data. The PCA scores plot of the average axonal layer spectra from the endstage animals (Figure 2A) shows that PSAPP mice are easily discriminated from CNT by using the first two components, PC1 and PC2, which account for 95% and 2% of the variance, respectively. Interestingly, the earlier time points or other ROIs did not show such discrimination (data not shown). To visualize the spectral regions displaying the most variability, loadings plots were used to highlight the contribution of each spectral frequency to each PC. In the PC1 loadings plots (Figure 2B), the major contributions to spectral variation between PSAPP and CNT endstage mice were the olefinic (=CH) stretching vibration (3012 cm−1), the asymmetric and symmetric C–H stretching vibrations from CH2 and CH3 groups (2865 – 2960 cm−1), the C=O stretching vibration of carbonyl esters (1740 cm−1), the Amide I (1654 cm−1) and Amide II (1550 cm−1) protein bands, and the symmetric C–H scissoring of CH2 (1465 cm−1). However, the C–H stretching region showed the largest differences.
Figure 2.
(A) Scores plot of average axonal layer spectra from 56 week-old PSAPP (●) and CNT (○) animals; (B) PC1 loadings of average axonal layer spectra from 56 week-old PSAPP and CNT animals.
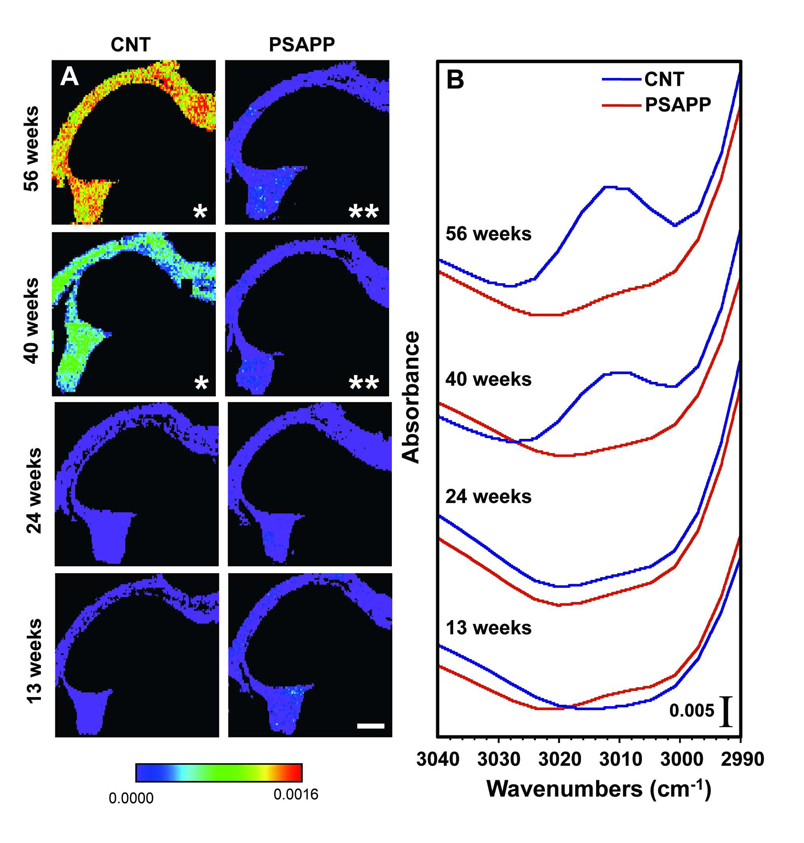
We then examined lipid unsaturation in each ROI by calculating the integrated area of the olefinic (=CH) peak (3000 – 3020 cm−1) divided by the integrated area of the total lipid region (2750 – 3020 cm−1). A lower ratio indicates a lower level of unsaturation in fatty acid chains, mainly arachidonic and docosahexaenoic acids 20,22. In both PSAPP and CNT animals, the olefinic peak at 3012 cm−1 was most easily detected in the axonal layer, while the content was below the detection limit in the other ROIs at all timepoints. Figure 3 shows that the intensity of the olefinic peak from the axonal layer average spectra increases in CNT mice, but not in PSAPP mice. The distribution of unsaturated lipid in the axonal layer is shown in Figure 3A, where red indicates a large olefinic peak area, demonstrating high unsaturated lipid content, whereas purple indicates a small olefinic peak area, demonstrating lower lipid unsaturation. In the 13 and 24 week-old CNT mice, a low olefinic content was observed throughout the axonal layer (Figure 3A, first column; Figure 3B). However, from 24 to 40 weeks-old, the level of unsaturated lipids increased, as evidenced by a significant increase in the median integrated area of the olefinic peak from 0 at 24 weeks to 0.0005 at 40 weeks (p < 0.01). The median integrated olefinic peak area increased even more at 56 weeks-old to 0.0013 (p < 0.01), indicating a further increase in unsaturated lipid content.
Figure 3.
(A) Olefinic content as a function of age in CNT mice (top row) and PSAPP (bottom row) in the axonal layer. Red indicates very high olefinic content while purple represents very low olefinic content. * = median is significantly different from both 13 and 24 week-old CNT mice (p < 0.01); ** = median is significantly different from CNT mice at the same time point (p < 0.01). Scale bar = 500 µm. (B) Average spectra from the axonal layer in CNT and PSAPP mice showing the olefinic 3012 cm−1peak.
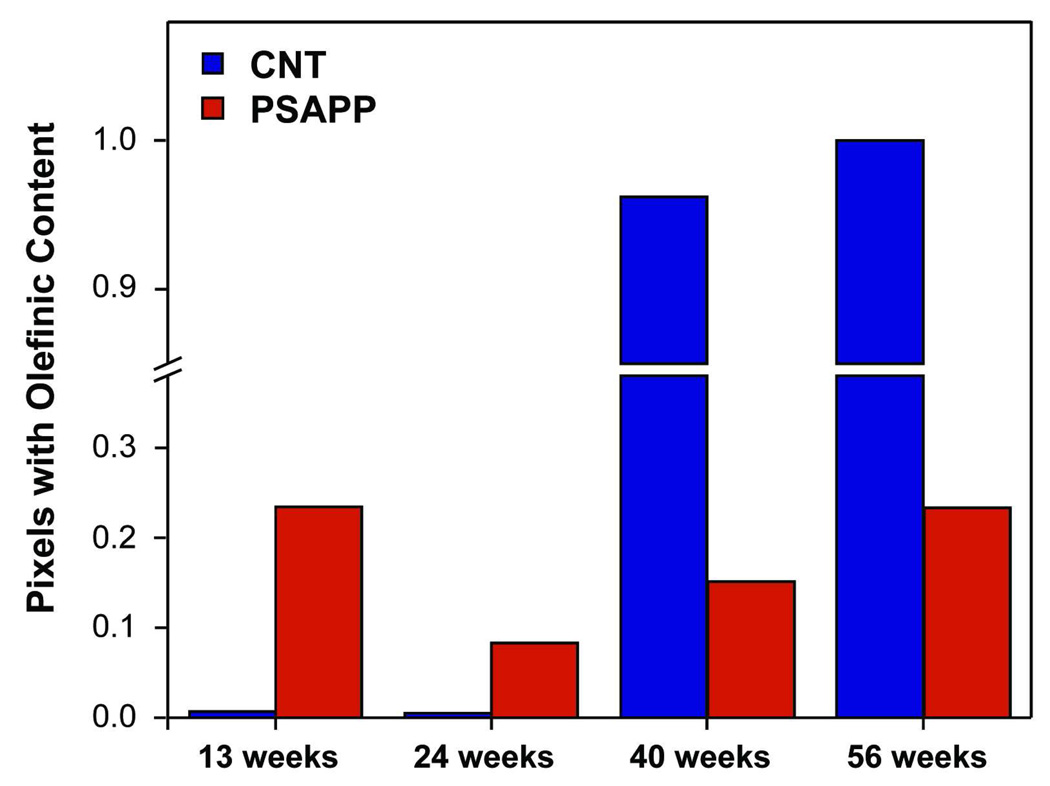
In the 13 and 24 week-old PSAPP mice, the unsaturated lipid content was similar to the CNT mice as the median integrated olefinic areas were identical (Figure 3A, second column; Figure 3B). However, at 40 and 56 weeks, the median integrated olefinic area remained low and relatively constant, indicating that plaque pathology was keeping the unsaturated lipid content low. Since the olefinic content was normalized to total lipid content, this implies an increase in unsaturation in the CNT mice with age, but not in the PSAPP mice. Likewise, the number of pixels in this region containing a non-zero value increased dramatically in CNT mice, but remained constant in PSAPP mice (Figure 4). Thus, while AD is thought to primarily affect the grey matter, these findings support evidence that white matter regions, including those connected to the hippocampus, are also affected 27.
Figure 4.
Fraction of pixels from the axonal layer ROI containing a non-zero value for integrated olefinic content.
Earlier studies have shown that the total lipid content in the myelin of the developing rat brain does not change, but the fatty acid composition of the lipid is altered28,29. For example, sphingomyelin, cerebroside, phosphatidic acid, and inositol plasmalogen are all increased30. In rodents, myelination is rapid during post-natal development, but steadily increases throughout aging30. Thus, it is possible that the observed increase in unsaturated lipid in the axonal layer of CNT mice represents an increase in myelination since myelin contains unsaturated fatty acids31. On the contrary, an increase in unsaturation in of the longest acyl chains of major myelin glycosphingolipids with aging could represent decreased myelin stability32 or possibly changes in the cellular makeup such as a loss of oligodendrocytes.
Since unsaturated lipid accumulated in the CNT mice, it is possible that accumulation in the PSAPP mice was inhibited. For example, the unsaturated class of lipids, dolichol, which increase membrane fluidity, increase substantially in the brain during aging33. In rats, it has been shown to increase 100-fold from birth to two years of age34 and appears to be a common feature of the aging process, though through unknown mechanisms35. However, in AD brain dolichol is markedly decreased36. A decrease in unsaturated lipids could also represent a deficiency in the uptake of the essential fatty acids linoleic and linolenic acids, both of which are required for biosynthesis of long-chain PUFAs37. Since mammals cannot produce either of these essential fatty acids, they must be introduced through the diet. Both the CNT and PSAPP mice in this study were fed identical, standard diets that contained linoleic acid. Thus, a dietary deficiency cannot explain the differences, but it is possible that the disease process impaired the normal processing pathways for the synthesis of PUFAs from dietary unsaturated fatty acids and/or the incorporation of them into the brain. For example, reduced desaturase enzyme activity has been implicated in AD 4.
The low unsaturated lipid content in the endstage PSAPP mice may also be a result of Aβ-mediated lipid peroxidation. Polyunsaturated lipids in the brain are readily attacked by free radicals because of their double bond content, becoming oxidized into lipid peroxides38. Aβ, the primary component of amyloid plaques, has been implicated in oxidative damage through metal ion reduction39, lipid peroxidation in synaptic plasma membranes40, 41, and production of HNE in hippocampal neurons42. Indeed, increased lipid peroxidation has been observed in human AD, mild cognitive impairment, and in transgenic mouse models of AD12,43,44, suggesting that antioxidant defenses are impaired in AD11. This is consistent with recent work by Petursdottir and colleagues18 who that demonstrated that 12 month-old senescence-accelerated mice had decreased α-tocopherol, a lipid soluble antioxidant, in the hippocampus and amygdala compared to young mice, which also coincided with a decreased proportion of docosahexaenoic acid and arachidonic acid in these regions.
In addition to lipid peroxidation, gliosis is a common occurrence in AD brain45,46. Glial cells are not only capable of producing free radicals47, but glial membranes also contain less unsaturated lipid than neuronal membranes48. In AD, astrocytes typically become reactive around amyloid plaques49; gliosis accompanied by tissue degeneration has also been observed in the dentate gyrus and subiculum of AD patients in response to lesions of the afferent projection sites50 and also in the white matter of AD patients51. Moreover, an increase in astrocytes is associated with a decrease in oligodendrocytes in the white matter of AD patients52. In addition, axonal and oligodendroglial loss is accompanied by deep white matter gliosis, which is independent from grey matter lesions53. It is possible that the decreased level of unsaturated lipid observed in the PSAPP mice in the present study may also represent astrocytic gliosis or oligodendroglial or axonal loss in response to the insults caused by plaque pathology.
CONCLUSION
In this study, we used FTIRI to demonstrate differences in unsaturated lipid accumulation in the hippocampus between control and a mouse model of Alzheimer’s disease. The PSAPP mouse model allowed us to examine unsaturated lipids as a function of age and plaque pathology. We found that unsaturated lipids increase during normal aging but this process is impaired in PSAPP mice beginning at early time points coincident with the formation of amyloid plaques. These results indicate that amyloid plaque formation may reduce lipid unsaturation through a number of routes, including the impairment of the normal processing pathways for PUFA synthesis, the mediation of lipid peroxidation, astrocytic gliosis, and oligodendroglial or axonal loss. While the exact mechanism is yet to be elucidated, it is clear that amyloid plaque formation reduces unsaturated lipid content in the brain, impacting brain growth, membrane fluidity, signal transduction, and cognitive development. Moreover, since these effects on white matter occur at the beginning stages of amyloid plaque formation, preventing the reduction of unsaturated lipids early may be critical in avoiding the onset of disease progression. Thus, FTIRI provides a direct approach for examining lipid composition as related to the progression of Alzheimer’s disease.
ACKNOWLEDGMENTS
The authors would like to thank Janelle Collins for her skillful technical assistance with the animal dissection and tissue preparation. We would also like to thank Alvin Acerbo and Randy Smith for assistance with the FTIR microscope. This work is funded by the National Institutes of Health Grants R01-GM66873 and S10-RR023782. The National Synchrotron Light Source is funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886.
Footnotes
There are no conflicts of interest associated with this work.
REFERENCES
- 1.Sastry PS. Prog. Lipid. Res. 1985;24:69–176. doi: 10.1016/0163-7827(85)90011-6. [DOI] [PubMed] [Google Scholar]
- 2.Salvati S, Attorri L, Di Benedetto R, Di Biase A, Leonardi F. Mini. Rev. Med. Chem. 2006;6:1201–1211. doi: 10.2174/138955706778742740. [DOI] [PubMed] [Google Scholar]
- 3.Lukiw WJ, Bazan NG. J. Nutr. 2008;138:2510–2514. doi: 10.3945/jn.108.096016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tully AM, Roche HM, Doyle R, Fallon C, Bruce I, Lawlor B, Coakley D, Gibney MJ. Br. J. Nutr. 2003;89:483–489. doi: 10.1079/BJN2002804. [DOI] [PubMed] [Google Scholar]
- 5.Calon F, Lim GP, Yang F, Morihara T, Teter B, Ubeda O, Rostaing P, Triller A, Salem N, Jr., Ashe KH, Frautschy SA, Cole GM. Neuron. 2004;43:633–645. doi: 10.1016/j.neuron.2004.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sahlin C, Pettersson FE, Nilsson LN, Lannfelt L, Johansson AS. Eur. J. Neurosci. 2007;26:882–889. doi: 10.1111/j.1460-9568.2007.05719.x. [DOI] [PubMed] [Google Scholar]
- 7.Lim GP, Calon F, Morihara T, Yang F, Teter B, Ubeda O, Salem N, Jr., Frautschy SA, Cole GM. J. Neurosci. 2005;25:3032–3040. doi: 10.1523/JNEUROSCI.4225-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oksman M, Iivonen H, Hogyes E, Amtul Z, Penke B, Leenders I, Broersen L, Lutjohann D, Hartmann T, Tanila H. Neurobiol. Dis. 2006;23:563–572. doi: 10.1016/j.nbd.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 9.Montine TJ, Neely MD, Quinn JF, Beal MF, Markesbery WR, Roberts LJ, Morrow JD. Free Radic. Biol. Med. 2002;33:620–626. doi: 10.1016/s0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 10.Butterfield DA, Perluigi M, Sultana R. Eur. J. Pharmacol. 2006;545:39–50. doi: 10.1016/j.ejphar.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Lovell MA, Ehmann WD, Butler SM, Markesbery WR. Neurology. 1995;45:1594–1601. doi: 10.1212/wnl.45.8.1594. [DOI] [PubMed] [Google Scholar]
- 12.Sayre LM, Zelasko DA, Harris PL, Perry G, Salomon RG, Smith MA. J. Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 13.Williams TI, Lynn BC, Markesbery WR, Lovell MA. Neurobiol. Aging. 2006;27:1094–1099. doi: 10.1016/j.neurobiolaging.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 14.Abdul HM, Sultana R, St Clair DK, Markesbery WR, Butterfield DA. Free Radic. Biol. Med. 2008;45:1420–1425. doi: 10.1016/j.freeradbiomed.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratico D, Uryu K, Leight S, Trojanoswki JQ, Lee VM. J. Neurosci. 2001;21:4183–4187. doi: 10.1523/JNEUROSCI.21-12-04183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, et al. J. Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 17.Braak H, Braak E. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 18.Petursdottir AL, Farr SA, Morley JE, Banks WA, Skuladottir GV. Neurobiol. Aging. 2007;28:1170–1178. doi: 10.1016/j.neurobiolaging.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 19.Soderberg M, Edlund C, Kristensson K, Dallner G. Lipids. 1991;26:421–425. doi: 10.1007/BF02536067. [DOI] [PubMed] [Google Scholar]
- 20.Sills RH, Moore DJ, Mendelsohn R. Anal. Biochem. 1994;218:118–123. doi: 10.1006/abio.1994.1149. [DOI] [PubMed] [Google Scholar]
- 21.Liu KZ, Bose R, Mantsch HH. Vib. Spectrosc. 2002;28:131–136. [Google Scholar]
- 22.Severcan F, Gorgulu G, Gorgulu ST, Guray T. Anal. Biochem. 2005;339:36–40. doi: 10.1016/j.ab.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 23.Garcia-Alloza M, Robbins EM, Zhang-Nunes SX, Purcell SM, Betensky RA, Raju S, Prada C, Greenberg SM, Bacskai BJ, Frosch MP. Neurobiol. Dis. 2006;24:516–524. doi: 10.1016/j.nbd.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 24.Jankowsky JL, Slunt HH, Ratovitski T, Jenkins NA, Copeland NG, Borchelt DR. Biomol. Eng. 2001;17:157–165. doi: 10.1016/s1389-0344(01)00067-3. [DOI] [PubMed] [Google Scholar]
- 25.Pidgeon C, Apostol G, Markovich R. Anal. Biochem. 1989;181:28–32. doi: 10.1016/0003-2697(89)90389-8. [DOI] [PubMed] [Google Scholar]
- 26.Roher AE, Weiss N, Kokjohn TA, Kuo YM, Kalback W, Anthony J, Watson D, Luehrs DC, Sue L, Walker D, Emmerling M, Goux W, Beach T. Biochemistry. 2002;41:11080–11090. doi: 10.1021/bi026173d. [DOI] [PubMed] [Google Scholar]
- 27.Salat DH, Tuch DS, van der Kouwe AJ, Greve DN, Pappu V, Lee SY, Hevelone ND, Zaleta AK, Growdon JH, Corkin S, Fischl B, Rosas HD. Neurobiol. Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norton WT, Poduslo SE. J. Neurochem. 1973;21:759–773. doi: 10.1111/j.1471-4159.1973.tb07520.x. [DOI] [PubMed] [Google Scholar]
- 29.Oulton MR, Mezei C. J. Lipid Res. 1976;17:167–175. [PubMed] [Google Scholar]
- 30.Wells MA, Dittmer JC. Biochemistry. 1967;6:3169–3175. doi: 10.1021/bi00862a026. [DOI] [PubMed] [Google Scholar]
- 31.Zabelinskii SA, Chebotareva MA, Kostkin VB, Krivchenko AI. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1999;124:187–193. doi: 10.1016/s0305-0491(99)00113-3. [DOI] [PubMed] [Google Scholar]
- 32.Malone MJ, Szoke MC. J Gerontol. 1982;37:262–267. doi: 10.1093/geronj/37.3.262. [DOI] [PubMed] [Google Scholar]
- 33.Soderberg M, Edlund C, Kristensson K, Dallner G. J. Neurochem. 1990;54:415–423. doi: 10.1111/j.1471-4159.1990.tb01889.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhang Y, Appelkvist EL, Kristensson K, Dallner G. Neurobiol. Aging. 1996;17:869–875. doi: 10.1016/s0197-4580(96)00076-0. [DOI] [PubMed] [Google Scholar]
- 35.Pallottini V, Marino M, Cavallini G, Bergamini E, Trentalance A. Biogerontology. 2003;4:371–378. doi: 10.1023/B:BGEN.0000006557.92558.60. [DOI] [PubMed] [Google Scholar]
- 36.Edlund C, Soderberg M, Kristensson K. Neurochem Int. 1994;25:35–38. doi: 10.1016/0197-0186(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 37.Youdim KA, Martin A, Joseph JA. Int. J. Dev. Neurosci. 2000;18:383–399. doi: 10.1016/s0736-5748(00)00013-7. [DOI] [PubMed] [Google Scholar]
- 38.Floyd RA. Proc. Soc. Exp. Biol. Med. 1999;222:236–245. doi: 10.1046/j.1525-1373.1999.d01-140.x. [DOI] [PubMed] [Google Scholar]
- 39.Huang X, Atwood CS, Hartshorn MA, Multhaup G, Goldstein LE, Scarpa RC, Cuajungco MP, Gray DN, Lim J, Moir RD, Tanzi RE, Bush AI. Biochemistry. 1999;38:7609–7616. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 40.Avdulov NA, Chochina SV, Igbavboa U, O'Hare EO, Schroeder F, Cleary JP, Wood WG. J. Neurochem. 1997;68:2086–2091. doi: 10.1046/j.1471-4159.1997.68052086.x. [DOI] [PubMed] [Google Scholar]
- 41.Murray IV, Sindoni ME, Axelsen PH. Biochemistry. 2005;44:12606–12613. doi: 10.1021/bi050926p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mark RJ, Lovell MA, Markesbery WR, Uchida K, Mattson MP. J. Neurochem. 1997;68:255–264. doi: 10.1046/j.1471-4159.1997.68010255.x. [DOI] [PubMed] [Google Scholar]
- 43.Markesbery WR, Kryscio RJ, Lovell MA, Morrow JD. Ann. Neurol. 2005;58:730–735. doi: 10.1002/ana.20629. [DOI] [PubMed] [Google Scholar]
- 44.Gu F, Zhu M, Shi J, Hu Y, Zhao Z. Neurosci. Lett. 2008;440:44–48. doi: 10.1016/j.neulet.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 45.Sturchler-Pierrat C, Abramowski D, Duke M, Wiederhold KH, Mistl C, Rothacher S, Ledermann B, Burki K, Frey P, Paganetti PA, Waridel C, Calhoun ME, Jucker M, Probst A, Staufenbiel M, Sommer B. Proc. Natl. Acad. Sci. U S A. 1997;94:13287–13292. doi: 10.1073/pnas.94.24.13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McGowan E, Sanders S, Iwatsubo T, Takeuchi A, Saido T, Zehr C, Yu X, Uljon S, Wang R, Mann D, Dickson D, Duff K. Neurobiol. Dis. 1999;6:231–244. doi: 10.1006/nbdi.1999.0243. [DOI] [PubMed] [Google Scholar]
- 47.Davis EJ, Foster TD, Thomas WE. Brain Res. Bull. 1994;34:73–78. doi: 10.1016/0361-9230(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 48.Norton WT, Abe T, Poduslo SE, DeVries GH. J. Neurosci. Res. 1975;1:57–75. doi: 10.1002/jnr.490010106. [DOI] [PubMed] [Google Scholar]
- 49.Pike CJ, Cummings BJ, Cotman CW. Exp. Neurol. 1995;132:172–179. doi: 10.1016/0014-4886(95)90022-5. [DOI] [PubMed] [Google Scholar]
- 50.Casanova MF, Stevens JR, Kleinman JE. Psychiatry Res. 1990;35:149–166. doi: 10.1016/0925-4927(90)90017-z. [DOI] [PubMed] [Google Scholar]
- 51.Chalmers K, Wilcock G, Love S. Neuropathol. Appl. Neurobiol. 2005;31:623–631. doi: 10.1111/j.1365-2990.2005.00678.x. [DOI] [PubMed] [Google Scholar]
- 52.Sjobeck M, Englund E. Neuropathol. Appl. Neurobiol. 2003;29:159–169. doi: 10.1046/j.1365-2990.2003.00456.x. [DOI] [PubMed] [Google Scholar]
- 53.Brun A, Englund E. Ann. Neurol. 1986;19:253–262. doi: 10.1002/ana.410190306. [DOI] [PubMed] [Google Scholar]