Abstract
To divide asymmetrically, the mitotic spindle is moved from the cell center to the cortex, a process that requires astral microtubules and the microtubule-based motor dynein. New work examining spindle positioning in large oocytes shows that in these cells actin and actin polymerization plays a key role.
Asymmetric cell division underlies fundamental developmental processes in all eukaryotes and serves to generate unequal daughter cells. In stem cells, for example, asymmetric division results in one daughter cell capable of self-renewal and a second daughter cell capable of differentiating. During the reductional division that leads to a mature egg, asymmetric division is used to ensure that the cytoplasmic components needed for early development are retained in the cell that will be fertilized. To divide asymmetrically, cells assemble a mitotic (or meiotic) spindle near the cell center and subsequently move the spindle to the cell cortex. Because spindle position dictates the location of the contractile ring, which is responsible for dividing the cytoplasm, proper orientation of the spindle relative to the cell is essential for asymmetric division.
So how does the spindle move to the appropriate location? Much of what we know about this process has been learned from studies of the first mitotic division in the worm Caenorhabditis elegans (Figure 1) [1]. In these cells, experiments utilizing a laser beam to sever the spindle show that pulling forces act on the two spindle poles, but that a greater net force acts on the posterior pole, resulting in spindle displacement [2]. Pulling forces require dynamic astral microtubules and cytoplasmic dynein, a minus-end-directed motor protein whose asymmetric activation, or localization, is regulated by cortical polarity factors [1]. Results in other systems, notably Drosophila, support this microtubule-dependent model for spindle positioning [3].
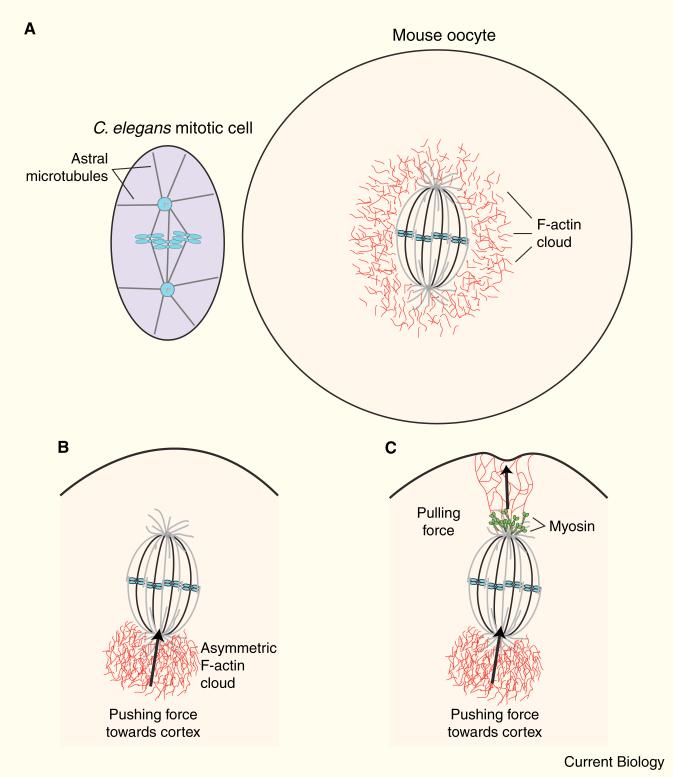
Figure 1. Actin and myosin power spindle positioning in mouse oocytes.
(A) Schematic diagrams of a mitotic spindle at first division in C. elegans (left) and the meiotic spindle in a mouse oocyte (right). Note that in the oocyte, few astral microtubules are present, and a cloud of F-actin surrounds the spindle. (B) Symmetry-breaking in an oocyte results in an asymmetric cloud of F-actin pushing the spindle toward the cortex. (C) Myosin, located at the spindle poles, pulls on actin filaments, buckling the cortex.
Meiotic spindles in the very large mammalian oocytes, however, lack centrosomes and have reduced numbers of astral microtubules, suggesting that a different mechanism may be responsible for spindle positioning. In fact, several studies have pointed to the involvement of actin in these cells [4–8]. Unfortunately, imaging actin in the interior of these large cells has been challenging. Dynamic actin has proved notoriously difficult to chemically fix and stain [9] and, although useful for imaging actin in some cell types [10,11], a probe comprising actin fused to the green fluorescent protein fails to incorporate into actin structures in other cell types [12]. Because of these technical challenges, definitive evidence for actin-mediated spindle positioning has remained elusive.
Two new reports [13,14] now show that dynamic actin and the actinpolymerizing factor formin (Fmn2) are required for spindle positioning during meiosis in mammalian oocytes. These studies both have taken advantage of new actin probes that enable imaging of actin structure and dynamics in live oocytes. Li et al. [13] used LifeAct, a probe that comprises the first 17 amino acids of ABP140 fused with enhanced green fluorescent protein (EGFP) and binds to actin without altering actin dynamics [12]. Schuh et al. [14], as reported in a recent issue of Current Biology, used the calponin homology domain of utrophin tagged with EGFP, which similarly binds F-actin without perturbing dynamics and has been used to examine actin in frog oocytes [15]. These new actin probes represent a breakthrough for imaging actin and have enabled detailed mechanistic studies of spindle positioning.
The Li study [13] observed a surprising finding in live oocytes — F-actin was organized as a ‘cloud’ that surrounded but didn't directly touch the chromosomes (Figure 1). As the chromosomes began to move, the actin cloud became asymmetric, with the bulk of F-actin behind the moving mass of chromosomes. Jasplakinolide, which stabilizes F-actin, prevented both chromosome motion and formation of the asymmetric actin cloud. By tracking the center of mass of the chromosomes, they showed that motion was pulsatile, and depolymerization of microtubules resulted in faster motion, suggesting that in these cells microtubules restrict, rather than mediate, movement. Oocytes from Fmn2–/– mutant mice lack cytoplasmic actin and fail to move their spindle. Full-length Fmn2 or the actin-elongating/nucleating FH1/FH2 domains of Fmn2 could rescue these phenotypes. The results favor a model in which forces generated by formin-dependent assembly of F-actin power spindle positioning, and that a symmetry-breaking event determines the direction of motion.
In agreement with Li et al. [13], Schuh et al. [14] report that actin and Fmn2 are required for spindle relocalization. They use confocal microscopy to show that short, dynamic actin filaments surround the spindle and extend into its interior. As the spindle approaches the cortex, the cortex dimples inward, and actin filaments are observed to move toward the spindle pole at the same rate that the spindle moves to the cortex. These observations suggest that myosin pulling on actin contributes to the motion. In support of this role for myosin, antibody staining showed that the activated, phosphorylated form of myosin light chain localizes to the spindle poles. Additionally, treatment with ML7, a kinase inhibitor that inhibits myosin light chain kinase, reduced the rate of spindle movement and blocked the poleward motion of actin filaments. From these results, the authors propose that a dynamic meshwork of actin filaments extends from the spindle poles to the cortex and that myosin pulls on these filaments to move the spindle. The model predicts, and experiments verify, that spindles will move end-on to the nearest cortex and that motion will speed up as the spindle nears the cortex.
Although both studies agree on the contribution of actin and Fmn2, Li et al. [13] and Schuh et al. [14] differ as to the role of myosin, based on studies using ML7. While Li et al. [13] tracked chromosome movement, Schuh et al. [14] observed the spindle. Perhaps actin polymerization works first to push the chromosomes and thus the spindle to the cortex (Figure 1), and then pole-localized myosin ‘steps in’ to finish the job and anchor the spindle to the cortex (Figure 1). It is possible that this latter step may not be as apparent when observing chromosomal movements. In addition, myosin may regulate dynamic turnover of actin, thus contributing to spindle positioning [11,16]. The existence of partially overlapping mechanisms is consistent with redundant pathways contributing to vital cellular processes.
Unlike the microtubule-based mechanisms for spindle positioning observed in other cells types, these studies provide evidence for a novel actin-based mechanism for spindle movement. Previous analysis of chromosome capture in early prometaphase cells showed that the ‘casting’ range of a microtubule is about 30 μm; chromosomes farther than this distance from a spindle pole are unlikely to interact with a microtubule [17]. In large oocytes with sparse astral microtubules, most microtubules are not long enough to reach the cortex and generate force. So large cells have devised another way — using actin polymerization and myosin pulling to generate force. These new insights raise additional questions, including: what triggers the relocalization of Fmn2 from the cortex to the interior cytoplasm? And what mediates the symmetry-breaking event required for the polymerization-dependent motility of the chromosomes? Answering these questions will provide additional insight into actin's role in diverse motility events.
References
- 1.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat. Rev. Mol. Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 2.Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the C. elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 3.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 4.Longo FJ, Chen DY. Development of cortical polarity in mouse eggs: involvement of the meiotic apparatus. Dev. Biol. 1985;107:382–394. doi: 10.1016/0012-1606(85)90320-3. [DOI] [PubMed] [Google Scholar]
- 5.Terada Y, Fukaya T, Yajima A. Localization of microfilametns during oocyte maturation and fertilization in vitro. Mol. Reprod. Dev. 1995;41:486–492. doi: 10.1002/mrd.1080410411. [DOI] [PubMed] [Google Scholar]
- 6.Kim NH, Chung HM, Cha KY, Chung KS. Microtubule and microfilament organization in maturing human oocytes. Hum. Reprod. 1998;13:2217–2222. doi: 10.1093/humrep/13.8.2217. [DOI] [PubMed] [Google Scholar]
- 7.Kim NH, Cho SK, Choi SH, Kim EY, Park SP, Lim JH. The distribution and requirements of microtubules and microfilaments in bovine oocytes during in vitro maturation. Zygote. 2000;8:25–32. doi: 10.1017/s0967199400000794. [DOI] [PubMed] [Google Scholar]
- 8.Sun QY, Lai L, Park KW, Kuhholzer B, Prather RS, Schatten H. Dynamic events are differently mediated by microfilaments, microtubules, and mitogen-activated protein kinase during porcine oocyte maturation and fertilization in vitro. Biol. Reprod. 2001;64:879–889. doi: 10.1095/biolreprod64.3.879. [DOI] [PubMed] [Google Scholar]
- 9.Lovy-Wheeler A, Wilsen KL, Baskin TI, Hepler PK. Enhanced fixation reveals the apical cortical fringe of actin filaments as a consistent feature of the pollen tube. Planta. 2005;221:95–104. doi: 10.1007/s00425-004-1423-2. [DOI] [PubMed] [Google Scholar]
- 10.Ballestrem C, Wehrle-Haller B, Imhof BA. Actin dynamics in living mammalian cells. J. Cell Sci. 1998;111:1649–1658. doi: 10.1242/jcs.111.12.1649. [DOI] [PubMed] [Google Scholar]
- 11.Murthy K, Wadsworth P. Myosin-II-dependent localization and dynamics of F-actin during cytokinesis. Curr. Biol. 2005;15:724–731. doi: 10.1016/j.cub.2005.02.055. [DOI] [PubMed] [Google Scholar]
- 12.Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. Lifeact: a versitile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Guo F, Rubinstein B, Li R. Actin-driven chromosomal motility leads to symmetry breaking in mammalian meiotic oocytes. Nat. Cell Biol. 2008;10:1301–1308. doi: 10.1038/ncb1788. [DOI] [PubMed] [Google Scholar]
- 14.Schuh M, Ellenberg J. A new model for asymmetric spindle positioning in mouse oocytes. Curr. Biol. 2008;18:1986–1992. doi: 10.1016/j.cub.2008.11.022. [DOI] [PubMed] [Google Scholar]
- 15.Burkel BM, von Dassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell Motil. Cytoskel. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guha M, Zhou M, Wang Y-L. Cortical actin turnover during cytokinesis requires myosin II. Curr. Biol. 2005;15:732–736. doi: 10.1016/j.cub.2005.03.042. [DOI] [PubMed] [Google Scholar]
- 17.Rieder CL. Mitosis: towards a molecular understanding of chromosome behavior. Curr. Opin. Cell Biol. 1991;3:59–66. doi: 10.1016/0955-0674(91)90166-v. [DOI] [PubMed] [Google Scholar]