Abstract
Objective
Chronic pancreatitis is a progressive inflammatory disorder of the pancreas characterized by permanent destruction of acinar cells. Mutations in the chymotrypsinogen C (CTRC) gene have been linked to the development of chronic pancreatitis. The aim of the present study was to explore whether CTRC mutants induce endoplasmic reticulum (ER) stress in pancreatic acinar cells.
Design
Dexamethasone-differentiated AR42J rat acinar cells and freshly isolated mouse acini were transfected with recombinant adenovirus carrying wild type CTRC or the p.A73T pancreatitis-associated mutant. ER stress markers were assessed by reverse transcription-PCR and western blotting. Apoptosis was characterized by caspase-3/7 activity and the TUNEL assay.
Results
Acinar cells transfected with the p.A73T mutant, but not those with wild-type CTRC, developed significant ER stress, as judged by elevated mRNA and protein levels of the ER chaperone immunoglobulin-binding protein (BiP), increased splicing of the X-box binding protein-1 (XBP1) mRNA and marked induction of the transcription factor C/EBP-homologous protein (CHOP), a mediator of ER stress-associated apoptosis. Consistent with higher CHOP expression, AR42J cells expressing the p.A73T mutant became detached over time and showed considerably increased caspase-3/7 activity and TUNEL staining.
Conclusions
Pancreatitis-associated CTRC mutations can markedly increase the propensity of chymotrypsinogen C to elicit ER stress in pancreatic acinar cells. Thus, carriers of CTRC mutations may be at a higher risk of developing ER stress in the exocrine pancreas, which may contribute to parenchymal damage through acinar cell apoptosis.
Keywords: chronic pancreatitis, protein misfolding, endoplasmic reticulum stress, unfolded protein response, apoptosis
Chronic pancreatitis is a persistent inflammatory disorder characterized by destruction of the pancreatic parenchyma, maldigestion, chronic pain and diabetes mellitus.[1] Identification of genetic risk factors and elucidation of their mechanism of action has allowed the formulation of a disease model in which increased trypsinogen activation and failure of protective mechanisms responsible for trypsin inactivation represent the major pathological pathways.[2–3] Four significant milestones contributed to the definition of this trypsin-dependent disease mechanism. First, cationic trypsinogen (PRSS1) mutations were identified in association with hereditary pancreatitis and the majority of these mutations were shown to stimulate autoactivation of trypsinogen to trypsin.[2–6] Second, mutations in the SPINK1 gene encoding pancreatic secretory trypsin inhibitor were detected in subjects with idiopathic, alcoholic and tropical pancreatitis.[7–9] Many of the SPINK1 mutations studied so far have been found to cause diminished trypsin inhibitor expression at the mRNA and/or protein level, although the main disease related variant (p.N34S) has yet to be shown to have a functional consequence. 10–13] Third, a mutation in anionic trypsinogen (PRSS2) was described, which promoted rapid autodegradation and afforded protection against chronic pancreatitis.[14] This finding was important conceptually, as it highlighted the protective effect of trypsinogen degradation against chronic pancreatitis. Finally, we recently demonstrated that the digestive enzyme chymotrypsin C (CTRC) promoted trypsinogen and trypsin degradation and mutations in the CTRC gene predisposed to chronic pancreatitis.[15–17] CTRC mutants exhibited diminished secretion and in some cases also loss of catalytic activity; therefore, we proposed that increased risk for chronic pancreatitis in mutation carriers is best explained by the reduced trypsin degrading activity within the pancreas.[16]
The trypsin-dependent disease model described above assumes that gain or loss of catalytic activity of the participant proteins is critical in disease pathogenesis. We hypothesized; however, that mutations in digestive enzymes might increase the risk of chronic pancreatitis by an alternative mechanism, which involves mutation-induced misfolding. Intracellular retention of misfolded proteins results in endoplasmic reticulum (ER) stress and activates a signaling pathway aimed at alleviating ER burden by increasing protein folding capacity and attenuating translation.[18–23] Potentially harmful consequences of this signaling process are the activation of the inflammatory transcription factor NFκB and the induction of apoptotic cell death. In the present study, we examined the effect of a representative CTRC mutant, p.A73T, on ER stress markers and apoptosis in the dexamethasone-differentiated rat acinar cell line AR42J and in primary mouse acini.
METHODS
Recombinant adenovirus construction
The cDNAs for the human wild type CTRC and the p.A73T mutant carrying a Glu-Glu epitope tag were excised from the previously constructed pcDNA3.1(−)_CTRC expression plasmids [15,16] with XhoI and EcoRI and were subcloned into the VQ Ad5CMV shuttle vector under the control of a CMV promoter. Recombinant adenovirus was custom-made by Viraquest, Inc. (North Liberty, Iowa). Adenovirus containing the enhanced green fluorescent protein (eGFP) cDNA was also purchased from Viraquest. Virus particles were stored in A195 buffer, at 1×1012 particles/mL (~1–4×1010 pfu/mL) concentration at −80 °C in aliquots. The infectious titer of the adenovirus stocks was confirmed with the Adeno-X Rapid Titer Kit (Clontech).
Cell culture and transfection of AR42J cells
Rat pancreatic AR42J acinar cells were purchased from ATCC (#CRL-1492) and were maintained as subconfluent cultures in D-MEM containing 20% fetal bovine serum, 2 mM glutamine and 1% penicillin/streptomycin solution at 37 °C in a humidified atmosphere containing 5% CO2. Prior to transfection cells were plated into 35-mm wells (106 cells per well) and were grown in the presence of 100 nM dexamethasone (final concentration; Sigma-Aldrich #D4902) for 48 h to induce differentiation.[24]
Unless indicated otherwise, transfections were performed with 2×108 pfu/mL final adenovirus concentrations in 1 mL OptiMEM supplemented with 2 mM glutamine and 1% penicillin/streptomycin in the presence of dexamethasone (100 nM final concentration).
Preparation and transfection of primary mouse pancreatic acini
Mice were treated according to the Federal Guidelines for Animal Care and the Institutional Animal Care and Use Committee of Boston University approved the animal research protocol. Pancreatic acini were prepared in D-MEM/F-12 medium supplemented with 0.1% BSA, 1 mM sodium pyruvate and 1% penicillin/streptomycin solution (final concentrations). Male Hsd:ICR (CD-1) mice (21–24 g; purchased from Harlan Sprague Dawley, Inc) were sacrificed by CO2 inhalation followed by cervical dislocation. The pancreata were excised, digested with collagenase (Sigma, #C5138; 0.25 mg/mL final concentration) at 37 °C in an atmosphere containing 100% O2, dispersed by pipetting and passed through a 160 µm nylon mesh (Sefar Filtration Inc., purchased from SmallParts, Inc.; part# CMN-LP160037-06). Acini were then washed with D-MEM/F-12 medium and plated into 35-mm wells (1 mL aliquots). Prior to transfection, acinar cells were incubated for 1 h at 37 °C in a humidified atmosphere containing 5% CO2. Transfections were performed with 2×108 pfu/mL final adenovirus concentrations in 1 mL D-MEM/F-12 medium supplemented with 0.1% BSA, 1 mM sodium pyruvate and 1% penicillin/streptomycin solution (final concentrations).
Preparation of cell lysates
Transfected cells were washed twice with phosphate buffered saline (PBS). Two-hundred µL reporter lysis buffer (Promega), 4 µL protease inhibitor cocktail (Sigma, #P8340) and 2 µL Halt phosphatase inhibitor cocktail (Thermo Scientific, #78420) was added and cells were briefly vortexed. After 15 min incubation at 4 °C the lysates were centrifuged for 5 min at 16,000g in a microcentrifuge and the pellet was discarded. The protein concentration of the supernatant was measured with the Micro BCA™ Protein Assay Kit (Thermo Scientific).
Reverse transcriptase (RT)-PCR analysis
RNA was isolated at 24 h after transfection using the RNAqueous kit (Ambion) and 1 µg RNA was reverse-transcribed with M-MLV reverse transcriptase (Ambion). Semi-quantitative measurements of cDNA levels for XBP1, XBP1s (spliced form), BiP and CHOP were performed by PCR using primers listed in Supplementary Table 1. As an internal control for mRNA integrity and equal loading, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was amplified, using pseudogene-free amplification conditions (Supplementary Table 1). PCR products were run on 1–2% agarose gels and stained with ethidium bromide; and band intensities were quantitated using Quantity One Software, version 4.3.1 (Bio-Rad).
Western blot analysis
Aliquots of conditioned media (100 µL per lane) or cell lysates (20 µg protein per lane) were electrophoresed on 15% Tris-glycine minigels and transferred onto Immobilon-P membranes (Millipore, Billerica, MA). For the analysis of BiP expression 10% gels were used. The membranes were blocked with 5% milk powder solution overnight and incubated with given primary and secondary antibodies for 1 h at room temperature. To detect the Glu-Glu-tag, a horseradish peroxidase (HRP)-conjugated goat polyclonal antibody (Abcam, Cambridge, MA, #ab1267) was used at a dilution of 1:10,000. BiP was detected with a rabbit polyclonal antibody (Abcam, #ab21685) used at a dilution of 1:5000. Phospho-eIF2α (Ser51) was detected with a rabbit monoclonal antibody (Epitomics, #1090-1) used at a dilution of 1:1000. IκBα was detected with a rabbit polyclonal antibody (Cell Signaling Technology, #9242) used at a dilution of 1:2000. Membranes probed with anti-BiP, anti-eIF2α and anti-IκBα primary antibodies were incubated with an HRP conjugated goat polyclonal anti-rabbit IgG secondary antibody (Pierce, #31460) at a dilution of 1:20,000. Actin was detected with a mouse monoclonal antibody (Sigma, #A4700, clone AC-40) used at a dilution of 1:1000 followed by HRP-conjugated goat polyclonal anti-mouse IgG (Abcam, #ab6789) at a dilution of 1:2500. HRP was detected using the SuperSignal West Pico Chemiluminescent Substrate (Pierce).
Cell death assays
Cell viability was determined based on the dehydrogenase activity of living cells using the Cell Counting Kit-8 from Dojindo Molecular Technologies, Inc. (Rockville, MD). Forty-eight hours after transfection both detached and attached AR42J cells were collected, washed with PBS, and total protein content was determined from an aliquot of the cells using the Micro BCA™ Protein Assay Kit. Approximately 1 × 105 cells per well were seeded into a 96-well flat bottom plate in 100 µL PBS. An aliquot (10 µL) of the tetrazolium substrate was added to each well and plates were incubated at 37 °C for 2 h, after which the absorbance at 450 nm was measured using a Spectramax Plus 384 microplate reader (Molecular Devices). Absorbance values were normalized to total protein concentrations.
Terminal deoxynucleotidyl transferase (TdT)-mediated dUTP nick-end labeling (TUNEL) detection of DNA fragmentation was carried out using a fluorescein-based detection kit (In Situ Cell Death Detection Kit, Roche Applied Science, Indianapolis, IN) according to the manufacturer’s instructions. Prior to transfection 1 × 105 AR42J cells per well were seeded into a 96-well flat bottom plate and allowed to grow for 48 h. Transfection was carried out in a final volume of 100 µL and the TUNEL assay was performed 48 h after transfection. Briefly, cultures were fixed with 4% paraformaldehyde in PBS (pH 7.4) for 1 h at room temperature, followed by 2 min incubation with 0.1% Triton X-100 in 0.1% sodium citrate at 4 °C to permeabilize the cells. TUNEL reagent (50 µL) was added to air dried cells and incubated for 60 min at 37 °C in a humidified atmosphere in the dark and the samples were analyzed under a fluorescence microscope. Cells were washed twice with PBS after each incubation step in this protocol.
Caspase-3/7 activity was measured using the Apo-ONE Homogenous Caspase-3/7 Assay (Promega, Madison, WI). AR42J cells were scraped and approximately 1 × 105 cells (in 50 µL PBS) were mixed with the same volume of the Apo-ONE Homogenous Caspase-3/7 reagent in a 96-well black plate. After incubation at room temperature for 2 h, the fluorescence emission was measured at 530 nm, using an excitation wavelength of 485 nm, in a Spectramax Gemini XS fluorescent microplate reader (Molecular Devices). PBS mixed with the same volume of Apo-one Homogenous Caspase-3/7 reagent served as a negative control. Caspase activity was corrected for the activity of the negative control and normalized to total protein concentrations.
Fluorescence microscopy
Cells expressing eGFP or subjected to the TUNEL assay were viewed under a Nikon Eclipse TE 300 inverted microscope with a TE-FM epifluorescence attachment.
Statistical analysis
The significance of changes was analyzed with the Tukey-Kramer multiple comparisons test; the differences were regarded significant when the P value was <0.05. Results are means ± standard error of the mean (SEM).
RESULTS
Expression of CTRC in pancreatic acinar cells via adenoviral transfection
To investigate whether the p.A73T CTRC mutant causes ER stress in pancreatic acinar cells, in the present study we used dexamethasone-differentiated AR42J rat pancreatic acinar cells and freshly prepared primary mouse acini for the transient expression of CTRC. Acinar cells were transfected with recombinant adenoviral vectors carrying wild type CTRC or the p.A73T mutant. As a negative control, an adenovirus carrying the eGFP gene was used. Efficiency of transfection exceeded 90% both in AR42J cells (Fig 1A) and in primary mouse acini (data not shown). Transfection of AR42J cells with increasing concentrations of recombinant CTRC-adenovirus resulted in secretion of CTRC into the growth medium as judged by SDS-PAGE analysis (Fig 1B). Secreted levels of CTRC increased as a function of the virus concentration and a robust CTRC band was detected on gels at 108 pfu/mL and above. Primary mouse acini transfected with CTRC-adenovirus secreted CTRC protein, which was readily detectable by western blots, but could not be visualized on Coomassie-stained SDS-PAGE gels due to the complex mixture of secretory proteins produced by the acini and the presence of high concentrations of bovine serum albumin (the Coomassie-stained gels were reviewed but are not shown). It is also important to note that in AR42J cells and mouse acini CTRC is expressed in its inactive, zymogen form and we detected no signs of spontaneous activation either in the conditioned medium or in cell lysates.
Figure 1.
Transfection of AR42J cells with enhanced green fluorescent protein (eGFP) and CTRC-adenovirus. (A) Efficiency of transfection as monitored by eGFP expression. Cells were analyzed under a fluorescence microscope 24 h after infection (200-fold magnification, scale bars: 50 µm). Left panel, phase-contrast image; right panel, fluorescent image. (B) CTRC secretion by AR42J cells transfected with increasing concentrations of CTRC-adenovirus (expressed in plaque-forming units per mL). Twenty-four hours post-transfection 100 µL aliquots of conditioned media were precipitated with 10% trichloroacetic acid, heat denaturated at 95 °C for 5 min in reducing Laemmli sample buffer and electrophoresed on 15% SDS-polyacrylamide gels. Gels were stained with Coomassie Blue.
Diminished secretion and intracellular retention/degradation of the p.A73T CTRC mutant
First, we compared spontaneous (basal) and secretagogue (cerulein) stimulated secretion of wild type CTRC and the p.A73T CTRC mutant from transfected AR42J cells and mouse acini. Secretion of the p.A73T mutant was markedly reduced compared to wild type CTRC in both cell types (Fig 2A). In AR42J cells, the p.A73T mutant was detected in cell lysates at levels that were comparable or higher than those of wild type, indicating that the p.A73T mutant is synthesized normally inside the cell but it undergoes degradation instead of secretion (Fig 2B). In lysates of mouse acini the p.A73T mutant was found in significantly lower amounts than wild type CTRC, suggesting that the mutant becomes degraded more efficiently in this cell type (Fig 2B).
Figure 2.
Diminished secretion and intracellular retention/degradation of the p.A73T CTRC mutant. Western blot analysis of conditioned media (A) and cell lysates (B) of pancreatic acinar cells expressing wild type CTRC, enhanced green fluorescent protein (control) or the p.A73T CTRC mutant. Twenty-four hours after transfection cells were washed twice with phosphate buffered saline and fresh OptiMEM medium was added. After 15 min incubation at 37 °C, the medium was collected (“basal”) and fresh OptiMEM containing 10 nM (AR42J) or 100 pM (mouse acini) concentration of cerulein was added to stimulate secretion. The “cerulein” medium was collected after 15 min incubation at 37 °C, and cells were also harvested for western blot analysis. Aliquots of conditioned media (100 µL) and cell lysates (20 µg total protein) were loaded onto a 15% SDS-polyacrylamide gel, transferred to Immobilon-P membrane and CTRC was detected with an antibody against the Glu-Glu epitope tag. Representative blots of three independent experiments are shown.
ER stress in AR42J cells expressing the p.A73T CTRC mutant
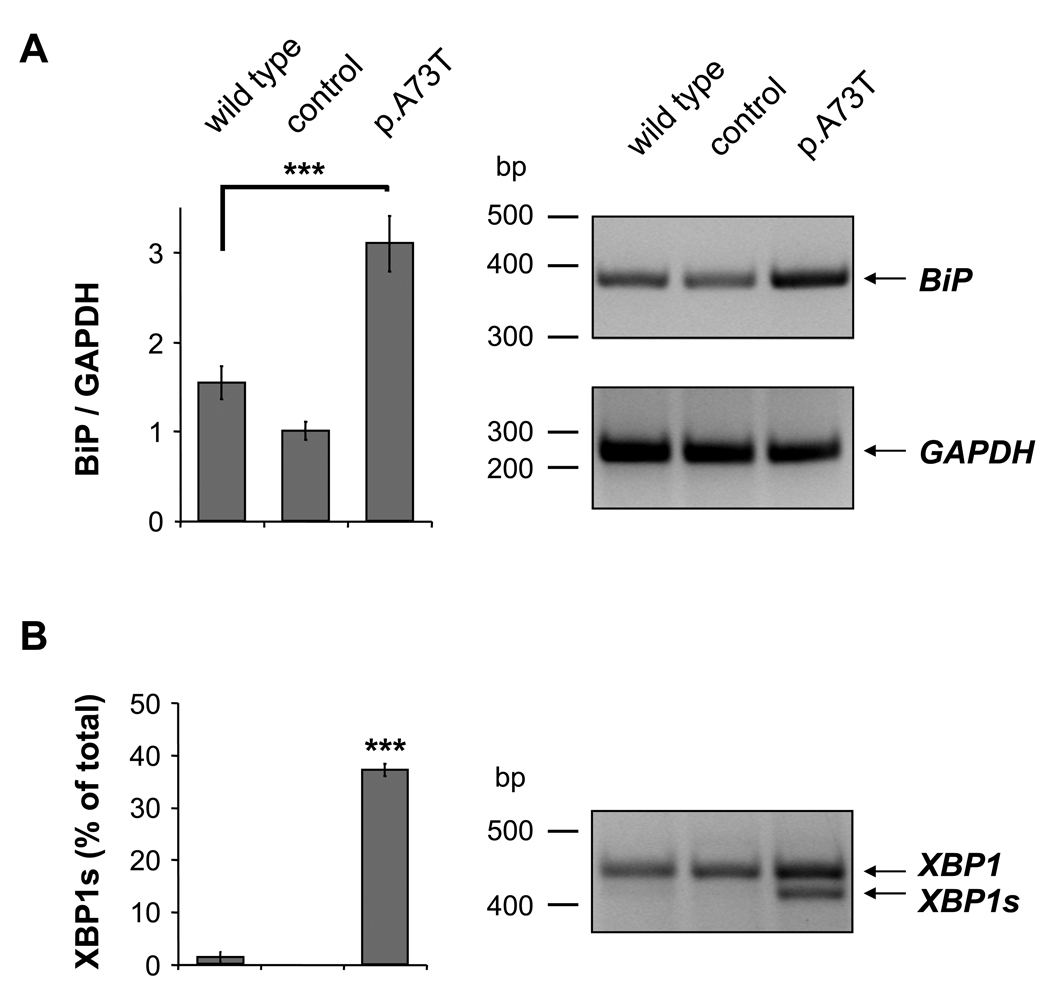
Intracellular retention of the p.A73T mutant most likely occurs in the ER due to mutation-induced misfolding, which then results in degradation through the ER-associated protein degradation (ERAD) pathway. If this were the case, the p.A73T mutant might cause ER stress and trigger the unfolded protein response, a signal transduction pathway aimed at alleviating ER protein burden and increasing ER folding capacity. The master regulator of the unfolded protein response is the chaperone immunoglobulin-binding protein (BiP), which becomes characteristically upregulated under conditions of ER stress. Therefore, to test whether expression of the p.A73T mutant in AR42J cells causes ER stress, we measured the protein and mRNA levels of BiP in cell lysates by western blotting (Fig 3A) and semi-quantitative RT-PCR analysis (Fig 3B). We found that BiP was significantly upregulated in acinar cells expressing the p.A73T mutant, relative to cells transfected with wild type CTRC or control adenovirus.
Figure 3.
ER stress markers in AR42J cells expressing wild type CTRC, enhanced green fluorescent protein (control) or the p.A73T CTRC mutant. (A) Levels of BiP protein (78 kDa) in cell lysates were analyzed by western blotting as described in Methods. Actin (42 kDa) was measured as a loading control. Band intensities were quantitated by densitometry and BiP/actin ratios are shown as bar graphs. (B) Semi-quantitative RT-PCR was performed to determine BiP (578 bp) mRNA levels. Expression of GAPDH (261 bp) was measured as a reference control. Band intensities were quantitated by densitometry and BiP/GAPDH ratios are depicted as bar graphs. (C) The extent of XBP1 splicing was determined by measuring the unspliced (XBP1; 447 bp) and spliced (XBP1s; 421 bp) forms by RT-PCR. Band intensities were quantitated by densitometry and the amount of the spliced form is shown as percentage of total XBP1 (unspliced and spliced). Values are means ± SEM, ***P<0.001, n=3.
In non-stressed mammalian cells BiP binds to the luminal domain of the membrane-embedded inositol-requiring enzyme 1 (IRE1) and keeps this ER stress sensor protein in an inactive state. During ER stress BiP dissociates from IRE1 and binds to misfolded proteins, thereby allowing the dimerization and activation of IRE1. IRE1 catalyzes the cytoplasmic splicing of the X-box binding protein-1 (XBP1) mRNA by removing a 26-nucleotide intron to create a shorter variant (XBP1s), which encodes a potent transcriptional activator of the unfolded protein response. We used RT-PCR to measure the extent of XBP1 splicing in AR42J cells expressing the p.A73T mutant. Fig 3C demonstrates that expression of the p.A73T mutant caused a significant increase in XBP1s compared to the lower levels observed in cells expressing wild type CTRC or infected with control virus.
ER stress in AR42J cells is proportional to intracellular levels of the p.A73T CTRC mutant
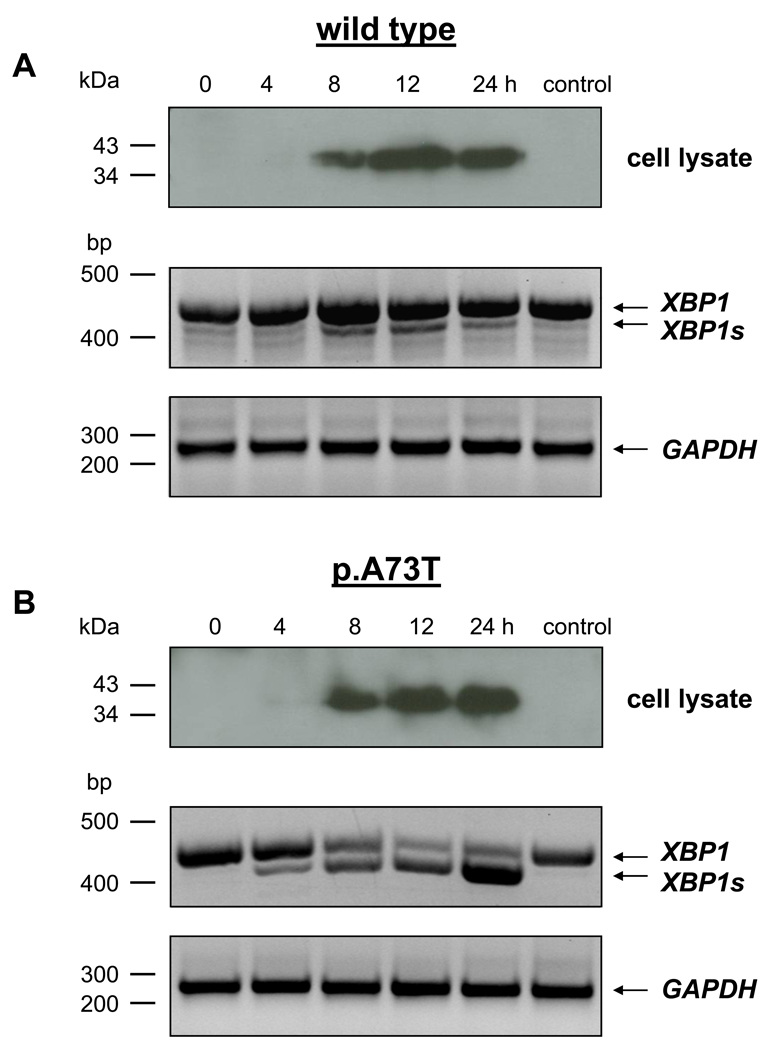
To demonstrate that ER stress is dependent on the intracellular accumulation of the misfolded p.A73T CTRC mutant, we measured the time-course of CTRC expression, for both wild type and mutant, and compared it to the time-course of XBP1 splicing. As shown in Fig 4, expression of CTRC in cell lysates became clearly detectable at 8 h; increased by 12 h and changed very little between 12 and 24 h. XBP1 splicing in cells expressing the p.A73T mutant showed a very similar kinetics. Thus, minimal splicing was detected already at 4 h; splicing became robust at 8 h and increased to near completion at 12 h. Although total XBP1 levels were somewhat higher at 24 h, the extent of splicing did not change significantly between 12 h and 24 h. In stark contrast to the p.A73T CTRC mutant, cells expressing wild type CTRC showed only negligible XBP1 splicing by 8 h which remained unchanged at 12 h and 24 h, despite the higher intracellular CTRC content at the later time points. These data clearly demonstrate a positive correlation between ER stress and the intracellular levels of the p.A73T CTRC mutant. Importantly, the results also confirm that heterologous overexpression of CTRC alone is insufficient to trigger significant ER stress.
Figure 4.
Time course of CTRC expression and XBP1 splicing in AR42J cells expressing wild type CTRC (A) or the p.A73T CTRC mutant (B). Control cells were infected with eGFP-adenovirus and were harvested at 24 h. Levels of CTRC protein in cell lysates were analyzed by western blotting as described in Fig 2 and Methods. XBP1 splicing was measured by RT-PCR as described in Fig 3 and Methods. Expression of GAPDH (261 bp) was measured as a control for mRNA integrity and equal loading.
ER stress in primary mouse acinar cells expressing the p.A73T CTRC mutant
We determined mRNA levels for BiP and XBP1s in freshly prepared mouse pancreatic acini using RT-PCR (Fig 5). Both ER stress markers were significantly elevated in acini expressing the p.A73T mutant compared to cells infected with wild type CTRC virus or control virus. We were unable to evaluate intracellular BiP protein levels in a reliable manner, as mouse acini exhibited robust BiP secretion, which resulted in variable BiP levels in cell lysates. The unusual phenomenon of BiP secretion by pancreatic acini has been reported previously, although its mechanism and physiological significance is unclear.[25]
Figure 5.
ER stress markers in mouse pancreatic acinar cells expressing wild type CTRC, enhanced green fluorescent protein (control) or the p.A73T CTRC mutant. (A) Semi-quantitative RT-PCR was performed to determine BiP (398 bp) mRNA levels. Expression of GAPDH (261 bp) was measured as a reference control. Band intensities were quantitated by densitometry and BiP/GAPDH ratios are depicted as bar graphs. (B) The extent of XBP1 splicing was determined by measuring the unspliced (XBP1; 447 bp) and spliced (XBP1s; 421 bp) forms by RT-PCR. Band intensities were quantitated by densitometry and the amount of the spliced form is shown as percentage of total XBP1 (unspliced and spliced). Values are means ± SEM, *** P<0.001, n=3.
Activation of the PERK pathway and NFκB is not observed in acinar cells expressing the p.A73T CTRC mutant
Translation attenuation in response to ER stress is mediated by the protein kinase RNA (PKR)-like endoplasmic reticulum kinase (PERK), an ER membrane protein. Dimerization of PERK leads to trans-autophosphorylation and activation of its cytoplasmic protein kinase domain, which in turn phosphorylates the α subunit of the eukaryotic translation initiation factor-2 (eIF2α) thereby impeding translation initiation. One of the consequences of translation attenuation is the activation of NFκB, due to the relative depletion of its short half-life inhibitor IκBα. To assess whether expression of the p.A73T mutant causes translation attenuation and NFκB activation, we measured levels of phosphorylated eIF2α and IκBα in lysates of AR42J and mouse pancreatic acinar cells at 24 h after transfection by western blot analysis (Supplementary Fig 1). No significant changes were detected in these markers in cells expressing the p.A73T mutant compared to cells transfected with wild type CTRC or control virus, indicating that the PERK pathway was not activated. To assess whether translation attenuation might be more prominent at earlier time points, we followed the time course of eIF2α phosphorylation in AR42J cells expressing the p.A73T CTRC mutant but found no increase in phosphorylation at 4, 8, and 12 h (reviewed but not shown).
Apoptotic cell death in AR42J cells expressing the p.A73T CTRC mutant
Chronic ER stress may trigger apoptosis and we observed that AR42J cells expressing the p.A73T mutant became almost completely detached from the tissue culture plates within 48 hours, whereas cells expressing the wild type CTRC or those infected with control virus remained attached to the plastic support and showed normal morphology (Fig 6A). Viability of the acinar cells expressing the p.A73T CTRC mutant was also significantly decreased when assessed by an assay based on the dehydrogenase activity of living cells (Fig 6B). To obtain evidence that AR42J cells expressing the p.A73T mutant undergo apoptosis we measured caspase 3/7 activity from cell lysates at 24 and 48 hours after transfection. Caspase activity increased between 24 h and 48 h by 2–3-fold, and at both time points it was significantly higher in cells expressing the p.A73T mutant, relative to cells transfected with wild type CTRC or control virus (Fig 7A). Furthermore, at 48 hours after transfection, TUNEL staining revealed a striking increase in the number of apoptotic nuclei in cells expressing the p.A73T mutant compared to cells expressing wild type CTRC (Fig 7B).
Figure 6.
Viability of AR42J cells expressing the p.A73T CTRC mutant. (A) Detachment of cells from the tissue culture plates. Cells expressing wild type CTRC or the p.A73T CTRC mutant were co-transfected with enhanced green fluorescent protein (eGFP) for better visualization. Control cells were transfected with eGFP adenovirus only. At the indicated times cells were washed twice with phosphate-buffered saline to remove detached cells and pictures were taken under a fluorescence microscope (100-fold magnification, scale bars: 100 µm). (B) The number of viable cells expressing wild type CTRC (WT) or the p.A73T mutant CTRC (M) was estimated 48 h after transfection by their dehydrogenase activity as described in Methods. In this assay both the detached and attached cells were included. Cell viability was expressed as percentage of the viability of non-transfected control (C) samples. Values are means ± SEM, *** P<0.001, n=3.
Figure 7.
ER stress-induced apoptosis in AR42J cells expressing the p.A73T CTRC mutant. (A) Caspase 3/7 activity was measured from cells expressing wild type CTRC (WT) or the p.A73T CTRC mutant (M) at 24 h and 48 h after transfection, as described in Methods. Caspase activities were expressed as percentage of the activity measured in the non-transfected control (C) cells at 24 h. (B) TUNEL assay. Cells expressing wild type CTRC or the p.A73T mutant were fixed at 48 h after transfection and apoptotic nuclei were detected by TUNEL staining. Upper panels show phase contrast images of cells and lower panels demonstrate TUNEL-positive apoptotic cells, which were seen in green color under the fluorescence microscope. Representative pictures are shown at 100-fold magnification (scale bars: 100 µm). The bar graphs present the number of TUNEL positive cells counted from 6 randomly chosen 1 mm2 fields in each well (means ± SEM). (C) Semi-quantitative RT-PCR was performed to determine CHOP (286 bp) mRNA levels. Expression of GAPDH (261 bp) was measured as reference control. Band intensities were quantitated by densitometry and CHOP/GAPDH ratios are depicted in bar graphs. Values are means ± SEM from 3 independent experiments (***P<0.001).
ER stress associated apoptosis is induced by the transcription factor C/EBP homologous protein (CHOP), which becomes highly expressed in ER stress and down-regulates expression of anti-apoptotic proteins. We measured the expression of CHOP mRNA in AR42J cells at 24 h after transfection by RT-PCR and found that CHOP levels were significantly elevated in cells expressing the p.A73T mutant, relative to cells transfected with wild type CTRC or control virus (Fig 7C). Elevated CHOP levels were also found in primary mouse acini expressing the p.A73T mutant at 24 h, indicating that activation of pro-apoptotic pathways by ER stress has occurred (Supplementary Fig 2). However, we did not observe signs of increased apoptosis at this early time point, as assessed by viability assays, caspase activity measurements or TUNEL staining (not shown). We were unable to evaluate apoptosis at longer culture times, as viability of primary cultures of acini kept beyond 24 h progressively deteriorated.
DISCUSSION
In the present study we demonstrate that mutations in human CTRC can increase the ability of chymotrypsinogen C to cause ER stress and subsequent cell death by a mechanism which is unrelated to the trypsin-degrading activity of CTRC but involves mutation-induced misfolding. Previous studies on the mechanism of genetic risk in chronic pancreatitis led to the formulation of a trypsin-dependent disease model, which posits that sustained, intrapancreatic conversion of trypsinogen to trypsin plays a central role in the development of chronic pancreatitis.[2–17] One of the defense mechanisms against unwanted trypsin activity in the pancreas is CTRC, which was recently shown to degrade trypsin and trypsinogen with high specificity.[15] Mutations in CTRC that abolish catalytic activity and/or decrease secretion have been found in association with chronic pancreatitis, suggesting that failure of the intrapancreatic trypsin-degrading mechanisms increases pancreatitis risk.[16,17] To date, three CTRC mutations have been confirmed statistically to increase the risk of chronic pancreatitis. Mutations p.R254W and p.K247_R254del are prevalent in European subjects whereas mutation p.A73T was found with higher frequency in India. When analyzed in transiently transfected HEK 293T cells, all three mutants exhibited a secretion defect apparently as a result of intracellular retention and degradation.[16] Secretion of mutant p.R254W was reduced by about 50%, whereas mutants p.K247_R254del and p.A73T were secreted only in trace amounts. This common phenotype led us to speculate that intracellular retention and degradation of CTRC mutants might cause ER stress, which can contribute to the development of chronic pancreatitis. This would represent a novel disease mechanism unrelated to the trypsin-degrading activity of CTRC and thus unrelated to intrapancreatic trypsin levels. We note, however, that loss of function mutations in CTRC might result in ER stress by other mechanisms as well, which might involve accumulation of trypsin or other malformed proteins which would be otherwise degraded by CTRC.
In the present study we established an adenovirus mediated transfection system for the expression of CTRC in pancreatic acinar cells. Using dexamethasone-differentiated AR42J pancreatic acinar cells and freshly isolated mouse acini we found that mutant p.A73T was intracellularly retained and degraded; and markers of ER stress were significantly elevated in cells expressing the p.A73T CTRC mutant relative to cells transfected with wild type CTRC or a control adenovirus. Furthermore, we observed that AR42J cells underwent apoptotic cell death as a result of expressing the p.A73T CTRC mutant. Apoptosis was related to ER stress, as evidenced by marked induction of the pro-apoptotic transcription factor CHOP. We also measured elevated levels of CHOP in primary mouse acinar cells expressing the p.A73T mutant; however, we could not detect apoptosis in these cells during short term culturing. Mouse acinar cells have been reported to exhibit higher resistance to apoptosis than rat acinar cells, which is consistent with our findings.[26]
Previously, we observed that the hereditary-pancreatitis associated p.R116C cationic trypsinogen mutant suffered intracellular misfolding and retention and caused ER stress in transiently transfected HEK 293T cells.[27] Taken together, the two studies propose a new paradigm for the mechanism of genetic risk in chronic pancreatitis, which involves ER stress caused by mutation-induced misfolding and ER retention of digestive (pro)enzymes or other secretory proteins. This model implies that the physiological function or catalytic activity of the mutant protein is immaterial and mutations in a variety of secretory proteins can increase pancreatitis risk as long as the protein is highly expressed and the mutation causes misfolding. ER stress induced apoptosis can accelerate the loss of functional acini and thus contribute to exocrine insufficiency, a hallmark of chronic pancreatitis. Apoptosis of pancreatic acinar cells was shown to be protective against necrosis and severe inflammation in acute models of experimental pancreatitis,[26] but in a chronic setting apoptosis would be clearly detrimental with respect to parenchymal cell loss. This notion is also supported by a recent study demonstrating that in pancreatic tissue of chronic pancreatitis patients the number of apoptotic acinar cells was 10-fold increased relative to cells from normal pancreata.[28]
Summary box
Loss-of-function mutations in the chymotrypsinogen C (CTRC) gene have been linked to the development of chronic pancreatitis.
Increased pancreatitis risk was so far explained by the loss of trypsin-degrading activity.
New findings indicate that CTRC mutations can also increase the propensity of chymotrypsinogen C to elicit endoplasmic reticulum (ER) stress in pancreatic acinar cells.
Carriers of CTRC mutations may be at a higher risk of developing ER stress in the exocrine pancreas.
ER stress may contribute to parenchymal damage in chronic pancreatitis through acinar cell apoptosis.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by US National Institutes of Health grants DK082412 and DK058088 (to M. S.-T) and a grant from the National Pancreas Foundation (to R. S.). The authors thank Rajinder Dawra and George Perides for helpful technical suggestions.
Footnotes
Competing interest: None to declare.
COPYRIGHT LICENCE STATEMENT
“The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non-exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd, and its Licensees to permit this article (if accepted) to be published in GUT and any other BMJPGL products and to exploit all subsidiary rights, as set out in our licence.”
REFERENCES
- 1.Witt H, Apte MV, Keim V, et al. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–1573. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Teich N, Rosendahl J, Tóth M, et al. Mutations of human cationic trypsinogen (PRSS1) and chronic pancreatitis. Hum Mutat. 2006;27:721–730. doi: 10.1002/humu.20343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sahin-Tóth M. Biochemical models of hereditary pancreatitis. Endocrinol Metab Clin North Am. 2006;35:303–312. doi: 10.1016/j.ecl.2006.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whitcomb DC, Gorry MC, Preston RA, et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat Genet. 1996;14:141–145. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 5.Howes N, Lerch MM, Greenhalf W, et al. Clinical and genetic characteristics of hereditary pancreatitis in Europe. Clin Gastroenterol Hepatol. 2004;2:252–261. doi: 10.1016/s1542-3565(04)00013-8. [DOI] [PubMed] [Google Scholar]
- 6.Sahin-Tóth M, Tóth M. Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem Biophys Res Commun. 2000;278:286–289. doi: 10.1006/bbrc.2000.3797. [DOI] [PubMed] [Google Scholar]
- 7.Witt H, Luck W, Hennies HC, et al. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat Genet. 2000;25:213–216. doi: 10.1038/76088. [DOI] [PubMed] [Google Scholar]
- 8.Pfützer RH, Barmada MM, Brunskill AP, et al. SPINK1/PSTI polymorphisms act as disease modifiers in familial and idiopathic chronic pancreatitis. Gastroenterology. 2000;119:615–623. doi: 10.1053/gast.2000.18017. [DOI] [PubMed] [Google Scholar]
- 9.Aoun E, Chang CC, Greer JB, et al. Pathways to injury in chronic pancreatitis: decoding the role of the high-risk SPINK1 N34S haplotype using meta-analysis. PLoS ONE. 2008;3:e2003. doi: 10.1371/journal.pone.0002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Király O, Boulling A, Witt H, et al. Signal peptide variants that impair secretion of pancreatic secretory trypsin inhibitor (SPINK1) cause autosomal dominant hereditary pancreatitis. Hum Mutat. 2007;28:469–476. doi: 10.1002/humu.20471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Király O, Wartmann T, Sahin-Tóth M. Missense mutations in pancreatic secretory trypsin inhibitor (SPINK1) cause intracellular retention and degradation. Gut. 2007;56:1433–1438. doi: 10.1136/gut.2006.115725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boulling A, Le Maréchal C, Trouvé P, et al. Functional analysis of pancreatitis-associated missense mutations in the pancreatic secretory trypsin inhibitor (SPINK1) gene. Eur J Hum Genet. 2007;15:936–942. doi: 10.1038/sj.ejhg.5201873. [DOI] [PubMed] [Google Scholar]
- 13.Kereszturi E, Király O, Sahin-Tóth M. Minigene analysis of intronic variants in common SPINK1 haplotypes associated with chronic pancreatitis. Gut. 2009;58:545–549. doi: 10.1136/gut.2008.164947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Witt H, Sahin-Tóth M, Landt O, et al. A degradation-sensitive anionic trypsinogen (PRSS2) variant protects against chronic pancreatitis. Nat Genet. 2006;38:668–673. doi: 10.1038/ng1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Szmola R, Sahin-Tóth M. Chymotrypsin C (caldecrin) promotes degradation of human cationic trypsin: identity with Rinderknecht's enzyme Y. Proc Natl Acad Sci USA. 2007;104:11227–11232. doi: 10.1073/pnas.0703714104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rosendahl J, Witt H, Szmola R, et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat Genet. 2008;40:78–82. doi: 10.1038/ng.2007.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masson E, Chen JM, Scotet V, et al. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum Genet. 2008;123:83–91. doi: 10.1007/s00439-007-0459-3. [DOI] [PubMed] [Google Scholar]
- 18.Marciniak SJ, Ron D. Endoplasmic reticulum stress signaling in disease. Physiol Rev. 2006;86:1133–1149. doi: 10.1152/physrev.00015.2006. [DOI] [PubMed] [Google Scholar]
- 19.Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- 20.Rutkowski DT, Kaufman RJ. That which does not kill me makes me stronger: adapting to chronic ER stress. Trends Biochem Sci. 2007;32:469–476. doi: 10.1016/j.tibs.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature. 2008;454:455–462. doi: 10.1038/nature07203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kubisch CH, Sans MD, Arumugam T, et al. Early activation of endoplasmic reticulum stress is associated with arginine-induced acute pancreatitis. Am J Physiol Gastrointest Liver Physiol. 2006;291:G238–G245. doi: 10.1152/ajpgi.00471.2005. [DOI] [PubMed] [Google Scholar]
- 23.Kubisch CH, Logsdon CD. Secretagogues differentially activate endoplasmic reticulum stress responses in pancreatic acinar cells. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1804–G1812. doi: 10.1152/ajpgi.00078.2007. [DOI] [PubMed] [Google Scholar]
- 24.Logsdon CD, Moessner J, Williams JA, et al. Glucocorticoids increase amylase mRNA levels, secretory organelles, and secretion in pancreatic acinar AR42J cells. J Cell Biol. 1985;100:1200–1208. doi: 10.1083/jcb.100.4.1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takemoto H, Yoshimori T, Yamamoto A, et al. Heavy chain binding protein (BiP/GRP78) and endoplasmin are exported from the endoplasmic reticulum in rat exocrine pancreatic cells, similar to protein disulfide-isomerase. Arch Biochem Biophys. 1992;296:129–136. doi: 10.1016/0003-9861(92)90554-a. [DOI] [PubMed] [Google Scholar]
- 26.Mareninova OA, Sung KF, Hong P, et al. Cell death in pancreatitis: caspases protect from necrotizing pancreatitis. J Biol Chem. 2006;281:3370–3381. doi: 10.1074/jbc.M511276200. [DOI] [PubMed] [Google Scholar]
- 27.Kereszturi E, Szmola R, Kukor Z, et al. Hereditary pancreatitis caused by mutation-induced misfolding of human cationic trypsinogen: a novel disease mechanism. Hum Mutat. 2009;30:575–582. doi: 10.1002/humu.20853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrader H, Menge BA, Schneider S, et al. Reduced pancreatic volume and beta-cell area in patients with chronic pancreatitis. Gastroenterology. 2009;136:513–522. doi: 10.1053/j.gastro.2008.10.083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.