Abstract
Molecular Dynamics simulations of a β-hairpin miniprotein, CLN025, were performed to examine the conformational stability of the peptide in H2O at 278 K, 300 K, 333 K and 363 K as well as in TFE, MeOH and DMSO at 300 K. CLN025 is a variant of the Chignolin miniprotein, in which the terminal Gly residues of Chignolin are replaced with Tyr residues, which leads to a 29.7 K increase in melting temperature. The energy of the intramolecular interactions were calculated using DFT quantum chemical calculations at the BH and HLYP/cc-pVTZ level of theory. CLN025 maintained a β-hairpin conformation in all environments. The β-hairpin is stabilized by hydrogen bonds, an electrostatic interaction between the charged termini of the peptide and weakly polar interactions. The interaction between the backbones of the N and C-terminal strands accounts for −97.32 to −120.87 kcal mol−1 of the stabilization energy. The energy of the CH-π interactions between Tyr2 and Pro4 were between −1.80 and −8.9 kcal mol−1 and the energy of the Tyr2-Trp9 Ar-Ar interaction was between −0.43 and −8.11 kcal mol−1. Increasing temperature caused the Tyr2-Pro4 CH-π and the Tyr2-Trp9 and Tyr2-Tyr10 Ar-Ar interactions to become less favorable but the Tyr1-Trp9 interaction became more favorable and played an important role in stabilizing the β-hairpin of CLN025 that resulted in the increased melting temperature. The weakly polar interactions play an important role in the structure and stability of CLN025 and other proteins.
Keywords: CLN025, Chignolin, beta-hairpins, miniprotein, Molecular Dynamics, weakly polar interactions, Ar-Ar interaction
Introduction
Miniproteins play an important role in the study of protein folding and structure, because they have properties that are normally associated only with proteins and include a well-defined structure and two-state folding but they are small enough to be studied using long-time Molecular Dynamics simulations and ab initio quantum chemical calculations. Numerous β-hairpin miniproteins that have high melting temperatures and are soluble in aqueous solutions have been designed recently. These β-hairpins are stabilized by cross-strand hydrogen bonds, disulfide bonds and weakly polar interactions.1–3 Miniproteins designed to take advantage of the weakly polar interactions include β-hairpins with aromatic-aromatic (Ar-Ar) interactions,4–8 cation-π interactions,7–9 and CH-π interactions.7,8
The weakly polar interactions play an important role in the stabilization of both secondary and tertiary structures due to the frequency with which they occur in the hydrophobic core of proteins and because they can be as strong as the hydrogen bonds in proteins.10–17 The Ar-Ar interactions can be modeled effectively by dispersion, electrostatics and short range repulsion.18 The dispersion energy accounts for 60% of the attractive interaction energy while 32% is due to electrostatics and 8% to polarization.19 Dispersion makes the largest contribution to the interaction energy but it is substantially canceled out by the short-range repulsion. Thus, the electrostatic energy component is the most important determinant of the orientation of the aromatic rings.20 The Ar-Ar interactions are attractive in two orientations. In the displaced-stack configuration, the aromatic rings are parallel and the centers of the aromatic rings are off-center. In the edge-to-face or T-shaped configuration, the aromatic rings are perpendicular. The face-to-face configuration, where the aromatic rings are parallel and directly over each other, is not stable due to enhanced electrostatic repulsion of the partial negative charges of the electron-rich atoms and the repulsion of the partial positive charges of the electron-deficient atoms.21 Thus, the displaced-stack and T-shaped configurations are more favorable.22, 23 The displaced-stack is typically more favorable than the T-shaped orientation in a polar environments such as the hydrophobic core of proteins.20
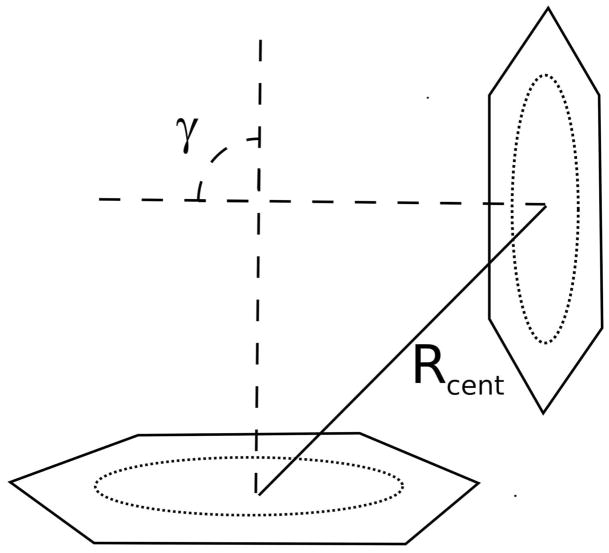
The geometry of the aromatic rings is described in terms of the distance between the centroids of the aromatic rings (Rcent) and the angle between the two normal vectors of the aromatic rings (γ) (Figure 1).24 The distance between aromatic rings is typically around 0.38 nm.25 The strength of the Ar-Ar interaction depends on the orientation and any substituents on the aromatic rings. The strength of the Ar-Ar interaction between benzene molecules at a distance of 0.38 nm was calculated to be −1.53 kcal mol−1 for the face-to-face orientation, −2.62 kcal mol−1 for the displaced-stack orientation and −2.77 kcal mol−1 for the T-shaped orientation.20 The energy of the Ar-Ar interaction in peptides is greater than that in the benzene dimer due to the substituent groups on the aromatic rings in proteins. Almost any substituent increases the strength of the interaction, because substituents that either donate or withdraw electron density from the aromatic rings increase the interaction energy, due to the interaction between the substituent and the other aromatic ring. The interaction between mono-substituted benzene and unsubstituted benzene can be 2.5 kcal/mol greater than the interaction energy of the benzene dimer.20 The strength of the Ar-Ar interaction in proteins was determined to be between −2.5 and −3.3 kcal mol−1 26 and −4.2 kcal mol−1.13
Figure 1.
The geometry of the Ar-Ar interaction showing the distance between the centroids of the aromatic rings (Rcent) and the angle between the two normal vectors of the aromatic rings (γ)
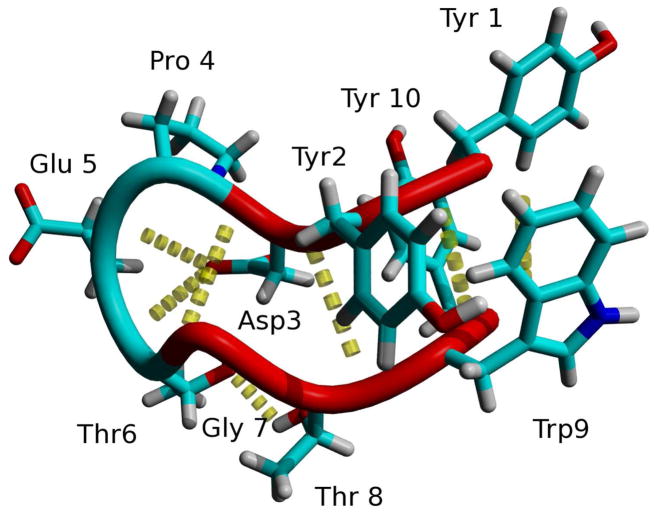
CLN025, Tyr1-Tyr2-Asp3-Pro4-Glu5-Thr6-Gly7-Thr8-Trp9-Tyr10 (Figure 2), is a miniprotein that forms a stable β-hairpin that undergoes a reversible cooperative transition upon thermal denaturation.27 The β-hairpin is stabilized by an electrostatic interaction between the charged termini, cross-strand hydrogen bonds, Ar-Ar interactions between Tyr2 and Trp9, Tyr1 and Tyr10, Tyr1 and Trp9 and Tyr2 and Tyr10 as well as a CH-π interaction between Try2 and Pro4.27 The difference in stability between CLN025 and Chignolin is due to the additional Ar-Ar interactions which involve the terminal tyrosyl residues and result in an increase in the stabilization energy of −4.83 kcal mol−1 and a 27.9 K increase in melting temperature.27
Figure 2.
The backbone and side chains of the NMR structure of CLN02527. Random meander is cyan, β-sheet is red, turn is green and the H-bonds are yellow dotted lines.
In this study, Molecular Dynamics simulations are used to investigate the conformational stability of the CLN025 β-hairpin at different temperatures and in TFE, MeOH and DMSO. The role of hydrogen bonds, the Ar-Ar interactions and the other weakly polar interactions in the stabilization of the peptide are investigated through geometric analysis of the molecular dynamics simulations and the determination of intramolecular interaction energies (ΔEint) using DFT quantum chemical calculations.
Methods
Molecular Dynamics Simulations
Molecular Dynamics simulations were performed with the GROMACS 3.2.1 software package28, 29 the GROMOS 96 force field and the 53a6 parameter set30, 31 using the X-ray structure of CLN02527 as the initial structure. CLN025 was solvated in a 45.61 nm3 dodecahedral box with the parameters listed (Table 1). The MeOH, TFE and DMSO models were optimized for the 53a6 parameter set.30 The compressibilities and dielectric constants used for the simulations were the experimentally determined values.32 One chloride and three sodium ions were used to neutralize the charge of the system. The solvated structures were energy minimized by the steepest descent method.33 An NVT simulation of the positionally restrained peptide was performed for 100 ps at the desired temperature. A 200.1 ns NPT simulation was performed at the desired temperature and 1 bar pressure by coupling the system to an external heat and pressure baths, respectively, using the method by Berendsen.34 The bonds were constrained35 using the Shake method36 in the TFE simulation and the Lincs method33 in all other simulations.
Table 1.
Molecular dynamics simulation parameters
| simulation | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| temperature/K | 278 | 300 | 333 | 363 | 300 | 300 | 300 |
| solvent | SPC water | SPC water | SPC water | SPC water | TFE | MeOH | DMSO |
| number of solvent molecules | 1438 | 1438 | 1438 | 1438 | 302 | 1866 | 286 |
| κ/bar−1 | 4.92 × 10−5 | 4.50 × 10−5 | 4.45 × 10−5 | 4.7 × 10−5 | 1.22 × 10−4 | 1.29 × 10−4 | 4.50 × 10−5 |
| εrel | 86 | 78 | 68 | 56 | 27 | 32 | 47 |
Trajectories were submitted to cluster analysis using the GROMOS method of clustering37 and a backbone RMSD cutoff of 0.1 nm using structures sampled in 10 ps intervals. The structure of CLN025 was examined using the middle structure of the largest cluster of structures from the trajectories of the simulations. The root-mean-square deviation (RMSD) of the backbone of the peptide structures were calculated with the VMD software package38 (http://www.ks.uiuc.edu/Research/vmd/). Secondary structure of CLN025 was analyzed with plots of the φ and ψ dihedral angles of the residues sampled every 100 ps along the trajectories. The width of the turn is determined by the distance between the α-carbons of the Asp3 and Thr8 residues sampled every 100 ps along the trajectories of the simulations. Hydrogen bond analysis was used to determine the number of hydrogen bonds in the structures of CLN025 in the presence of the solvent sampled every 100 ps along the trajectory of the simulations. Radial distribution functions of the distance between the center-of-mass of the aromatic rings of Tyr1, Tyr2, Trp9 and Tyr10 and the oxygen atoms of the solvents were calculated every 100 ps along the trajectories of the simulations using a binwidth of 0.01 nm.
The percentage of interacting structures was determined by the percentage of structures, sampled every 10 ps along the trajectories of the simulations, in which the distance between the center-of-mass of the aromatic side chains or the prolyl ring were below a cutoff distance of 0.75 nm, which was determined previously39 to indicate a probable weakly polar interaction between the residues. Geometric analysis of the Ar-Ar interactions (Figure 1) was examined by the distances between the center-of-mass of the aromatic rings (Rcent) and the angle between the planes (γ) determined by the CG, CE1 and CE2 atoms of Tyr aromatic side chains and the CD1, CZ3 and C atoms of the Trp aromatic side chain, sampled every 100 ps along the trajectories of the simulations.
Interaction energy calculations
The interaction energies were calculated as described previously13–17 using the X-ray27 and NMR27 structures as well as the middle structures of the largest clusters from the trajectories of the simulations. Hydrogens were added to the crystal structure and the united atoms in the middle structure of CLN025. The energies of the interactions between Tyr2 and Trp9, Tyr1 and Tyr10, Tyr1 and Trp9 and Tyr2 and Tyr10 as well as between the backbones of these residues and between the backbones of the N- and C-terminal strands of the β-hairpin were calculated. To calculate the energies of interactions between residues, the chain of the polypeptide was fragmented into sets of interacting molecules that were capped as described previously.40 Non-terminal fragments were capped41, 42 by acetylation and methyl amidation of their α-amino and carboxyl groups, respectively, in order to preserve the original local electronic environment40 For the backbone-backbone (Bb-Bb) interactions, the side chains of the amino acid residues were replaced with hydrogens. The backbones of the N- and C-terminal strands were modeled as the capped Tyr1-Tyr2-Asp3-Pro4 and Gly7-Thr8-Trp9-Tyr10 fragments, respectively, with the side chains replaced with hydrogens. The strength of the Tyr2-Pro4 interaction was determined using the rotation method.15–17
Intramolecular interaction energy calculations
The added hydrogens were optimized at the B3LYP/6–31G** level of theory. Interaction energies of the molecular fragments were calculated at the BH and HLYP/cc-pVTZ level of theory. The BH and HLYP functional used is expressed as:
is the exact Hartree-Fock exchange, is the exact Slater exchange, and is the Lee-Yang-Parr correlation. This functional was chosen because it reliable reproduces high level ab initio results for weakly polar interactions in polypeptides and systems in which dispersion plays an important role.10, 12, 14, 15 The Boys and Bernardi method was used to correct the basis set superposition error (BSSE).43
Calculation of the Ar-Ar interactions
The energy of the Ar-Ar interactions was determined by subtracting the interaction energy of the backbone atoms of the residues from the interaction energy of the residues as is shown by the equation:
where ΔEint (Re-ReXxx-Yyy) is the energy of the interaction between residues Xxx and Yyy and ΔEint (Bb-BbXxx-Yyy) is the energy of the interaction between the backbones of the Xxx and Yyy residues.
Programs
The Jaguar V5.5 release 11 (Schrödinger LLC, Portland, OR) program was used for calculations and the YASARA (http://www.yasara.org) program was used for visualization of the peptide and the preparation of figures.
Results and Discussion
MD simulations
Analysis of the clusters generated from the trajectories of CLN025 in the various conditions is given in Table 2. The large contribution made by largest cluster in all environments studied indicates that CLN025 forms a stable structure. The β-hairpin appears to be destabilized at 363 K as indicated by the larger number of clusters and the lower population of the largest cluster.
Table 2.
Analysis of trajectories of CLN025
| Simulation |
|||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Number of clusters | 8 | 15 | 76 | 369 | 3 | 19 | 6 |
| % of structures in most populated cluster | 92.3 | 94.4 | 72.0 | 40.0 | 99.8 | 83.9 | 98.2 |
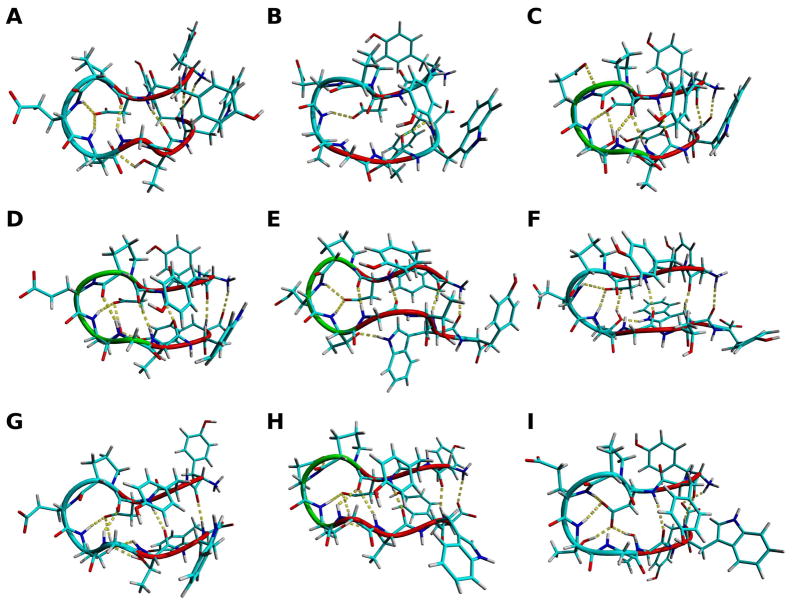
The middle structures (Figure 3) indicate that CLN025 maintains a β-hairpin conformation in all environments studied. In H2O at 333 K (simulation 3), the turn is a β-bend and the φ and ψ dihedral angles of Gly7 change. This causes the side chain of Gly7 to rotate away from the turn and the C-terminal strand to elongate and shift the hydrogen bonding pattern. The Trp9 residue moves 0.13 nm further from the Tyr2 residue, which causes the Ar-Ar interaction to break and form a new Ar-Ar interaction with the Tyr1 residue. The turn of the β-hairpin is also a β-bend in TFE (simulation 5) and DMSO (simulation 7). In DMSO (simulation 7), the Asp3 side chain moves closer to the opposite strand and rotates so that the oxygen atoms point towards the turn and strand. This allows the side chain to participate in more hydrogen bonds. The backbone structure of CLN025 from the Molecular Dynamics simulation at 300 K (simulation 2) are consistent with the experimentally determined X-ray and NMR structures27 (Table 3). The backbone RMSD values between the middle structures from the simulations and the X-ray structure are lower than the RMSD value of the first NMR structure and only the RMSD values between the backbone of the peptide at 300 K and 363 K (simulations 3, 4) and the NMR structure are greater than the RMSD value of the X-ray structure. ECD and VCD spectra of CLN02544 confirm that the peptide maintains a β-hairpin in the environments studied and that in TFE and MeOH the turn of the peptide changes.
Figure 3.
The backbone and side chains of CLN025. A, X-ray structure27; B, the first NMR structure27 and the middle structures of the most populated clusters of the trajectories of the MD simulations of CLN025 in C, H2O (278 K); D, H2O (300 K); E, H2O (333 K); F, H2O (363 K); G, TFE; H, MeOH and I, DMSO. Random meander is cyan, β-sheet is red, turn is green and the H-bonds are yellow dotted lines.
Table 3.
Backbone RMSD (Å) of the structures of CLN025 compared to the X-ray and first NMR structure
| Reference structure | Structure |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| X-ray | NMR | Simulation |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| X-ray | 0 | 1.843 | 1.021 | 1.034 | 1.538 | 1.600 | 1.055 | 1.086 | 1.075 |
| NMR | 1.843 | 0 | 1.213 | 1.265 | 2.284 | 2.347 | 1.211 | 1.232 | 1.217 |
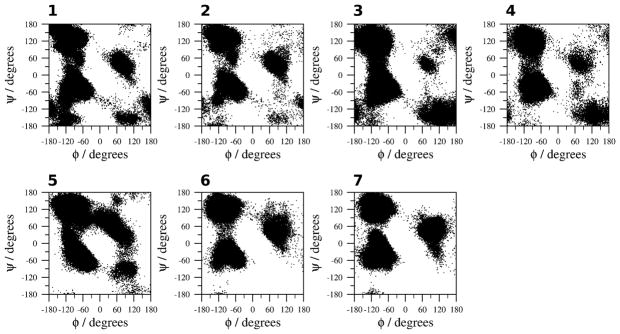
The plots of the φ and ψ dihedral angles of the residues of CLN025 over the 200 ns trajectories (Figure 4) indicate that the backbone conformations of the peptide are similar in all solvents, although the φ and ψ dihedral angles of the backbone are less restricted in TFE (simulation 5). The region around (−140°, 140°) is indicative of anti-parallel β-sheet, the regions around (−60°, −30°) and (−90°, 0°) are indicative of a type I β-turn and the regions at (−60°, 120°) and (80°, 0°) are indicative of a type II β-turn. In H2O at 333 and 363 K (simulations 3, 4) and to a lesser extent in TFE (simulation 5), the region at (90°, 30°) shifts to (120°, −150°) which indicates a change from type II to type II′ β-turn in a region of the plot that is populated primarily by glycyl residues.45
Figure 4.
φ and ψ dihedral angles of the residues of CLN025 along the trajectories of the MD simulations. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
The distance between the α-carbons of the Asp3 and Thr8 residues (Table 4) indicate that the turn is wider in the experimental X-ray and NMR structures than in the middle structure of CLN025 in H2O at 300 K (simulation 2). The width of the turn increases from 0.534 nm to 0.606 nm at 333 K and 0.704 nm at 363 K (simulations 3, 4). The turn is wider in DMSO (simulation 7) with a width of 0.548 nm. In MeOH (simulation 6), the turn is narrower with a width of 0.527 nm.
Table 4.
Effect of the turn width on the Cα-Cα Asp3-Thr8 distance (nm)
| Structure |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| X-ray | NMR | Simulation |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Distance | 0.550 | 0.545 | 0.534 | 0.534 | 0.606 | 0.704 | 0.535 | 0.527 | 0.548 |
| Std. Dev. | 0.03 | 0.03 | 0.05 | 0.26 | 0.03 | 0.02 | 0.05 | ||
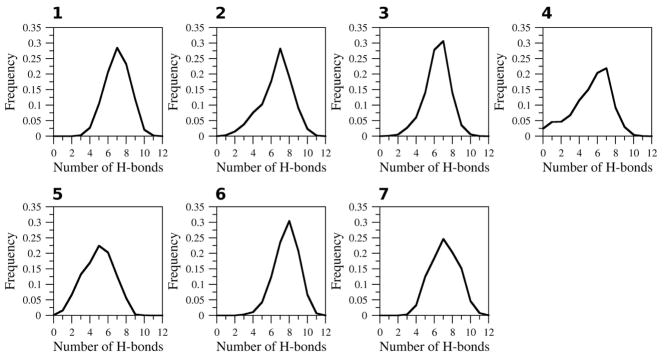
The number of hydrogen bonds in CLN025 for the different simulations is shown (Figure 5). In H2O at 278 K, 300 K and 333 K (simulation 1–3) the peptide has an average of 7.03 and 6.60 and 6.35 hydrogen bonds, respectively. Upon heating to 363 K (simulation 4) the average number of hydrogen bonds decreases to 5.36. In TFE (simulation 5), the number of hydrogen bonds in CLN025 reduced to an average of 4.93 hydrogen bonds with greater variability in the number of hydrogen bonds than seen in any of the other solvents. The number of hydrogen bonds in the peptide in MeOH (simulation 6) increases to an average of 7.69. The average number of hydrogen bonds in CLN025 in DMSO (simulation 7) is 7.13.
Figure 5.
Frequency of the number of hydrogen bonds in CLN025. CLN025 in 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
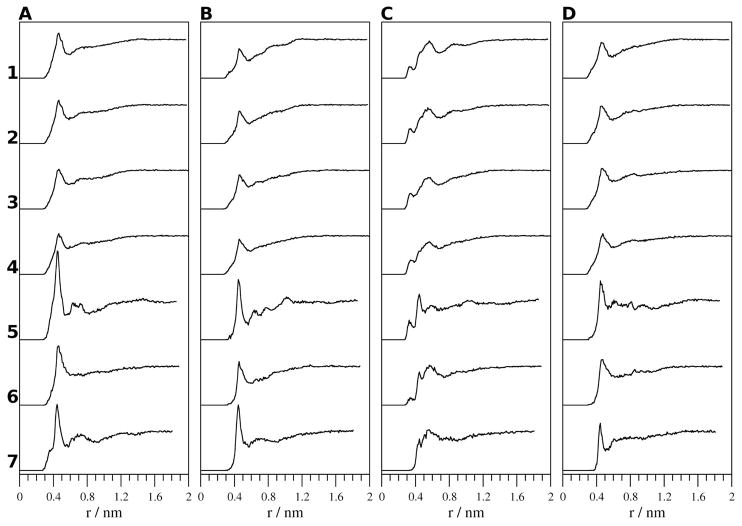
The radial distribution functions (Figure 6) of the distance between the center-of-mass of the aromatic rings of the Tyr residues in the peptide and the oxygen atoms in the solvent show a peak at 0.46 nm in H2O and MeOH (simulations 1–4, 6) and 0.44 nm in TFE and DMSO (simulations 5, 7). In the radial distribution functions between the center of mass of the indole ring of Trp9 and the oxygen atoms of the solvent, peaks are seen at 0.33 and 0.57 nm in simulations where either H2O or MeOH is the solvent (simulations 1–4, 6). In TFE (simulation 5), the peak at 0.33 nm is present but the other peak is shifted from 0.57 nm to 0.45 nm. In DMSO (simulation 7), a single broad peak has a maximum at 0.57 nm. The heights of the peaks in the radial distribution functions indicate that as the temperature increases the solvation decreases around the Tyr1 and Trp9 aromatic rings and increases around the aromatic rings of Tyr2 and Tyr10. This indicates that the Tyr1 and Trp9 residues are shielded from the solvent more at higher temperatures, possibly due to a more energetically favorable Ar-Ar interaction between them. The heights of the peaks also indicate that in H2O (simulations 1–4) Tyr1 and Tyr10 are more solvated than Tyr2, while in TFE and MeOH (simulations 5, 6) Tyr1 is more solvated than Tyr2 and Tyr10. In DMSO (simulation 7), Tyr1 and Tyr2 are more solvated than Tyr10.
Figure 6.
Radial distribution functions between the center of mass of the aromatic rings of A, Tyr1; B, Tyr2; C, Trp9; and D, Tyr10 and the oxygen atoms in the solvent from the simulations of 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH; and 7, DMSO.
Analysis of the percentage of interacting structures (Table 5) indicates that in H2O at 333 K and 363 K (simulations 3, 4) the Tyr2-Trp9 and Tyr1-Tyr10 Ar-Ar interactions are lost and a new Ar-Ar interaction between the side chains of Tyr1 and Trp9 is formed. In TFE (simulation 5), the Tyr1-Tyr10 Ar-Ar interaction is reduced, while in MeOH (simulation 6) the Tyr2-Pro4 CH-π and the Tyr2-Trp9 and Tyr1-Tyr10 Ar-Ar interactions are reduced. In DMSO (simulation 7), the Tyr1-Tyr10 Ar-Ar interaction is lost and the Tyr2-Trp9 Ar-Ar interaction and the Tyr2-Pro4 CH-π interactions are reduced.
Table 5.
Structures (%) in the trajectories of the MD simulations of CLN025 which have weakly polar interactions
| Interaction | Simulation |
||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| Ar-ArTyr2-Trp9 | 97 | 95 | 10 | 25 | 93 | 84 | 71 |
| Ar-ArTyr1-Tyr10 | 86 | 88 | 15 | 22 | 58 | 47 | 12 |
| Ar-ArTyr1-Trp9 | 2 | 1 | 65 | 32 | 1 | 5 | 1 |
| CH-πTyr2-Pro4 | 95 | 97 | 93 | 88 | 98 | 86 | 71 |
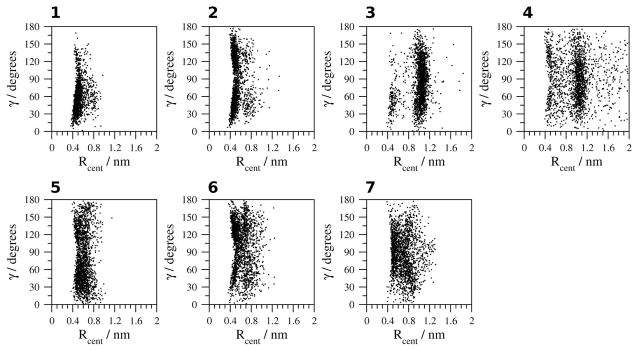
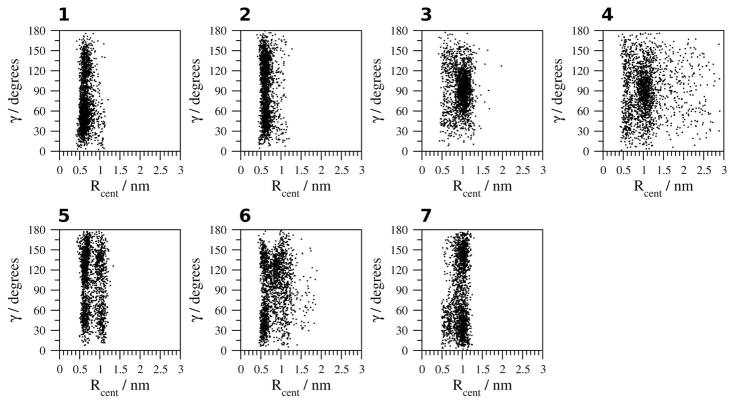
Geometric analysis of the Ar-Ar interactions of CLN025 are shown (Figures 7–11). The geometric analysis of the Tyr2 and Trp9 aromatic side chains (Figure 7) indicates that the aromatic rings are 0.4 nm apart, with no angle dependence in H2O at 278 K or 300 K (simulations 1, 2). In H2O at 333 K and 363 K (simulation 3, 4), however, the distance between the aromatic rings increases to 1 nm, which is too long for weakly polar interactions. In TFE, MeOH and DMSO (simulations 5–7), the distance between the aromatic rings varies from 0.4 to 1 nm. The smaller population of structures with 90° to 180° angles between the aromatic rings in H2O at 278 K (simulation 1) may indicate that at this temperature the aromatic ring of the Tyr2 residue does not flip as readily as it does in the other simulations.
Figure 7.
Geometric analysis of the relative orientation of the aromatic rings of Tyr2 and Trp9. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
Figure 11.
Geometric analysis of the relative orientation of the aromatic rings of Tyr1 and Tyr2. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
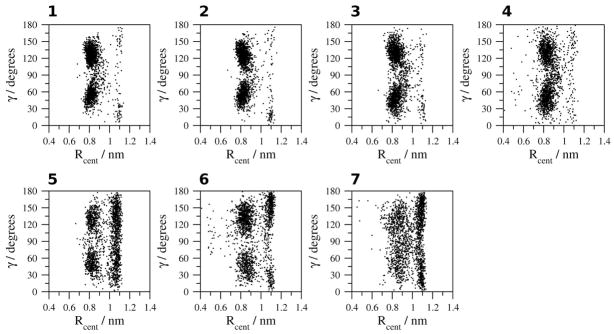
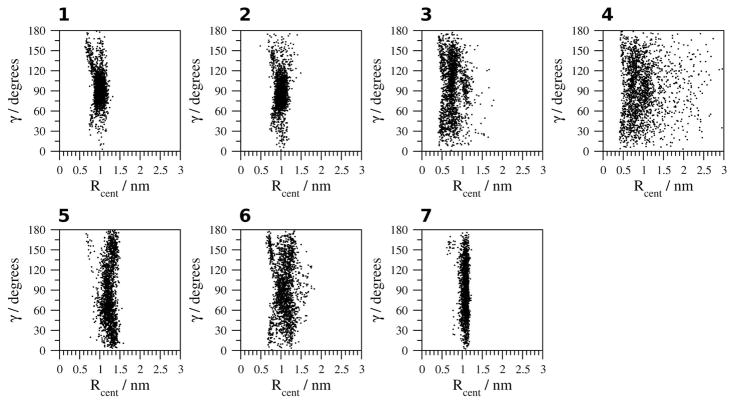
The geometry of the Ar-Ar interaction between the side chains of Tyr1 and Tyr 10 (Figure 8) indicates an orientation in which no angle between the aromatic rings is favored and a distance between 0.4 and 0.8 nm in H2O at 278 K and 300 K (simulations 1,2). In TFE and MeOH (simulations 5, 6), a group of structures in which the distance between the aromatic rings is 0.8 to 1.2 nm indicate a non-interacting population of structures. In H2O at 333 K and 363 K and in DMSO (simulations 3, 4, 7), the non-interacting population dominates, favoring a 60° to 120° edge-to-face orientation between the aromatic rings of Tyr1 and Tyr10 in H2O at 333 K (simulation 3) and a 0° to 60° or 120° to 180° orientation in DMSO (simulation 7).
Figure 8.
Geometric analysis of the relative orientation of the aromatic rings of Tyr1 and Tyr10. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
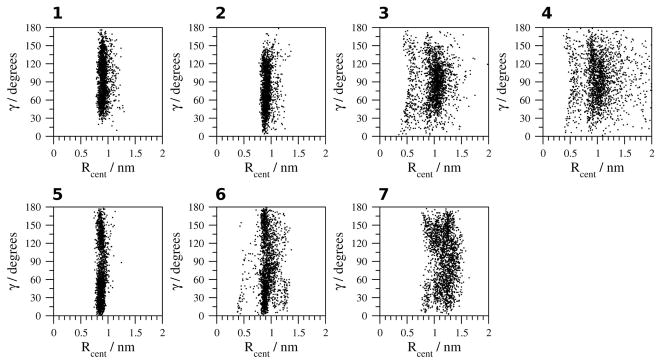
The geometric analysis of the orientation between the aromatic rings of the Tyr1 and Trp9 side chains (Figure 9) indicates a distance of 0.8 to 1.2 nm between aromatic rings and an edge-to-face orientation in H2O at 278 K and 300 K (simulations 1, 2). At 333 K (simulation 3), however, the distance between the aromatic rings is shorter, ranging from 0.4 to 0.8 nm, which indicates a more energetically favorable Ar-Ar interaction between the side chains of Tyr1 and Trp9. In H2O at 363 K (simulation 4), the distance between the rings varies from 0.4 to 3 nm. In TFE and MeOH (simulations 5, 6), the distance ranges from 0.7 to 1.5 nm with no angle dependence. In DMSO (simulation 7), the distance between the aromatic rings is constrained between 1.0 to 1.2 nm with no angle dependence.
Figure 9.
Geometric analysis of the relative orientation of the aromatic rings of Tyr1 and Trp9. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
The geometry of the Tyr2 and Tyr10 aromatic rings (Figure 10) indicates the absence of an Ar-Ar interaction with a distance of 0.8 to 1.0 nm between the rings. In H2O at 333 K and 363 K and MeOH (simulations 3, 4, 6), however, a population of structures with distances of 0.4 to 0.7 nm are likely to be involved in Ar-Ar interactions. In DMSO (simulation 7), the aromatic rings are further apart with distances between 0.8 and 1.5 nm.
Figure 10.
Geometric analysis of the relative orientation of the aromatic rings of Tyr2 and Tyr10. 1, H2O (278 K); 2, H2O (300 K); 3, H2O (333 K); 4, H2O (363 K); 5, TFE; 6, MeOH and 7, DMSO.
The geometry of the aromatic rings of the Tyr1 and Tyr2 side chains indicate two orientation groupings (Figure 11). One grouping at 0.8 nm has either a 30° to 75° or 105° to 150° edge-to-face orientation, while a second at 1.1 nm has no favored angle. In H2O (simulations 1–4), the first grouping is favored, accounting for 95% of the structures, while in TFE, MeOH and DMSO (simulations 5–7) the first cluster is less populated, contributing 49%, 66% and 51% of the sampled structures, respectively.
Intramolecular interaction energy calculations
The energies of the interactions between residues and of the interactions between the backbones of the residues are shown (Table 6). The energy of the interactions between the Tyr2 and Trp9 residues is diminished in the NMR structure and the middle structure of CLN025 in H2O at 333 K and 363 K (simulations 3, 4) due to a loss of stabilization energy from the backbone cross-strand hydrogen bonds. The Tyr1-Tyr10 interaction has energies of −70 kcal mol−1 to −100 kcal mol−1 contributed by the electrostatic interaction between the charged termini, hydrogen bonds and the Ar-Ar interaction. The energy of this interaction is 14 kcal mol−1 lower in the middle structure of CLN025 in H2O at 333 K (simulation 3) than in the middle structure of the peptide in H2O at 300 K (simulation 2). The energy of the interaction is reduced to −66.06 kcal mol−1 in the middle structure of CLN025 in TFE (simulation 5).
Table 6.
ΔEint (kcal mol−1) of interacting residues of CLN025
| Interaction | Structure |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| X-ray | NMR | Simulation |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Re-ReTyr2-Trp9 | −29.86 | −11.23 | −30.05 | −28.08 | −7.07 | −8.61 | −23.25 | −24.52 | −19.44 |
| Bb-BbTyr2-Trp9 | −23.56 | −6.18 | −21.94 | −23.42 | −4.71 | −8.18 | −20.65 | −21.81 | −15.49 |
| Re-ReTyr1-Tyr10 | −90.3 | −88.14 | −89.72 | −78.87 | −78.68 | −71.03 | −66.06 | −94.74 | −94.16 |
| Bb-BbTyr1-Tyr10 | −87.11 | −93.22 | −94.18 | −82.04 | −79.14 | −71.17 | −69.48 | −93.51 | −95.26 |
| Re-ReTyr1-Trp9 | −26.94 | −5.05 | −13.05 | −9.46 | −28.84 | −26.55 | −6.53 | −8.3 | −2.34 |
| Bb-BbTyr1-Trp9 | −7.45 | −2.77 | −5.99 | −9.02 | −27.11 | −23.13 | −5.91 | −7.21 | −3.04 |
| Re-ReTyr2-Tyr10 | −5.61 | −16.27 | −3.84 | −3.28 | 2.53 | 2.03 | −1.87 | −8.1 | −1.31 |
| Bb-BbTyr2-Tyr10 | −2.8 | −12.58 | −2.02 | −2.29 | 2.86 | 1.99 | −0.04 | −5.12 | −0.65 |
| Re-ReTyr2-Pro4 | 1.72 | −12.47 | −8.9 | −3.16 | −3.42 | −1.80 | −2.41 | −2.83 | −3.6 |
| Bb-BbTyr1Tyr2Asp3Pro4-Gly7Thr8Trp9Tyr10 | −114.93 | −110.67 | −120.87 | −109.4 | −99.97 | −87.32 | −92.75 | −116.16 | −105.4 |
The energy of the Tyr1-Trp9 interaction is greatest in the X-ray structure and the simulated structure in H2O at 333 K (simulation 3). The energy is diminished in the middle structure of the peptide in DMSO (simulation 7). The Tyr2-Tyr10 interaction is weak except in the NMR structure where it is 10 kcal mol−1 stronger than the interaction in the other structures. The energy of the Tyr2-Pro4 interaction in the X-ray and NMR structures is 1.72 kcal mol−1 and −12.47 kcal mol−1 respectively. The difference between these values is attributed to the twisting of the aromatic ring of Try2 away from the Pro4 ring and towards the Trp9 aromatic ring in the X-ray structure while in the NMR structure the aromatic ring is directly over the prolyl ring (Figure 3A, B).
The energy of the Bb-Bb interaction between the N- and C-terminal strands ranges from −90 kcal mol−1 to −120 kcal mol−1 and is attributed primarily to the electrostatic interaction between the charged termini. The energy of the Bb-Bb interaction between the strands is greatest in the middle structures of CLN025 in MeOH (simulation 6) and H2O (simulations 1–4) and weakest in the middle structures of the peptide in TFE and DMSO (simulations 5, 7). The energy of the interaction between the backbones of the strands is 20 kcal mol−1 lower in the middle structure of the peptide in H2O at 333 K (simulation 3) than in H2O at 300 K (simulation 2).
The energies of the Ar-Ar interactions were determined by subtracting the energies of the interactions between the backbones of the residues from the energies of the interactions between the residues (Table 7). The energy of the Tyr2-Trp9 Ar-Ar interaction ranges from −0.43 kcal mol−1 to −8.11 kcal mol−1. The energy of the Ar-Ar interaction is lower in the simulated middle structures of CLN025 in H2O at 333 K and 363 K, TFE, MeOH and DMSO (simulations 3–7).
Table 7.
ΔEint (kcal mol−1) of the Ar-Ar interactions
| Interaction | Structure |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| X-ray | NMR | Simulation |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| Ar-ArTyr2-Trp9 | −6.3 | −5.05 | −8.11 | −4.66 | −2.36 | −0.43 | −2.6 | −2.71 | −3.95 |
| Ar-ArTyr1-Tyr10 | −3.19 | 5.08 | 4.46 | 3.17 | 0.46 | 0.14 | 3.2 | −1.23 | 1.1 |
| Ar-ArTyr1-Trp9 | −19.49a | −2.28 | −7.03a | −0.44 | −1.73 | −3.42 | −0.62 | −1.09 | 0.7 |
| Ar-ArTyr2-Tyr10 | −2.81 | −3.69 | −1.82 | −0.99 | −0.33 | 0.04 | −1.83 | −2.98 | −0.66 |
The aromatic ring of Trp9 is also involved in a cation-π interaction with the positively charged Tyr1 residue.
In the NMR structure and the middle structures from the H2O, TFE and DMSO simulations (simulations 1–5, 7), no Ar-Ar interaction between Tyr1 and Tyr10 are found because of the large distance between the aromatic rings. The energy of interaction is positive due to a repulsive interaction between the β-carbon methylene groups of the tyrosyl side chains In structures where the energy of the interaction is greater than +1 kcal mol−1, the Cβ-Cβ distances are between 0.46 and 0.61 nm and the centroids of the aromatic rings are more than 0.73 nm apart, indicating no Ar-Ar interaction (Table 8). In the middle structures of CLN025 in H2O at 333 K and 363 K (simulations 3, 4) a greater Cβ-Cβ distance of 0.72 nm and 0.75 nm, respectively, leads to less repulsion. The energy of the Tyr1-Tyr10 Ar-Ar interaction is attractive in the X-ray structure and the middle structure of the peptide in MeOH (simulation 6), due to a shorter distance of 0.57 and 0.58 nm, respectively, between the aromatic rings.
Table 8.
The Tyr1-Tyr10 Ar-Ar interaction
| Structure |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| X-ray | NMR | Simulation |
|||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||
| aΔEint/kcal mol−1 | −3.9 | 5.08 | 4.46 | 3.17 | 0.46 | 0.14 | 3.2 | −1.23 | 1.1 |
| b number of structures | 1 | 20 | 2000 | 2000 | 2000 | 2000 | 2000 | 2000 | 2000 |
| c Cβ-Cβ/nm | 0.59 | 0.57 | 0.46 | 0.50 | 0.72 | 0.75 | 0.51 | 0.53 | 0.61 |
| d Cβ-Cβ/nm | - | 0.68 ± 0.12 | 0.48 ± 0.08 | 0.47 ± 0.07 | 0.74 ± 0.14 | 0.89 ± 0.44 | 0.57 ± 0.07 | 0.61 ± 0.19 | 0.56 ± 0.06 |
| c Rcent/nm | 0.57 | 0.90 | 0.73 | 0.74 | 0.97 | 1.06 | 0.98 | 0.58 | 1.02 |
| d Rcent/nm | - | 0.83 ± 0.13 | 0.65 ± 0.12 | 0.64 ± 0.12 | 0.97 ± 0.19 | 1.08 ± 0.44 | 0.78 ± 0.19 | 0.83 ± 0.26 | 0.95 ± 0.15 |
Energy of the interaction between the aromatic rings of Tyr1 and Tyr10.
Number of structures used to determine the average distances.
The distance in the X-ray structure, the first of the NMR structures and the middle structure from the largest cluster from the MD simulations.
The average and standard deviation of the distance.
The Cβ-Cβ distance in the first NMR structures is 0.11 nm shorter than the average distance, indicating that the interaction in the average structure is less repulsive. The Rcent of the middle structures from the simulations in H2O at 278 K and 300 K (simulation 1, 2) are 0.73 and 0.74 nm, respectively, while the average distances are 0.65 and 0.64 nm. This indicates that an Ar-Ar interaction is present in most of the structures but is not present in the middle structures. The middle structure of CLN025 from the simulation in H2O at 333 K (simulation 3) is an suitably representative structure because the Rcent and Cβ-Cβ distances are within 0.02 nm of the average values. The Cβ-Cβ distance in the middle structure from the TFE simulation (simulation 5) is 0.06 nm lower than the average distance and the Rcent is 0.20 nm greater. While the Tyr1-Tyr10 Ar-Ar interaction is attractive in the middle structure from the MeOH simulation (simulation 6), the average Rcent is 0.83 nm, which indicates that an attractive Ar-Ar interaction is not present in the average structure. In the middle structure from the DMSO simulation (simulation 7) the Rcent and Cβ-Cβ distances, respectively, are 0.07 and 0.05 nm longer than the average values.
The energy of the Tyr1-Trp9 Ar-Ar interaction (Table 7) is greatest in the X-ray structure, −19.49 kcal mol−1 and in the middle structure of CLN025 in H2O at 278 K (simulation 1), −7.03 kcal mol−1, due to the presence of a cation-π interaction between the aromatic ring of Trp9 and the positively charged amide in the Tyr1 residue which is present only in these two structures. The energy of the Tyr1-Trp9 Ar-Ar interaction is greater in the middle structure from the simulation in H2O at 363 K (simulation 4) than it is in the middle structures from the simulations in H2O at 300 K and 333 K (simulations 2, 3). The energy of the Tyr2-Tyr10 Ar-Ar interaction ranges from 0.04 kcal mol−1 to −3.69 kcal mol−1 and is weakest in the middle structures of the peptide in H2O at 333 K and 363 K and in DMSO (simulations 3, 4, 7).
The calculated energies of the weakly polar interactions in the CLN025 miniprotein are consistent with energies determined previously for the weakly polar interactions. The energy of the Ar-Ar interaction in CLN025 is between −0.43 and −8.11 kcal mol−1. Energies of −1 to −2 kcal mol,−1 46 −2 to −4 kcal mol−1 12, 13, 26 and −3 to −7 kcal mol−1 47 were calculated previously for the Ar-Ar interaction. The energies of the CH-π interactions between Tyr2 and Pro4 were between 1.72 and −12.47 kcal mol−1 which is consistent with previously calculated energies for Ar-Pro interactions of −1.41 to −8.05 kcal mol−1 13 and individual CH-π interactions of −1.5 to −3 kcal mol.−1 13, 46 The energies of the cation-π interaction in the X-ray structure and the middle structure of CLN025 from the simulation in H2O at 278 K (simulation 1) were −19.49 and −7.03 kcal mol−1, respectively, and these are consistent with previously determined energies of the cation-π interaction of −10.6 kcal mol−1 48 and −8 to −23 kcal mol.−1 47
CLN025 is a β-hairpin variant of Chignolin, having the terminal Gly replaced by Tyr, with −4.83 kcal mol−1 more stabilization energy and a 27.9 K higher melting temperature.27 This increase in stabilization energy is attributed to the additional Ar-Ar interactions in which the Tyr1 and Tyr10 residues participate in mutually and with the Tyr2 and Trp9 residues. The increase in the energy of the Tyr1-Trp9 Ar-Ar interaction at higher temperatures plays an important role in the stability of CLN025 and in the increase of the melting temperature.
Conclusions
In H2O (simulations 1–4), CLN025 forms a β-hairpin with a mixture of type I and type II β-turns. The β-hairpin is stabilized by 7 hydrogen bonds and an electrostatic interaction between the charged termini of the peptide which account for 110 kcal mol−1 stabilization energy of which 80 kcal mol−1 is due to the hydrogen bonds and the electrostatic interaction between Tyr1 and Tyr10. The β-hairpin is also stabilized by CH-π interactions between the aromatic ring of Tyr2 and the aliphatic hydrogens of the Pro4 ring of −1.80 to −8.9 kcal mol−1 as well as Ar-Ar interactions between Tyr2 and Trp9 of −0.43 to −8.11 kcal mol−1, Tyr1 and Tyr9 of −0.44 to −3.42 kcal mol−1 and between Tyr2 and Tyr10 of 0.04 to −1.82 kcal mol−1. As the temperature increases, the Tyr2-Trp9 and Tyr2-Tyr10 Ar-Ar interactions and the Tyr2-Pro4 CH-π interactions decrease, due to greater solvation around the Tyr2 and Tyr10 aromatic rings and the Tyr1-Trp9 interaction intensifies as the Tyr1 and Trp9 residues are shielded from the solvent. In TFE (simulation 5), the β-hairpin has a β-bend and the interaction between the backbones of the N- and C-terminal strands of the peptide is destabilized by 17 kcal mol−1 due to the loss of 3 hydrogen bonds, a weaker electrostatic interaction and a reduction in the energy of the Ar-Ar interactions. In MeOH (simulation 6), the conformation of the β-hairpin is similar to that in H2O, except that the width of the turn is narrower. In DMSO (simulation 7), the turn widens to allow the Asp3 side chain to move closer to the C-terminus of the peptide and the orientation of the oxygen atoms of the side chain to form additional hydrogen bonds.
Analysis of the MD simulations and the interaction energies of intramolecular interactions in CLN025 in H2O at 278 K, 300 K, 333 K and 363 K and in TFE, MeOH and DMSO at 300 K (simulations 1–7) indicates that CLN025 forms a stable β-hairpin in all environments studied. The β-hairpin is stabilized primarily by hydrogen bonds and the electrostatic interaction between the charged termini as well as the CH-π interactions between Tyr2 and Pro4, the Ar-Ar interactions and a cation-π interaction evident in the X-ray structure and the middle structure of CLN025 from the simulation in H2O at 278 K (simulation 1). The weakly polar interactions contribute between −4 and −30 kcal mol−1 to the stabilization of the β-hairpin. The Tyr1-Trp9 and Tyr2-Tyr10 Ar-Ar interactions that involve the terminal aromatic residues which are not present in Chignolin are responsible for the added stability of CLN025. The ability of the Tyr1-Trp9 Ar-Ar interaction to intensify as the temperature increases plays an important role in the higher melting temperature of CLN025 than Chignolin. The Ar-Ar interactions have an important role in maintaining the structure and the stability of CLN025. This reflects the importance of the weakly polar interactions in proteins. This study shows how the weakly polar interactions vary in contributing to a stable β-hairpin conformation when solvents and temperatures are changed.
Acknowledgments
This work was supported by the NIH-INBRE grant P20 RR016469.
References
- 1.Ramirez-Alvarado M, Kortemme T, Blanco FJ, Serrano L. Bioorg Med Chem. 1999;7:93–103. doi: 10.1016/s0968-0896(98)00215-6. [DOI] [PubMed] [Google Scholar]
- 2.Maynard AJ, Sharman GJ, Searle MS. J Am Chem Soc. 1998;120:1996–2007. [Google Scholar]
- 3.Andersen NH, Dyer RB, Fesinmeyer RM, Gai F, Liu Z, Neidigh JW, Tong H. J Am Chem Soc. 1999;121:9879–9880. [Google Scholar]
- 4.Cochran AG, Skelton NJ, Starovasnik MA. Proc Natl Acad Sci U S A. 2001;98:5578–5583. doi: 10.1073/pnas.091100898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu Y, Oyola R, Gai F. J Am Chem Soc. 2003;125:15388–15394. doi: 10.1021/ja037053b. [DOI] [PubMed] [Google Scholar]
- 6.Takekiyo T, Wu L, Yoshimura Y, Shimizu A, Keiderling TA. Biochemistry. 2009;48:1543–1552. doi: 10.1021/bi8019838. [DOI] [PubMed] [Google Scholar]
- 7.Eidenschink LA, Kier BL, Huggins KNL, Andersen NH. Proteins. 2009;75:308–322. doi: 10.1002/prot.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andersen NA, Olsen KA, Fesinmeyer Rm, Tan X, Hudson FM, Eidenschink LA, Farazi SR. J Am Chem Soc. 2006;128:6101–6110. doi: 10.1021/ja054971w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Riemen AJ, Waters ML. Biochemistry. 2009;48:1525–1531. doi: 10.1021/bi801706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Palermo NY, Csontos J, Owen MC, Murphy RF, Lovas S. J Comput Chem. 2007;28:1208–1214. doi: 10.1002/jcc.20578. [DOI] [PubMed] [Google Scholar]
- 11.Borics A, Murphy RF, Lovas S. Protein Pept Lett. 2007;14:353–359. doi: 10.2174/092986607780363907. [DOI] [PubMed] [Google Scholar]
- 12.Csontos J, Palermo NY, Murphy RF, Lovas S. J Comput Chem. 2008;29:1344–1352. doi: 10.1002/jcc.20898. [DOI] [PubMed] [Google Scholar]
- 13.Hatfield MPD, Palermo NY, Csontos J, Murphy RF, Lovas S. J Phys Chem B. 2008;112:3503–3508. doi: 10.1021/jp077674h. [DOI] [PubMed] [Google Scholar]
- 14.Csontos J, Murphy RF, Lovas S. Biopolymers. 2008;89:1002–1011. doi: 10.1002/bip.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palermo NY, Csontos J, Murphy RF, Lovas S. Int J Quantum Chem. 2008;108:814–819. doi: 10.1002/qua.21521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Csontos J, Murphy RF, Lovas S. Adv Exp Med Biol. 2009;611:79–80. doi: 10.1007/978-0-387-73657-0_35. [DOI] [PubMed] [Google Scholar]
- 17.Palermo NY, Csontos J, Murphy RF, Lovas S. Adv Exp Med Biol. 2009;611:89–90. doi: 10.1007/978-0-387-73657-0_40. [DOI] [PubMed] [Google Scholar]
- 18.Gervasio FL, Chelli R, Procacci P, Schettino V. Proteins. 2002;48:117–125. doi: 10.1002/prot.10116. [DOI] [PubMed] [Google Scholar]
- 19.Ribas J, Cubero E, Luque J, Orozco M. J Org Chem. 2002;67:7057–7065. doi: 10.1021/jo0201225. [DOI] [PubMed] [Google Scholar]
- 20.Lee EC, Kim D, Jurecka P, Tarakeshwar P, Hobza P, Kim KS. J Phys Chem A. 2007;111:3446–3457. doi: 10.1021/jp068635t. [DOI] [PubMed] [Google Scholar]
- 21.Tsuzuki S, Honda K, Uchimaru T, Mikami M, Tanabe K. J Am Chem Soc. 2002;124:104–112. doi: 10.1021/ja0105212. [DOI] [PubMed] [Google Scholar]
- 22.Hobza P, Selzle HL, Schlag EW. J Chem Phys. 1990;93:5893–5897. [Google Scholar]
- 23.Gwench O, Brooks CL., III J Am Chem Soc. 2005;127:4668–4674. doi: 10.1021/ja043492e. [DOI] [PubMed] [Google Scholar]
- 24.McGaughey GB, Gagne M, Rappe AK. J Biol Chem. 1998;273:15458–15463. doi: 10.1074/jbc.273.25.15458. [DOI] [PubMed] [Google Scholar]
- 25.Thomas A, Meurisse R, Charloteaux B, Brasseur R. Proteins. 2002;48:628–634. doi: 10.1002/prot.10190. [DOI] [PubMed] [Google Scholar]
- 26.Vondrasek J, Bendova L, Klusak V, Hobza P. J Am Chem Soc. 2005;127:2615–2619. doi: 10.1021/ja044607h. [DOI] [PubMed] [Google Scholar]
- 27.Honda S, Akiba T, Kato YS, Sawada Y, Sekijima M, Ishimura M, Ooishi A, Watanabe H, Odahara T, Harata K. J Am Chem Soc. 2008;130:15327–15331. doi: 10.1021/ja8030533. [DOI] [PubMed] [Google Scholar]
- 28.Lindahl E, Hess B, Van der Spoel D. J Mol Mod. 2001;7:306–317. [Google Scholar]
- 29.Berendsen HJC, Van der Spoel D, Van Drunen R. Comp Phys Comm. 1995;91:43–56. [Google Scholar]
- 30.Oostenbrink C, Villa A, Mark AE, van Gunsteren WF. J Comp Chem. 2004;25:1656–1676. doi: 10.1002/jcc.20090. [DOI] [PubMed] [Google Scholar]
- 31.Oostenbrink C, Soares TA, van der Vegt NF, van Gunsteren WF. Eur Biophys J. 2005;34:273–284. doi: 10.1007/s00249-004-0448-6. [DOI] [PubMed] [Google Scholar]
- 32.Weast RC. CRC Handbook of Chemistry and Physics. 1. CRC Pres, Inc; Boca Raton, FL: 1988. Student Edition. [Google Scholar]
- 33.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. J Comp Chem. 1997;18:1463–1472. [Google Scholar]
- 34.Berendsen HJC, Postma JPM, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 35.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comp Phys. 1977;23:327–341. [Google Scholar]
- 36.Miyamoto S, Kollman PA. J Comp Chem. 1992;13:952–962. [Google Scholar]
- 37.Daura X, Gademann K, Juan B, Seebach D, van Gunsteren WF, Mark AE. Agnew Chem Int Ed. 1999;38:236–240. [Google Scholar]
- 38.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 39.Toth G, Murphy RF, Lovas S. Protein Eng. 2001;14:543–547. doi: 10.1093/protein/14.8.543. [DOI] [PubMed] [Google Scholar]
- 40.Hatfield MPD, Palermo NY, Csontos J, Murphy RF, Lovas S. Int J Quantum Chem. 2008;108:1017–1021. doi: 10.1002/qua.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D, Zhang JZH. J Chem Phys. 2003;119:3599–3605. [Google Scholar]
- 42.Zhang DW, Chen XH, Zhang JZH. J Comput Chem. 2003;24:1846–1852. doi: 10.1002/jcc.10346. [DOI] [PubMed] [Google Scholar]
- 43.Boys SF, Bernardi F. Mil Phys. 1970;19:553–566. [Google Scholar]
- 44.Hatfield MPD, Murphy RF, Lovas S. Biopolymers. 2009 doi: 10.1002/bip.21356. [DOI] [Google Scholar]
- 45.Creighton TE. Protein Folding. Chapter 1 W. H. Freeman and Company; New York: 1992. [Google Scholar]
- 46.Burley SK, Petsko GA. Science. 1985;229:23–28. doi: 10.1126/science.3892686. [DOI] [PubMed] [Google Scholar]
- 47.Singh NJ, Min SK, Kim DY, Kim KS. J Chem Theory Comput. 2009;5:515–529. doi: 10.1021/ct800471b. [DOI] [PubMed] [Google Scholar]
- 48.Gallivan JP, Dougherty DA. Proc Natl Acad Sci U S A. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]