Abstract
Objectives
XPD is a major player in nucleotide excision repair, which is one of the basic pathways of DNA repair. The objective of this study was to investigate the association of XPD single nucleotide polymorphisms (SNPs) and the risk of squamous cell carcinoma of the head and neck (SCCHN) in Koreans.
Methods
We performed XPD +23591G>A and +35931A>C genotyping in 290 SCCHN patients and 358 controls.
Results
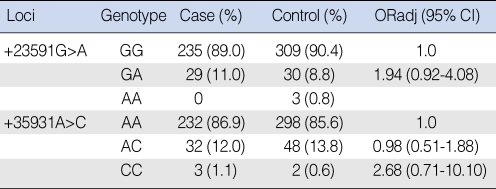
The frequencies of the XPD +23591G>A (GG/GA/AA) genotypes were 89.0%/11.0%/0% in the patients and 90.3%/8.8%/0.9% in the controls, respectively. The odds ratio (OR) of the XPD +23591 GA genotype was 1.94 (0.92 to 4.08) in reference to the GG genotype. The frequencies of the XPD +35931A>C (AA/AC/CC) genotypes were 86.9%/12.0%/1.1% in the patients and 85.6%/13.8%/0.6% in the controls, respectively. The OR of the XPD +35931 AC and CC genotypes were 0.98 (0.51 to 1.88) and 2.68 (0.71 to 10.1), respectively, in reference to the AA genotype. On the subgroup analyses according to the smoking and drinking statuses, the SNPs and haplotypes of XPD showed no statistically significant association with the risk of SCCHN.
Conclusion
The results of this study suggest that the XPD +23591G>A and +35931A>C SNPs are not associated with the risk of SCCHN in Koreans; however, a further study with a larger number of subjects is necessary to verify this conclusion.
Keywords: Polymorphism, XPD, Head and neck cancer, Squamous cell carcinoma
INTRODUCTION
Squamous cell carcinoma of the head and neck (SCCHN) accounted for 3.5% of all the registered cancers in Korea during 2001 (1). SCCHN is believed to be induced by environmental carcinogens. Tobacco smoking and alcohol consumption are recognized as major risk factors of SCCHN, yet only a small fraction of individuals exposed to tobacco or alcohol actually develop SCCHN. (2). Therefore, the possibility of genetic susceptibility due to the polymorphisms and phenotypic variations in the DNA repair enzymes or the metabolic enzymes that protect against the carcinogens in tobacco or alcohol have received much attention in attempts to determine the causes of SCCHN (3-6). The higher frequency of SCCHN in the first-degree relatives of SCCHN patients (7) and the early onset SCCHN in a subgroup of patients (8) lends support for such genetic susceptibility.
Accumulating evidence has shown that genetic differences in the DNA repair capacity and that are the result of genetic polymorphisms influence the risk of environmental carcinogenesis (9, 10). The importance of DNA repair for modulating the cancer risk of humans originated from the observation that individuals with the skin cancer-susceptible human disease xeroderma pigmentosum (XP) have defective nucleotide excision repair (NER). XP is a genetically complex disease that involves eight different complementation groups (A-G) (11).
XPD protein is one of nine subunits that compose transcription factor IIH (TFIIH), and TFIIH is a basal transcription factor that participates in NER and transcription initiation. The role of TFIIH in NER is to open up the damaged DNA to permit damage-specific nucleases to cleave both sides of the damage site. XPD protein has single strand DNA-dependent ATPase and 5'-3' DNA helicase activities and this protein is thought to participate in DNA unwinding during NER. Mutations in the XPD gene can give rise to repair and transcription defects (12).
In addition to these point mutations, variations in the XPD sequence are found in the general population. These variations are called single nucleotide polymorphisms (SNPs) with a highly variable frequency above 1%. To date, 17 polymorphisms in the XPD gene have been identified, and it is thought that certain XPD polymorphisms may be associated with the susceptibility to developing cancer (13).
Polymorphisms in the XPD gene have been studied in relation to the risk for head and neck cancer or lung cancer. Sturgis et al. (14, 15) reported that the XPD +35931A>C and +23591G>A SNPs are associated with a slightly increased risk of head and neck cancer, although this was not statistically significant. In a study on larynx, oral cavity and lung cancer, Buch et al. (16) reported that XPD +23591G>A and XPD +35931A>C SNPs are associated with a statistically significant increased risk of SCCHN; however, in another study the variant genotype of XPD +35931A>C showed no association with the risk of head and neck cancer (17). Similar contradictory results have been reported by studies on lung cancer (18-22).
No genetic study of XPD polymorphism in SCCHN has been performed in a sample of the Korean population. In this study, we investigated the frequencies of XPD SNPs in the SCCHN patients and controls in a sample of the Korean population and we evaluated the estimated risk of SCCHN.
MATERIALS AND METHODS
Study population
This case-control study was performed at the Department of Otolaryngology-Head and Neck Surgery, Hanyang University Hospital, Seoul, Korea from 1997 to 2004. The patient group contained 290 cases with pathologically verified SCCHN; there were 148 cases of larynx cancer, 56 cases of oral cancer, 42 cases of oropharynx cancer, 40 cases hypopharyx cancer and four cases of cancer at other sites. The control group consisted of 358 patients with chronic otitis media, chronic sinusitis or chronic tonsillitis, and they had no history of previous malignant disease or genetic disease. The patient group contained 252 males and 38 females with a mean age of 62.6 yr (range, 28 to 90 yr), and the control group contained 339 males and 19 females with a mean age of 38.8 yr (range, 21 to76 yr). All the participants provided us their written informed consent. The study protocol was approved by The Institutional Review Board of Hanyang University Hospital. Peripheral blood specimens were taken from all the participants and the samples were stored at -80℃ for DNA isolation. All the participants completed a questionnaire on personal information and lifestyle, including their smoking and drinking history. All the participants were ethnic Koreans.
Genotyping
We extracted the DNA from the peripheral blood using the Wizard™ Genomic DNA purification kit (Promega, Madison, WI, USA).
Two SNPs of XPD, +23591G>A and +35931A>C, were analyzed using the single base extension (SBE) technique. Polymerase chain reaction (PCR) using a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA) was performed on the samples, and the samples contained 1.25 pM of each primer, 5 ng of genomic DNA, 250 µM of the dNTPs and 0.15U Taq DNA polymerase. The primers used in this study are listed in Table 1. The primer extension reaction was performed using the SNaPshot ddNTP primer extension kit (Applied Biosystems). One unit of shrimp alkaline phosphatase (Amersham Life Science, Cleveland, OH, USA) was added to the reaction mixture to clean up the primer extension reaction products, and the mixture was incubated at 37℃ for one hour, followed by 15 min at 72℃ for enzyme inactivation. The amplified material and Genescan 120 Liz size-standard solution (Applied Biosystems) were added to Hi-Di formamide (Applied Biosystems) and this was all reacted at 95℃ for five minutes, and then it was incubated on ice for five minutes. Electrophoresis was performed on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems) and the genes were analyzed using ABI Prism GeneScan and Genotyper software.
Table 1.
Sequences of the primers used for XPD single nucleotide polymorphism genotyping by the single-base extension method
Statistical analysis
Statistical analysis of the XPD polymorphism frequencies between the SCCHN and normal control groups was performed using the chi square test. Odds ratios (OR), adjusted for age and gender, and the 95% confidence intervals (95% CI) of a genetic polymorphism and its associations were obtained using a logistic regression model. All the statistical data was obtained using SPSS ver. 15.0 (SPSS Inc., Chicago, IL, USA).
RESULTS
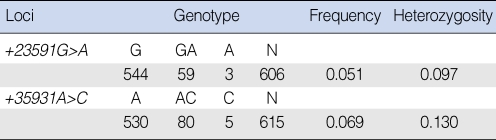
The allele and genotype frequencies are summarized in Tables 2 and 3. For the XPD +23591G>A SNP, genotyping was available in 606 of 648 subjects and the frequency of the variant allele A was 5.1%. The genotype distributions of XPD +23591 GG, GA, and AA were 89.0%, 11.0%, and 0% in the patient group and 90.4%, 8.8%, and 0.8% in the control group, respectively. The odds ratio and 95% confidence interval of the genotype GA in reference to GG was 1.94 (0.92 to 4.08). For the XPD +35931A>C SNP, genotyping was available in 615 of 648 subjects and the frequency of the variant allele C was 6.9%. The genotype distributions of XPD +35931 AA, AC, and CC were 86.9%, 12.0%, and 1.1% in the patient group and 85.6%, 13.8%, and 0.6% in the control group, respectively. The odds ratios and 95% confidence intervals of the genotypes AC and CC in reference to AA were 0.98 (0.51 to 1.88) and 2.68 (0.71 to 10.10), respectively.
Table 2.
Allele frequencies of the XPD single nucleotide polymorphisms in the Korean head and neck squamous cell carcinoma patients and the controls
Table 3.
Genotype frequencies of the XPD single nucleotide polymorphisms in the Korean head and neck squamous cell carcinoma patients and the controls
ORadj: Odds ratio adjusted for age and gender; CI: confidence interval.
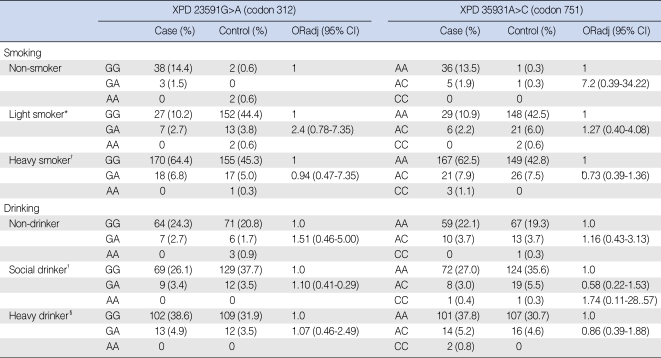
The risk estimates of SCCHN in the subgroups according to the smoking and alcohol consumption status are summarized in Table 4. The patient and control groups were classified as non-smokers, light smokers or heavy smokers. The light smoker group was defined as having a smoking history of <20 pack-year (number of packs smoked/day multiplied by the number of years of smoking) and the heavy smoker group was defined as having ≥20 packyear. There were no statistically significant differences in the risk of SCCHN according to the smoking status.
Table 4.
Genotype frequencies of the XPD single nucleotide polymorphisms and risk estimates according to the smoking and alcohol drinking statuses
*<20 pack-year; †≥20 pack-year; ‡alcohol consumption <2 times per week; §alcohol consumption ≥2 times per week.
ORadj: odds ratio adjusted for age and gender; CI: confidence interval.
The patient and control groups were also classified according to alcohol consumption into non-drinkers, social drinkers or heavy drinker. The social drinker group was defined as those who consumed alcohol <2 times per week and the heavy drinker group was defined as those who consumed alcohol ≥2 times per week. There were no statistically significant differences in the risk of SCCHN according to the alcohol drinking status.
We also analyzed the XPD polymorphisms and the risk of SCCHN according to the tumor subsite (oral cavity, larynx, hypopharynx and oropharynx). There were no statistically significant differences according to the tumor subsite.
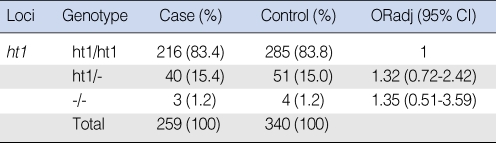
The most common haplotype (abbreviated as ht1) for XPD +23591G>A, +35931A>C was analyzed and the haplotype distribution is summarized in Table 5. There were no significant differences in the distribution of the XPD haplotype between the SCCHN and normal control groups.
Table 5.
Analysis of the haplotype of the XPD gene in the Korean head and neck squamous cell carcinoma patients and the controls
ORadj: odds ratio adjusted for age and gender; CI: confidence interval.
DISCUSSION
DNA repair systems play an important role not only in ensuring cellular survival, but also in preventing the development of cancer. At least four DNA repair pathways, including nucleotide excision repair, base excision repair, mismatch repair and double strand DNA break repair are involved in the repair of specific types of DNA damage. Aberrant DNA repair is associated with the development of several cancer types (13).
The NER pathway is the most versatile and ubiquitous mechanism for DNA repair and it is primarily involved in protection against the genotoxic damage induced by UV-irradiation or chemical carcinogens. In NER, the damaged part of a DNA strand is excised and then the gap is filled by repair replication using the complementary strand as a template. This "cut and patch" mode consists of five major steps: 1) recognition of the damaged DNA site, 2) incision of the damaged DNA strand on both sides of the defect, 3) removal of the damaged strand containing the lesion, 4) DNA replication to replace the excised region using the complementary strand as a template and 5) ligation to join the 3' end of the repair patch to the contiguous parental DNA strand (13, 23). Defects in the NER pathway are responsible for several human syndromes, including Cockayne syndrome, trichothiodystrophy and xeroderma pigmentosum, which are all characterized by defective repair of UV-damaged DNA and an increased risk of skin cancer (11).
Many of the studies on XPD SNPs have focused on +23591G>A, which is located at codon 312 of exon 10 and Asn is substituted for Asp, and +35931A>C, which is located at codon 751 of exon 2 and Gln is substituted for Lys. Several authors have suggested that these amino acid substitutions might alter XPD activity and so contribute to cancer development (13, 24).
This is the first study that has examined the relationship between XPD SNPs and the risk of SCCHN in a Korean cohort. We analyzed the XPD +23591G>A and +35931A>C SNPs in SCCHN patients and a control group. Overall, we could not find statistically significant differences in the XPD polymorphisms +23591G>A and +35931A>C between the SCCHN patients and the controls, although the XPD +23591 GA genotype showed a slightly increased risk of SCCHN (OR, 1.94; 95% CI, 0.92 to 4.08). We believe that further studies with larger numbers of subjects are warranted to confirm the role of these SNPs in SCCHN.
Several studies have examined the significance of XPD polymorphisms in head and neck cancer or lung cancer. Sturgis et al. reported that the XPD +35931 CC genotype is associated with a borderline increased risk for SCCHN (OR, 1.55; 95% CI, 0.96 to 2.52) (14). Furthermore, the XPD +23591 GA/AA genotype was associated with a slightly increased risk of SCCHN (OR, 1.28; 95% CI, 0.93 to 1.76), and subsequent studies showed that the risk was higher in combination with the ERCC1 8092 CC genotype (OR, 1.78; 95% CI, 0.99 to 3.17) (15), but they could not demonstrate statistical significance. In a study on larynx, oral cavity and lung cancer, Buch et al. (16) demonstrated that both the XPD +23591 GA/AA and XPD +35931 AC/CC genotypes were associated with a significantly increased risk of cancer (OR, 1.3, 95% CI, 1.0 to 1.8; OR, 1.5, 95% CI, 1.3 to 2.0, respectively). Furthermore, they found that the XPD +35931 AC/CC genotype in combination with the CCND1 870 GA/AA genotype markedly increased the risk of cancer (OR, 7.09; 95% CI, 4.03 to 12.5). In contrast, Huang et al. (17) reported that the XPD +39591A>C SNP was not associated with an increased risk of head and neck cancer.
There is similar controversy concerning studies on XPD SNPs and lung cancer. Spitz et al. (18) demonstrated that the combination of the XPD +23591 AA and +35931 CC genotypes showed an increased risk of lung cancer (OR, 1.84; 95% CI, 1.11 to 3.04), although each variant genotype of XPD +23591G>A and +35931A>C was not associated with increased cancer risk by itself. Two studies on Chinese lung cancer patients also reported that the XPD +23591 AA and +35931 CC genotype was associated with an increased risk of lung cancer (19, 20). However, several other authors have reported different results. Park et al. (21) reported that the distribution of the XPD +35931 AA/AC/CC genotype was not significantly different between the patients and controls, and they concluded that the XPD +35931A>C SNP was not associated with the risk of lung cancer in Koreans. Ryu et al. (22) also reported that the XPD +23591G>A and +35931A>C SNPs had no association with the chemotherapy response and prognosis of Korean lung cancer patients.
In the present study using a Korean cohort, the frequency of the variant A allele of the XPD +23591G>A SNP was 5.1%, and the frequency of the GA and AA genotypes was 9.6% and 0.5%, respectively. The frequency of the variant C allele of the XPD +35931A>C SNP was 6.9%, and the frequency of the AC and CC genotypes was 13.0% and 0.8%, respectively. The variant allele frequencies of the XPD +23591G>A SNP and the +35931A>C SNP in this study were markedly lower than those of previous studies in western countries, which reported the frequency of the variant A allele of XPD +23591G>A to be 33-38% and frequencies of the GA and AA genotypes to be 37-47% and 10-19%, respectively. They also reported the frequency of the variant C allele of XPD +35931A>C to be 36-41%, and frequencies of the AC and CC genotypes to be 37-52% and 10-17%, respectively (25-27). However, our results are similar to several studies conducted in East Asian populations. In one study on lung cancer in a Chinese sample, the frequency of the A allele in XPD +23591G>A was 6.9% and the frequencies of the GA and AA genotypes were 12.6% and 0.6%, respectively; the frequency of the C allele of XPD +35931A>C was 8.8% and the frequencies of the AC and CC genotype were 15.7% and 1.0%, respectively (19). Another study of a Japanese cohort reported the frequency of the C allele of XPD +35931A>C to be 5.2% and the frequencies of the AC and CC genotypes to be 8.8% and 0.8% (28). To date, there are two reports on XPD polymorphisms in Korean lung cancer patients. One study showed that the frequency of the C allele of XPD +35931A>C was 6.7% and the frequency of the AC and CC genotypes was 11.6% and 0.4%, respectively (21). In the other study, the frequency of the A allele in XPD +23591G>A was 3.7%, and the frequency of the GA and AA genotypes was 7.4% and 0%, respectively; the frequency of the C allele of XPD +35931A>C was 5.6% and the frequency of the AC and CC genotypes was 11.1% and 0%, respectively (22). We suggest that a low variant allele frequency of XPD SNPs in the Korean population might explain the difference between our results and those derived from Caucasian samples.
The XPD +23591G>A and +35931A>C SNPs were not associated with the risk of SCCHN in a Korean sample. We observed markedly lower frequencies of the variant alleles XPD +23591G>A and +35931A>C SNPs in this Korean sample, as compared with those of Caucasians. The low frequency of variant alleles might explain our failure to find a statistically significant association between the XPD SNPs and the risk of SCCHN. Further studies with larger numbers of cases are necessary to clarify the exact relationship between XPD SNPs and the risk of SCCHN.
Footnotes
No potential conflict of interest relevant to this article was reported.
References
- 1.National Central Cancer Center Registry. 2001 Annual report of the Korea Central Cancer Registry. Goyang: National Cancer Center; 2003. [Google Scholar]
- 2.Landis SH, Murray T, Bolden S, Wingo PA. Cancer statistics, 1998. CA Cancer J Clin. 1998 Jan–Feb;48(1):6–29. doi: 10.3322/canjclin.48.1.6. [DOI] [PubMed] [Google Scholar]
- 3.Yang M, Kim WH, Choi Y, Lee SH, Kim KR, Lee HS, et al. Effects of ERCC1 expression in peripheral blood on the risk of head and neck cancer. Eur J Cancer Prev. 2006 Jun;15(3):269–273. doi: 10.1097/01.cej.0000195709.79696.0c. [DOI] [PubMed] [Google Scholar]
- 4.Yang M, Kang MJ, Choi Y, Kim CS, Lee SM, Park CW, et al. Associations between XPC expression, genotype, and the risk of head and neck cancer. Environ Mol Mutagen. 2005 May;45(4):374–379. doi: 10.1002/em.20097. [DOI] [PubMed] [Google Scholar]
- 5.Tae K, Lee HS, Park BJ, Park CW, Kim KR, Cho HY, et al. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004 Sep 20;111(5):805–808. doi: 10.1002/ijc.20338. [DOI] [PubMed] [Google Scholar]
- 6.Shin CS, Ahn KS, Tae K, Lee HS, Kim HJ, Kong G. Genetic susceptibilities of cytochrome P4501A1 and glutathione S-transferase M1 to the risk for Korean head and neck squamous cell carcinoma patients. Korean J Otolaryngol - Head Neck Surg. 1999 Feb;42(2):202–208. [Google Scholar]
- 7.Foulkes WD, Brunet JS, Sieh W, Black MJ, Shenouda G, Narod SA. Familial risks of squamous cell carcinoma of the head and neck: retrospective case-control study. BMJ. 1996 Sep 21;313(7059):716–721. doi: 10.1136/bmj.313.7059.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schantz SP, Byers RM, Goepfert H, Shallenberger RC, Beddingfield N. The implication of tobacco use in the young adult with head and neck cancer. Cancer. 1988 Oct 01;62(7):1374–1380. doi: 10.1002/1097-0142(19881001)62:7<1374::aid-cncr2820620723>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Shen H, Zheng Y, Sturgis EM, Spitz MR, Wei Q. P53 codon 72 polymorphism and risk of squamous cell carcinoma of the head and neck: a case-control study. Cancer Lett. 2002 Sep 26;183(2):123–130. doi: 10.1016/s0304-3835(02)00117-9. [DOI] [PubMed] [Google Scholar]
- 10.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001 May 17;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 11.Lehmann AR. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 2001 Jan 01;15(1):15–23. doi: 10.1101/gad.859501. [DOI] [PubMed] [Google Scholar]
- 12.Evans E, Moggs JG, Hwang JR, Egly JM, Wood RD. Mechanism of open complex and dual incision formation by human nucleotide excision repair factors. EMBO J. 1997 Nov 03;16(21):6559–6573. doi: 10.1093/emboj/16.21.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002 Dec;11(12):1513–1530. [PubMed] [Google Scholar]
- 14.Sturgis EM, Zheng R, Li L, Castillo EJ, Eicher SA, Chen M, et al. XPD/ERCC2 polymorphisms and risk of head and neck cancer: a case-control analysis. Carcinogenesis. 2000 Dec;21(12):2219–2223. doi: 10.1093/carcin/21.12.2219. [DOI] [PubMed] [Google Scholar]
- 15.Sturgis EM, Dahlstrom KR, Spitz MR, Wei Q. DNA repair gene ERCC1 and ERCC2/XPD polymorphisms and risk of squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg. 2002 Sep;128(9):1084–1088. doi: 10.1001/archotol.128.9.1084. [DOI] [PubMed] [Google Scholar]
- 16.Buch S, Zhu B, Davis AG, Odom D, Siegfried JM, Grandis JR, et al. Association of polymorphisms in the cyclin D1 and XPD genes and susceptibility to cancers of the upper aero-digestive tract. Mol Carcinog. 2005 Apr;42(4):222–228. doi: 10.1002/mc.20086. [DOI] [PubMed] [Google Scholar]
- 17.Huang WY, Olshan AF, Schwartz SM, Berndt SI, Chen C, Llaca V, et al. Selected genetic polymorphisms in MGMT, XRCC1, XPD, and XR CC3 and risk of head and neck cancer: a pooled analysis. Cancer Epidemiol Biomarkers Prev. 2005 Jul;14(7):1747–1753. doi: 10.1158/1055-9965.EPI-05-0162. [DOI] [PubMed] [Google Scholar]
- 18.Spitz MR, Wu X, Wang Y, Wang LE, Shete S, Amos CI, et al. Modulation of nucleotide excision repair capacity by XPD polymorphisms in lung cancer patients. Cancer Res. 2001 Feb 15;61(4):1354–1357. [PubMed] [Google Scholar]
- 19.Liang G, Xing D, Miao X, Tan W, Yu C, Lu W, et al. Sequence variations in the DNA repair gene XPD and risk of lung cancer in a Chinese population. Int J Cancer. 2003 Jul 10;105(5):669–673. doi: 10.1002/ijc.11136. [DOI] [PubMed] [Google Scholar]
- 20.Chen S, Tang D, Xue K, Xu L, Ma G, Hsu Y, et al. DNA repair gene XRCC1 and XPD polymorphisms and risk of lung cancer in a Chinese population. Carcinogenesis. 2002 Aug;23(8):1321–1325. doi: 10.1093/carcin/23.8.1321. [DOI] [PubMed] [Google Scholar]
- 21.Park JY, Lee SY, Jeon HS, Park SH, Bae NC, Lee EB, et al. Lys751Gln polymorphism in the DNA repair gene XPD and risk of primary lung cancer. Lung Cancer. 2002 Apr;36(1):15–16. doi: 10.1016/s0169-5002(01)00447-0. [DOI] [PubMed] [Google Scholar]
- 22.Ryu JS, Hong YC, Han HS, Lee JE, Kim S, Park YM, et al. Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer. 2004 Jun;44(3):311–316. doi: 10.1016/j.lungcan.2003.11.019. [DOI] [PubMed] [Google Scholar]
- 23.Wood RD. Nucleotide excision repair in mammalian cells. J Biol Chem. 1997 Sep 19;272(38):23465–23468. doi: 10.1074/jbc.272.38.23465. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Y, Spitz MR, Shen H, Guo Z, Shete S, Hedayati M, et al. Modulation of repair of ultraviolet damage in the host-cell reactivation assay by polymorphic XPC and XPD/ERCC2 genotypes. Carcinogenesis. 2002 Feb;23(2):295–299. doi: 10.1093/carcin/23.2.295. [DOI] [PubMed] [Google Scholar]
- 25.Zhou W, Liu G, Miller DP, Thurston SW, Xu LL, Wain JC, et al. Gene-environment interaction for the ERCC2 polymorphisms and cumulative cigarette smoking exposure in lung cancer. Cancer Res. 2002 Mar 01;62(5):1377–1381. [PubMed] [Google Scholar]
- 26.Misra RR, Ratnasinghe D, Tangrea JA, Virtamo J, Andersen MR, Barrett M, et al. Polymorphisms in the DNA repair genes XPD, XRCC1, XRC C3, and APE/ref-1, and the risk of lung cancer among male smokers in Finland. Cancer Lett. 2003 Mar 10;191(2):171–178. doi: 10.1016/s0304-3835(02)00638-9. [DOI] [PubMed] [Google Scholar]
- 27.Benhamou S, Sarasin A. ERCC2 /XPD gene polymorphisms and lung cancer: a HuGE review. Am J Epidemiol. 2005 Jan 01;161(1):1–14. doi: 10.1093/aje/kwi018. [DOI] [PubMed] [Google Scholar]
- 28.Hamajima N, Saito T, Matsuo K, Suzuki T, Nakamura T, Matsuura A, et al. Genotype frequencies of 50 polymorphisms for 241 Japanese non-cancer patients. J Epidemiol. 2002 May;12(3):229–236. doi: 10.2188/jea.12.229. [DOI] [PMC free article] [PubMed] [Google Scholar]