Abstract
Summary
The development of the central nervous system (CNS) is governed by networks of extrinsic and intrinsic molecular programs that together orchestrate precise gene regulation. For the past few years, significant progress has been made in the characterization of histone modifying enzymes and the roles they play in transcriptional control by affecting chromatin structure. Importantly, recent studies have revealed dynamic changes in histone modifications over the course of neural cell fate specification. Further understanding of physiological functions of histone modifying enzymes and their molecular mechanisms of action in CNS development will provide crucial insights into the process of generating neural cell types with tremendous diversity. Here we discuss the recent advancement in understanding the roles of enzymes involved in histone acetylation and methylation during neural cell type specification.
Introduction
During CNS development, neural progenitor cells (NPCs) sequentially produce neurons, oligodendrocytes and astrocytes, while maintaining NPC population for the generation of later born cell types. The precise progress of neural development is controlled by a complex network of transcription factors, epigenetic regulators, and extrinsic developmental cues [1,2]. One important issue in neurobiology is to understand how the neural cell-type specific profiles of gene expression are established at right time and space in response to extracellular signals and cell intrinsic programs to produce a myriad of cell types constituting the CNS.
Many families of transcription factors, including Sox, basic helix-loop-helix (bHLH), and LIM homeodomain or homeodomain-containing proteins, have been shown to control neurogenesis and neural cell-fate specification [3-7]. These factors are expressed in a highly cell-type specific manner and play instructive roles in cell-fate decision. They either stimulate or suppress transcription of their target genes in specific developmental contexts, likely through affecting chromatin landscapes. Nonetheless, little is known about the chromatin modifying factors that enable these transcription factors to control the expression of their target genes during CNS development.
For the last decade, rapid progress has been made in elucidating epigenetic regulation of gene expression. Epigenetic modifications include DNA methylation, post-translational modification of histone tails, nucleosomal remodeling, and modifications by small non-coding RNAs [8,9]. Historically, “epigenetics” has been defined as meiotically or mitotically heritable changes in gene expression that are not encoded in the primary DNA sequences. However, the recent studies expanded the definition of “epigenetics”, as many epigenetic modifications considered to be stable in the past turned out to be reversible.
The vast majority of eukaryotic genomic DNA is wrapped around a histone octamer core and compacted to form chromatin [10]. Chromatin is a highly dynamic environment that can alternate between transcriptionally repressive/structurally condensed and transcriptionally active/structurally accessible states, influencing gene expressions directly. The changes in chromatin architecture are often evoked by protein complexes that carry out covalent post-translational modifications of histones. The amino- and carboxy-terminal tails of histones H3, H4, H2A, and H2B are particularly susceptible to a variety of post-translational modifications, such as phosphorylation, acetylation, methylation, ubiquitylation, sumoylation, ADP-ribosylation, and glycosylation [11].
Given the indispensable functions of histone modifications in transcription, it is reasonable to speculate that histone modifying complexes play an important role in CNS development. However, a major gap still exists in the mechanistic understanding of how these histone modifiers cooperate with the neural-specific transcription factors in orchestrating CNS development. Here, we aim to briefly review the recent progress on the role of histone modifying complexes in neurogenesis and neural cell-fate specification, particularly focusing on the modifiers controlling histone acetylation and methylation. We also attempt to highlight important issues to be resolved in the future.
Histone acetylation and deacetylation
Many lysine residues in histone tails of H3, H4, H2A and H2B are subject to acetylation, which decreases the interaction of the positively charged histone tails with the negatively charged phosphate backbone of DNA. This results in relaxation of the higher order of chromatin [11] (Fig. 1A). Thus, upon acetylation of histone tails, DNA transiently becomes accessible to transcription factors and the RNA pol II complex, facilitating transcription. Conversely, histone deacetylation leads to transcriptionally inactive chromatin structures by packaging the DNA into condensed chromatin. Histone acetylation is conducted by histone acetyltransferases (HATs), a group of enzymes that add an acetyl group to histones, including CBP (CREB-binding protein), p300, GCN5, PCAF (p300/CBP-associating factor), Tip60, and MOF (males absent on the first) [12]. Histone deacetylation is catalyzed by histone deacetylases (HDACs), which are grouped into four classes: class I (HDAC1, 2, 3 and 8), class II (HDAC4, 5, 6, 7, 9, and 10), class III (Sirt1-7), and class IV (HDAC11) [13]. Intriguingly, a recent study revealed that both HATs and HDACs associate with transcriptionally active genes occupied by RNA pol II, indicating that HATs and HDACs control transcription dynamically by adding or removing acetyl groups to/from target histones, respectively, thereby allowing an adequate level of transcription [14].
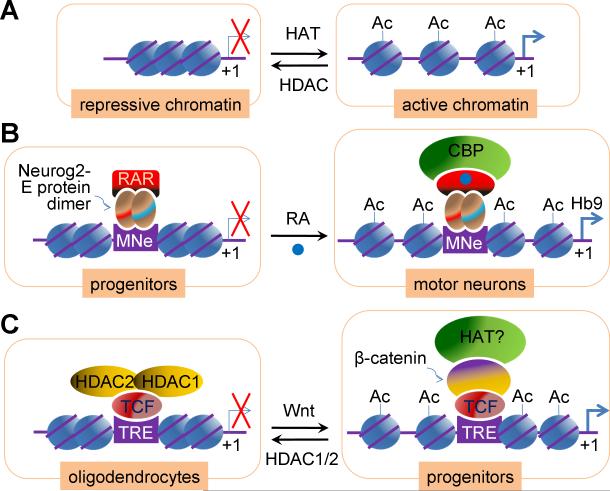
Figure 1.
Regulation of neural cell fate specification by histone acetylation and deacetylation.
(A) Histone acetylation by histone acetyltransferases (HATs) leads to the relaxation of chromatin, enabling transcription factors and RNA pol II complex to bind DNA and activate transcription. Conversely, histone deacetylation catalyzed by histone deacetylases (HDACs) results in transcriptionally inactive condensed chromatin, repressing gene expression.
(B) CBP is recruited to a motor neuron enhancer (MNe) of a motor neuron-specific gene Hb9 by a complex of Neurog2, a proneural bHLH factor, and RAR (RA receptor), when RA (retinoic acid) signal arrives. This induces histone acetylation of Hb9 gene and Hb9 expression, promoting motor neuron specification.
(C) HDAC1/2 play a key role in oligodendrocyte differentiation by associating with TCF7L2 (TCF), which binds TCF-responsive element (TRE), and repressing target genes of TCF7L2. β-catenin competes with HDAC1/2 for binding TCF7L2, inhibiting oligodendrocyte differentiation.
Mouse mutants deficient in several HAT genes, including CBP, p300 and GCN5, exhibited severe neural tube closure defects, including excencephaly [15-17]. Recently we found that the CNS-specific deletion of CBP and p300 results in neural tube closure defects [18] (S. Lee and S. Lee, unpublished results), indicating the essential role of CBP and p300 in the embryonic NPCs. Accumulating evidence suggest that CBP and p300 are critical components of specific networks of multiple transcription factors and extrinsic signals in directing neural cell-fates during development. First, CBP is required for the specification of motor neuron fate in the developing spinal cord [18]. In this context, the complex of Neurog2 (Neurogenin 2, Ngn2) and retinoic acid receptor (RAR) recruits CBP to motor neuron genes when retinoid signal arrives (Fig. 1B). This triggers histone acetylation in motor neuron genes, subsequently activating their expression. Second, p300 mediates the synergistic signaling of LIF (leukemia inhibitory factor) and BMP2 (bone morphogenetic protein 2) to trigger the differentiation of NPCs into astrocytes [19]. p300 bridges the formation of a complex between STAT3 and Smad1, downstream transcription factors of LIF and BMP2, respectively, when both signaling pathways are activated. Third, CBP enables Neurog1 (Neurogenin 1, Ngn1) to inhibit astrocyte differentiation [20]. CBP is a common coactivator for Neurog1-mediated neurogenic pathway and STAT3-mediated astrogenic pathway. When Neurog1 is highly expressed it prevents STAT3 from activating astrocytic genes, because Neurog1 sequesters the CBP-Smad1 transcription complex away from STAT3. These reports indicate that CBP and p300 regulate differentiation of NPCs by mediating the crosstalks among extrinsic signals and transcription factors.
Several reports show that increasing histone acetylation with class I and II HDAC inhibitors, such as valproic acid (VPA) and trichostatin A (TSA), profoundly affects the proliferation and differentiation of NPCs in the embryonic and adult brain. First, VPA triggers neuronal differentiation of adult hippocampal NPCs by inducing proneural transcription factors, such as Neurog1, Atoh1 (Math1), and NeuroD, while inhibiting NPC differentiation into astrocyte and oligodendrocyte lineages [21]. Notably, the VPA-triggered neuronal differentiation is accompanied by induction of histone H4 acetylation in the Neurog1 promoter [22]. Second, in developing mouse embryos, HDAC inhibition by TSA modestly increased neurogenesis in the cortex, but led to a dramatic reduction in neurogenesis accompanied by an increase in the production of immature astrocytes in the ganglionic eminences [23]. Third, perinatal HDAC inhibition in rodent brain resulted in delayed differentiation of oligodendrocytes and significant hypomyelination [24]. These studies with HDAC inhibitors suggest that the level of histone acetylation is tightly regulated by HDACs for the timely activation and/or inactivation of neurogenic and gliogenic programs during CNS development. How does HDAC inhibition result in pleiotropic outcomes in proliferation and differentiation of NPCs depending on the developmental timing and context? Considering that HDAC inhibitors, such as VPA and TSA, suppress the activity of many HDACs, it is possible that each HDAC has unique functions in developing CNS and HDAC inhibitors block the activity of different groups of HDACs depending on the timing and cellular context of treatment. Genetic analyses to inactivate each HDAC gene in specific neural cell-types would provide better insights into the role of an individual HDAC in neural cell-fate determination.
Loss-of-function studies in mice begun to uncover the role of HDAC1 and HDAC2 in neural development. First, CNS-specific deletion of both Hdac1 and Hdac2 results hippocampal abnormalities, absence of cerebellar foliation, disorganization of cortical neurons, accompanied by reduced neuronal differentiation and increased cell death [25]. Second, removal of Hdac1 and Hdac2 in oligodendrocyte lineage cells abolished oligodendrocyte differentiation in the brain and spinal cord [26]. In both cases, a single gene deletion of either Hdac1 or Hdac2 did not cause any apparent phenotype. These reports demonstrate critical but redundant functions of HDAC1 and HDAC2 in neuronal and oligodendrocyte differentiation.
What are the molecular mechanisms by which HDACs control the neurogenic and gliogenic genes? Recent studies have provided evidence that HDACs participate actively in networks of transcription factors and extrinsic signals in developing CNS, as was the case with CBP and p300. First, HDAC1/2 are recruited to TCF7L2 and mediate TCF7L2-dependent transcriptional repression, which promotes oligodendrocyte differentiation (Fig. 1C). Interestingly, HDAC1/2 compete with β-catenin for binding TCF7L2, thereby suppressing Wnt/β-catenin signaling, an inhibitory pathway for oligodendrocyte differentiation (Fig. 1C). This provides the mechanisms by which HDAC1/2 promote oligodendrocyte differentiation. Second, S-nitrosylation of HDAC2 is induced by BDNF (brain-derived neurotrophic factor) and regulates dendritic growth and branching in embryonic cortical neurons [27]. S-nitrosylation of HDAC2 triggers release of HDAC2 from chromatin, permitting acetylation of histones in neurotrophin-dependent genes and facilitating their transcription. Third, HDAC3 and HDAC5 interact with TLX, an important regulator of NPC proliferation and self-renewal in the adult brain, and suppress TLX-target genes, such as p21cip1 and pten, in adult NPCs [28]. Finally, Sirt1, a class III NAD+-dependent HDAC, also controls differentiation of NPCs in response to intrinsic and extrinsic signals. In embryonic NPCs, Sirt1 is localized in the cytoplasm. Upon the arrival of the differentiation signal, Sirt1 becomes transiently translocated to the nucleus, where Sirt1 binds and suppresses the promoter of Hes1 [29]. By antagonizing Notch1-mediated activation of Hes1, Sirt1 facilitates neuronal differentiation. In addition, Sirt1 has also shown to function as a sensor of the intracellular redox state of NPCs, and to regulate proliferation and differentiation of NPCs as a co-repressor of Hes1, an inhibitor of neurogenesis [30].
Collectively, these reports demonstrate that HATs and HDACs are involved in neurogenesis and neural cell-fate specification as integrators of multiple extrinsic signals and intrinsic transcription factors.
Histone methylation and demethylation
Methylation occurs on multiple lysine (K) and arginine (R) residues of histones H3 and H4, including histone H3-R2, K4, K9, R17, R26, K27, and K36 as well as histone H4-R3 and K20 [11]. Histone methylation is linked to both transcriptional activation and repression, and can occur combinatorially with each other or with other types of histone modifications, providing a tremendous degree of complexity in chromatin configuration. Although histone methylation was once treated as stable ‘marks’ of chromatin, recent discovery of histone demethylases (HDMs) changed this traditional view and suggested that histone methylation can be reversibly and dynamically regulated.
Among the numerous types of histone methylation, we will mainly discuss methylation of histone H3-K4, K9 and K27 to illustrate the general theme for the role of histone methylation in neural development (Fig. 2A). High levels of trimethylation of H3-K4 (H3K4me3) are associated with virtually all active genes and concomitantly occur with RNA pol II occupancy and histone acetylation [31]. In contrast, methylation in H3-K9 and H3-K27 are hallmarks of a condensed/transcriptionally repressed chromatin state [11]. H3-K4 methylation is performed by a group of HMTs that contain the catalytic SET domain, including Mixed Lineage Leukemia 1 (MLL1), MLL2, MLL3/HALR, MLL4/ALR, SET1A, and SET1B [31]. The MLL/SET1-family HMTs exist in multiprotein complexes that are related to Drosophila Trithorax complex, and share common subunits, such as WDR5, RBBP5 and ASH2. H3-K27 methylation is catalyzed by the polycomb repressive complex 2 (PRC2) that consists of Eed, Suz12 (Suppressor of zeste 12), RbAp46/48 and Ezh2, a catalytic component [32]. The HMTs for H3-K9 residue include G9a, SUV39H1 and SUV39H2 [11]. Several enzymes that erase histone methylation marks have also been discovered [33,34]. Lysine specific deaminase 1 (LSD1) mediates demethylation of mono- and dimethylated H3-K4, and form a complex with a corepressor CoREST (corepressor of REST). More recently, a family of the Jumonji (Jmj) domain-containing proteins was found to function as HDMs. The X-linked mental retardation gene SMCX/JARID1C reverses H3K4me3 to di- and monomethylated H3-K4, whereas UTX and JMJD3 demethylate H3K27me3.
Figure 2.
Regulation of neural cell fate specification by histone methylation and demethylation.
(A) Trimethylation (3me, red dots) of H3-K9 and H3-K27 are hallmarks of a condensed/transcriptionally repressed chromatin state, whereas trimethylation of H3-K4 (H3K4me3) marks transcriptionally active chromatin.
(B) In neural progenitors, the promoter of GFAP, an astrocytes-specific marker gene, exhibits high level of H3K9me3, transcriptionally repressive chromatin mark. FGF2 signal removes H3K9me3 and triggers H3K4me3, transcriptionally active chromatin mark, on the GFAP promoter. This facilitates access of the STAT-CBP complex to the STAT-response element (SRE) in the GFAP promoter upon CNTF signal, leading to efficient differentiation of neural progenitors to astrocytes.
(C) MLL1, a H3-K4 methyltransferase, is recruited to the promoter of Dlx2 by an unknown transcription factor (X), and promotes neuronal differentiation as well as Dlx2 expression. In the absence of MLL1, H3K27me3 levels on Dlx2 gene were markedly increased, suppressing Dlx2 expression and neuronal differentiation. The identity of H3-K27 demethylases (H3K27DMs) involved in the removal of H3K27me3 on Dlx2 gene remains unclear.
(D) During the neurogenic-to-astrogenic fate switch of neural progenitors, the chromatin status of the Neurog1 promoter changes from the acetylated open chromatin to the transcriptionally repressive chromatin marked by H3K27me3. These changes are mediated by Polycomb repressor complexes (PRCs).
The presence of multiple HMTs and HDMs decorating distinct residues of histones led to the prediction that combinatorial action of HMTs and HDMs at specific loci results in the coordinated modifications of histone tails, leading to either transcriptional activation or repression. Indeed, UTX, a HDM removing H3K27me3 repressive mark, forms a complex with MLL3 and MLL4 that trigger H3K4me3 [35]. In addition, the H3K4me3 demethylase Rbp2/JARID1A associates with the PRC2 complex containing a H3K27me3 methyltransferase [36]. Similarly, the H3K4me3 demethylase SMCX assembles a complex with the H3K9 methyltransferase G9a and HDAC1/2, and binds H3K9me3 [37,38]. This likely results in efficient transcriptional repression through concomitant execution of H3-K4 demethylation, H3-K9 methylation, and histone deacetylation. Together, these reports strongly suggest a synchronized action of multiple histone modifying enzymes to establish either transcriptionally active or inactive chromatin.
It is noteworthy that in Drosophila, the Trithorax and polycomb complexes are well studied as epigenetic activator and silencer of the homeotic (Hox) gene clusters, respectively [39]. This implies an antagonistic relationship between H3K4me3 and H3K27me3 in development. In vertebrates, imbalance between H3-K4 methylation and H3-K27 methylation caused by either removal or overexpression of MLLs, WDR5, JMJD3, UTX, or Ezh2 results in mis-regulation of Hox genes and various developmental defects, indicating that the role of MLLs and PRC in regulating Hox genes is well conserved evolutionarily [32,39-42].
An important question to be resolved is whether the histone methylation/demethylation complexes play roles in vertebrate CNS development. During differentiation of embryonic NPCs, the promoter of GFAP, an astrocytes-specific marker gene, undergoes drastic changes in histone lysine methylations [43] (Fig. 2B). When NPCs differentiate into astrocytes, originally high levels of H3K9me3 on the GFAP promoter of NPCs decrease, whereas H3K4me3 levels increase. Interestingly, FGF2 (fibroblast growth factor 2) triggers H3K4me3 and suppresses H3K9me3 on the GFAP promoter. This facilitates access of the STAT-CBP complex to the GFAP promoter when CNTF (ciliary neurotrophic factor) signal arrives. Consistent with these chromatin changes, FGF2 synergizes with CNTF to induce astrocyte differentiation. This study suggests that extrinsic signals regulate histone methylation status in cell-type specific genes, thereby controlling developmental competence of neural progenitors. While the identity of HMTs and HDMs that respond to FGF2 is yet to be discovered, recent loss- and gain-of-function studies uncovered several HMTs and HDMs involved in neural development. First, MLL1-deficient NPCs exhibit specific impairment of neuronal differentiation in the subventricular zone of brain [44], suggesting a requirement of MLL1 in neurogenesis in the mouse postnatal brain. This phenotype is attributed to the markedly reduced expression of Dlx2, a homeodomain-containing transcription factor whose promoter recruits MLL1 specifically (Fig. 2C). Interestingly, in the absence of H3-K4 methylating MLL1, H3K27me3 levels in Dlx2 gene were strongly increased, although H3K4me3 levels did not decrease (Fig. 2C). This indicates that a complex relationship between HTMs and HDMs exists to coordinate chromatin modifications at specific loci to promote neurogenesis. Second, deletion of Ptip (pax transcription activation domain interacting protein), a component of MLL3/4 complexes, in developing CNS results in a marked reduction of H3K4me3 level in the embryonic spinal cord [45]. This suggests that MLL3/4/PTIP complexes are important in establishing and/or maintaining the level of H3K4me3 in the neural tube. Third, the expression of JMJD3, a H3K27 demethylase, is induced by retinoid signal during neuronal differentiation of embryonic NPCs [46], as well as during neural lineage commitment of embryonic stem cells (ES cells) [47]. Overexpression of JMJD3 in NPCs stimulates expression of several neuronal genes, including Dlx5, Gad1/2 and Dcx, in a catalytic domain-dependent manner [46]. On the other hand, knockdown of JMJD3 suppresses neural lineage differentiation of ES cells [47]. These suggest that JMJD3 plays a key role in removing the H3K27me3 repressive mark in neuronal genes during neurogenesis. Fourth, SMCX, a H3K4me3 demethylase, suppresses neuronal genes by associating with REST (RE1-silencing transcription factor) [37]. SMCX has also been reported to play important roles in neuronal survival during zebrafish development and in dendritic morphogenesis of cerebellar granular neurons [38]. Fifth, PRC proteins have emerged as key players in the self-renewal of stem cell populations [48-50]. In ES cells, genome wide profiling techniques to map PRC target genes revealed that PRC proteins occupy and directly repress a large cohort of neuro-developmental regulators, which become de-repressed during neural lineage commitment [51-54]. During ES cell differentiation into neural lineages, PRC proteins and H3K27me3 repressive marks decrease at PRC-target neural-specific genes [47,51]. Sixth, PRC proteins promote the transition of NPC fate from neurogenic to astrogenic in the developing neocortex [55] (Fig. 2D). During the neurogenic-to-astrogenic switch of NPCs, histone acetylation of the Neurog1 promoter declines while H3K27me3 level elevates. This induction of the transcriptionally repressive chromatin in the Neurog1 promoter leads to the suppression of Neurog1 expression. PRC proteins, Ring 1B, Eed, and Ezh2, appear to mediate these chromatin changes and repression of Neurog1 in late stage NPCs. Finally, Ezh2 is needed for the efficient differentiation of mouse embryonic NPCs to oligodendrocytes in vitro [56]. High levels of Ezh2 expression in NPCs are maintained in the oligodendrocyte lineage during in vitro differentiation of embryonic NPCs, whereas Ezh2 becomes reduced during neuronal and astrocytic differentiation, further supporting the role of Ezh2 in oligodendrocyte differentiation.
Together, the aforementioned reports strongly suggest that HMTs and HDMs are required for the proper development of the nervous system and regulate specific cohort of genes depending on developmental timing and cellular context. This raises the important question of what controls the targeting of HMTs and HDMs to particular genomic loci. The report that FGF2 influences histone modification of the astrocyte-specific promoter [43] (Fig. 2B) points to the possibility that extrinsic signals control the recruitment of HMTs and HDMs to their target genes. In addition, given that HMTs and HDMs exist as multiprotein complexes, one or more components of each complex may determine the recruitment of HMTs and HDMs to target loci. Perhaps the best example is the binding of LSD1, G9a, and SMCX to RE-1 sites (restriction element-1) by forming a complex with REST and/or CoREST [37,57].
Conclusions and Future Perspectives
Changes in chromatin structure are expected to be important for coordinately activating and repressing large cohorts of genes according to intrinsic cellular programs and extrinsic signals during CNS development. Recent identification of many histone modifying enzymes led to the realization that these enzymes would function as key components of gene regulatory networks for neural cell-type specification. An important issue to be addressed is to what extent histone modifying enzymes are involved in regulating neural cell-fate decision; i.e. do they play instructive roles to dictate cell-type specific patterns of gene expression or permissive roles to enable instructive factors to establish cell-type specific gene expression? Currently, most evidence support the permissive roles of histone modifying enzymes for cell-type determination in that they mediate the actions of instructive transcription factors in making a decision on NPC proliferation and differentiation. Given that histone modifying enzymes are much fewer than neural cell-type specific transcription factors and are expressed relatively widely, histone modifying enzymes are likely to be shared by multiple transcription factors in many cell-types. This raises numerous questions for the future studies. First, how do histone modifying enzymes find appropriate target genes in a given developmental context? We speculate that emerging networks of extrinsic signals and intrinsic transcription factors orchestrate the recruitment of histone modifying complexes to specific sets of target genes, conferring the cell-type specific actions on histone modifying complexes. The identity of relevant histone modifying complexes that each instructive transcription factor associates with is largely unexplored. Also, how extrinsic signals control the function of histone modifying complexes remains to be determined. The extrinsic signals may modify the histone modifying enzymes directly or affect the association between the histone modifying enzymes and transcription factors. Carefully-designed investigation with combined genetic and biochemical tools is needed to address these questions. Second, a better understanding of the role of chromatin modifiers for neural cell-type specification will require comprehensive analyses of histone modifications in genomic loci over the course of neural development. One major challenge to this effort is the fact that the developing CNS consists of heterogeneous cell populations under various developmental stages. Thus, it is difficult to determine the dynamic changes of histone modifications in a specific cell lineage from the CNS. ES cells might serve as a useful model to circumvent this difficulty, because the differentiation of ES cells can be directed into specific neural cell-types in a relatively synchronous pattern, and the differentiation of ES cells has been shown to mimic embryonic development [58]. Genome-wide profiling studies, such as chromatin immunoprecipitation-sequencing (ChIP-Seq) analyses with multiple modified histone-specific antibodies, would reveal global chromatin changes in histone modifications over the course of neural cell differentiation, yielding novel insights into the relationship between histone modifications and neural cell-type specification. Studies in ES cells to define chromatin changes and the histone modifiers mediating such changes will need to be recapitulated in vivo during development. Finally, given the presence of multiple histone modifying enzymes that make distinct marks on histones, it would be interesting to investigate how their actions are coordinated to either promote or suppress transcription of neural genes synchronously.
Acknowledgements
We apologize to all the researchers whose important contributions could not be acknowledged due to space constraints. We thank Joanna Asprer and Jae W. Lee for stimulating discussions and comments on this manuscript. This work was supported by NIH/NINDS (R01 NS054941), PEW Scholars Program, and March of Dimes Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 2.Guillemot F. Cellular and molecular control of neurogenesis in the mammalian telencephalon. Curr Opin Cell Biol. 2005;17:639–647. doi: 10.1016/j.ceb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Wegner M, Stolt CC. From stem cells to neurons and glia: a Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 4.Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- 5.Hobert O, Westphal H. Functions of LIM-homeobox genes. Trends Genet. 2000;16:75–83. doi: 10.1016/s0168-9525(99)01883-1. [DOI] [PubMed] [Google Scholar]
- 6.Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Supp 1):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- 7.Guillemot F. Cell fate specification in the mammalian telencephalon. Prog Neurobiol. 2007;83:37–52. doi: 10.1016/j.pneurobio.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 9.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Luger K. Structure and dynamic behavior of nucleosomes. Curr Opin Genet Dev. 2003;13:127–135. doi: 10.1016/s0959-437x(03)00026-1. [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 12.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanaka Y, Naruse I, Hongo T, Xu M, Nakahata T, Maekawa T, Ishii S. Extensive brain hemorrhage and embryonic lethality in a mouse null mutant of CREB-binding protein. Mech Dev. 2000;95:133–145. doi: 10.1016/s0925-4773(00)00360-9. [DOI] [PubMed] [Google Scholar]
- 16.Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R. Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 17.Bu P, Evrard YA, Lozano G, Dent SY. Loss of Gcn5 acetyltransferase activity leads to neural tube closure defects and exencephaly in mouse embryos. Mol Cell Biol. 2007;27:3405–3416. doi: 10.1128/MCB.00066-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18••.Lee S, Lee B, Lee JW, Lee SK. Retinoid signaling and neurogenin2 function are coupled for the specification of spinal motor neurons through a chromatin modifier CBP. Neuron. 2009;62:641–654. doi: 10.1016/j.neuron.2009.04.025. [This paper shows that the histone acetyltransferase CBP is a key player to integrate the extrinsic retinoid signaling and the cell intrinsic pronerual bHLH factor Neurog2 during neurogenesis and motor neuron specification. Furthermore, the paper provides the mechanisms by which CBP establishes the transcriptionally active chromatin in motor neuron genes in response to retinoid signal.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Nadal-Vicens M, Misono S, Lin MZ, Zubiaga A, Hua X, Fan G, Greenberg ME. Neurogenin promotes neurogenesis and inhibits glial differentiation by independent mechanisms. Cell. 2001;104:365–376. doi: 10.1016/s0092-8674(01)00224-0. [DOI] [PubMed] [Google Scholar]
- 21.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu IT, Park JY, Kim SH, Lee JS, Kim YS, Son H. Valproic acid promotes neuronal differentiation by induction of proneural factors in association with H4 acetylation. Neuropharmacology. 2009;56:473–480. doi: 10.1016/j.neuropharm.2008.09.019. [DOI] [PubMed] [Google Scholar]
- 23.Shaked M, Weissmuller K, Svoboda H, Hortschansky P, Nishino N, Wolfl S, Tucker KL. Histone deacetylases control neurogenesis in embryonic brain by inhibition of BMP2/4 signaling. PLoS One. 2008;3:e2668. doi: 10.1371/journal.pone.0002668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone deacetylases 1 and 2 control the progression of neural precursors to neurons during brain development. Proc Natl Acad Sci U S A. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26••.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, Bu H, Hu T, Taketo MM, van Es JH, Clevers H, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [This paper describes a novel crosstalk between HDAC1/2 and the canonical Wnt signaling pathway mediated by TCF7L2 during oligodendrocyte differentiation. HDAC1/2 facilitate oligodendrocyte formation by competing with •-catenin for TCF7L2 interaction. This is a good example for the selective developmental function for chromatin modifiers.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S-Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- 28.Sun G, Yu RT, Evans RM, Shi Y. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc Natl Acad Sci U S A. 2007;104:15282–15287. doi: 10.1073/pnas.0704089104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hisahara S, Chiba S, Matsumoto H, Tanno M, Yagi H, Shimohama S, Sato M, Horio Y. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A. 2008;105:15599–15604. doi: 10.1073/pnas.0800612105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prozorovski T, Schulze-Topphoff U, Glumm R, Baumgart J, Schroter F, Ninnemann O, Siegert E, Bendix I, Brustle O, Nitsch R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 31.Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 32.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 33.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007;8:829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 34.Shi Y, Whetstine JR. Dynamic regulation of histone lysine methylation by demethylases. Mol Cell. 2007;25:1–14. doi: 10.1016/j.molcel.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Roeder RG, Lee J. Progress in Molecular Biology and Translational Science. Vol. 87. Academic Press; 2009. Roles of Histone H3 - Lysine 4 methyltransferase complexes in NR-mediated gene transcription. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37•.Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [This paper describes an identification of a complex containing the H3K4 demethylase SMCX, HDAC1/2, G9a and REST, which plays critical roles in REST-mediated neuronal gene regulation.] [DOI] [PubMed] [Google Scholar]
- 38••.Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [The authors demonstrate that SMCX is a H3K4me3 demethylase and specifically binds to H3K9me3, suggesting the coordinated changes in H3K4 and H3K9 methylation. Moreover, they show the roles of SMCX in zebrafish and mammalian neurons.] [DOI] [PubMed] [Google Scholar]
- 39.Kiefer JC. Epigenetics in development. Dev Dyn. 2007;236:1144–1156. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- 40.Wysocka J, Swigut T, Milne TA, Dou Y, Zhang X, Burlingame AL, Roeder RG, Brivanlou AH, Allis CD. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121:859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 41.Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 42.Lan F, Bayliss PE, Rinn JL, Whetstine JR, Wang JK, Chen S, Iwase S, Alpatov R, Issaeva I, Canaani E, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 43.Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- 44••.Lim DA, Huang YC, Swigut T, Mirick AL, Garcia-Verdugo JM, Wysocka J, Ernst P, Alvarez-Buylla A. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458:529–533. doi: 10.1038/nature07726. [By analyzing CNS-specific Mll1-mutant mice, the authors show that MLL1 promotes neurogenesis in the mouse postnatal brain, at least in part, by regulating Dlx2 expression. In Mll1-deficient NPCs, the Dlx2 gene is marked by both H3K4me3 and H3K27me3 and Dlx2 expression is suppressed.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patel SR, Kim D, Levitan I, Dressler GR. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell. 2007;13:580–592. doi: 10.1016/j.devcel.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46•.Jepsen K, Solum D, Zhou T, McEvilly RJ, Kim HJ, Glass CK, Hermanson O, Rosenfeld MG. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [In this paper, JMJD3, a H3K27me3 demethylase, is identified as a direct target of retinoid acid receptor. This paper further shows that JMJD3 induces neuronal gene expression and binds to the promtoer of the neuronal gene Dlx5.] [DOI] [PubMed] [Google Scholar]
- 47.Burgold T, Spreafico F, De Santa F, Totaro MG, Prosperini E, Natoli G, Testa G. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3:e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 49.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 50.Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 51.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 52.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ringrose L. Polycomb comes of age: genome-wide profiling of target sites. Curr Opin Cell Biol. 2007;19:290–297. doi: 10.1016/j.ceb.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 55••.Hirabayashi Y, Suzki N, Tsuboi M, Endo TA, Toyoda T, Shinga J, Koseki H, Vidal V, Gotoh Y. Polycomb limits the neurogenic competence of neural precursor cells to promote astrogenic fate transition. Neuron. 2009;63:600–613. doi: 10.1016/j.neuron.2009.08.021. [The authors demonstrate that PRC proteins suppress neurogenic competence of NPCs by inhibiting Neurog1 expression in late stage NPCs. Thus, PRC promotes the neurogenic-to-astrogenic switch of NPC fate, limiting the duration of the neurogenic phase of NPCs.] [DOI] [PubMed] [Google Scholar]
- 56.Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells. 2008;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- 57.Ballas N, Mandel G. The many faces of REST oversee epigenetic programming of neuronal genes. Curr Opin Neurobiol. 2005;15:500–506. doi: 10.1016/j.conb.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 58.Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell. 2008;132:661–680. doi: 10.1016/j.cell.2008.02.008. [DOI] [PubMed] [Google Scholar]