Abstract
The processing of N-linked oligosaccharides by α-mannosidases in the endoplasmic reticulum and Golgi is a process conserved in plants and animals. After the transfer of a GlcNAc residue to Asn-bound Man5GlcNAc2 by N-acetylglucosaminyltransferase I, an α-mannosidase (EC 3.2.1.114) removes one α1,3-linked and one α1,6-linked mannose residue. In the present study, we have identified the relevant α-mannosidase II gene (aman-2; F58H1.1) from Caenorhabditis elegans and have detected its activity in both native and recombinant forms. For comparative studies, the two other cDNAs encoding class II mannosidases aman-1 (F55D10.1) and aman-3 (F48C1.1) were cloned, which encode, respectively, a putative lysosomal α-mannosidase and a Co(II)-activated α-mannosidase. The analysis of the N-glycan structures of an aman-2 mutant strain demonstrates that the absence of α-mannosidase II activity results in a shift to structures not seen in wild-type worms (e.g., N-glycans with the composition Hex5-7HexNAc2-3Fuc2Me) and an accumulation of hybrid oligosaccharides. Paucimannosidic glycans are almost absent from aman-2 worms, indicative also of a general lack of α-mannosidase III activity. We hypothesise that there is a tremendous flexibility in the glycosylation pathway of C. elegans which does not impinge, under standard laboratory conditions, on the viability of worms with glycotypes very unlike the wild-type pattern.
Keywords: Caenorhabditis, mannosidase, N-glycans
During the biosynthesis of N-glycans in eukaryotes a number of trimming and extension steps occur - the relative contributions of these steps then determine the final nature of the oligosaccharides present on a glycoprotein. As part of this process, a number of α-mannosidases are required to remove first a number of α1,2-linked mannose residues (1,2); after attainment of the Man5GlcNAc2 structure, N-acetylglucosaminyltransferase I (GlcNAc-TI1) may, depending on the species, the cell or the glycoprotein, act to facilitate the formation of hybrid, paucimannosidic and complex N-glycans (3). Thereafter, according to the ‘classical’ pathway a Golgi α1,3/6-mannosidase (α-mannosidase II) will, dependent on the action of GlcNAc-TI, remove two mannose residues - the result being so-called MGn (GlcNAc1Man3GlcNAc2), which is a substrate for other enzymes, such as GlcNAc-TII, core fucosyltransferases and, in insects and nematodes, a processing Golgi hexosaminidase (4,5). The effect on glycan structure of ‘knocking-out’ α-mannosidase II genes in invertebrates has been previously unknown, but an α-mannosidase III has been characterised in insects (6), which could in theory act in an alternative route of N-glycan processing. However, even though GlcNAc-TI-independent Golgi α1,2/3/6-mannosidases similar to mannosidase III have also been found by assaying crude mammalian extracts (7), recent data indicate that mammals have two GlcNAc-TI dependent α-mannosidases (mannosidases II and IIx); knocking-out both these genes is necessary and sufficient to abolish complex N-glycan biosynthesis in embryos (8).
In addition to its demonstrated presence in a number of mammalian cell extracts, α-mannosidase II activity has been recently reported in Caenorhabditis elegans microsomes (5), but otherwise no data exists about this enzyme in the nematode. Considering that the complete genome of C. elegans has been sequenced, an obvious aid in identifying any α-mannosidase II gene is that various cDNA sequences encoding this enzyme have been cloned from insects (9) and mammals (10,11); phylogenetic and biochemical comparisons indicate that these are members of the class II mannosidase family. As such their sequences display similarity to those of lysosomal mannosidase (12), α-mannosidase III (6), α-mannosidase IIx (11), a cytosolic/ER α-mannosidase (13) and a lysosomal α1,6-specific mannosidase (14). On the other hand, mannosidase II is unlike the ‘classical’ ER α-mannosidase (15) and Golgi α-mannosidases IA, IB and IC (16) which are class I mannosidases. Most recently, the Drosophila α-mannosidase II has been crystallized and its 3D-structure determined (17).
In this study, we identified three class II mannosidase genes from Caenorhabditis elegans, cloned the corresponding cDNAs, expressed these in Pichia pastoris and compared the properties of the encoded proteins; on the basis of the various analyses, we conclude that we have identified the α-mannosidase II (aman-2) gene from the nematode. Furthermore, examination of a relevant mutant indicated that this enzyme has a major role in “normal” N-glycan biosynthesis in the worm.
Experimental Procedures
Cloning of the nematode class II mannosidase cDNAs
Open reading frames for putative C. elegans class II mannosidases were identified by BLAST searching and cDNA fragments encoding truncated forms of these enzymes (lacking the cytosolic and transmembrane regions) were amplified by RT-PCR using Trizol-purified RNA from N2 worms, Expand polymerase and the primer pairs Ce_ManI/1/ClaI (ccatcgatccaagattgtgcatggaata) and Ce_ManI/2/SacII (tccccgcggttataattttgacattttgcgttc), Ce_ManII/1 (ggaattgaaaccggagctg) and Ce_ManII/2/SacII (tccccgcggttaaaatgatacaagaatact), and Ce_ManIII/1/AclI (ccggaacgtttcatcaaagattaggacagca) and Ce_ManIII/2/SacII (tccccgcggtcatcgataaagaatcaaagtc) with an annealing temperature of 58 °C and an extension time of 4′30″. The fragments were purified using a GFX DNA purification kit (GE Healthcare), cut with ClaI (a site being naturally present in the aman-2 cDNA and in the primer for the aman-1 cDNA) or with AclI (for the aman-3 cDNA fragment) and SacII, prior to ligation into the pPICZαC vector (Invitrogen) cut with ClaI and SacII. Positive clones were sequenced, linearised and transformed into the Pichia pastoris GS115 strain.
Characterisation of recombinant α-mannosidases
Transformed Pichia were grown at 16 or 30 °C in the presence of 1% methanol and culture supernatants were collected on the second, third, fourth, fifth and sixth days after commencement of induction. Supernatants were initially screened using p-nitrophenyl-α-mannopyranoside as a substrate in assays containing a final concentration of 5 mM substrate (stock 100 mM in dimethylsulphoxide), McIlvaine buffers of varying pH (18) and 20 μl of culture supernatant; tests with cations were performed at 10 mM final concentration of the chloride salts, whereas swainsonine (Sigma) and deoxymannojirimycin (Industrial Research Ltd, New Zealand) were employed at concentrations of, respectively, 0.01 - 1 μM and 0.1 - 10 mM. Incubations (total volume 50 μl) were allowed to proceed for 2-16 hours at various temperatures prior to termination with 250 μl 0.4 M glycine-NaOH, pH 10.4; absorbances were measured using a 96-well plate reader at 405 nm and corrected by subtracting the values of control incubations lacking substrate in order to calculate ΔA405. Furthermore, other controls with substrate and either no supernatant or a supernatant of yeast not expressing recombinant mannosidase were also performed.
For assays with pyridylaminated GlcNAc1Man5GlcNAc2 (Man5Gn-PA; see Scheme I for an explanation of glycan nomenclature) or Man5GlcNAc2 (Man5-PA), 0.1 nmol substrate was incubated with 1 μl unconcentrated yeast culture supernatant and 2 μl 0.4 M 2-morpholinoethanesulfonic acid buffer (pH 6) in a final volume of 5 μl for 14 hours in a PCR tube at 30 °C, prior to RP-HPLC (Hypersil ODS) using a gradient of 4.2 - 5.7 % MeOH (95.8 - 94.3% 0.1 M ammonium acetate, pH 4); the pyridylamino-labeled glycans were detected by fluorescence (320/400 nm). Control assays were performed using the supernatant of yeast transformed with the pPICZαC vector lacking an insert. Fractions were collected, dried and subjected to either MALDI-TOF MS using 2,5-dihydroxybenzoic acid as matrix or normal phase HPLC using a Palpak Type N column (4.6 × 250 mm; Takara). In the latter case, mixtures of 25 (buffer A) or 50 (buffer B) parts 3% acetic acid (adjusted to pH 7.3 with triethylamine) and 10% acetonitrile with, respectively, 75 or 50 parts 100 % acetonitrile were prepared. The gradient was then as follows: 0 - 5 minutes, 10% B; 5 - 45 minutes, 10 - 100% B; 45 - 55 minutes, 100%, 55 - 56 minutes, 10% (total run time, 65 minutes; flow rate, 1 ml/min). Samples were taken up in 80% acetonitrile prior to injection and peaks detected by fluorescence (310 nm excitation, 380 nm emission).
Scheme I.
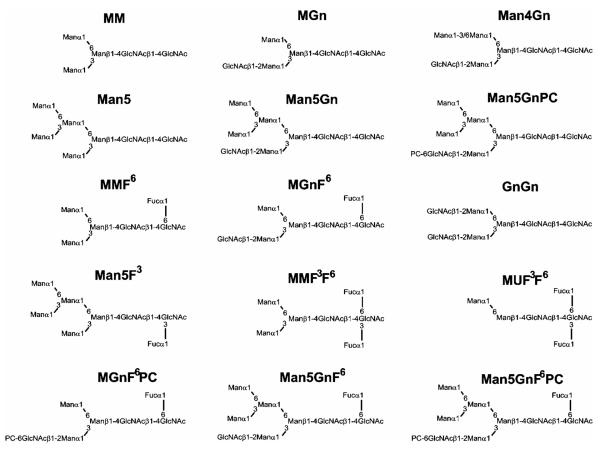
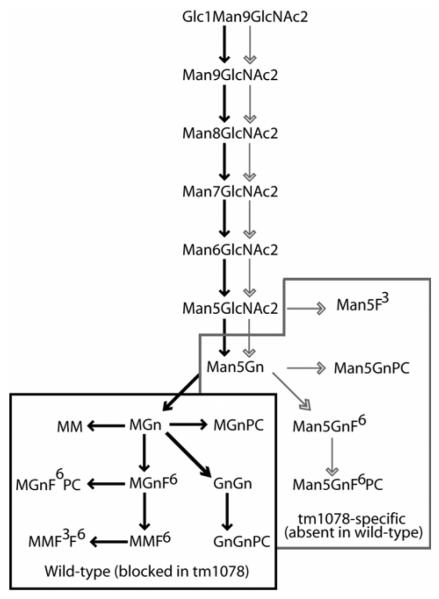
Structures of N-glycans referred to in this study. Abbreviations are based on the system of Schachter (ref. 3). For the smaller structures, the order of the letters M (mannose), Gn (N-acetylglucosamine) and U (non-substituted) indicates the antenna on which the sugar is the terminal residue; thus, MGn indicates a structure for which mannose is the terminal residue on the ‘upper’ α1,6-arm and GlcNAc is on the α1,3-arm, whereas GnGn indicates there are two non-reducing terminal GlcNAc residues. For fucose modifications of the core, F3 indicates a core α1,3-fucose and F6 a core α1,6-fucose. Based on other studies, the terminal phosphorylcholine (PC) is assumed to be 6-linked to GlcNAc.
Mannosidase assays of wild-type and mutant worms
Extracts of worms grown in liquid culture with E. coli OP50 were prepared as previously described (19). The tm1078 strain (displaying a 304 bp deletion including parts of the fifth and sixth exons of the F58H1.1 gene) was acquired from the National Bioresource Project for the Experimental Animal Nematode C. elegans (Tokyo Women’s Medical University, Japan). The yield of tm1078 worms (9 g wet weight from 500 ml culture) was comparable to that of the wild-type N2. α-Mannosidase II activity in worm extracts was determined using Man5Gn-PA as substrate and analysed by RP-HPLC as described above.
Western blotting
Crude whole extracts of worms were analysed by Western blotting using anti-horseradish peroxidase and anti-phosphorylcholine (TEPC-15) antibodies after separation by SDS-PAGE on 12.5% gels and transfer to nitrocellulose. After blocking with 0.5% (w/v) BSA, membranes were incubated with either rabbit anti-horseradish peroxidase (Sigma; 1:12,500) or mouse TEPC-15 (1:170). To detect the bound primary antibody or lectin, alkaline phosphatase-conjugated anti-rabbit IgG (1:2000) or horseradish peroxidase-conjugated anti-mouse IgA (1:200) were used together with the relevant chromogenic substrate (5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium or chloronaphthol/hydrogen peroxide).
Glycan analysis of wild-type and mutant worms
N-glycans were prepared from worms according to our previously-published procedures by PNGase A digestion of peptic peptides, prior to MALDI-TOF MS analysis in both linear and reflectron modes on respectively a ThermoBioanalysis Dynamo or a Bruker Ultraflex instrument (19). Furthermore, portions of N-glycans of wild-type and mutant strains were subject to pyridylamination and RP-HPLC also as previously described (20,21); collected fractions were analysed further by MALDI-TOF MS on a Perspective Biosystems Voyager DE using either 2,5-dihydroxybenzoic acid or 6-aza-2-thiothymine as matrix. Selected fractions were analysed further after treatment overnight either with jack bean β-hexosaminidase (5 mU; in the presence or absence of 5 mU bovine α-fucosidase) in 0.1 M sodium citrate, pH 5, buffer at 37 °C (22) or with 48% aqueous hydrofluoric acid at 0 °C (23). In the latter case, the reagent was removed by evaporation under a stream of nitrogen.
Phosphorylcholine-substituted glycans were also analysed by MALDI-TOF-MS (Ultraflex Tof/Tof, BrukerDaltonics, Bremen) using a dihydroxybenzoic acid matrix (10 mg/ml DHB in 50% aqueous acetonitrile, 1% phosphoric acid) either in the MS or MS/MS mode. Normally 100 - 500 shots were summed. For external calibration, a peptide standard mixture (Bruker) was used, whereas for MS/MS calibration the theoretical monoisotopic masses of the phosphorylcholine-substituted glycans were applied.
Tissue-specific promoter analysis
The reporter plasmid used was a form of the pPD95.67 (L2459) promoterless gfp vector2, which was the kind gift of Dr. Andrew Fire. This vector was then modified by insertion of a HindIII/XbaI fragment carrying the unc-119 gene into its multiple cloning site to generate pPD95.67/unc-119. Then, a genomic fragment corresponding to the 2000 bp upstream of the aman-2 gene was isolated by PCR using the primers ManII_prom/1/XbaI gctctagacggacgagaagtacaaat and ManII_prom/2/XbaI gctctagacccatgctttatcttgccat. Both the fragment and the pPD95.67/unc-119 vector were cut with XbaI and ligated. A selected clone was sequenced and verified to contain the expected aman-2 upstream region and was used to transform unc-119 (ed3) mutants with a particle gun (24). Individuals from integrated lines were examined by confocal laser scanning microscopy (Leica TCS SP2) with Ar-laser excitation at 488 nm and emission 500-540 nm using an HC Plans 10×/25 occular and HC PL Fluotar 10×/0.30 objective.
Results
Identification of the nematode class II mannosidase genes
Databank searches using the BLAST program, as well as examination of the CaZy database3, indicate that the C. elegans genome encodes three class II mannosidases, which are members of glycohydrolase family 38 (25). These have the Wormbase designations F48C1.1, F55D10.1 and F58H1.14. The predicted F48C1.1 cDNA encodes a protein of 1064 amino acids with 30% identity to the human α-mannosidase II (MAN2A1) (11) and 28% to insect (Sf9) α-mannosidase III (6), whereas F55D10.1 encodes a protein of 955 residues with 38% identity to human lysosomal mannosidase (MAN2B1) (12) and F58H1.1 encodes a protein of 1143 residues with 36% identity to human α-mannosidase II, 38% to human α-mannosidase IIx (MAN2A2) (11), 29% to C. elegans F55D10.1 and 25% to the human lysosomal mannosidase. A recent phylogenetic analysis (14) also suggests that F55D10.1 and F58H1.1 are, respectively, within the lysosomal and Golgi clades of the class II mannosidase family, whereas F48C1.1 is not closely associated with any other mannosidase sequence.
All worm class II mannosidase homologues contain an Asp-Phe-Pro motif, conserved in comparison to mammalian lysosomal and Golgi mannosidases; the aspartate residue of this motif has been predicted to be the catalytic nucleophile of the Drosophila Golgi α-mannosidase II (17), jack bean α-mannosidase (26) and bovine lysosomal α-mannosidase (27). The predicted F58H1.1 protein sequence contains three predicted N-glycosylation sites, one of which (Asn320) has been confirmed to be occupied in vivo (28); the F48C1.1 sequence also contains three predicted N-glycosylation sites, whereas the F55D10.1 sequence possesses seven. On the basis of the in silico analysis and its length, we postulated that F58H1.1 is the closest relative to Golgi α-mannosidase II. Thus, this gene, which we designate aman-2, and the corresponding mutant were the major focus of our later studies. The aman-1 (F55D10.1) and aman-3 (F48C1.1) cDNAs were also cloned so as to facilitate a comparative enzymatic characterisation to verify that the AMAN-2 protein is the sole Golgi α-mannosidase II in the worm.
Characterisation of nematode class II mannosidases
The cloned partial cDNAs encoding AMAN-2 and the two other class II mannosidases were engineered into vectors suitable for expression in Pichia pastoris. In the case of aman-2, an expression vector was constructed containing nt 366-3432 (corresponding to residues 123-1143 of the predicted protein) in frame with the yeast α-mating factor secretion signal. Truncation of the first 122 residues was performed on the basis of two criteria: first, due to the relative lack of suitable restriction sites, use was made of the natural ClaI site within the F58H1.1 sequence which was fortuitously suitable for in-frame ligation into the pPICZαC vector; second, the first 106 amino acids of mammalian α-mannosidase II were previously shown to be absent from an active chymotrypsin-released form of the enzyme (10). Indeed residue 124 of the worm enzyme corresponds to residue 113 of the murine enzyme and is prior to the first significant region of homology displayed by a DXQMLDXY sequence conserved between the mammalian, fruitfly and worm sequences. The aman-1 and aman-3 constructs, generated so as to facilitate comparative characterisation, merely lacked the regions encoding the putative cytoplasmic and transmembrane domains.
Initial screening of supernatants of yeast transfected with the aman-2 cDNA construct indeed showed that a mannosidase activity, not present in control yeast supernatants, hydrolysed p-nitrophenyl-α-mannoside at a pH value of 6, but not at pH 4.5, regardless of whether the expression was performed at 16 or 30 °C. On the other hand, yeast transformed with aman-1 expressed a secreted α-mannosidase with activity at pH 4.5, which corresponds to the optimum found for the recombinant human lysosomal α-mannosidase (12). In the case of yeast transformed with the aman-3 construct, we considered the possibility that the encoded enzyme may be similar to the Sf9 Co(II)-activated mannosidase III (6); therefore, initial assays were performed in the absence and presence of Co(II) and indeed a mannosidase activity not found in control supernatants was detected overnight only in the presence of the cation at pH 6. Furthermore, control incubations of supernatants of mannosidase-transformed yeast with no substrate yielded results comparable to incubations of supernatants of yeast transformed with empty vector (data not shown).
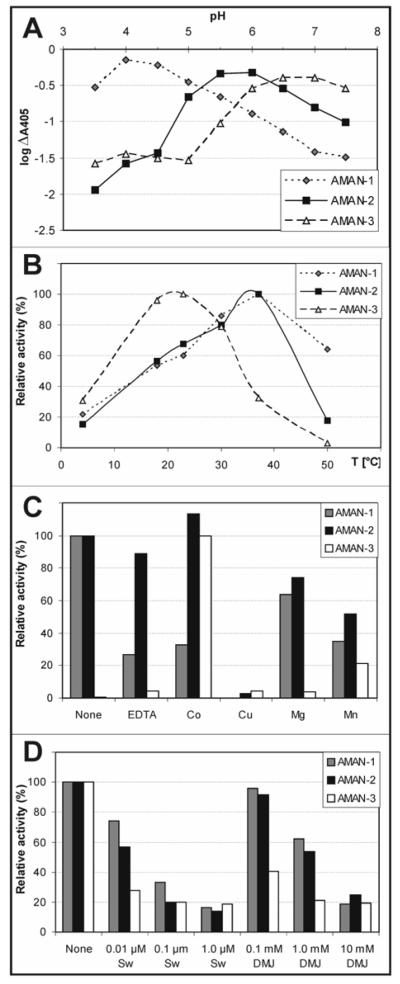
A fuller analysis with p-nitrophenyl-α-mannoside as substrate verified that the pH optimum of AMAN-2 was indeed at pH 5.5-6, compatible with other studies on Golgi α-mannosidase II from other species; based on a method described by Dixon (29), extrapolation of the linear regions of the plot of pH against log10ΔA405 (as a measure of log V) suggests the presence of titratable side chains with pKa values of 5.3 and 6.0 (Figure 1A) and so is potentially consistent with the putative roles of Asp204 and Asp341 as being, respectively, the catalytic nucleophile and the catalytic acid base in the Drosophila α-mannosidase II (17). The optima of AMAN-1 and AMAN-3 were at, respectively, pH 4-4.5 and pH 6.5 (Figure 1A) and buffers of these pH values were used in the subsequent assays. The temperature optima for AMAN-1 and AMAN-2 were in the region of 30 - 37 °C; AMAN-3, on the other hand, displayed optimal activity at 23 °C (Figure 1B). Taking the ΔA405 as a measure of V, an Arrhenius plot (not shown) suggested an activation energy for the AMAN-2 reaction of 28 - 30 kJ/mol (7 kcal/mol; data not shown), as compared to the 46 kJ/mol (11 kcal/mol) determined, also using V rather than an actual kcat, for the native rat liver α-mannosidase II (30).
Figure 1.
Characterisation of Caenorhabditis class II mannosidases. Supernatants of yeast expressing Caenorhabiditis mannosidases were assayed with p-nitrophenyl-α-mannoside for either two (AMAN-1), four (AMAN-2) or sixteen (AMAN-3) hours either using McIlvaine buffers with differing pH values (A), at different temperatures (B), in the presence of 10 mM of various cations or EDTA (C) or in the absence or presence of different concentrations of swainsonine (Sw) or deoxymannojirimycin (DMJ) (D). The amount of enzyme activity was calculated after subtracting the values of incubations lacking substrate from the observed A405 data, thus yielding the ΔA405 values used to calculate relative activity.
Of various cations tested, only Cu(II) was observed to completely inhibit AMAN-2 - such a result has been previously shown with the Drosophila and murine forms of α-mannosidase II (31). On the other hand, Co(II), Mg(II), Mn(II) and EDTA had either no or only a minor effect on the activity of AMAN-2 in comparison to incubations performed in the absence of these reagents (Figure 1C); this is again consistent with previous data on mannosidase II from other species. The EDTA-insensitivity of AMAN-2 would indicate that, if the Caenorhabditis enzyme is also a zinc-containing protein like the Drosophila form, the Zn(II) is tightly bound. The Co(II)-insensitivity of both AMAN-1 and AMAN-2 is in stark contrast to the properties of AMAN-3 (Figure 1C), which, like its presumed ‘orthologue’ from Sf9 cells (6), only showed significant activity in the presence of Co(II) and, to a lesser extent, Mn(II); indeed, for all other tests shown, AMAN-3 was assayed in the presence of 10 mM CoCl2. The inhibitors swainsonine and deoxymannojirimycin were also tested; approximately 45% inhibition of AMAN-2 was achieved at 10 nM of the former and 1 mM of the latter (Figure 1D). These results are in line with the respective IC50 values of 20 nM and 0.4 mM determined for inhibition of Drosophila Golgi mannosidase II (17). AMAN-1 and AMAN-3 were also similarly sensitive to these reagents, although AMAN-3 appeared to be somewhat more sensitive to deoxymannojirimycin.
To confirm the specificity of the enzymes, assays were performed using pyridylaminated forms of Man5Gn (referred to by others sometimes as M5Gn3 or M5Gn) and Man5 as substrates. The subsequent RP-HPLC chromatogram for AMAN-2 showed, after 14 hours, the conversion of around 80% of the Man5Gn substrate to a peak co-eluting with an MGn standard; this activity is absent from a control supernatant (Figure 2A and 2B). In contrast, no digestion of Man5 was observed as judged by RP-HPLC and MALDI-TOF MS (data not shown), demonstrating the requirement of the enzyme for the prior action of N-acetylglucosaminyltransferase I. Furthermore, no digestion of Man5Gn was observed when using supernatants of yeast expressing AMAN-1 and AMAN-3, whereas with AMAN-3 some degradation of Man5 was observed as expected (data not shown).
Figure 2.
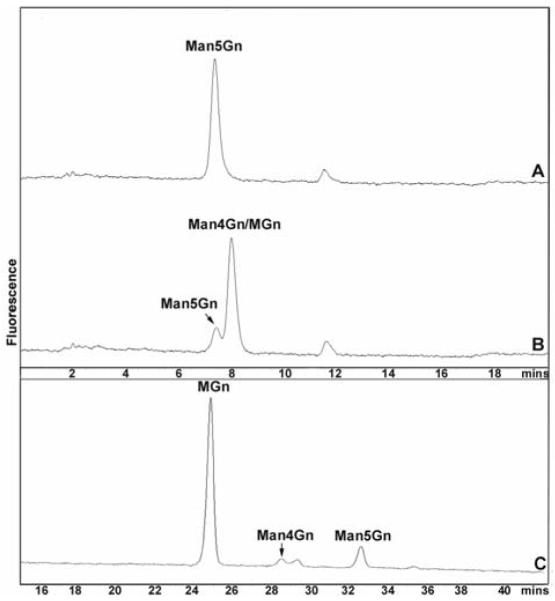
N-glycan digestion products of Caenorhabditis AMAN-2. Pyridylaminated Man5Gn was incubated with supernatant of yeast either transformed with empty vector (A) or expressing Caenorhabiditis AMAN-2 (B) for 14 hours prior to RP-HPLC. The two products MGn and Man4Gn co-elute, whereas the small peak is due to residual Man5Gn. The combined fractions from chromatogram B were then subject to normal phase HPLC (C) and were also subject to MALDI-TOF MS (not shown).
The product peak from the 14 hour incubation of AMAN-2 with Man5Gn was collected and subject to MALDI-TOF MS; this analysis showed the presence of both MGn (m/z 1194; [M+H]+) and Man4Gn (m/z 1356) as products. The purified incubation products were also analysed by normal phase HPLC, which more easily differentiates MGn, Man4Gn and Man5Gn; this verified that the vast majority of product at this time-point co-eluted with an MGn standard (Figure 2C). A time course analysis was also performed (data not shown); samples were removed after 1, 2, 4 and 12 hours, subject to HPLC as well as MALDI-TOF MS of the combined substrate and product peaks. These data confirmed Man4Gn as being an intermediate product of the enzyme reaction throughout the time course.
Biochemical analysis of a Golgi α-mannosidase II mutant
In order to test the in vivo function of the aman-2 gene, we made use of a publicly-available mutant C. elegans strain (tm1078), which has a deletion within the F58H1.1 gene5. First enzyme assays were performed on extracts of the mutant and these tests, using pyridylaminated Man5Gn, indicated that there was no α-mannosidase II activity in the tm1078 worms, in contrast to the wild-type (Figure 3). The wild-type extract indeed also digested the MGn α-mannosidase II product partly to MM, presumably due to the presence of a putative Golgi hexosaminidase (5). As a control, to show that other enzymes were indeed active in the mutant worm extract, the activity of hexosaminidase was tested with pyridylaminated GnGn, which was indeed degraded primarily to GnM in a manner similar to that seen in wild-type extract (data not shown).
Figure 3.
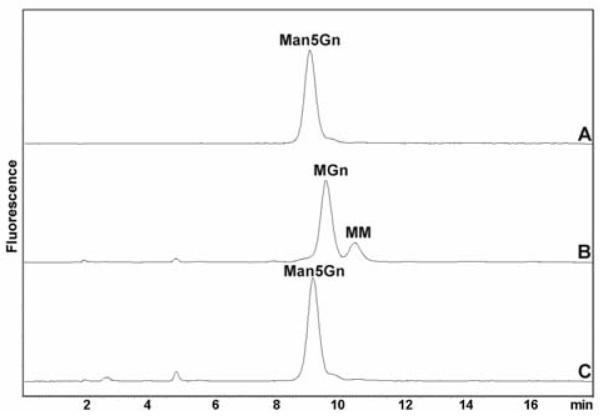
Mannosidase assays of wild-type and mutant Caenorhabditis. Pyridylaminated Man5Gn was incubated either with no enzyme (A) or with either wild-type (B) or tm1078 (C) extracts overnight at 30 °C prior to RP-HPLC. The tm1078 extract displays no α-mannosidase II activity compatible with deletion of part of the aman-2 gene in this strain.
Considering the importance of α-mannosidase II in the ‘classical’ N-glycan processing pathway, changes in the occurrence of carbohydrate-relevant epitopes may be expected to occur. Previously wild-type N2 worm extracts have been shown to display binding, upon Western blotting, to anti-horseradish peroxidase (19,32) and to TEPC-15 (an anti-phosphorylcholine IgA) (33). Anti-horseradish peroxidase stains neural cells in C. elegans and other invertebrates, whereas TEPC-15 recognises phosphorylcholine on bacterial cell surfaces (34,35) and on the N-glycans of the immunomodulatory excretory-secretory glycoprotein ES-62 from the nematode Acanthocheilonema viteae (36). Western blotting was therefore performed with these reagents: this showed a reduced, but still visible, presence of both the anti-horseradish peroxidase and phosphorylcholine epitopes as compared to the wild-type (Figure 4). The data with the former can be compared to the complete abolition of anti-HRP staining in the fut-1 mutant (19).
Figure 4.
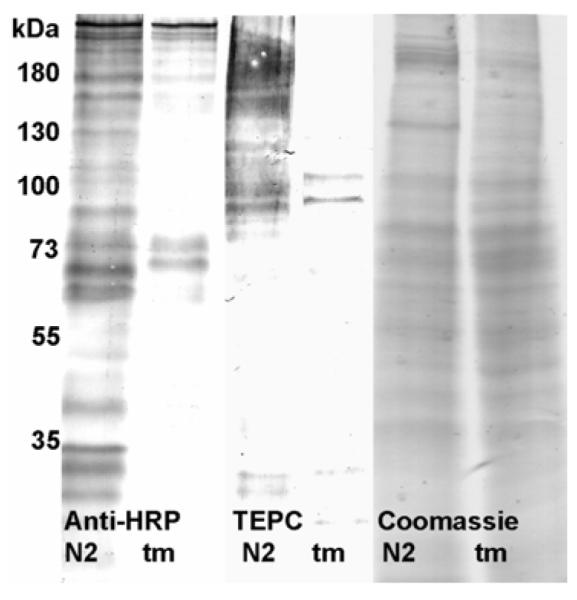
Western blotting of wild-type and mutant Caenorhabditis. Extracts of wild-type (N2) and aman-2 (tm1078; tm) worms were subject to SDS-PAGE and Western blotting using either anti-horseradish peroxidase (anti-HRP) or anti-phosphorylcholine (TEPC); Coomassie blue staining indicates that comparable amounts of protein were applied.
Comparative glycan analysis of wild-type and mutant nematodes
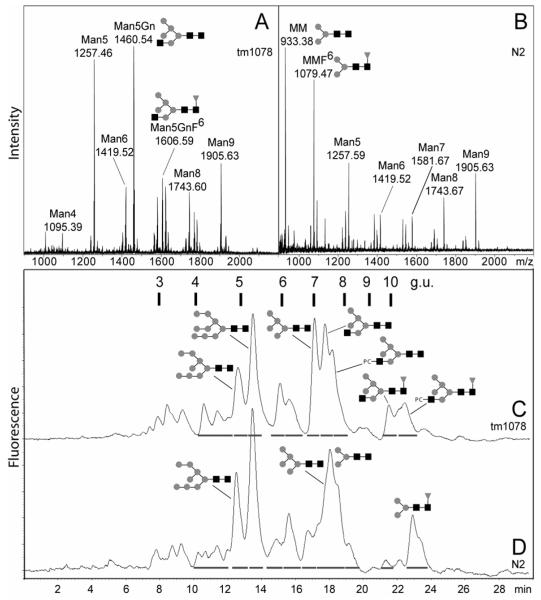
In order to show the importance of AMAN-2 in glycan processing in vivo, we performed N-glycan analysis by MS and HPLC. As shown in Figure 5 and Table I, a radical shift in the N-glycan MS profile was observed in comparison to wild-type. Rather than Hex3HexNAc2 (putatively MM) and Fuc1Hex3HexNAc2 (putatively MMF6) being major peaks, the dominant species were Hex5HexNAc2 (putatively Man5, also found in the wild-type) and Hex5HexNAc3 (putatively Man5Gn, not detectable in the wild-type). Also significant amounts of novel structures of the form Me0-2Fuc1-2Hex5-7HexNAc2-3, as well as PC1Fuc0-1Hex5HexNAc3, were found. On the other hand, in addition to the reduced occurrence and number of paucimannosidic and trifucosylated structures, tetrafucosylated species were completely absent.
Figure 5.
N-glycan analysis of Caenorhabditis elegans aman-2 mutant. N-glycans from either the tm1078 (A, C) or wild-type N2 (B, D) strains were isolated and subject to either monoisotopic MALDI-TOF MS (A, B) or, after pyridylamination, RP-HPLC (C, D). In the mass spectra the m/z values for seven selected species are shown, whereas the RP-HPLC chromatograms are annotated with retention times in terms of both minutes and glucose units (g.u.). Selected identifiable structures are depicted using the symbolic nomenclature of the Consortium for Functional Glycomics (http://www.functionalglycomics.org). For further details see Tables 1 and 2.
Table 1. Summary of MS data for underivatised glycans from the N2 and tm1078 strains.
Monoisotopic [M+Na+] m/z values from the spectra presented in Figure 5 (panels A and B) and the corresponding hypothesised compositions are shown for PNGase A-released N-glycans from wild-type N2 and mutant tm1078 aman-2 strains. ND, not detected. Abundances for detected glycans refer to percentage intensities relative to the peak of highest intensity (for N2 this is the m/z 933 peak and for tm1078 the m/z 1460 peak).
| Putative composition | N2 | tm1078 | ||
|---|---|---|---|---|
| m/z | abundance | m/z | abundance | |
| Fuc1Hex2HexNAc2 | 917.27 | 10 | ND | |
| Fuc1Hex2HexNAc2Me1 | 931.31 | 13 | ND | |
| Hex3HexNAc2 | 933.39 | 100 | 933.52 | 3 |
| Fuc2Hex2HexNAc2 | 1063.44 | 13 | ND | |
| Fuc2Hex2HexNAc2Me1 | 1077.27 | 14 | ND | |
| Fuc1Hex3HexNAc2 | 1079.46 | 81 | ND | |
| Fuc1Hex3HexNAc2Me1 | 1093.37 | 7 | ND | |
| Hex4HexNAc2 | 1095.46 | 33 | 1095.64 | 10 |
| Hex3HexNAc3 | 1136.54 | 22 | ND | |
| Fuc2Hex3HexNAc2 | 1225.59 | 16 | ND | |
| Fuc2Hex3HexNAc2Me1 | 1239.61 | 11 | 1239.73 | 8 |
| Fuc1Hex4HexNAc2 | 1241.56 | 17 | 1241.73 | 5 |
| Fuc1Hex4HexNAc2Me1 | 1255.62 | 10 | 1255.71 | 28 |
| Hex5HexNAc2 | 1257.59 | 43 | 1257.74 | 94 |
| Fuc1Hex3HexNAc3 | 1282.61 | 8 | ND | |
| Hex3HexNAc3PC1 | 1301.45 | 6 | ND | |
| Hex3HexNAc4 | 1339.62 | 6 | ND | |
| Fuc3Hex3HexNAc2 | 1371.66 | 7 | ND | |
| Fuc3Hex3HexNAc2Me1 | 1385.67 | 8 | ND | |
| Fuc2Hex4HexNAc2 | 1387.66 | 17 | ND | |
| Fuc2Hex4HexNAc2Me1 | 1401.63 | 14 | 1401.80 | 5 |
| Fuc1Hex5HexNAc2 | 1403.55 | 9 | 1403.84 | 9 |
| Fuc1Hex5HexNAc2Me1 | 1417.58 | 5 | 1417.84 | 15 |
| Hex6HexNAc2 | 1419.62 | 20 | 1419.84 | 28 |
| Fuc1Hex5HexNAc2Me2 | ND | 1431.86 | 5 | |
| Hex6HexNAc2Me1 | ND | 1433.85 | 4 | |
| Fuc1Hex3HexNAc3PC1 | 1447.54 | 3 | ND | |
| Fuc1Hex4HexNAc3Me1 | ND | 1458.86 | 27 | |
| Hex5HexNAc3 | ND | 1460.87 | 100 | |
| Fuc1Hex3HexNAc4 | 1485.32 | 6 | ND | |
| Hex3HexNAc4PC1 | 1504.56 | 3 | ND | |
| Fuc3Hex4HexNAc2 | 1533.69 | 13 | ND | |
| Fuc3Hex4HexNAc2Me1 | 1547.40 | 9 | ND | |
| Fuc2Hex5HexNAc2 | 1549.75 | 8 | ND | |
| Fuc2Hex5HexNAc2Me1 | 1563.66 | 5 | 1563.92 | 10 |
| Fuc1Hex6HexNAc2 | ND | 1565.91 | 7 | |
| Fuc2Hex5HexNAc2Me2 | ND | 1577.94 | 5 | |
| Fuc1Hex6HexNAc2Me1 | ND | 1579.92 | 13 | |
| Hex7HexNAc2 | 1581.68 | 19 | 1581.92 | 22 |
| Fuc2Hex4HexNAc3Me1 | ND | 1604.93 | 11 | |
| Fuc1Hex5HexNAc3 | ND | 1606.95 | 34 | |
| Fuc1Hex5HexNAc3Me1 | ND | 1620.93 | 11 | |
| Hex6HexNAc3 | ND | 1622.95 | 32 | |
| Hex5HexNAc3PC1 | ND | 1625.95 | 10 | |
| Fuc1Hex5HexNAc3Me2 | ND | 1634.96 | 4 | |
| Hex6HexNAc3Me1 | ND | 1636.97 | 9 | |
| Fuc4Hex4HexNAc2 | 1679.79 | 5 | ND | |
| Fuc4Hex4HexNAc2Me1 | 1693.65 | 7 | ND | |
| Fuc3Hex5HexNAc2 | 1695.61 | 14 | ND | |
| Fuc3Hex5HexNAc2Me1 | 1709.33 | 6 | ND | |
| Fuc2Hex6HexNAc2 | 1711.53 | 4 | ND | |
| Fuc2Hex6HexNAc2Me1 | ND | 1726.00 | 8 | |
| Fuc1Hex7HexNAc2 | ND | 1728.01 | 4 | |
| Fuc1Hex7HexNAc2Me1 | ND | 1741.99 | 10 | |
| Hex8HexNAc2 | 1743.68 | 25 | 1743.99 | 23 |
| Fuc1Hex6HexNAc3 | ND | 1769.03 | 15 | |
| Fuc1Hex5HexNAc3PC1 | ND | 1772.02 | 6 | |
| Fuc2Hex5HexNAc3Me2 | ND | 1781.02 | 5 | |
| Fuc1Hex6HexNAc3Me1 | ND | 1783.04 | 15 | |
| Fuc1Hex6HexNAc3Me2 | ND | 1797.09 | 5 | |
| Fuc4Hex5HexNAc2 | 1841.70 | 4 | ND | |
| Fuc4Hex5HexNAc2Me1 | 1855.61 | 6 | ND | |
| Fuc3Hex6HexNAc2 | 1857.82 | 5 | ND | |
| Fuc2Hex7HexNAc2Me1 | ND | 1888.09 | 3 | |
| Fuc1Hex8HexNAc2Me1 | 1903.42 | 3 | 1904.05 | 10 |
| Hex9HexNAc2 | 1905.63 | 32 | 1906.06 | 35 |
| Fuc2Hex6HexNAc3Me1 | ND | 1929.11 | 9 | |
| Fuc1Hex7HexNAc3 | ND | 1931.10 | 4 | |
| Fuc4Hex6HexNAc2Me1 | 2017.65 | 2 | ND | |
| Hex10HexNAc2 | 2067.54 | 2 | 2068.13 | 2 |
| Fuc3Hex6HexNAc3Me2 | ND | 2089.15 | 2 | |
| Fuc2Hex7HexNAc3Me1 | ND | 2091.18 | 3 | |
Subsequent RP-HPLC analysis of pyridylaminated glycans derived from tm1078 also showed a large-scale change in the glycan profile. MALDI-TOF MS of the collected fractions, estimation of glucose units, co-elution with known standards and/or hexosaminidase/fucosidase treatment were used to aid the identification of the major peaks. These analyses indicated that Man5, Man5Gn, Man5GnF6, Man8 and Man9 are the major N-glycans in the mutant (Figure 5C and Table 2), whereas MM, MMF6, Man5, Man8 and Man9 predominate in the wild-type (Figure 5D). Peaks from the tm1078 mutant eluting between 10 and 12 minutes (3.8-4.5 glucose units) were, in part, absent after hydrofluoric acid treatment of the whole glycan pool and included, as judged by MALDI-TOF MS, singly-fucosylated species of the form Fuc1Hex5-7HexNAc2-3 (Table 2). By comparison to the properties of MMF3, a structure previously shown to elute at 5 glucose units (21), it is concluded that these structures may be core α1,3-fucosylated glycans. The presence of such species would explain the anti-HRP staining in tm1078 worms; however, the low amount and complex mixture of glycans in this region precluded a final structural identification. It is also to be noted that some Fuc1Hex5-7HexNAc2-3 structures also elute in the region above 10 glucose units and so the latter are presumed to be core α1,6-fucosylated. The general accumulation of hybrid and oligomannosidic N-glycans was also demonstrated by the apparently complete digestion of the whole pyridylaminated tm1078 glycan pool by endoglycosidase H (data not shown).
Table 2. Summary of HPLC data for pyridylaminated glycans from the tm1078 and N2 strains.
Major peaks collected from the RP-HPLC run shown in Figure 5C were analysed by reflectron MALDI-TOF MS (increase of ca. m/z 78 as compared to underivatised glycans); retention times are expressed in both minutes and glucose units (g.u.); designations with defined hexoses or hexosamines are based on the retention times described in previous studies (21,22).
| Retention time | Tm1078 glycans | N2 glycans | ||
|---|---|---|---|---|
|
| ||||
| Putative glycans | m/z [M+Na+] | Putative glycans | m/z [M+Na+] | |
|
| ||||
| 10-12 min (3.8-4.5 g.u.) | Hex5HexNAc2 | 1336.03 | Hex4HexNAc2 | 1173.63 |
| Fuc1Hex5HexNAc2 | 1482.45 | Fuc1Hex4HexNAc2 | 1319.98 | |
| Hex6HexNAc2 | 1498.38 | Fuc2Hex4HexNAc2 | 1466.32 | |
| Fuc1Hex6HexNAc2 | 1644.8 | Fuc1Hex5HexNAc2 | 1482.5 | |
| Hex7HexNAc2 | 1660.84 | Hex6HexNAc2 | 1498.37 | |
| Hex6HexNAc3 | 1701.9 | Fuc2Hex5HexNAc2 | 1628.48 | |
| Fuc1Hex6HexNAc3a | 1846.93 | |||
| Fuc1Hex7HexNAc3 | 2008.77 | |||
|
| ||||
| 12.5 min (4.8 g.u.) | Fuc1Hex5HexNAc2Me1 | 1496.01 | Man8GlcNAc2 | 1823.09 |
| Man7GlcNAc2 (isomer M7.1) | 1660.22 | |||
| Man8GlcNAc2 | 1822.64 | |||
|
| ||||
| 14 min (5.2 g.u.) | Man9GlcNAc2 | 1986.87 | Man9GlcNAc2 | 1985.9 |
|
| ||||
| 15 min (6.0 g.u.) | Man7GlcNAc2 (isomer M7.2) | 1660.61 | Man7GlcNAc2 (isomer M7.2) | 1661.05 |
| Hex6HexNAc3 | 1700.93 | |||
|
| ||||
| 15.5-16 min (6.3 g.u.) | Man6GlcNAc2 | 1498.25 | Man6GlcNAc2 | 1497.46 |
| Glc1Man9GlcNAc2 | 2147.77 | |||
| Fuc1Hex6HexNAc3Me1 | 1862.17 | |||
|
| ||||
| 16.5 min | No corresponding peak | N/A | Fuc2Hex6HexNAc2 | 1790.47 |
|
| ||||
| 17 min (7.0 g.u.) | Man5GlcNAc3 (Man5Gn) b | 1538.67 | Shoulder not collected separately | N/A |
|
| ||||
| 18 min (7.5 g.u.) | Man5GlcNAc2 (Man5), major component c | 1335.06 | Man5GlcNAc2 (Man5) | 1335.09 |
| Fuc1Hex5HexNAc3 | 1684.65 | |||
| Fuc1Hex6HexNAc3Me2 | 1875.3 | |||
|
| ||||
| 18.5-19 min (8.0 g.u.) | Man3GlcNAc2 (MM) | 1011.1 | Man3GlcNAc2 (MM), major component | 1011.63 |
| Man4GlcNAc2 | 1173.46 | Fuc2Man2GlcNAc2 (MUF3F6) | 1140.82 | |
| Man5GlcNAc3PC1 d, e | 1704.45 | Fuc2Man3GlcNAc2 (MMF3F6) | 1313.19 | |
| Hex6HexNAc3Me1 | 1715.73 | |||
|
| ||||
| 21.5 min (10 g.u.) | Fuc1 Man5GlcNAc3 (Man5GnF6) f | 1685.6 | Fuc4Hex5HexNAc2 | 1920.4 |
|
| ||||
| 22-23 min | Fuc1Hex4HexNAc2 | 1319.84 | Fuc1Man3GlcNAc2 (MMF6) | 1157.62 |
| Fuc1Hex5HexNAc2 | 1482.16 | |||
| Fuc1Man5GlcNAc3PC1(Man5GnF6PC) e, g | 1850.5 | |||
| Fuc1Hex7HexNAc3 | 2010.44 | |||
| Fuc2Hex7HexNAc3Me1 | 2170.83 | |||
Confirmatory data from further analyses of selected major peaks are summarised as follows:
Hydrofluoric acid treatment of the relevant sub-fraction (10.5 min, 3.8 g.u.) generated a structure with same retention time (15 min, 6.0 g.u.) as an isoform of Hex6HexNAc3, suggesting the presence of core α1,3-fucose
Reinjected fraction has same retention time as a previously-generated Man5Gn standard; hexosaminidase treatment generated a structure with same retention time as Man5
Re-injected fraction has same retention time as a Man5 standard generated from fungal amylase
Hydrofluoric acid treatment generated a structure with same retention time and m/z as Man5Gn, compatible with the removal of phosphorylcholine from a portion of the glycans in this fraction
Presence of terminal PC-HexNAc and PC-HexNAc-Hex shown by MALDI-TOF MS-MS (see Figure 6)
Fucosidase and hexosaminidase treatment generated a structure with same retention time as Man5
Treatment of the whole glycan pool with either hydrofluoric acid or fucosidase resulted in the disappearance of the putative Man5GnF6PC RP-HPLC peak and respective increases in the heights of the putative Man5GnF6 and Man5GnPC peaks
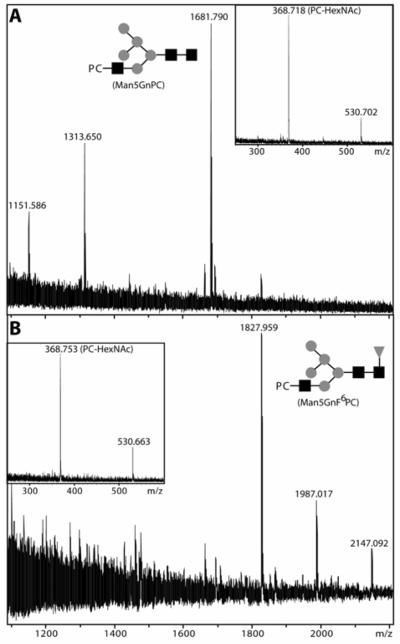
The two phosphorylcholine-containing glycans (Man5GnPC and Man5GnF6PC) detected by MALDI-TOF MS of both the underivatised glycans and collected HPLC fractions were further analysed. Consistent with the known sensitivity of phosphorylcholine-containing glycans (23), treatment with hydrofluoric acid of the fraction containing the putative Man5GnPC structure resulted in a shift of a major portion of this peak to a species with the same retention time and m/z as Man5Gn (see Table 2). The proof that the phosphorylcholine residue is located on the outer GlcNAc residues of both the putative Man5GnPC and Man5GnF6PC was given by MALDI-TOF MS-MS which resulted in fragments of 368.7 and 530.7 (PC-HexNAc and PC-HexNAc-Hex; see Figure 6). This is consistent with the presence of the phosphorylcholine on the α1,3-antennae and with the requirement of the prior action of GlcNAc-TI before modification by phosphorylcholine can take place. Furthermore, as in other studies (37-43), both the mass spectrometric and HPLC analyses underline the complexity of the N-glycome of C. elegans.
Figure 6.
MALDI-TOF MS analysis of phosphorylcholine-substituted N-glycans. RP-HPLC-purified fractions 18.5-19 min (A) and 22-23 min (B) were subject to MALDI-TOF MS. The [M+H]+ species of m/z 1681 and m/z 1827 putatively corresponding to Man5GnPC and Man5GnF6PC were then examined by MALDI-TOF MS-MS; as shown in the inserts of both panels, the two experiments revealed fragments characteristic of PC-HexNAc (m/z 368) and PC-HexNAc-Hex (m/z 530). The putative structures Man5GnPC and Man5GnF6PC are depicted using the symbolic nomenclature of the Consortium for Functional Glycomics.
Tissue and stage specific expression
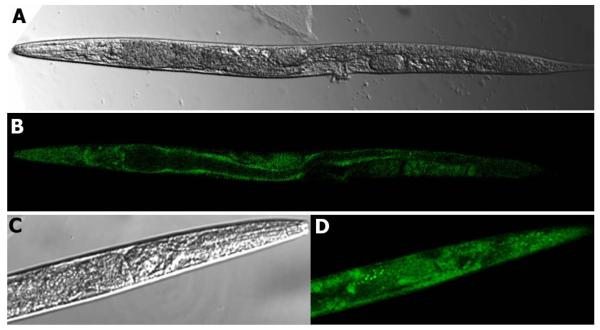
In order to test the tissue-specificity of the expression of Caenorhabditis mannosidase II, a promoter-gfp reporter construct was designed to generate a translational fusion. In order to select easily for low copy number transformants carrying the aman-2::gfp reporter fusion, the 2000 bp upstream of the aman-2 gene were ligated into a vector carrying not only a gfp sequence, but also the unc-119 gene and stably integrated into unc-119 (ed3) mutants. Two independent low copy number transgenic lines were generated and analysed. Confocal microscopy indicated that the aman-2 promoter is most active in the gut wall, pharynx and grinder, hypodermal cells, cells of the nervous system (ventral nerve cord cells, neurons surrounding the pharyngeal bulb and in the head) in adult animals; also rather ubiquitous expression was seen in larvae (Figure 7). The relatively abundant expression is consistent with the major effect on the N-glycan spectrum of deletion of part of the aman-2 gene and our results are also consistent with the results of a large-scale expression pattern screen4 which indicate expression in the intestine and nervous system.
Figure 7.
aman-2 promoter-driven GFP expression. Confocal transmission (A) and fluorescence (B) microscopy of an aman-2::gfp transgenic Caenorhabditis adult worm indicates relatively ubiquitous expression. The confocal transmission (C) and fluorescence (D) micrographs of the head region of an aman-2::gfp transgenic L3 worm are shown at higher magnification.
Discussion
Class II mannosidase homologues in the worm
The in silico analysis in this study, as well as that of Park et al. (14), suggested the presence of three class II mannosidases in C. elegans. Even based upon the substrate specificity of the recombinant enzyme, we have strong evidence to hypothesise that AMAN-2 is a Golgi α-mannosidase II. This designation is further confirmed by the previous use of GFP or YFP-fusions with the transmembrane and stalk regions of C. elegans AMAN-2, which were found to localise in a punctate pattern consistent with those of Golgi proteins in other invertebrates (44,45). Homologues of AMAN-2 undoubtedly play a key role in N-glycan biosynthesis in mammals and its activity was first demonstrated in rat liver by Dewald and Touster (46), who found an enzyme which cleaved of p-nitrophenyl-α-mannoside with an optimum of pH 5.5 - thereby distinguishing it from lysosomal and cytoplasmic activities. Afterwards, this enzyme was confirmed to be the major Golgi p-nitrophenyl-α-mannosidase in rat liver and found to specifically cleave Man5Gn (47); this latter activity was also found in the laboratories of Kornfeld (in CHO cells) and of Schachter (in hen oviduct) (48,49). The enzyme was also immunolocalised to the Golgi in rat liver (50); the intracellular localization (31) and the same specificity (51) have also been shown for the orthologous enzyme in insects.
In contrast to aman-2, the aman-1 gene encodes an enzyme with optimal activity at pH 4-4.5, the same as that previously reported for the putative lysosomal mannosidase in nematode extracts (52); furthermore, it has length and homology characteristics compatible with it being the worm’s lysosomal mannosidase. The Km for p-nitrophenyl-α-mannoside (~ 2.5 mM)6 is also similar to the value obtained for the recombinant human orthologue (12). However, AMAN-1 expressed in yeast appeared not to digest natural substrates such as Man5GlcNAc2, Man9GlcNAc2 and the endoglycosidase product Man9GlcNAc1 (data not shown), even though the latter substrate is degraded by the human enzyme expressed in Pichia (12). One explanation is that, since the bovine, human and feline lysosomal enzymes consist of five polypeptides (53-56), the worm AMAN-1 may require proteolytic and/or pH-dependent activation in the lysosome before it is capable of accepting such oligosaccharide substrates.
The aman-3 gene, in contrast to the other two worm class II mannosidases, appears to encode a Co(II)-dependent enzyme with a pH optimum of 6.5; preliminary data indicated that it could digest Man5GlcNAc2 to Man4GlcNAc2. Thus, AMAN-3 may be an orthologue of the α-mannosidase III found in insects which is activated 23-fold by Co(II) and has a pH optimum of 6.5-6.7. Sensitivity to Co(II) is also displayed by, e.g., the human lysosomal α1,6-mannosidase, which is more-or-less inactive in the absence of divalent cations (14), as well as by the MAN2C1 cytosolic mannosidase (57). The importance of AMAN-3, however, is not especially clear, because the worm aman-2 mutant is nearly devoid of paucimannosidic structures. Furthermore, even though AMAN-3 has a potential transmembrane domain, any hypothesis as to its function in vivo will require determination, for instance, of its actual intracellular localisation and tissue expression.
Altered N-glycan biosynthesis in worm glycosylation mutants
In order to show the function of AMAN-2 in vivo, we analysed the enzyme activity and the N-glycans of the corresponding mutant and conclude that the natural spectrum of N-glycans in the nematode C. elegans is dependent on the activity of mannosidase II. Analyses of the pyridylaminated N-glycans from the mannosidase II mutant confirm the mass spectrometric data and indicate that the major N-glycans present in the mutant are Man5 and Man5Gn, the latter not detected in the wild-type. On the other hand, MM is a major N-glycan in the wild-type. Interestingly, our own RP-HPLC analyses suggested that the major species in a mutant lacking all three GlcNAc-TI genes co-elutes with a Man5 standard (data not shown), a result consistent with previous MS data (42). The results from both the mannosidase II and GlcNAc-TI mutants, therefore, indicate that a major proportion of the N-glycans of wild-type worms are normally modified by GlcNAc-TI and then are rapidly subject to the action of mannosidase II; thus they can be considered ‘complex’ in the broadest sense of the word. The data are also in accordance with previous biochemical analyses (5) indicating that Man5Gn is not a substrate for the putative Golgi hexosaminidase in the worm.
A lack of the tetrafucosylated N-glycans previously found to be novel to C. elegans (19,37,38,41) was also observed in the mannosidase II mutant, as was a reduction in the presence of the anti-horseradish peroxidase epitope. Since FUT-1 (also known as CEFT1) requires for its activity the prior removal of the GlcNAc transferred by GlcNAc-TI (19,58), but that this GlcNAc is still present on a large proportion of glycans in the aman-2 mutant that would otherwise be ‘complex’, a reduced amount of anti-horseradish peroxidase binding in the aman-2 mutant could be expected. However, core α1,3-fucosylation pathway is not completely blocked, but probably takes a different route. Indeed, we have recently found that FUT-1 does transfer fucose to Man5, albeit at a fraction of the rate at which it utilises MM and MMF6 as a substrate7. Furthermore, as shown in a study to generate substrates for Drosophila FucTA (59), the core α1,6-fucosyltransferase FUT-8 can still transfer to Man5Gn, as well as to MGn, thus accounting for the presence of Man5GnF6 in the aman-2 worm as well as, after the action of the Golgi hexosaminidase, of MMF6 in the wild-type (see Figure 8). However, the action of at least one other fucosyltransferase, with an unknown substrate specificity, necessary to generate tri- and tetrafucosylated structures in the wild-type, is presumably blocked in the aman-2 worm.
Figure 8.
Biosynthetic pathways in wild-type and aman-2 worms. Structures with Glc1Man9GlcNAc2 and Man5-9GlcNAc2 are present in both N2 and tm1078 strains and the pathways between them are shown with both solid and non-filled arrows; however, the absence of Golgi α-mannosidase II (AMAN-2) blocks the left-hand “solid arrow” route to the paucimannosidic-type mono- and difucosylated species and to the complex (in part phosphorylcholine-substituted) N-glycans found in wild-type N2 worms (shown within the black box). As judged by the accumulation of Man5Gn in aman-2 worms, the Golgi hexosaminidase is obviously unable to digest this structure in the absence of the mannosidase. On the other hand, the absence of the AMAN-2 mannosidase pushes the N-glycan biosynthesis into the right-hand “non-filled arrow” route resulting in many “non-wild type” hybrid glycans that retain the GlcNAc residue transferred by GlcNAc-TI (structures within the grey box). Furthermore, this probably allows the retention of a pool of Man5GlcNAc2 which is a substrate for the FUT-1 core α1,3-fucosyltransferase at least in vitro (the occurrence of the Man5F3 structure in vivo is putative, but is suggested by MALDI-TOF MS analysis of RP-HPLC fractions). The reduced transfer by FUT-1 to Man5GlcNAc2 (i.e., Man5) shown in vitro and the putatively reduced transfer by phosphorylcholinyltransferases to Man5Gn and Man5GnF6 would be compatible with the lower reactivity with anti-horseradish peroxidase and TEPC-15 as shown in Figure 4. The hybrid structure Man5Gn is still a substrate for the FUT-8 core α1,6-fucosyltransferase. For simplicity and due to lack of data on the exact pathways, the biosynthesis of tri- and tetrafucosylated glycans in the wild-type as well as methylated and fucosylated Hex6-8HexNAc2-3 species in the mutant is not shown. The glycan nomenclature is based on that of Schachter (3); see also Scheme I.
Additionally, a reduction in TEPC-15 reactivity was seen in aman-2 extracts. This is compatible with the inability of GlcNAc-TII (GLY-20) and GlcNAc-TV (GLY-2) to act (due to blocking of the acceptor site by the mannose residues still present in the aman-2 mutant) and accounts for the reduced amount of protein-bound phosphorylcholine in the aman-2 worm. Indeed the C. elegans phosphorylcholine transferase has an apparent preference for tetra-antennary complex N-glycans as substrates in vitro (60). As confirmed by electrospray-MS8, wild-type worms express putatively multiantennary glycans carrying one or two phosphorylcholine residues. However, the phosphorylcholination pathway is not completely blocked in aman-2 worms, but as indicated in Figure 8 takes a different route, since both residual staining and relevant N-glycans (PC1Man5GlcNAc3Fuc0-1) are present. Thus, we assume that the relevant phosphorylcholine transferase(s) display activity towards Man5Gn. Indeed, swainsonine does not affect the transfer of this modification to a nematode secreted glycoprotein (36); on the other hand, Western blotting suggests that worms lacking all three GlcNAc-TI genes have drastically-reduced levels of protein-bound phosphorylcholine epitopes9.
Another feature of both the tm1078 (aman-2) and the VC378 (fut-1) worms is the presence of dimethylated N-glycans not found in the wild-type. Certainly monomethylated species (40,42,61), as well as Fuc3Hex4HexNAc2Me1-2 (38), Fuc1Hex4-5HexNAc2Me2 (62) and Fuc2-4Hex4-6HexNAc2Me2 (43), are included in the summaries of previous analyses of N2 glycans. The novelty of the apparently methylated aman-2 glycans, however, is that the methylation is not necessarily associated with fucosylation, since species are present with an m/z consistent with glycans with the compositions Hex6HexNAc2-3Me1-2. The presence of such structures is an indication that, in addition to the ‘obvious’ results of the aman-2 deletion (i.e., the presence of structures of the general form Fuc0-1Hex5HexNAc3Me0-2PC0-1 which can be easily deduced as resulting from accumulated Man5Gn), there are other novel structures in the mutant worm: this is unlike the case with the mannosidase II knock-out plant in which only ‘expected’ hybrid and oligomannosidic structures are seen (63). Indeed, due to the presence of Fuc0-3Hex6-7HexNAc2-3Me0-2 structures, it may be that either Man6-7GlcNAc2 is a target for the worm GlcNAc-TI, fucosyltransferases and methyltransferases and/or that Man5GlcNAc2-3 (Man5Gn/Man5) can be acted upon by hexosyltransferases prior to further modification. Thus, as also shown in other studies (19,42), it appears that otherwise ‘hidden’ glycosylation pathways may be revealed in mutant worms since substrates generally absent may then be accessible for enzymes whose products are not normally visible by the analytical methods employed or which then display abnormal activities.
Although the paucimannosidic glycans occur at a very low level in aman-2 worms, an explanation for the low residual levels of Fuc0-1Hex3-4HexNAc2-3Me0-1 has to be considered. It may be that either lysosomal or other α-mannosidase activities (possibly either AMAN-1 or AMAN-3) present in the tm1078 strain act on Man5 and/or on the more ‘complex’ structures; however, the glycan analyses and enzyme assays would certainly indicate that the normal levels of α-mannosidase(s) capable of processing Man5Gn are hugely reduced or abolished in this mutant. Based, therefore, on its in vitro and in vivo activity, we conclude that the C. elegans aman-2 gene encodes a ‘classical’ Golgi α-mannosidase II, which converts Man5Gn to Man4Gn (as an intermediate) and MGn and which, based on the promoter analysis, is expressed in most cells of the worm.
Biological repercussions of abnormal N-glycan biosynthesis
Our data are the first on the effect on glycosylation of an ablation of the mannosidase II gene in any invertebrate; however, the mutant strain appears perfectly viable under normal laboratory conditions. Furthermore, wild-type phenotypes were also described for RNAi experiments for all three class II mannosidases4. This is not particularly surprising, since the worm lacking all three GlcNAc-TI enzymes (required to generate the substrate for α-mannosidase II) also has no obvious viability defect under standard conditions (42), although recently preliminary data indicated an altered sensitivity to bacteria for this mutant (64). Also, the Arabidopsis cgl mutant, lacking GlcNAc-TI and so not expressing plant complex N-glycans has only a marginal phenotype (65). On the other hand, the ‘molecular phenotype’ in terms of N-glycosylation is very profound with Man5 accounting for 75% of the structures in cgl mutants (66); furthermore, an Arabidopsis Golgi mannosidase II mutant, containing like the aman-2 worm predominantly hybrid N-glycans, is also viable and fertile under standard conditions (63). This indicates that, at least under non-stressful conditions, there exists a tremendous flexibility in terms of a glycosylation pattern compatible with viability. However, in mammals, a defect in GlcNAc-TI, as in the mgat1 knockout mouse, results in embryonic lethality (67,68). Also, very recent data indicate that mannosidase II/IIx double knock-out mice expressing no detectable complex N-glycans display severe phenotypes (8); indeed the same data argues against an earlier hypothesis (69,70) that a mannosidase III route is significant in mammals.
In contrast to mammals, worms and plants lack a second mannosidase II isoform and so any mannosidase III route is also either very minor or absent in the aman-2 worm and in the Arabidopsis mannosidase II mutant. However, a gene-trap line affecting the closest homologue in the Drosophila genome to Sf9 α-mannosidase III (CG4606; α-Man-IIb) has a recessive lethal phenotype (71), whereas overexpression of the fruitfly α-mannosidase II (CG18474) results in a ‘rough eye’ phenotype and increased life span (72). However, neither the enzymatic function of CG4606 nor the effect on the glycosylation profile of an ablation or overexpression of either fly mannosidase has been examined. Overall, the data from knock-outs would suggest that, during evolution, glycosylation has accumulated a set of roles, in which ‘decorating’ glycozymes aid survival under stress or infection, but are not absolutely required for life. The importance of these functions has increased during evolution; indeed, plants and invertebrates are still viable when major changes in N-glycosylation occur. This, in turn, means that these organisms are far more flexible in terms of their glycogenomic requirements. Despite this, the fact that pathways involving a requirement for GlcNAc-TI and α-mannosidase II have survived suggests that ‘complex’ N-linked oligosaccharide biosynthesis is still needed in order to have a competitive advantage in the ‘wild’ and is thus an indication of this metabolic pathway’s importance in eukaryotic biology.
ACKNOWLEDGEMENTS
The authors thank Denise Kerner for help with maintenance of worms and glycan purification, Josef Voglmeir for pyridylamination of N-glycans, Dr. Dubravko Rendić for reading the manuscript and Dr. Andrew Fire for the pPD95.67 vector. The tm1078 strain was obtained from the National Bioresource Project for the Experimental Animal Nematode C. elegans (Tokyo Women’s Medical University, Japan).
Footnotes
Abbreviations: GlcNAc-TI, N-acetylglucosaminyltransferase I (EC 2.4.1.101); GlcNAc-TII, N-acetylglucosaminyltransferase II (EC 2.4.1.143); MALDI-TOF, matrix-assisted laser desorption ionisiation/time-of-flight; MS, mass spectrometry: RP-HPLC, reverse phase high pressure liquid chromatography; glycan abbreviations are based on those of Schachter (Ref. 3) and are shown in Scheme I.
A. Fire, S. Xu, J. Ahnn and G. Seydoux, personal communication
M.H. and I.B.H.W., unpublished data
I.B.H.W., unpublished data
G. Pöltl and I.B.H.W., unpublished data
K.P. and I.B.H.W., unpublished data
This work was supported in part by grants from the Fonds zur Förderung der wissenschaftlichen Forschung (P15475 and P18447 to I.B.H.W.).
REFERENCES
- 1.Moremen KW, Trimble RB, Herscovics A. Glycobiology. 1994;4:113–125. doi: 10.1093/glycob/4.2.113. [DOI] [PubMed] [Google Scholar]
- 2.Herscovics A. Biochimie. 2001;83:757–762. doi: 10.1016/s0300-9084(01)01319-0. [DOI] [PubMed] [Google Scholar]
- 3.Schachter H. Biochem Cell Biol. 1986;64:163–181. doi: 10.1139/o86-026. [DOI] [PubMed] [Google Scholar]
- 4.Altmann F, Schwihla H, Staudacher E, Glössl J, März L. J. Biol. Chem. 1995;270:17344–17349. doi: 10.1074/jbc.270.29.17344. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Cao P, Chen S, Spence AM, Zhu S, Staudacher E, Schachter H. Biochem. J. 2003;372:53–64. doi: 10.1042/BJ20021931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kawar Z, Karaveg K, Moremen KW, Jarvis DL. J Biol Chem. 2001;276:16335–16340. doi: 10.1074/jbc.M100119200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monis E, Bonay P, Hughes RC. Eur J Biochem. 1987;168:287–294. doi: 10.1111/j.1432-1033.1987.tb13419.x. [DOI] [PubMed] [Google Scholar]
- 8.Akama TO, Nakagawa H, Wong NK, Sutton-Smith M, Dell A, Morris HR, Nakayama J, Nishimura S-I, Pai A, Moremen KW, Marth JD, Fukuda MN. Proc Natl Acad Sci U S A. 2006 doi: 10.1073/pnas.0603248103. 0603248103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster JM, Yudkin B, Lockyer AE, Roberts DB. Gene. 1995;154:183–186. doi: 10.1016/0378-1119(94)00867-r. [DOI] [PubMed] [Google Scholar]
- 10.Moremen KW, Robbins PW. J Cell Biol. 1991;115:1521–1534. doi: 10.1083/jcb.115.6.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misago M, Liao YF, Kudo S, Eto S, Mattei MG, Moremen KW, Fukuda MN. Proc. Natl. Acad. Sci. USA. 1995;92:11766–11770. doi: 10.1073/pnas.92.25.11766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liao Y-F, Lal A, Moremen KW. J Biol Chem. 1996;271:28348–28358. doi: 10.1074/jbc.271.45.28348. [DOI] [PubMed] [Google Scholar]
- 13.Bischoff J, Moremen K, Lodish HF. J Biol Chem. 1990;265:17110–17117. [PubMed] [Google Scholar]
- 14.Park C, Meng L, Stanton LH, Collins RE, Mast SW, Yi X, Strachan H, Moremen KW. J. Biol. Chem. 2005;280:37204–37216. doi: 10.1074/jbc.M508930200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. J Biol Chem. 1999;274:21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- 16.Tremblay LO, Hersovics A. J Biol Chem. 2000;275:31655–31660. doi: 10.1074/jbc.M004935200. [DOI] [PubMed] [Google Scholar]
- 17.van den Elsen JM, Kuntz DA, Rose DR. EMBO J. 2001;20:3008–3017. doi: 10.1093/emboj/20.12.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McIlvaine TC. J Biol Chem. 1921;49:183–186. [Google Scholar]
- 19.Paschinger K, Rendic D, Lochnit G, Jantsch V, Wilson IBH. J. Biol. Chem. 2004;279:49588–49598. doi: 10.1074/jbc.M408978200. [DOI] [PubMed] [Google Scholar]
- 20.Hase S, Ibuki T, Ikenaka T. J Biochem (Tokyo) 1984;95:197–203. doi: 10.1093/oxfordjournals.jbchem.a134585. [DOI] [PubMed] [Google Scholar]
- 21.Wilson IBH, Altmann F. Glycoconjugate Journal. 1998;15:1055–1070. doi: 10.1023/a:1006960401562. [DOI] [PubMed] [Google Scholar]
- 22.Fabini G, Freilinger A, Altmann F, Wilson IBH. J. Biol. Chem. 2001;276:28058–28067. doi: 10.1074/jbc.M100573200. [DOI] [PubMed] [Google Scholar]
- 23.Haslam SM, Khoo KH, Houston KM, Harnett W, Morris HR, Dell A. Mol Biochem Parasitol. 1997;85:53–66. doi: 10.1016/s0166-6851(96)02807-1. [DOI] [PubMed] [Google Scholar]
- 24.Praitis V, Casey E, Collar D, Austin J. Genetics. 1998;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henrissat B, Bairoch A. Biochem. J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howard S, He S, Withers SG. J Biol Chem. 1998;273:2067–2072. doi: 10.1074/jbc.273.4.2067. [DOI] [PubMed] [Google Scholar]
- 27.Numao S, He S, Evjen G, Howard S, Tollersrud O-K, Withers SG. FEBS Lett. 2000;484:175–178. doi: 10.1016/s0014-5793(00)02148-7. [DOI] [PubMed] [Google Scholar]
- 28.Kaji H, Saito H, Yamauchi Y, Shinkawa T, Taoka M, Hirabayashi J, Kasai K.-i., Takahashi N, Isobe T. Nat. Biotech. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- 29.Dixon M. Biochem. J. 1953;55:161–170. doi: 10.1042/bj0550161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tulsiani DRP, Opheim DJ, Touster O. J Biol Chem. 1977;252:3227–3233. [PubMed] [Google Scholar]
- 31.Rabouille C, Kuntz DA, Lockyer A, Watson R, Signorelli T, Rose DR, van den Heuvel M, Roberts DB. J. Cell. Sci. 1999;112:3319–3330. doi: 10.1242/jcs.112.19.3319. [DOI] [PubMed] [Google Scholar]
- 32.van Die I, Gomord V, Kooyman FNJ, van der Berg TK, Cummings RD, Vervelde L. FEBS Lett. 1999;463:189–193. doi: 10.1016/s0014-5793(99)01508-2. [DOI] [PubMed] [Google Scholar]
- 33.Gerdt S, Dennis RD, Borgonie G, Schnabel R, Geyer R. Eur J Biochem. 1999;266:952–963. doi: 10.1046/j.1432-1327.1999.00937.x. [DOI] [PubMed] [Google Scholar]
- 34.Leon MA, Young NM. Biochemistry. 1971;10:1424–1429. doi: 10.1021/bi00784a024. [DOI] [PubMed] [Google Scholar]
- 35.Weiser JN, Shchepetov M, Chong ST. Infect. Immun. 1997;65:943–950. doi: 10.1128/iai.65.3.943-950.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houston KM, Cushley W, Harnett W. J. Biol. Chem. 1997;272:1527–1533. doi: 10.1074/jbc.272.3.1527. [DOI] [PubMed] [Google Scholar]
- 37.Altmann F, Fabini G, Ahorn H, Wilson IBH. Biochimie. 2001;83:703–712. doi: 10.1016/s0300-9084(01)01297-4. [DOI] [PubMed] [Google Scholar]
- 38.Haslam SM, Gems D, Morris HR, Dell A. Biochem. Soc. Symp. 2002;69:117–134. [PubMed] [Google Scholar]
- 39.Natsuka S, Adachi J, Kawaguchi M, Nakakita S, Hase S, Ichikawa A, Ikura K. J. Biochem. (Tokyo) 2002;131:807–813. doi: 10.1093/oxfordjournals.jbchem.a003169. [DOI] [PubMed] [Google Scholar]
- 40.Hirabayashi J, Hayama K, Kaji H, Isobe T, Kasai K. J Biochem (Tokyo) 2002;132:103–114. doi: 10.1093/oxfordjournals.jbchem.a003186. [DOI] [PubMed] [Google Scholar]
- 41.Cipollo JF, Awad AM, Costello CE, Hirschberg CB. J Biol Chem. 2004;279:52893–52903. doi: 10.1074/jbc.M409557200. [DOI] [PubMed] [Google Scholar]
- 42.Zhu S, Hanneman A, Reinhold V, Spence A, Schachter H. Biochem. J. 2004;382:995–1001. doi: 10.1042/BJ20040793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hanneman AJ, Rosa JC, Ashline D, Reinhold V. Glycobiology. 2006 doi: 10.1093/glycob/cwl011. in press. [DOI] [PubMed] [Google Scholar]
- 44.Treusch S, Knuth S, Slaugenhaupt SA, Goldin E, Grant BD, Fares H. Proc Natl Acad Sci U S A. 2004;101:4483–4488. doi: 10.1073/pnas.0400709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolls MM, Hall DH, Victor M, Stelzer EHK, Rapoport TA. Mol. Biol. Cell. 2002;13:1778–1791. doi: 10.1091/mbc.01-10-0514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewald B, Touster O. J Biol Chem. 1973;248:7223–7233. [PubMed] [Google Scholar]
- 47.Tulsiani DRP, Hubbard SC, Robbins PW, Touster O. J. Biol. Chem. 1982;257:3660–3668. [PubMed] [Google Scholar]
- 48.Tabas I, Kornfeld S. J Biol Chem. 1978;253:7779–7786. [PubMed] [Google Scholar]
- 49.Allen SD, Tsai D, Schachter H. J. Biol. Chem. 1984;259:6984–6990. [PubMed] [Google Scholar]
- 50.Novikoff PM, Tulsiani DRP, Touster O, Yam A, Novikoff AB. Proc Natl Acad Sci U S A. 1983;80:4364–4368. doi: 10.1073/pnas.80.14.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Altmann F, März L. Glycoconjugate Journal. 1995;12:150–155. doi: 10.1007/BF00731359. [DOI] [PubMed] [Google Scholar]
- 52.Bolanowski M, Jacobson LA, Russell RL. Mech. Aging Dev. 1983;21:295–319. doi: 10.1016/0047-6374(83)90048-9. [DOI] [PubMed] [Google Scholar]
- 53.Nilssen Ø, Berg T, Riise HMF, Ramachandran U, Evjen G, Hansen GM, Malm D, Tranebjaerg L, Tollersrud O-K. Hum. Mol. Genet. 1997;6:717–726. doi: 10.1093/hmg/6.5.717. [DOI] [PubMed] [Google Scholar]
- 54.Berg T, Tollersrud OK, Walkley SU, Siegel D, Nilssen O. Biochem J. 1997;328:863–870. doi: 10.1042/bj3280863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tollersrud O-K, Berg T, Healy P, Evjen G, Ramachandran U, Nilssen Ø. Eur J Biochem. 1997;246:410–419. doi: 10.1111/j.1432-1033.1997.00410.x. [DOI] [PubMed] [Google Scholar]
- 56.Heikinheimo P, Helland R, Leiros H.-K. Schrøder, Leiros I, Karlsen S, Evjen G, Ravelli R, Schoehn G, Ruigrok R, Tollersrud O-K, McSweeney S, Hough E. J. Mol. Biol. 2003;327:631–644. doi: 10.1016/s0022-2836(03)00172-4. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki T, Hara I, Nakano M, Shigeta M, Nakagawa T, Kondo A, Funakoshi Y, Taniguchi N. Biochem J. 2006 doi: 10.1042/BJ20060945. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IBH. Glycobiology. 2005;15:463–474. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 59.Rendic D, Linder A, Paschinger K, Borth N, Wilson IBH, Fabini G. J. Biol. Chem. 2006;281:3343–3353. doi: 10.1074/jbc.M508334200. [DOI] [PubMed] [Google Scholar]
- 60.Cipollo JF, Awad A, Costello CE, Robbins PW, Hirschberg CB. Proc Natl Acad Sci U S A. 2004;101:3404–3408. doi: 10.1073/pnas.0400384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haslam SM, Dell A. Biochimie. 2003;85:25–32. doi: 10.1016/s0300-9084(03)00041-5. [DOI] [PubMed] [Google Scholar]
- 62.Cipollo JF, Awad A, Costello CE, Hirschberg CB. J Biol Chem. 2005;280:26063–26072. doi: 10.1074/jbc.M503828200. [DOI] [PubMed] [Google Scholar]
- 63.Strasser R, Schoberer J, Jin C, Glössl J, Mach L, Steinkellner H. Plant J. 2006;45:789–803. doi: 10.1111/j.1365-313X.2005.02648.x. [DOI] [PubMed] [Google Scholar]
- 64.Schachter H, Shi H, Spence A. Glycobiology. 2005;15:1191. (Abs. 1117) [Google Scholar]
- 65.von Schaewen A, Sturm A, O’Neill J, Chrispeels MJ. Plant Physiol. 1993;102:1109–1118. doi: 10.1104/pp.102.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strasser R, Stadlmann J, Svoboda B, Altmann F, Glössl J, Mach L. Biochem. J. 2005;387:385–391. doi: 10.1042/BJ20041686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Metzler M, Gertz A, Sarkar M, Schachter H, Schrader JW, Marth JD. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ioffe E, Stanley P. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chui D, Oh-Eda M, Liao YF, Panneerselvam K, Lal A, Marek KW, Freeze HH, Moremen KW, Fukuda MN, Marth JD. Cell. 1997;90:157–167. doi: 10.1016/s0092-8674(00)80322-0. [DOI] [PubMed] [Google Scholar]
- 70.Chui D, Sellakumar G, Green RS, Sutton-Smith M, McQuistan T, Marek KW, Morris HR, Dell A, Marth JD. Proc Natl Acad Sci U S A. 2001;98:1142–1147. doi: 10.1073/pnas.98.3.1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lukacsovich T, Asztalos Z, Awano W, Baba K, Kondo S, Niwa S, Yamamoto D. Genetics. 2001;157:727–742. doi: 10.1093/genetics/157.2.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Landis G, Bhole D, Lu L, Tower J. Genetics. 2001;158:1167–1176. doi: 10.1093/genetics/158.3.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]