Summary
Multiple excitatory and inhibitory interneurons form the motor circuit with motor neurons in the ventral spinal cord. Notch signaling initiates the diversification of immature V2-interneurons into excitatory V2a-interneurons and inhibitory V2b-interneurons. Here we provide a transcriptional regulatory mechanism underlying their balanced production. LIM-only protein LMO4 controls this binary cell fate choice by regulating the activity of V2a- and V2b-specific LIM-complexes inversely. In the spinal cord, LMO4 induces GABAergic V2b-interneurons in collaboration with SCL and inhibits Lhx3 from generating glutamatergic V2a-interneuons. In LMO4;SCL compound mutant embryos, V2a-interneurons increase markedly at the expense of V2b-interneurons. We further demonstrate that LMO4 nucleates the assembly of a novel LIM-complex containing SCL, Gata2 and NLI. This complex activates specific enhancers in V2b-genes consisting of binding sites for SCL and Gata2, thereby promoting V2b-interneuron fate. Thus, LOM4 plays essential roles in directing a balanced generation of inhibitory and excitatory neurons in the ventral spinal cord.
Introduction
A proportional production of excitatory and inhibitory neuronal subtypes is important, as the balance between these two opposing activities is critical to establish functional neuronal circuits. In the ventral spinal cord, interneurons and motor neurons form a neural circuit that coordinates locomotion. Four classes of ventral interneurons, V0, V1, V2 and V3, emerge from progenitors in distinct progenitor domains, termed p0, p1, p2 and p3, respectively (Jessell, 2000). These interneurons acquire characteristics of either excitatory neurons that use glutamate as neurotransmitters or inhibitory neurons that utilize GABA (gamma-aminobutyrate) and/or glycine (Lanuza et al., 2004; Alvarez et al., 2005; Kimura et al., 2006). However, mechanisms that govern the alternative fate choices between excitatory and inhibitory neurons in the ventral spinal cord are poorly understood.
The p2 progenitor cells produce immature V2-interneruons (V2-INs) that express combinations of transcription factors; LIM homeodomain (LIM-HD) factor Lhx3, zinc finger protein Gata2, basic helix-loop-helix (bHLH) factor Mash1 and forkhead protein FoxN4 (Del Barrio et al., 2007; Karunaratne et al., 2002; Li et al., 2005; Parras et al., 2002; Thaler et al., 2002; Zhou et al., 2000). These cells diversify into two distinct cell types, V2a-INs and V2b-INs. While V2a and V2b-INs share several properties such as dorso-ventral position and ipsilateral axonal projection, they differ in the expression of marker genes and the choice of neurotransmitters. Notch-Delta interactions initiate this binary cell fate choice in immature V2-INs (Fig. 1A) (Del Barrio et al., 2007; Peng et al., 2007; Yang et al., 2006). Delta4+ signal-sending V2a-INs maintain Lhx3 while suppressing Gata2, whereas Notch1+ signal-receiving V2b-INs upregulate a bHLH factor SCL (also known as Tal1) and Gata2 while silencing Lhx3. V2a-INs mature to become Lhx3+Chx10+ excitatory neurons, whereas V2b-INs develop into inhibitory neurons labeled by Gata2/3 and SCL (Kimura et al., 2006; Lundfald et al., 2007; Peng et al., 2007). Thus, cell-cell interactions through Delta4 and Notch1 set up distinct transcription factor profiles in V2a and V2b cells, thereby generating two distinct V2-IN subtypes from a pool of genetically homogenous p2 progenitors. Forced expression of Gata2 in the dorsal spinal cord triggers Gata3+ V2b-INs, while suppressing the development of other interneurons, including V2a-INs (Karunaratne et al., 2002). Gata2-null mutants are deficient in V2a-INs as monitored by Chx10 expression (Zhou et al., 2000), but remain undetermined for V2b-INs. Expression of SCL also induces ectopic formation of Gata2/Gata3+ V2b-INs in the neural tube and simultaneously suppresses Chx10+ V2a-IN formation (Muroyama et al., 2005). Conversely, deletion of SCL gene in the spinal cord leads to downregulation of Gata2 and loss of Gata3+ V2b-INs, accompanied by increased V2a-INs (Muroyama et al., 2005). These results indicate that Gata2 and SCL are capable of directing transcription pathways to specify V2b-INs, bypassing the initial diversification step by Notch-Delta signaling, and that V2 cells remain plastic between V2a and V2b fates even after adopting cell identities via Notch-Delta signaling. A majority of Chx10+ V2a-INs are glutamatergic, whereas GATA3+ V2b-INs become mainly GABAergic although a small fraction of V2b-derived cells display a glycinergic phenotype (Al-Mosawie et al., 2007; Batista et al., 2008; Kimura et al., 2006; Lundfald et al., 2007). Consistently, ablation of Chx10+ V2a-INs results in a substantial reduction of ventral glutamatergic neurons (Crone et al., 2008). Key questions, however, remain to be answered; first, what is the mechanism that segregates V2a and V2b fate after the initial binary cell identity selection by Notch-Delta signaling; and second, how are immature V2-INs transcriptionally directed to either glutamatergic or GABAergic cell fates.
Figure 1. LMO4 suppresses formation of glutamatergic V2a-INs and cooperates with SCL to promote GABAergic V2b-IN generation.
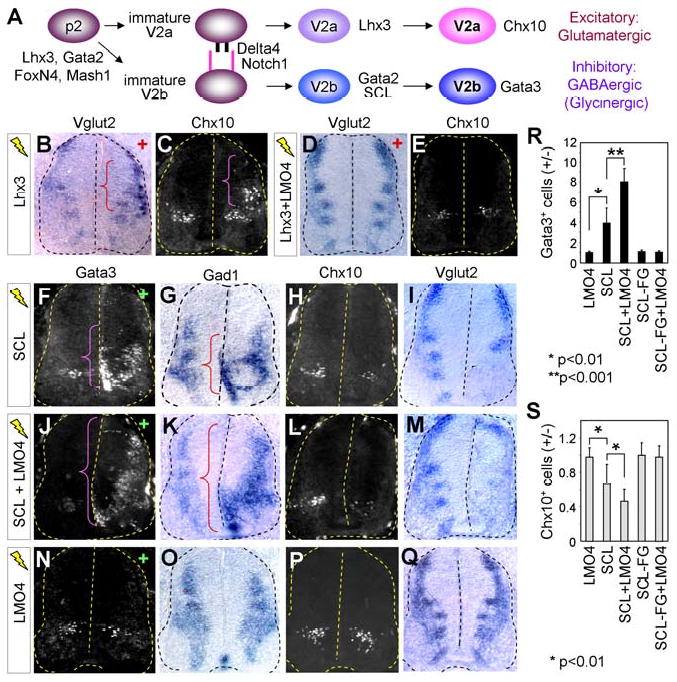
(A) A schematic model shows the diversification process of p2 progenitors to V2a-INs or V2b-INs. (B-E) In situ hybridization for Vglut2 and immunostaining with Chx10 antibody in chicks electroporated (+ side) with Lhx3 alone or Lhx3 plus LMO4. Ectopic Chx10+Vglut2+ V2a-INs in the dorsal spinal cord are marked by brackets. (F-Q) Cell differentiation analyses on chick embryos electroporated with constructs listed on left, using immunostaining with Gata3 or Chx10 antibodies and in situ hybridization for Gad1 or Vglut2. Brackets mark ectopic Gata3+Gad1+ V2b-INs on the electroporated side (+). (R, S) The effect on V2a-IN and V2b-IN formation was quantified by the ratio of Gata3+ or Chx10+ cells on the electroporated side (+) over the control side (-). The error bars represent the standard deviation.
The nuclear LIM proteins are composed of LIM-HD transcription factors and LIM-only proteins (LMOs) (Hobert and Westphal, 2000). LIM-HD factors, which contain LIM domains for protein-protein interactions and the DNA-binding homeodomain, play important roles in establishing neuronal subtype identities in the CNS (Lee and Pfaff, 2001). Structural and in vivo functional studies in vertebrates and invertebrates have revealed that LIM-HD factors form multi-protein complexes with a cofactor NLI (Ldb, CLIM, Chip), which consists of a LIM domain interaction region (NLI-LID) and a self dimerization domain (NLI-DD) (Jurata et al., 1998; Thaler et al., 2002; van Meyel et al., 1999). These NLI-based LIM-complexes vary considerably in their components and organization depending on cell types, leading to cell context-specific gene expression. Lhx3 specifies V2a-IN identity by assembling a tetrameric complex of 2NLI:2Lhx3 (denoted as V2-tetramer) that regulates V2a-IN genes by binding the V2-tetramer-specific response elements (Thaler et al., 2002; Lee et al., 2008). Mice deficient in Lhx3 (and its redundant factor Lhx4) are impaired in V2a-IN formation, whereas misexpression of Lhx3 induces ectopic Chx10+ V2a-INs in the dorsal spinal cord (Lee and Pfaff, 2001; Sharma et al., 1998; Tanabe et al., 1998; Thaler et al., 2002). The role of Lhx3 in establishing neurotransmitter characteristics of V2a-INs remains to be determined. Unlike LIM-HD proteins, LMO proteins have LIM domains but lack any functional DNA-binding motif. Among the four vertebrate LMO genes, LMO4 has been shown to suppress the V2a-IN differentiation pathway by inhibiting the assembly of the V2-tetramer by competing with Lhx3 for binding NLI (Lee et al., 2008). As LIM-HD factors are not expressed in V2b-INs, V2b-specification is unlikely directed by a conventional LIM-complex containing LIM-HD proteins and NLI. Interestingly, however, the identification of the hematopoietic complex containing NLI, LMO2, SCL and Gata1 (Wadman et al., 1997), along with aforementioned roles of SCL and Gata2 in V2b-IN formation, raises the possibility that a similar complex may be involved in V2b-IN-specification.
In this report, we set out to understand the mechanistic basis of the balance between excitatory V2a-INs and inhibitory V2b-INs that are initially set up by Notch-Delta signaling. Our results reveal an essential function for LMO4 in directing a balanced generation of V2a- and V2b-INs in the ventral spinal cord. While suppressing glutamatergic V2a fate by inhibiting the formation of the V2-tetramer, LMO4 simultaneously promotes GABAergic V2b-IN identity by assembling a novel LIM-complex with SCL, Gata2 and NLI. The opposite action of LMO4 in two distinct LIM-complexes directing V2a-IN and V2b-IN fates provides a novel molecular mechanism by which a transcriptional modulator such as LMO4 contributes to striking a balance between two parallel developmental pathways following the initial alternate cell fate determination.
Results
LMO4 inhibits glutamatergic differentiation by Lhx3
We tested whether Lhx3 is capable of promoting a glutamatergic cell fate in the developing chick spinal cord. Ectopic expression of Lhx3 using electroporation induced the glutamatergic neuronal marker, vesicular glutamate transporter 2 (Vglut2), concomitant with the upregulation of Chx10, in the dorsal neural tube (parenthesis in Fig. 1B, C). Interestingly, coelectroporation of LMO4 with Lhx3 suppressed the glutamatergic neuronal differentiation by Lhx3 (Fig. 1D, E), consistent with the previous report that LMO4 antagonizes the formation of the V2-tetramer complex that contains Lhx3 (Lee et al., 2008). These findings suggest that Lhx3 is sufficient to induce glutamatergic cell fates in the neural tube and LMO4 is an efficient blocker of glutamatergic differentiation by Lhx3.
LMO4 cooperates with SCL to reciprocally regulate development of V2a- and V2b-INs
At E11.5, LMO4, Gata2 and SCL are expressed in overlapping areas in the ventricular zone where V2b-INs are being specified (S-Fig. 1A, B). To investigate the possible function of LMO4 in GABAergic V2b-IN development, we misexpressed LMO4, in combination with the V2b-IN-specific factor SCL, using in ovo electroporation of chick embryos. As reported (Muroyama et al., 2005), misexpressed SCL led to the ectopic formation of Gata3+ V2b-INs, resulting in four times more Gata3+ V2b-INs in the electroporated side compared to the control side (Fig. 1F, R). Interestingly, while LMO4 did not induce the formation of V2b-INs, coexpression of LMO4 along with SCL dramatically increased the number of V2b-INs to eight-fold (Fig. 1J, N, R). To determine whether SCL regulates the GABAergic cell fate, we monitored the expression of the Gad1 gene encoding glutamic acid decarboxylase that syhthesizes GABA. SCL induced Gad1 expression modestly (Fig. 1G). Coexpression of LMO4 with SCL resulted in a robust upregulation of Gad1 throughout dorso-ventral spinal cord (parenthesis in Fig. 1K), whereas LMO4 alone is not sufficient for triggering Gad1 expression (Fig. 1O). These results suggest that LMO4 promotes the activity of SCL in generating GABAergic V2b-INs.
These results led us to test whether LMO4 also facilitates the ability of SCL to antagonize V2a-IN differentiation (Muroyama et al., 2005). While forced expression of SCL reduced Chx10+ endogenous V2a-INs to 65% of the control side, the combination of SCL and LMO4 further suppressed V2a-IN formation to 44% (Fig. 1H, L, P, S). Similarly, SCL suppressed Vglut2 expression, which was markedly exacerbated by coexpression of LMO4, leaving only a small number of glutamatergic neurons in the electroporated side (Fig. 1I, M, Q). These results indicate that LMO4 cooperates with SCL in inhibiting glutamatergic V2a-IN differentiation.
To understand the functional basis underlying the cooperativity of SCL and LMO4, we employed SCL-F238G bearing a point mutation in helix2 of the bHLH domain. This mutant is unable to transcriptionally synergize with LMO2 in mammalian cells and ineffective in triggering ectopic V2b-IN generation (Muroyama et al., 2005; Schlaeger et al., 2004). SCL-F238G did not cooperate with LMO4 either in induction of V2b-INs or in suppression of V2a-INs (S-Fig. 1C, Fig. 1R, S), suggesting that the transcriptional synergy between SCL and LMO4 is necessary for their cooperative action in promoting V2b-fate and blocking V2a-differentiation.
These results demonstrate that LMO4 plays opposing roles in the determination of excitatory V2a and inhibitory V2b fates. LMO4 functionally collaborates with SCL in promoting GABAergic V2b-IN fate and inhibiting glutamatergic V2a-IN differentiation.
LMO4 and SCL are required for GABAergic V2b-IN generation
To further define the role of LMO4 in V2b-IN development, we investigated LMO4 mutant and LMO4;SCL compound mutant embryos (Elefanty et al., 1998; Lee et al., 2005). Due to the early embryonic lethality of SCL-null mutants (Porcher et al., 1996), we analyzed LMO4-heterozygote and LMO4-null mutations in a SCL-heterozygous background. We used a previously reported SCL-LacZ allele, which deletes all SCL coding sequences, in our analyses (Elefanty et al., 1998). First, we monitored V2b-IN differentiation by measuring the level of Gata3 mRNA, a V2b-IN-specific marker, from dissected spinal cords using quantitative RT-PCR (qRT-PCR). The level of Gata3 mRNA relative to the internal control cyclophilin was highest in the wild-type and progressively decreased to 50% in LMO4+/- and 17% in LMO4-/- embryos at E14.5 (Fig. 2A). In contrast to the marked downregulation of Gata3, the expression of Gata2 and SCL did not significantly reduce in LMO4-/- embryos (S-Fig. 2). The reduction of Gata3 expression was further augmented by the loss of one allele of SCL gene, resulting in 11% in LMO4+/-;SCL+/- and only 5% in LMO4-/-;SCL+/- embryos (Fig. 2A), consistent with the cooperative action between LMO4 and SCL in the chick spinal cord. Second, to determine whether the loss of LMO4 and/or SCL affects the inhibitory neuronal phenotypes of V2b-INs, we examined GABAergic and glycinergic neurons in the ventral spinal cord of LMO4 and SCL mutants at E13.5 using immunostaining with anti-Glycine and anti-GABA antibodies. While the small number of Glycine+ cells in the ventral spinal cord did not vary among wild-type and mutant embryos (data not shown), the number of GABA+ cells within the ventral spinal cord of LMO4 and SCL mutants was observed to change in comparison to wild-type animals. LMO4+/-;SCL+/- embryos exhibited a 26% decrease in GABA+ cells, whereas neither LMO4+/- nor SCL+/- showed any observable change in GABA+ ventral interneurons (Fig. 2B, C). Given the specific expression of SCL in V2b-INs (Muroyama et al., 2005; Smith et al., 2002), the change in GABA+ ventral interneurons in LMO4+/-;SCL+/- embryos likely reflects a defective acquisition of appropriate neurotransmitter profiles by the V2b-INs. Similarly to Gata3 expression, the greatest reduction in GABA+ ventral interneurons was seen in LMO4-/-;SCL+/- embryos to 46% of wild-type controls (Fig. 2A, B), indicating that the loss of LMO4 and SCL genes leads to a progressive loss of ventral GABAergic interneurons. Considering the dramatic downregulation of Gata3 in LMO4-/-;SCL+/- embryos (Fig. 2A) and the presence of multiple types of GABAergic interneurons in the ventral spinal cord in addition to V2b-INs (Lanuza et al., 2004; Alvarez et al., 2005; Kimura et al., 2006), it is likely that the majority of remaining GABAergic interneurons in LMO4-/-;SCL+/- mutants are non-V2b-INs. These results establish that LMO4 and SCL cooperate to specify the GABAergic fate in V2b-INs. Lastly, we examined the production of V2a-INs in mutant embryos of various genotypes using immunostaining with anti-Chx10 antibody. Chx10+ V2a-INs were significantly increased in LMO4-null embryos to ∼134% of wild-type at E13.5 (Fig. 2B, C). Interestingly, LMO4+/-;SCL+/- embryos displayed increased V2a-IN generation to 157% of wild-type control littermate, although neither LMO4+/- nor SCL+/- showed any significant change in the number of V2a-INs (Fig. 2B, C), indicating a genetic interaction between LMO4 and SCL. In LMO4-/-;SCL+/- embryos, glutamatergic Chx10+ V2a-INs were further increased to 193% of wild-type control (Fig. 2B, C, S-Fig. 3). Combined with the loss of Gata3+GABA+ V2b-INs, these results suggest that the prospective Gata3+GABA+ V2b-INs changed their fate to glutamatergic Chx10+ V2a-INs in LMO4;SCL mutants. Together, these data establish that LMO4 is required to enhance GABAergic V2b-differentiation and suppress glutamatergic V2a-fate, playing an important role in maintaining a balance between V2a-INs and V2b-INs, in part by collaborating with SCL.
Figure 2. Suppression of Gata3+ GABAergic V2b-INs and increase in Chx10+ V2a-INs in LMO4 and SCL mutant embryos.
(A) qRT-PCR to monitor Gata3 mRNA levels in E14.5 littermate embryos. (B) Quantification of GABA+ and Chx10+ cells in E13.5 embryos by the percentage of Chx10+ or GABA+ cells in each embryo over the wild-type littermate control. * indicates significant differences for both Chx10 and GABA, and ** indicates a significant difference for Chx10. * & **, p<0.001 in the two-tailed t-test. (C) Immunohistochemical analyses with Chx10 and GABA antibodies in E13.5 littermate embryos at thoracic spinal cord. The ventral quadrant spinal cord is outlined. The error bars represent the standard deviation (A, B).
LMO4 nucleates the assembly of a multi-protein complex including SCL and Gata2
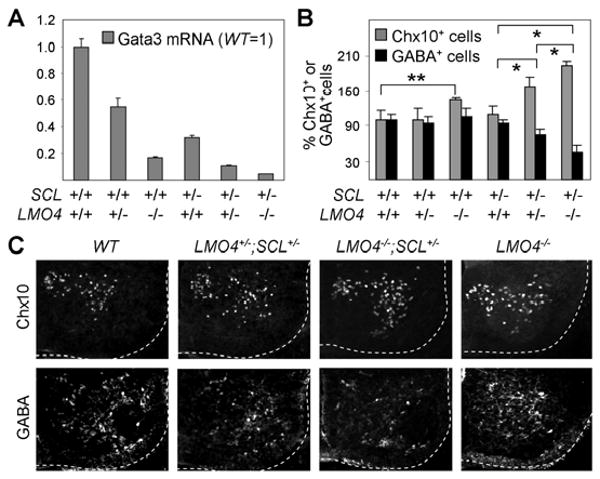
To understand the molecular basis of the cooperative action between LMO4 and SCL in promoting V2b fate, we considered the possibility that LMO4 mediates the formation of a higher-order LIM-complex with SCL, Gata2 and NLI during V2b-IN development. To test this idea, we assessed interactions among LMO4, SCL, Gata2 and NLI in yeast two hybrid and GST-pull down assays. LMO4 interacts with not only a well-known LIM-interactor NLI but also Gata2 and SCL, whereas SCL binds neither Gata2 nor NLI (Fig. 3A-D). NLI interacts with Gata2 as well as LMO4 and NLI (Fig. 3A-C). The interaction of LMO4 with SCL, Gata2 and NLI prompted us to test whether LMO4 enables SCL to be indirectly tethered to Gata2 and/or NLI. In support of this idea, GST-pull down assays revealed that LMO4 bridges SCL to Gata2 and NLI (Fig. 3C, D). Moreover, the addition of NLI strengthened the LMO4-mediated association between Gata2 and SCL (Fig. 3D), raising the possibility that all four proteins assemble into a multi-protein complex. To map the Gata2-interaction domain in NLI, we tested the binding of Gata2 to either NLI-DD, which contains the dimerization domain (DD) of NLI, or NLI-LID consisting of the LIM-interaction domain (LID) of NLI (S-Fig. 4) (Thaler et al., 2002). Gata2 preferentially bound NLI-DD in GST-pull down and yeast two hybrid tests, whereas LMO4 interacted with NLI-LID (Fig. 3B, data not shown), suggesting that NLI:Gata2 and NLI:LMO4 interactions could occur simultaneously allowing the multi-protein complex to form. The binding between Gata2 and NLI was also strengthened by the addition of LMO4 (Fig. 3C), further supporting the high-order complex model. Consistent with this in vitro interaction profile, coimmunoprecipiation assays in HEK293 cells revealed that LMO4 associates with SCL and Gata2 in cells (Fig. 3E). Together, our results suggest that LMO4 and NLI play critical roles for nucleating the multi-protein complex of NLI:Gata2:LMO4:SCL and that two of these units could be assembled into a higher-order LIM-complex of 2NLI:2Gata2:2LMO4:2SCL through NLI-dimerization (Fig. 3G).
Figure 3. Formation of a high-order LIM-complex mediated by LMO4 and NLI.
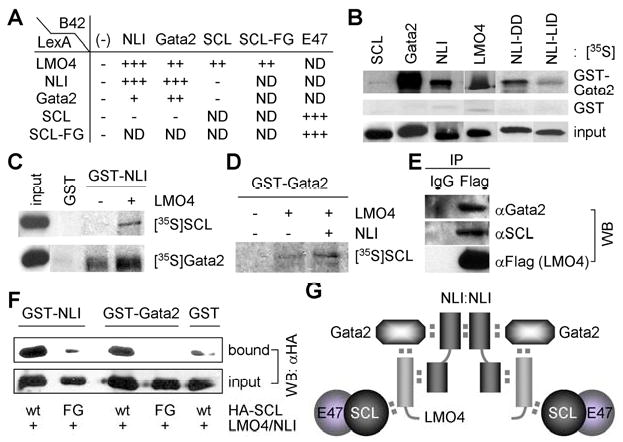
(A) Protein-protein interactions monitored by the yeast two hybrid assays. − represents no interaction and + shows the interaction intensity determined by the color intensity on the X-gal containing plates. ND, not determined. (B-D) In vitro GST-pull down assays using in vitro translated, 35S-labeled proteins and bacterially purified GST-fusion proteins. (E) LMO4 associates with SCL and Gata2 in cells, as assessed by co-IP assays in HEK293 cells transfected with SCL, Gata2 and Flag-tagged LMO4. (F) In vivo GST-pull down assays in HEK293 cells expressing LMO4, NLI, and HA-SCL or HA-SCL-F238G (FG), along with GST-NLI or GST-Gata2. NLI- or Gata2-bound proteins were purified with glutathione sepharose beads and probed with HA-antibody. (G) The proposed model for the V2b-complex consisting of 2NLI:2Gata2:2LMO4:2SCL-E47.
While SCL can transcriptionally synergize with LMO2 and induce hematopoietic differentiation of embryonic stem cells, SCL-F238G is inert (Muroyama et al., 2005; Schlaeger et al., 2004). SCL-F238G is also inactive in V2b-specification in the developing spinal cord, and does not cooperate with LMO4 for V2b-IN differentiation (S-Fig. 1C, Fig. 1R). Thus, understanding the mechanism of the functional deficiency of SCL-F238G is likely to provide crucial insights into the cooperation of SCL and LMO4 in V2b-IN specification. In yeast two hybrid assays, both SCL and SCL-F238G heterodimerized with E47 effectively (Fig. 3A). Contrary to the expectation that SCL-F238G would be unable to bind LMOs, SCL-F238G interacted with LMO1, LMO2 and LMO4 as strongly as wild-type SCL (Fig. 3A, data not shown), suggesting that SCL-F238G is intact in an one-to-one interaction with LMOs. Next, we investigated whether SCL-F238G is incorporated into the complex of NLI:Gata2:LMO4:SCL in cells using in vivo GST-pull down assays. GST-NLI and LMO4 were expressed along with either SCL or SCL-F238G in HEK293 cells and NLI-associated proteins were purified with glutathione sepharose beads. Consistent with the in vitro results, NLI associated with SCL, presumably through a bridging molecule LMO4 (Fig. 3F). Interestingly, NLI failed to co-purify SCL-F238G under the same condition (Fig. 3F), suggesting that SCL-F238G is incapable of forming a complex with NLI. We also investigated the LMO4-mediated association between Gata2 and SCL in cells. GST-Gata2 associated with SCL, but not with SCL-F238G (Fig. 3F). Thus, SCL-F238G, which is defective in V2b-IN induction, is incapable of forming a complex with NLI, Gata2 and LMO4 in cells. These data strongly suggest that formation of a complex containing SCL, NLI, Gata2 and LMO4 in cells is needed for V2b-IN induction.
Dimerization of NLI is required for V2b-IN differentiation
Using the N-terminal self-dimerization and C-terminal LIM-interaction domains (NLI-DD and NLI-LID), NLI mediates the formation of two LIM-complexes that induce the specification of motor neurons and V2a-INs (Lee and Pfaff, 2003; Thaler et al., 2002). As NLI is highly upregulated in differentiating neurons in the developing spinal cord (Thaler et al., 2002), we predicted that NLI-dimerization would bring two NLI:Gata2:LMO4:SCL units into a higher-order LIM-complex that we named the V2b-complex (Fig. 3G), and that the V2b-complex is responsible for directing V2b-IN specification. To test this hypothesis, we used a dominant negative NLI-DD, which blocks the dimerization of endogenous NLI (S-Fig. 4) (Thaler et al., 2002). NLI-DD effectively inhibited V2b-IN specification triggered by SCL expression in the chick neural tube (Fig. 4A, B). These results establish that the dimerization of NLI is required for SCL to induce ectopic V2b-IN generation and support our V2b-complex model.
Figure 4. NLI dimerization is required for SCL-induced V2b-IN formation.
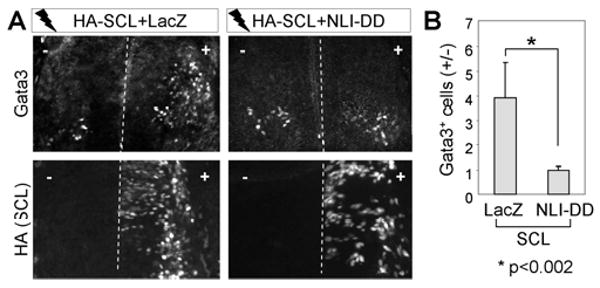
(A) Immunostaining analyses in chicks electroporated with HA-SCL along with either LacZ or NLI-DD. (B) The efficiency of V2b-IN induction was quantified by the ratio of Gata3+ V2b-INs on the electroporated side (+) over the control side (-). The error bars represent the standard deviation.
The transcriptional synergy among components of the V2b-complex and SSDP1
Our data establish the biochemical basis underlying the assembly of the V2b-complex and its critical role in V2b-IN differentiation. However, the transcriptional regulatory mechanism by which the components of the V2b-complex cooperate with each other to specify V2b-INs remains unclear. It is possible that the formation of the V2b-complex results in transcriptional synergy by facilitating recruitment of coactivators. This could involve new coactivator-interacting interfaces resulting from formation of the complex and/or a combined action of multiple interaction interfaces provided by individual subunits of the complex. Interestingly, single stranded DNA-binding proteins (SSDPs) have been found as NLI-interacting proteins (Chen et al., 2002; van Meyel et al., 2003) and to function as coactivators for the hematopoietic complex consisting of SCL, Gata1, LMO2 and NLI (Xu et al., 2007). Thus, NLI (and possibly other subunits of the V2b-complex) may recruit SSDPs to the V2b-complex, which subsequently enhances the transcriptional activating potential of the V2b-complex and thus promotes V2b-IN specification. To test this possibility, we first tested whether LMO4 associates with SSDP1, which is abundantly expressed in spinal neurons (unpublished data). HA-LMO4 and Flag-SSDP1 were expressed without or with NLI in HEK293 cells and the complex containing SSDP1 was immunopurified with Flag-antibody. Interestingly, overexpressed NLI stabilized LMO4 protein (Fig. 5A), suggesting that formation of a complex between LMO4 and NLI increases the stability of LMO4 protein. This is consistent with the report that the expression of dLMO protein requires Chip, a NLI ortholog, during Drosophila wing development (Milan and Cohen, 2000). SSDP1 efficiently immunoprecipitated LMO4 as well as its known interactor NLI in coimmunoprecipitation assays (co-IP) (Fig. 5A). These results suggest that NLI and LMO4 are capable of recruiting SSDP1 to the V2b-complex.
Figure 5. The recruitment of SSDP1 to the V2b-complex leads to transcriptional synergy.
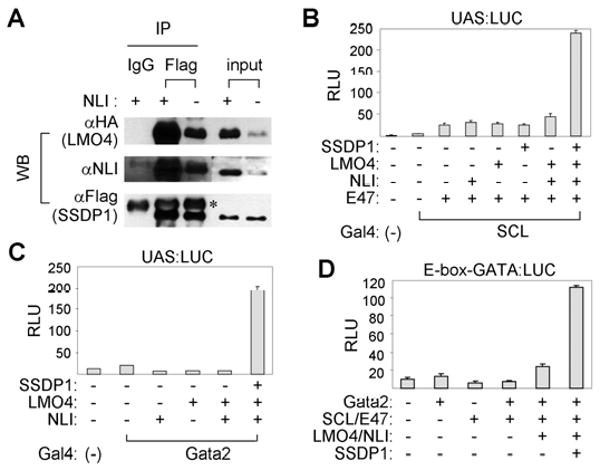
(A) Co-IP assays in HEK293 cells transfected with HA-LMO4, Flag-SSDP1 and NLI. * shows the heavy chain bands. (B-D) Luciferase reporter assays in HEK293 cells. UAS:LUC with Gal4-SCL (B) or Gal4-Gata2 (C) or E-box-GATA:LUC (D) were used as reporters. The error bars represent the standard deviation.
Next, we tested whether LMO4, NLI and SSDP1 potentiate the transcriptional activity of SCL or Gata2 using cell-based luciferase reporter assays. SCL and GATA2 are mild transcriptional activators when their transcriptional activity was measured as Gal4-DNA binding domain (Gal4-DBD) fusions in Gal4-binding UAS:Luciferase reporter (Fig. 5B, C). Interestingly, coexpression of LMO4, NLI and SSDP1 led to a strongly synergistic stimulation of the SCL- or Gata2-mediated transcription, while expression of LMO4, NLI, or SSDP1 alone was not effective (Fig. 5B, C, data not shown). These results suggest that recruitment of SSDP1 to SCL or Gata2 through NLI and LMO4 augments the transcriptional activity of SCL and Gata2. Next, we wanted to monitor the transcriptional activity of the V2b-complex that contains both DNA-binding components SCL and Gata2. To this end, we utilized an E-box-GATA composite element, which was identified as a binding site of the hematopoietic complex containing Gata1, SCL, NLI and LMO2 (Wadman et al., 1997). This is a bipartite DNA response element composed of a bHLH-binding E-box and a Gata protein-binding GATA-site separated by nine base-pairs. We measured the transcriptional activation of a luciferase reporter linked to multimerized E-box-GATA elements, upon expression of SCL, Gata2, LMO4, NLI and SSDP1, alone and in combinations. Remarkably, coexpression of the V2b-complex components along with SSDP1 led to a strongly synergistic activation of this reporter (Fig. 5D). Together, these data indicate that the V2b-complex recognizes and transactivates E-box-GATA composite elements and demonstrate that the formation of the V2b-complex results in transcriptional synergy at least in part by recruiting the coactivator SSDP1.
Identification of enhancers in Gata2 and Gata3 genes responsive to the V2b-complex
To characterize the in vivo function of the V2b-complex in transcriptional regulation during spinal cord development, we sought the physiological target genes of the V2b-complex. Based on the architecture of the V2b-complex (Fig. 3G), we reasoned that the V2b-complex-response elements would consist of at least two reiterated GATA-E-box composite elements and have a capacity to direct gene expression to V2b-INs, in which the endogenous V2b-complex is expected to be assembled. Notably, the screen for binding elements of the SCL/Gata1-containing hematopoietic complex isolated not only the consensus E-box sequences (CAnnTG) and GATA site (GATA) but also non-canonical forms of the E-box motif (CAnnnTG) and GATA site (GATT) (Wadman et al., 1997). Thus, we also considered the possibility that the V2b-complex-response elements consist of canonical and/or non-canonical E-boxes and GATA sites. A 190 bp enhancer region in intron 5 of Gata2 has previously been shown to be sufficient for directing the reporter gene expression to V2-INs in transgenic mouse embryos (Zhou et al., 2000). However, the specific DNA sequences critical for this V2-specific enhancer activity remain unclear. Intriguingly, we noted that a pair of bipartite DNA sequences consisting of non-canonical E-boxes (CAnnnTG) and GATA sites is present in this enhancer region (Fig. 6A). This feature prompted us to ask whether the V2b-complex recognizes the V2-IN-specific Gata2-enhancer in vivo. To this end, we performed chromatin immunoprecipitation (ChIP) analyses in mouse embryonic P19 cells transfected with constructs expressing Gata2, SCL and LMO4, which are expected to form the V2b-complex with endogenous NLI. Gata2, SCL and LMO4-containing chromatin-protein complexes were purified by Gata2, SCL and LMO4 antibodies, respectively, and the presence of Gata2-enhancer in the chromatin-protein complexes was determined by PCR. The ChIP assays revealed that Gata2, SCL and LMO4 are recruited to the genomic Gata2-enhancer in cells (Fig. 6C). Moreover, consistent with the in vivo recruitment of the V2b-complex, the Gata2-enhancer linked to GFP reporter directs expression of GFP to Gata3+ V2b-INs with ipsilateral axonal projections in chick embryos (Fig. 6D). We then asked whether ectopic expression of the V2b-complex activates Gata2-enhancer in the dorsal spinal cord. Co-injection of SCL and LMO4, along with Gata2-enhancer:GFP reporter, triggered GFP expression in the dorsal spinal cord and dorsal root ganglion (Fig. 6H, S-Fig. 5). Collectively, these data suggest that the V2b-complex regulates Gata2 expression during V2-IN development by binding to the Gata2-enhancer.
Figure 6. The V2b-complex binds the intronic enhancers of Gata2 and Gata3 genes.
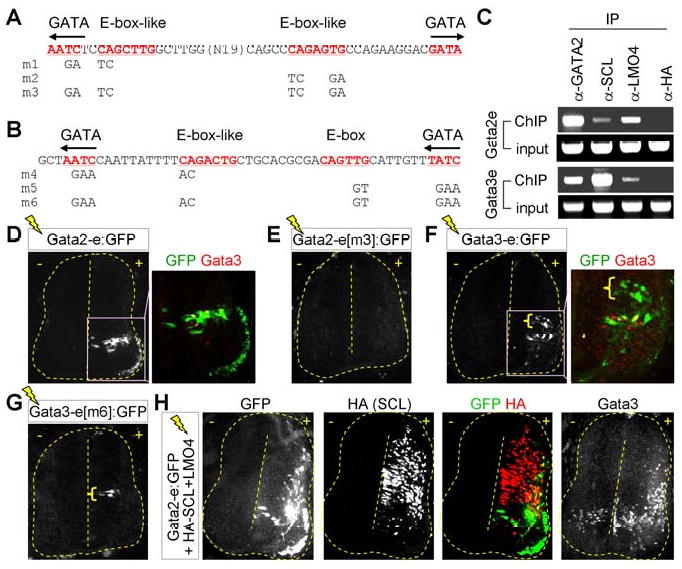
(A, B) E-box/E-box-like and GATA sites within Gata2-enhancer (A) or Gata3-enhancer (B) are underlined. Mutations in E-box/E-box-like or GATA sites are as indicated. (C) ChIP assays using P19 cells expressing Gata2, SCL and LMO4. (D-H) Immunohistochemical analyses in HH stage 24 chick embryos following electroporation of indicated constructs. GFP expression in the dorsal interneurons is marked by brackets (F, G).
The preceding observations indicate that repeated bipartite DNA sequences consisting of E-boxes and GATA sites are likely to be the signature motif recognized by the V2b-complex in vivo. Thus, we searched for similar motifs in Gata3 gene, a well-established marker for V2b-INs. Our bioinformatics approach discovered a 300 bp region in intron 3 of Gata3 gene, whose sequences are highly conserved across multiple species (S-Fig. 6). Intriguingly, this intronic region includes an evolutionarily conserved pair of bipartite E-box-GATA elements, which is highly homologous to the Gata2-enhancer (Fig. 6B). ChIP experiments using P19 cells expressing Gata2, SCL and LMO4 demonstrated that the V2b-complex specifically binds the genomic bipartite E-box-GATA elements of the Gata3 gene (Fig. 6C). To determine the functional responsiveness of this 300 bp intronic region of Gata3 gene to the endogenous V2b-complex, we asked whether this can direct reporter gene expression to V2b-INs in the embryonic spinal cord. To this end, we electroporated Gata3 genomic region linked to GFP reporter to chick embryos and monitored GFP expression in the neural tube. The strongest GFP expression was detected in Gata3+ V2b-INs, but a small population of interneurons positioned medially along the dorso-ventral axis also expressed GFP (Fig. 6F). These data establish that the 300 bp intronic region of Gata3 gene functions as an enhancer to stimulate transcription in V2b-INs by recruiting the V2b-complex.
The presence of a pair of E-box-GATA composite elements in Gata2/3-enhancers along with their in vivo recruitment of SCL, Gata2 and LMO4 leads to the prediction that E-boxes and GATA sites are required for the activity of Gata2/3-enhancers in V2b-INs by serving as binding sites for SCL and Gata2 within the endogenous V2b-complex. To test this, we mutated either one or both of the E-box-GATA elements in Gata2/3-enhancers and monitored their activity to drive gene transcription in V2b-INs using chick embryo electroporations. For both Gata2 and Gata3-enhancer:GFP reporters, mutation of each pair of E-box-GATA unit resulted in weaker but significant GFP expression in V2-INs (m1, m2, m4 and m5 in Fig. 6A, B, data not shown). However, simultaneous mutation of both E-box-GATA composite elements completely abolished GFP expression in V2-INs by Gata2 and Gata3-enhancer:GFP reporters (m3 and m6 in Fig. 6A, B, Fig. 6E, G), indicating that a pair of E-box-GATA composite elements are needed for Gata2/3-enhancers to activate the gene transcription in V2 cells. In contrast to V2-INs, GFP expression in dorsal interneurons still persisted when the Gata3-enhancer was mutated on both E-box-GATA elements (bracket in Fig. 6G), suggesting that expression in dorsal interneurons is regulated independently of E-box-GATA composite elements.
Collectively, we identified enhancers in Gata2 and Gata3 genes targeted by the V2b complex. Each enhancer contains a pair of E-box and GATA composite elements and directs gene expression to V2b-INs.
The V2b-complex is responsible for transactivation of the Gata2/3-enhancers
To examine the requirement for the endogenous V2b-complex in stimulating Gata3-enhancer in V2b-INs, we assessed the transcriptional activity of the Gata3-enhancer in the neural tube upon disruption of the V2b-complex assembly. To inhibit the formation of the V2b-complex, we utilized NLI-DD, which blocks the self-association of NLI, and NLI-LID, which binds LIM domains but lacks the dimerization domain (S-Fig. 4) (Thaler et al., 2002). The Gata3-enhancer:GFP reporter was electroporated into chick embryos along with vectors encoding LacZ control, NLI-DD or NLI-LID. Overexpressed NLI-DD or NLI-LID suppressed the V2b-IN-specific enhancer activity of the Gata3-enhancer, whereas NLI-DD did not suppress the GFP expression in the dorsal interneurons (Fig. 7A). These results suggest that the assembly of the V2b-complex, mediated by NLI through its ability to self-dimerize and to interact with LIM-domains, is required for the Gata3-enhancer to stimulate transcription in V2b-INs.
Figure 7. The assembly of the V2b-complex is required to recognize and transactivate the Gata2/3-enhancers.
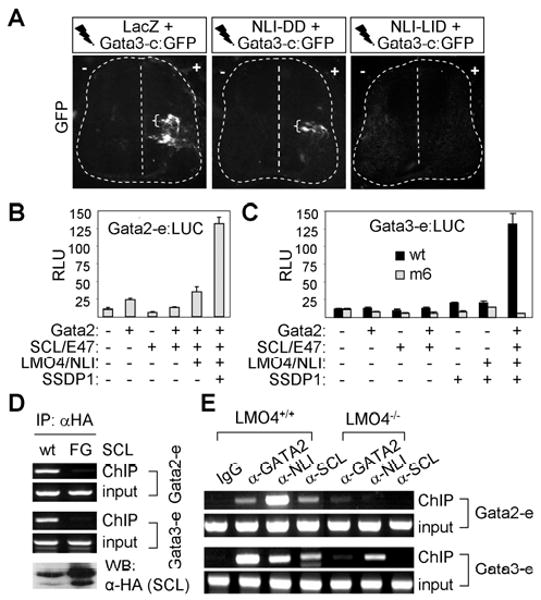
(A) GFP expression in chicks electroporated with Gata3-enhancer:GFP along with LacZ, NLI-DD or NLI-LID. Brackets mark GFP expression in the dorsal interneurons. (B, C) Luciferase reporter assays with Gata2-enhancer:LUC (B) and Gata3-enhancer:LUC (C) in HEK293 cells. (D) ChIP assays in P19 cells transfected with Gata2, LMO4 and NLI along with HA-SCL or HA-SCL-F238G (FG). The bottom panel shows western blot analyses with HA-antibody. (E) ChIP assays using spinal cord extracts from E14.5 wild-type or LMO4-null embryos.
Next, we examined whether SCL and Gata2 synergize to activate the Gata2/3-enhancers, physiological target elements of the V2b-complex, using luciferase reporter assays. SCL and GATA2, alone or in combination, failed to strongly activate the Gata2-enhancer:LUC or Gata3-enhancer:LUC reporters (Fig. 7B, C). Interestingly, the coexpression of LMO4, NLI and SSDP1 along with SCL and Gata2 resulted in a robust stimulation of transcription in Gata2/3-enhancer:LUC reporters, but not in Gata3-enhancer[m6]:LUC reporter in which a pair of E-box-GATA composite elements are mutated (Fig. 6B, 7B, C). These results suggest that formation of the V2b-complex and the subsequent recruitment of SSDP1 to E-box-GATA composite elements are necessary to transactivate the Gata2/3-enhancers effectively in cells.
It is possible that formation of the V2b-complex facilitates efficient binding of SCL and Gata2 to the combinatorial E-box-GATA elements in Gata2/3-enhancers, and this enhancement in DNA binding may underlie the transcriptional synergy among components of the V2b-complex. To test this idea, we transfected P19 cells with Gata2 and LMO4 along with either HA-SCL or HA-SCL-F238G mutant, which is defective in forming the V2b-complex in cells (Fig. 3F), and performed ChIP analyses by immunopurifying the chromatin-protein complex with HA-antibody. Because SCL-F238G has an intact DNA-binding basic domain and efficiently heterodimerizes with E47, it is likely capable of binding conventional E-boxes as a SCL/E47 heterodimer. Interestingly, although SCL-F238G was expressed no less than SCL wild-type (Fig. 7D), SCL-F238G was not recruited to the genomic Gata2/3-enhancers under the condition in which wild-type SCL readily bound the Gata2/3-enhancers (Fig. 7D). These results support the idea that assembly of the V2b-complex promotes the efficient recruitment of SCL and Gata2 to the V2b-complex responsive enhancers.
LMO4 is required for the efficient binding of SCL and Gata2 in the V2b-complex to Gata2/3-enhancers
Given the essential role of LMO4 in nucleating the V2b-complex formation (Fig. 3), we reasoned that inefficient assembly of the V2b-complex in LMO4-null embryos would compromise the binding of SCL and Gata2 to the V2b-complex response elements thereby contributing to reduced Gata3 expression and GABAergic neurons (Fig. 2). To test this possibility, we monitored the occupancy of the Gata2/3-enhancers by SCL and Gata2 in the spinal cord of wild-type and LMO4-null embryos using ChIP assays. The spinal cords of E14.5 mouse embryos were dissected and then SCL, Gata2, or NLI-containing chromatin-protein complexes were immunopurified by SCL-, Gata2-, or NLI-antibodies, respectively. The SCL, Gata2, and NLI proteins in the spinal cord cells from wild-type embryos bound the Gata2/3-enhancers (Fig. 7E), in agreement with the recruitment pattern in P19 cells (Fig. 6C). This establishes that the endogenous V2b-complex directly interacts with the Gata2/3-enhancers. Interestingly, in contrast to wild-type embryos, the recruitment of Gata2, NLI and SCL to the Gata2/3-enhancers was severely attenuated in LMO4-null spinal cords (Fig. 7E), indicating that LMO4 is needed for the efficient binding of SCL, Gata2 and NLI to the Gata2/3-enhancers. Because LMO4 lacks any DNA-binding domain, this function of LMO4 is likely to reflect its role in promoting the assembly and stabilization of the V2b-complex. The greatly attenuated binding of SCL and Gata2, two distinct DNA-binding components of the V2b-complex, to the Gata3-enhancer in LMO4-null embryos is consistent with reduced expression of Gata3 (Fig. 7E, 2A). Together, these results demonstrate that assembly of the V2b-complex mediated by LMO4 appears to be a prerequisite for the efficient recognition of the Gata2/3-enhancers by SCL and Gata2.
Discussion
A majority of neurons in vertebrates make a developmental choice between excitatory and inhibitory neurotransmitter phenotypes, predominantly glutamatergic versus GABAergic (Bennett and Balcar, 1999). As unbalanced activity of inhibitory and excitatory neurons can result in various neuronal disorders, the generation of neuronal subtypes with different neurotransmitter phenotypes is tightly controlled during CNS development. In this report, we identified LMO4 as a critical factor, which controls this intricate balance during spinal V2-IN development and refines the binary cell fate choices initiated by Notch-Delta signaling. We further defined the biochemical basis of the dual action of LMO4 in promoting GABAergic V2b fate and simultaneously inhibiting glutamatergic V2a fate.
Transcriptional regulatory pathway to control excitatory and inhibitory V2-IN identities
In the ventral spinal cord, four classes of interneurons are further subdivided into functionally distinct subtypes on the basis of neurotransmitter choices, expression of marker genes, and axonal projections (Al-Mosawie et al., 2007; Lanuza et al., 2004; Lundfald et al., 2007). Immature V2-INs expressing both Lhx3 and Gata2 diversify into excitatory V2a-INs and inhibitory V2b-INs over the same period of time (Fig. 8) (Peng et al., 2007). Notch-Delta cell-signaling orchestrates the initial diversification step of V2-INs (Del Barrio et al., 2007; Kimura et al., 2008; Peng et al., 2007). However, little is known about the transcription regulators which control neurotransmitter identity in the ventral spinal cord. We showed that LMO4 inhibits Lhx3 from ectopically inducing the glutamatergic fate and cooperates with SCL to suppress endogenous glutamatergic neuronal differentiation and promote GABAergic neurotransmitter expression. Correspondingly, in LMO4-/-;SCL+/- compound mutant embryos, the number of glutamatergic Chx10+ V2a-INs increases dramatically at the expense of GABAergic Gata3+ V2b-INs, suggesting that prospective V2b-INs are converted to V2a cells. Together, LMO4 promotes GABAergic neuron identity and simultaneously suppresses glutamatergic neuron identity during the diversification of V2-INs, keeping the necessary balance between excitatory and inhibitory activities in the motor circuit (Fig. 8). This dual regulatory role for LMO4 involves its positive role in assembling the V2b-IN-specifying V2b-complex with SCL and Gata2 as well as its negative action in disrupting the assembly and activity of the V2a-IN-specifying 2NLI:2Lhx3 V2-tetramer complex (Fig. 8) (Lee et al., 2008). Our results also raised the possibility that the LIM-complexes directing V2a and V2b identities function as postmitotic selectors for maintaining the desired neurotransmitter phenotypes.
Figure 8. The working model.
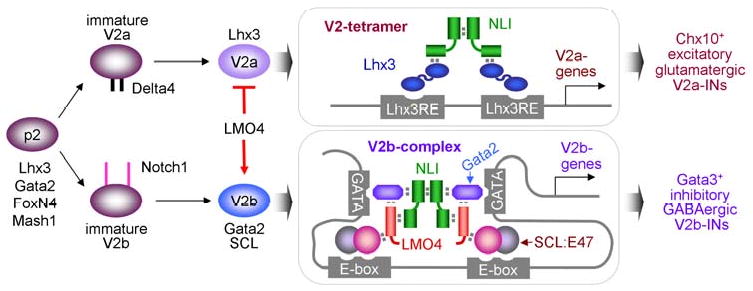
LMO4 stimulates V2b-IN development by mediating the assembly of the V2b-complex that recognizes E-box-GATA composite elements in V2b-IN genes, while it inhibits differentiation of p2 progenitors to excitatory V2a-IN by suppressing the formation of Lhx3-containing V2-tetramer complex.
The similarity in the process of choosing GABAergic over glutamatergic neuronal cell fate between ventral and dorsal spinal cord is noteworthy. In the dorsal spinal cord, another bHLH factor Ptf1a (also known as p48) plays a central role in the generation of inhibitory GABAergic neurons while suppressing the production of excitatory glutamatergic neurons (Glasgow et al., 2005; Hori et al., 2008). Both Ptf1a and SCL form a multi-protein complex to fulfill their function in neurotransmitter specification, rather than functioning through a conventional heterodimer with E-proteins. The formation of a complex between Ptf1a and Rbpj, the vertebrate ortholog of the Drosophila Suppressor of Hairless, is required for Ptf1a to promote GABAergic neuronal identity (Hori et al., 2008). Ptf1a/Rbpj-complex has been shown to recognize DNA sequences consisting of Ptf1a-binding E-box and Rbpj-binding T/C-box (Masui et al., 2007). Unlike Ptf1a, Ptf1a-W298A mutant, which fails to cooperate with Rbpj in activating transcription, is inert in inducing GABAergic dILA neurons and suppressing glutamatergic dILB neurons (Hori et al., 2008). Similarly, the SCL-F238G mutant, defective in forming the V2b-complex, was unable to induce GABAergic V2b-INs and suppress glutamatergic V2a-INs. Thus, it is tempting to speculate that complex formation with multiple DNA-binding components provides bHLH factors with higher specificity in the recognition of target elements and subsequently target gene selections. In support of this idea, we found that the assembly of the V2b-complex is required for the efficient binding of SCL to the enhancers of V2b-IN genes Gata2/3. However, it is unclear whether the V2b-complex regulates genes involved in GABAergic neurotransmission directly. Thus, it will be interesting to test whether the V2b-complex directly controls expression of Gad1 gene in the ventral spinal cord through an evolutionarily conserved E-box-GATA element that we have found in Gad1 gene (unpublished data). Different bHLH factors appear to be required for GABAergic neurons in distinct regions of the nervous system. For instance, Ptf1a and Mash1 mediate GABAergic neuronal specification in dorsal spinal cord/cerebellum and telencephalon/midbrain, respectively (Fode et al., 2000; Glasgow et al., 2005; Hoshino et al., 2005; Nakatani et al., 2007). We report that SCL is important to establish GABAergic identity of V2b-INs in the ventral spinal cord. Thus, genes involved in GABAergic neurotransmission are likely to be regulated by multiple bHLH factors in different regions of the vertebrate CNS, rather than by universal regulators that govern all GABAergic neuronal subtypes.
LMO4 mediates the assembly of the V2b-complex
In higher vertebrates, twelve LIM-HD and four LMO proteins have been implicated in the development of multiple cell types (Hobert and Westphal, 2000). The LIM-HD factors not only label distinct cell populations within the nervous system (Tsuchida et al., 1994) but also function in combinations by forming cell type-specific complexes with a self-dimerizing adaptor NLI protein (Thaler et al., 2002). Based on the fact that LMOs lack a functional DNA-binding domain, they have been proposed to disrupt the formation of conventional LIM-complexes consisting of LIM-HD factors and NLI by competing with LIM-HD proteins for NLI-interaction, thus acting as negative regulators of LIM-complexes (Lee et al., 2008; Milan and Cohen, 1999; Milan et al., 1998). In contrast, here we presented that LMO4 functions as a positive transcriptional regulator by nucleating, along with NLI, the assembly of a novel-type of LIM-complex comprised of SCL and Gata2 during V2b-IN development. It is noteworthy that a similar DNA-binding complex consisting of SCL, Gata1, LMO2 and NLI has been proposed in hematopoietic cells (Wadman et al., 1997), although its exact stoichiometry remains unknown. This indicates that formation of multi-protein complexes bridged by LMOs may be a recurring strategy to achieve cellular diversity in multiple tissues including the CNS.
The one-to-one interaction profiles among SCL, Gata2, LMO4 and NLI suggest at least four possible models for multi-protein complexes. One possibility is that Gata2 forms a complex with NLI, which then homodimerizes to form a 2Gata2:2NLI tetramer (S-Fig. 7A). Alternatively, SCL can interact with NLI via LMO4 and this complex then homodimerizes through NLI-dimerization thereby forming a 2SCL:2LMO4:2NLI complex (S-Fig. 7B). Since these interactions are occurring in the same cell co-expressing SCL and Gata2, it is also possible that Gata2:NLI associates with SCL:LMO4:NLI, forming a heteromeric Gata2:NLI-NLI:LMO4:SCL complex that may operate to co-regulate V2b-targets by SCL and Gata2 (S-Fig. 7C). However, a paradox emerges in these complex assembly models. Considering that all three complexes can be formed in a single cell that expresses SCL, Gata2, LMO4 and NLI, only a fraction of the complexes will have both SCL and Gata2. In a fourth model, an NLI:LMO4 module binds Gata2 and SCL simultaneously forming a subcomplex of Gata2:NLI:LMO4:SCL and NLI-dimerization subsequently results in the multiprotein complex that we denoted as the V2b-complex (Fig. 3G, S-Fig. 7D). This model would ensure an enrichment of both SCL and Gata2 in a single complex, thereby avoiding the generation of multiple homomeric and heteromeric complexes within a single cell. Our biochemical analyses support this model, as incubation of multiple components of the putative V2b-complex tends to stabilize associations among the subunits, rather than disrupting each interaction (Fig. 3). The composition of the V2b-complex is also mirrored in the cis-regulatory elements of the V2b-specific genes, Gata2 and Gata3, which consist of a pair of bipartite E-box-GATA elements (Fig. 6A, B). Our results do not exclude the possibility that complexes which have either SCL or Gata2, but not both, could still be instrumental in regulation of genes responsive to SCL or Gata2 alone during V2b-IN differentiation. For example, Gata2 expression precedes SCL expression in p2 progenitors differentiating to V2-INs, where it may regulate a discrete set of genes by forming a complex with NLI, but not with SCL.
The “LIM code” in V2b-IN specification
LIM-HD codes are crucial in implementing cell-type-specific transcription by directing different types of LIM-complexes in a cell context-dependent manner (Thaler et al., 2002). Our studies expand the LIM codes to include bHLH and Gata proteins as these two factors form an atypical LIM-complex via a non-DNA binding LIM factor LMO4. Unlike typical LIM-complexes such as the V2-tetramer complex, which utilize LIM-HD proteins for recognition of specific DNA response elements (Fig. 8) (Lee et al., 2008), SCL and Gata2 serve as the major DNA-binding components in the V2b-complex. A couple of unique advantages of assembling the V2b-complex can be proposed in cell fate specification.
First, our results suggest that the V2b-complex allows integration of SCL and Gata2 functions by selecting a group of target genes that bear both SCL- and GATA-recognition sites. This should ensure the expression of V2b-target genes specifically in cells coexpressing SCL and Gata2. We found that the enhancers of Gata2 and Gata3 genes display striking similarity in that they contain reiterated bipartite elements composed of E-box (CAnnTG) and/or atypical E-box (CAnnnTG) for SCL-binding and GATA sites for recruiting Gata proteins. E-boxes and GATA sites occur relatively often in the genome due to their short sequences and serve as binding motifs for multiple bHLH and Gata factors. Thus, simultaneous recognition of paired E-box-GATA composite elements by the V2b-complex is expected to provide the required stringency in choosing the target genes coregulated by SCL and Gata2.
Second, we found that formation of the V2b-complex facilitates the transcriptional synergy among its components by enabling the recruitment of coactivators including SSDP1. Coexpression of SSDP1 allowed a potent transcriptional activation by the V2b-complex on its physiological targets, Gata2/3-enhancers. Given that SCL and Gata2 are relatively weak transcriptional activators in Gal4-DBD fusion transcription assays, the transcriptional synergy between SCL and Gata2 resulting from forming a complex may be due, at least in part, to the recruitment of SSDP1. The facilitated recruitment of SSDP1 and possibly other coactivators may account for the necessity of the V2b-complex formation for inducing the V2b-IN genes.
Experimental Procedures
Mice
The SCL and LMO4 mutant mice have been previously described (Elefanty et al., 1998; Lee et al., 2005). Embryos were collected at stages E11.5-E15.5, fixed in 4% paraformaldehyde, sunk in 30% sucrose embedded in OCT for cryosectioning and subsequent analyses.
In ovo electroporation, immunohistochemistry and in situ hybridization
In ovo electroporation was performed as described (Thaler et al., 2002). Briefly, plasmid DNA was injected into the lumen of the neural tube of HH stage 13 chick embryos which were then electroporated. A mammalian expression vector expressing dsRed or GFP (Clonthech) or nuclear lacZ was co-injected to monitor electroporation efficiency. The embryos were harvested at HH stage 24-26, fixed in 4% paraformaldehyde, embedded in OCT and cryosectioned at 12-18 μm. Immunohistochemistry and in situ hybridization were performed as described (Thaler et al., 2002). Digoxigenin-labeled riboprobe complementary to mouse or chick Vglut2 and Gad1; mouse Gata2, SCL, and LMO4 were used for in situ hybridization.
Quantification and statistical analysis
Cell counts were obtained from 12 μm sections of embryos and represented as a ratio to cells in the control side in chicks and a ratio to cells in the wild-type littermate in mice with standard deviation as error bars. The significance was determined using two-tailed Student's t-test.
Cell culture and luciferase assays
Cell culture and luciferase assays were used as described (Lee et al., 2005). Histograms show mean normalized luciferase units and error bars represent standard deviation. All transfections were repeated multiple times.
CoIP, ChIP, GST-pull down and yeast two hybrid assays
These assays were performed as described previously (Lee et al., 2008; Lee et al., 2005). In yeast two hybrid assays, at least eight independently derived colonies were tested for each protein interaction experiment.
Quantitative Reverse Transcriptase PCR (qRT-PCR)
Total RNA was isolated from mouse embryonic spinal cord tissue using BioRad mini-kit. Superscript II (Invitrogen) was used for reverse transcription. qRT-PCR were performed using SYBR-Green kit (Invitrogen) and Mx3000P (Stratagene). Three different litters have been analyzed and the representative results are shown. The primers used were: mouse Gata3, forward 5′-CTT ATC AAG CCC AAG CGA AG, reverse 5′-TTT GCA CTT TTT CGA TTT GC; Cyclophilin, forward 5′-GTC TCC TTC GAG CTG TTT GC, reverse 5′-GAT GCC AGG ACC TGT ATG CT.
Supplementary Material
Acknowledgments
We thank Margaret Goodell and Huda Zoghbi for critically reading the manuscript, Stacey Glassgow for technical assistance, and David Curtis for SCL mutant mice. This research was supported by grants from NINDS (R01 NS054941), PEW, March of Dimes Foundations and MRDDRC P30 HD24064.
Footnotes
Additional experimental procedures are provided as supplements.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Al-Mosawie A, Wilson JM, Brownstone RM. Heterogeneity of V2-derived interneurons in the adult mouse spinal cord. Eur J Neurosci. 2007;26:3003–3015. doi: 10.1111/j.1460-9568.2007.05907.x. [DOI] [PubMed] [Google Scholar]
- Alvarez FJ, Jonas PC, Sapir T, Hartley R, Berrocal MC, Geiman EJ, Todd AJ, Goulding M. Postnatal phenotype and localization of spinal cord V1 derived interneurons. J Comp Neurol. 2005;493:177–192. doi: 10.1002/cne.20711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Bennett MR, Balcar VJ. Forty years of amino acid transmission in the brain. Neurochem Int. 1999;35:269–280. doi: 10.1016/s0197-0186(99)00068-6. [DOI] [PubMed] [Google Scholar]
- Chen L, Segal D, Hukriede NA, Podtelejnikov AV, Bayarsaihan D, Kennison JA, Ogryzko VV, Dawid IB, Westphal H. Ssdp proteins interact with the LIM-domain-binding protein Ldb1 to regulate development. Proc Natl Acad Sci U S A. 2002;99:14320–14325. doi: 10.1073/pnas.212532399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crone SA, Quinlan KA, Zagoraiou L, Droho S, Restrepo CE, Lundfald L, Endo T, Setlak J, Jessell TM, Kiehn O, Sharma K. Genetic ablation of V2a ipsilateral interneurons disrupts left-right locomotor coordination in mammalian spinal cord. Neuron. 2008;60:70–83. doi: 10.1016/j.neuron.2008.08.009. [DOI] [PubMed] [Google Scholar]
- Del Barrio MG, Taveira-Marques R, Muroyama Y, Yuk DI, Li S, Wines-Samuelson M, Shen J, Smith HK, Xiang M, Rowitch D, Richardson WD. A regulatory network involving Foxn4, Mash1 and delta-like 4/Notch1 generates V2a and V2b spinal interneurons from a common progenitor pool. Development. 2007;134:3427–3436. doi: 10.1242/dev.005868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elefanty AG, Begley CG, Metcalf D, Barnett L, Kontgen F, Robb L. Characterization of hematopoietic progenitor cells that express the transcription factor SCL, using a lacZ “knock-in” strategy. Proc Natl Acad Sci U S A. 1998;95:11897–11902. doi: 10.1073/pnas.95.20.11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fode C, Ma Q, Casarosa S, Ang SL, Anderson DJ, Guillemot F. A role for neural determination genes in specifying the dorsoventral identity of telencephalic neurons. Genes Dev. 2000;14:67–80. [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, et al. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, et al. Ptf1a, a bHLH transcriptional gene, defines GABAergic neuronal fates in cerebellum. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- Jurata LW, Pfaff SL, Gill GN. The nuclear LIM domain interactor NLI mediates homo- and heterodimerization of LIM domain transcription factors. J Biol Chem. 1998;273:3152–3157. doi: 10.1074/jbc.273.6.3152. [DOI] [PubMed] [Google Scholar]
- Karunaratne A, Hargrave M, Poh A, Yamada T. GATA proteins identify a novel ventral interneuron subclass in the developing chick spinal cord. Dev Biol. 2002;249:30–43. doi: 10.1006/dbio.2002.0754. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Okamura Y, Higashijima S. alx, a zebrafish homolog of Chx10, marks ipsilateral descending excitatory interneurons that participate in the regulation of spinal locomotor circuits. J Neurosci. 2006;26:5684–5697. doi: 10.1523/JNEUROSCI.4993-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y, Satou C, Higashijima S. V2a and V2b neurons are generated by the final divisions of pair-producing progenitors in the zebrafish spinal cord. Development. 2008;135:3001–3005. doi: 10.1242/dev.024802. [DOI] [PubMed] [Google Scholar]
- Lanuza GM, Gosgnach S, Pierani A, Jessell TM, Goulding M. Genetic identification of spinal interneurons that coordinate left-right locomotor activity necessary for walking movements. Neuron. 2004;42:375–386. doi: 10.1016/s0896-6273(04)00249-1. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee B, Joshi K, Pfaff SL, Lee JW, Lee SK. A regulatory network to segregate the identity of neuronal subtypes. Dev Cell. 2008;14:877–889. doi: 10.1016/j.devcel.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Jurata LW, Nowak R, Lettieri K, Kenny DA, Pfaff SL, Gill GN. The LIM domain-only protein LMO4 is required for neural tube closure. Mol Cell Neurosci. 2005;28:205–214. doi: 10.1016/j.mcn.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4 1:1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Synchronization of neurogenesis and motor neuron specification by direct coupling of bHLH and homeodomain transcription factors. Neuron. 2003;38:731–745. doi: 10.1016/s0896-6273(03)00296-4. [DOI] [PubMed] [Google Scholar]
- Li S, Misra K, Matise MP, Xiang M. Foxn4 acts synergistically with Mash1 to specify subtype identity of V2 interneurons in the spinal cord. Proc Natl Acad Sci U S A. 2005;102:10688–10693. doi: 10.1073/pnas.0504799102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundfald L, Restrepo CE, Butt SJ, Peng CY, Droho S, Endo T, Zeilhofer HU, Sharma K, Kiehn O. Phenotype of V2-derived interneurons and their relationship to the axon guidance molecule EphA4 in the developing mouse spinal cord. Eur J Neurosci. 2007;26:2989–3002. doi: 10.1111/j.1460-9568.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- Masui T, Long Q, Beres TM, Magnuson MA, MacDonald RJ. Early pancreatic development requires the vertebrate Suppressor of Hairless (RBPJ) in the PTF1 bHLH complex. Genes Dev. 2007;21:2629–2643. doi: 10.1101/gad.1575207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Regulation of LIM homeodomain activity in vivo: a tetramer of dLDB and apterous confers activity and capacity for regulation by dLMO. Mol Cell. 1999;4:267–273. doi: 10.1016/s1097-2765(00)80374-3. [DOI] [PubMed] [Google Scholar]
- Milan M, Cohen SM. Temporal regulation of apterous activity during development of the Drosophila wing. Development. 2000;127:3069–3078. doi: 10.1242/dev.127.14.3069. [DOI] [PubMed] [Google Scholar]
- Milan M, Diaz-Benjumea FJ, Cohen SM. Beadex encodes an LMO protein that regulates Apterous LIM-homeodomain activity in Drosophila wing development: a model for LMO oncogene function. Genes Dev. 1998;12:2912–2920. doi: 10.1101/gad.12.18.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroyama Y, Fujiwara Y, Orkin SH, Rowitch DH. Specification of astrocytes by bHLH protein SCL in a restricted region of the neural tube. Nature. 2005;438:360–363. doi: 10.1038/nature04139. [DOI] [PubMed] [Google Scholar]
- Nakatani T, Minaki Y, Kumai M, Ono Y. Helt determines GABAergic over glutamatergic neuronal fate by repressing Ngn genes in the developing mesencephalon. Development. 2007;134:2783–2793. doi: 10.1242/dev.02870. [DOI] [PubMed] [Google Scholar]
- Parras CM, Schuurmans C, Scardigli R, Kim J, Anderson DJ, Guillemot F. Divergent functions of the proneural genes Mash1 and Ngn2 in the specification of neuronal subtype identity. Genes Dev. 2002;16:324–338. doi: 10.1101/gad.940902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CY, Yajima H, Burns CE, Zon LI, Sisodia SS, Pfaff SL, Sharma K. Notch and MAML signaling drives Scl-dependent interneuron diversity in the spinal cord. Neuron. 2007;53:813–827. doi: 10.1016/j.neuron.2007.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86:47–57. doi: 10.1016/s0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- Schlaeger TM, Schuh A, Flitter S, Fisher A, Mikkola H, Orkin SH, Vyas P, Porcher C. Decoding hematopoietic specificity in the helix-loop-helix domain of the transcription factor SCL/Tal-1. Mol Cell Biol. 2004;24:7491–7502. doi: 10.1128/MCB.24.17.7491-7502.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. LIM homeodomain factors Lhx3 and Lhx4 assign subtype identities for motor neurons. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- Smith E, Hargrave M, Yamada T, Begley CG, Little MH. Coexpression of SCL and GATA3 in the V2 interneurons of the developing mouse spinal cord. Dev Dyn. 2002;224:231–237. doi: 10.1002/dvdy.10093. [DOI] [PubMed] [Google Scholar]
- Tanabe Y, William C, Jessell TM. Specification of motor neuron identity by the MNR2 homeodomain protein. Cell. 1998;95:67–80. doi: 10.1016/s0092-8674(00)81783-3. [DOI] [PubMed] [Google Scholar]
- Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. LIM factor Lhx3 contributes to the specification of motor neuron and interneuron identity through cell-type-specific protein-protein interactions. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- Tsuchida T, Ensini M, Morton SB, Baldassare M, Edlund T, Jessell TM, Pfaff SL. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–970. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- van Meyel DJ, O'Keefe DD, Jurata LW, Thor S, Gill GN, Thomas JB. Chip and apterous physically interact to form a functional complex during Drosophila development. Mol Cell. 1999;4:259–265. doi: 10.1016/s1097-2765(00)80373-1. [DOI] [PubMed] [Google Scholar]
- van Meyel DJ, Thomas JB, Agulnick AD. Ssdp proteins bind to LIM-interacting co-factors and regulate the activity of LIM-homeodomain protein complexes in vivo. Development. 2003;130:1915–1925. doi: 10.1242/dev.00389. [DOI] [PubMed] [Google Scholar]
- Wadman IA, Osada H, Grutz GG, Agulnick AD, Westphal H, Forster A, Rabbitts TH. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. Embo J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Meng X, Cai Y, Liang H, Nagarajan L, Brandt SJ. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 2007;21:942–955. doi: 10.1101/gad.1528507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Tomita T, Wines-Samuelson M, Beglopoulos V, Tansey MG, Kopan R, Shen J. Notch1 signaling influences v2 interneuron and motor neuron development in the spinal cord. Dev Neurosci. 2006;28:102–117. doi: 10.1159/000090757. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Yamamoto M, Engel JD. GATA2 is required for the generation of V2 interneurons. Development. 2000;127:3829–3838. doi: 10.1242/dev.127.17.3829. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


