Abstract
Evidence for genetic linkage to alcohol and other substance dependence phenotypes in areas of the human and mouse genome have now been reported with some consistency across studies. However, the question remains as to whether the genes that underlie the alcohol-related behaviors seen in mice are the same as those that underlie the behaviors observed in human alcoholics. The aims of the current set of analyses were to identify a small set of alcohol-related phenotypes in humans and in mouse by which to compare quantitative trait locus (QTL) data between the species using syntenic mapping. These analyses identified that QTLs for alcohol consumption and acute and chronic alcohol withdrawal on distal mouse chromosome 1 are syntenic to a region on human chromosome 1q where a number of studies have identified QTLs for alcohol-related phenotypes. Additionally, a QTL on human chromosome 15 for alcohol dependence severity/withdrawal identified in two human studies was found to be largely syntenic with a region on mouse chromosome 9 where two groups have found QTLs for alcohol preference. In both of these cases while the QTLs were found to be syntenic the exact phenotypes between humans and mice did not necessarily overlap. These studies demonstrate how this technique might be useful in the search for genes underlying alcohol-related phenotypes in multiple species. However, these findings also suggest that trying to match exact phenotypes in humans and mice may not be necessary or even optimal for determining whether similar genes influence a range of alcohol-related behaviors between the two species.
Keywords: Alcohol dependence, phenotypes, linkage analyses, genome scan, mouse models of alcoholism, QTL, genetics
1. Introduction
Alcohol dependence is characterized by the emergence of a set of specific cognitive, physiological, and behavioral phenomena in which the use of alcohol eventually takes on greater importance than other salient activities and often results in physical and psychological harm (Ehlers and Chester, 2009). There is consistent evidence from family, twin, and adoption studies for the existence of a genetic contribution to the etiology of alcohol dependence (see Cotton, 1979; McGue, 1994, 1999; Heath et al., 1997). The individual differences seen in risk for alcohol dependence are almost certainly due to the combined effects of many genes, each exerting a specific effect on protection from and risk for the disorder, as well as gene-gene and gene-environment interactions (see Dick et al., 2006a; Kohnke, 2008). Over the last decade there have been several large studies in which investigators have identified families with alcoholism, collected DNA and family relationships, and conducted linkage and association analyses with the goal of identifying the genes associated with alcohol dependence (see Long et al., 1998; Reich et al., 1998; Ehlers et al., 2004; Hill et al., 2004; Vieten et al., 2004; Wilhelmsen et al., 2005; Dick and Bierut, 2006; Edenberg and Foroud, 2006; Prescott et al., 2006).
One of the most consistent findings among the several genetic linkage studies is evidence for a protective association for alcohol dependence and related behaviors on chromosome 4q near the alcohol dehydrogenase (ADH) gene cluster. In one genetic linkage study evaluating large families who were members of a Southwest Indian tribe, three loci near the ADH gene cluster showed evidence for linkage (Long et al., 1998). Additionally, genome screens for both the “unaffected by alcoholism” (Reich et al., 1998) and “maximum drinks ever consumed in a 24 hour period” (Saccone et al., 2001) phenotypes, provided evidence for linkage on chromosome 4 in the Collaborative Study of the Genetics of Alcoholism (COGA). Linkage at this site to various alcohol-related phenotypes has been confirmed by a genome screen in Mission Indian families (Ehlers et al., 2004), in Irish sib-pairs (Prescott et al., 2006), and in a population ascertained for smoking behaviors (Wilhelmsen et al., 2005). Linkage to this chromosome 4 site has also been found for use of tobacco phenotypes (Duggirala et al., 1999; Straub et al., 1999; Ehlers and Wilhelmsen, 2006).
Evidence for genetic linkage to alcohol and other substance dependence in other areas of the human genome has also been reported with some consistency across studies. At least five studies have reported evidence for linkage of substance dependence to a broad area on distal chromosome 1 (Reich et al., 1998; Foroud et al., 2000; Nurnberger et al., 2001; Dick et al., 2002; Hill et al., 2004; Lappalainen et al., 2004; Guerrini et al., 2005; Ehlers and Wilhelmsen, 2006). A site on chromosome 4 near a GABAA receptor gene cluster was first identified as being associated with alcohol dependence by Long et al. (1998) in a southwest Indian tribe, and also by COGA (Reich et al., 1998). This chromosomal region, which contains genes coding for the GABAA receptor subunits GABRG1, GABRA2, GABRA4, and GABRB1, was also linked to an electrophysiological phenotype (EEG beta activity) that characterizes individuals at risk for alcoholism in the COGA study (Porjesz et al., 2002), heavy drinking in the Framingham study (Wyszynski et al., 2003), cannabis dependence in the nicotine addiction genetics project (Agrawal et al., 2008), and haplotypes of “addiction phenotypes” in substance abusers (Drgon et al., 2006). Additional sites in the genome where there have been duplications of findings for linkage to alcohol and other substance dependence phenotypes within a broad support interval can be found on chromosome 2 (Reich et al., 1998; Dick et al., 2004; Ehlers et al., 2008), chromosome 5 (Hill et al., 2004; Ehlers and Wilhelmsen, 2005; Gelernter et al., 2006), chromosome 6 (Cantor and Lanning, 1999; Hill et al., 2004; Swan et al., 2006; Ehlers and Wilhelmsen, 2007), chromosome 15 (Dick et al., 2002; Ehlers et al., 2004), and chromosome 16 (Foroud et al., 1998, Ehlers et al., 2004). Strong evidence for a site on chromosome 7 has also been found by the COGA study; however, it has not as yet been duplicated by other studies (see Saccone et al., 2005; Dick et al., 2008).
The complexity of finding the genes for alcohol dependence derives both from its multigenic and polygenic character but also from the fact that alcohol dependence is co-morbid with other psychiatric disorders and other drug dependencies. Additionally, a host of environmental factors influence the development of alcoholism, and genetic and environmental risk factors may interact substantially (see Schuckit, 1995; Johnson et al., 1998; Goldman et al., 2005). Animal models of alcoholism have an advantage in that they allow for the control of a number of characteristics of the animal's genetic background, prior drug exposure, and to a large extent, the environment (for recent reviews see Rodd et al., 2004; Lovinger and Crabbe, 2005; Bell et al., 2006, Bennett et al 2006; Sanchis-Segura and Spanagel, 2006; Treadwell, 2006). Invertebrates such as C. elegans (Crowder, 2004) and D. melanogaster (Heberlein, 2000) have been used and been found to be informative. Notably, genetic selection studies have established high drinking lines of mice and rats (see McBride and Li, 1998; Bell et al., 2006; Green and Grahame, 2008), and some models have also been developed in non-human primates (see Grant and Bennett, 2003; Barr and Goldman, 2006).
Mice have some distinct advantages for use as animal models in alcohol research, particularly in genetic analyses. Over 90% of the mouse and human genomes can be partitioned into corresponding regions of conserved synteny, reflecting segments in which the gene order in the most recent common ancestor has been conserved in both species. The proportion of mouse genes without any homologue currently detectable in the human genome (and vice versa) seems to be less than 1% (Waterston et al., 2002). The first mouse genome that was fully sequenced was the C57BL/6J (B6) inbred strain, for which the physical map is readily available (http://www.ensembl.org/Mus_musculus/). Currently an additional 17 key mouse strains are being sequenced in their entirety by the Sanger Center (http://www.sanger.ac.uk/modelorgs/mousegenomes/), and all single nucleotide polymorphism (SNP) calls are publically available. For example, the QTL region of mouse distal chromosome 1 was completely sequenced for the B6 and DBA/2J (D2) strains commonly used to study alcohol related phenotypes (Walter et al., 2009). In a 3 megabase (Mb) region surveyed, 11,824 SNPs were identified between just these two strains. Additionally, mice can be genetically manipulated by knockout, knockin, RNAi, transgenic, and mutagenic technologies more easily than other mammalian species (see Bennett et al., 2006). Disadvantages of the use of mice, and other rodents, are that they cannot model the entire psychobiological diagnostic construct of alcoholism seen in humans (Lovinger and Crabbe, 2005). However, what has been developed are genetic models that robustly interrogate key features of alcoholism including preferential drinking of alcohol vs. water, conditioned preference for (or avoidance of) locations or flavors conditioned by ethanol, sensitivity and neuroadaption (sensitization and tolerance) to ethanol's stimulating and depressant effects, and the severity of alcohol physiological dependence and associated withdrawal following acute or chronic alcohol exposure (Crabbe, 2008).
One method that has been used in the search for genes contributing to alcohol-related behaviors in these “partial” models of alcoholism in mice is quantitative trait locus (QTL) analyses. QTL analyses can identify regions of the genome linked to the trait of interest. QTL analyses require one or more informative mapping populations (e.g., recombinant inbred strains [e.g., RI, RIX], F2 or advanced intercrosses, selectively bred lines, heterogeneous stocks) and informative genetic markers. Much of the early QTL mapping data in mice were obtained in panels of recombinant inbred (RI) lines of mice that were derived from crosses between D2 and B6 mice; these inbred strains have been extensively studied and differ in many alcohol-related phenotypes such as: voluntary ethanol consumption and ethanol preference (Phillips et al., 1994; Rodriguez et al., 1995; Melo et al., 1996; Gehle and Erwin, 1998; Tarantino et al., 1998; Fernandez et al., 1999; Whatley et al.,1999; Ruf et al., 2004; Belknap and Atkins, 2001), ethanol-induced locomotor responses (Cunningham, 1995; Demarest et al., 1999), loss of righting reflex (Browman and Crabbe, 2000; Bennett et al., 2008), withdrawal following acute and chronic ethanol exposure (Crabbe et al, 1983, Buck et al., 1997, 2002, Crabbe, 1998), tolerance (Kirstein et al., 2002; Tabakoff et al., 2003), responses conditioned by ethanol injections (Cunningham, 1995; Risinger and Cunningham, 1998), and metabolism (Grisel et al., 2002). Using B6D2-derived populations, significant QTLs for ethanol preference and/or withdrawal have been confirmed on mouse chromosomes 1, 2, 3, 4, 9, 11, and 19. In some cases, the QTLs have been fine-mapped, e.g., using interval-specific congenic animals (Kozell et al., 2008, 2009; Fehr et al., 2002; Shirley et al., 2004). These congenic models possess a short interval of DNA surrounding (or flanking) the QTL that has been introduced from a donor strain onto the distinct genetic background of a recipient background strain. Narrowing the QTL interval to a small region (~1-2 Mb) also substantially reduces the number of potential candidate genes remaining within the finely mapped interval. A data base is available that summarizes these alcohol related QTLs, as well as additional significant (and suggestive) QTLs for a variety of ethanol response traits, and information about the size of the confidence interval surrounding the QTL and/or its definitive limits derived from subsequent analyses with congenic strains from fine mapping studies (http://www.ohsu.edu/parc/data/qtl/by_phen.shtml). QTL fine-mapping has been crucial to reduce the number of potential candidate genes within the QTL interval and, when coupled with detailed molecular analyses of candidate gene allelic variation in sequence and/or expression, has been key to identifying high-quality quantitative trait gene [QTG] candidates. In at least one case, studies have actually identified a QTG for alcohol withdrawal severity, i.e., Mpdz, which encodes the multi-PDZ domain protein (MPDZ, also called MUPP1) (see Shirley et al., 2004).
Using short term selective breeding from a more genetically diverse population (heterogeneous stock [HS] animals) derived from four inbred strains (B6, D2, BALB/cJ, LP/J), Hitzemann et al (2009) recently identified seven significant, coincident QTLs on mouse chromosomes 1 (two), 3, 6, 11, 16 and 17 with apparent reciprocal effects on ethanol consumption and acute ethanol withdrawal severity. Previous work using B6D2 populations detected putative reciprocal QTLs for these two phenotypes on chromosomes 1, 2, 4 and 15 that may contribute to the negative genetic relationship between ethanol consumption and acute withdrawal (Metten et al., 1998). Although the selection from HS and B6D2 QTL data appear discordant, this is not necessarily the case, since a QTL detected in one genotype may be ‘silent’ in another, or the loci may have strong epistatic effects (Hitzemann et al., 2009).
While these studies in mouse models have been successful in identifying regions in the genome associated with alcohol phenotypes of interest, and in some cases have actually identified a QTG or promising QTG candidates, the question remains as to whether the genes that underlie the alcohol-related behaviors seen in mice are the same as those that underlie the behaviors observed in human alcoholics. One procedure that may assist in answering this question is to use mouse QTG data to inform human studies. For example, MPDZ, the human homolog of Mpdz (identified as a QTG for alcohol withdrawal in mice; Shirley et al., 2004), is currently being studied in populations of human alcoholics by NIH-NIAAA intramural scientists with encouraging preliminary results (Dr. David Goldman, personal communication). Additionally, although limited to a small population thus far, Karpyak et al. (2009) recently reported a potential role of MPDZ in alcoholism, though a particular role in the withdrawal syndrome was not apparent. Also, Tabakoff et al. (2009) found that an MPDZ SNP is significantly associated with alcohol consumption based on data from a WHO/ISBRA Montreal study. In some cases a QTG will be the same in mouse and man, while, in other cases, animal models may identify a relevant gene network operating in both species, within which many drugable targets may exist (Hopkins, 2008). Another procedure that may be useful in this regard is to make a direct comparison of QTL data obtained from mouse studies with those from human studies using synteny mapping. Syntenic conservation makes it possible to locate the homologous regions of the genome from mice to particular chromosomal regions in humans, and vice versa.
The present report is part of a larger effort to develop greater consilience between phenotypes for alcohol dependence used in human studies and those used in animal models. The aim of the current set of analyses was to identify a limited set of alcohol-related phenotypes in humans and in mouse by which to compare QTL data between the species using syntenic mapping. The intent of this effort was not meant to be exhaustive but rather to demonstrate how the technique might be applied to a few phenotypes and their QTLs and to identify specific phenotypes in both mice and humans where future studies might improve both data collection and analyses in order to optimize the eventual success of this endeavor.
2. Materials and Methods
2.1 Identification of human phenotypes and QTLs
Most studies that have conducted linkage analyses for alcohol related traits have used DSM based alcohol dependence as the phenotype of interest. However, there have also been genome screens conducted on a number of other alcohol-related traits including: number of dependence symptoms (Ehlers et al., 2004), alcohol withdrawal severity (Ehlers et al., 2004), antisocial or conduct disorder symptoms or traits co-occurring with alcohol dependence (Dick et al., 2004; Kendler et al., 2006; Ehlers et al., 2008), personality factors co-occurring with alcohol dependence (Nurnberger et al., 2001; Dick et al., 2002), maximum drinks in 24 hours (Saccone et al., 2000), craving for alcohol (Ehlers and Wilhelmsen, 2005; Foroud et al., 2007), alcohol and tobacco usage (Ehlers and Wilhelmsen, 2006), latent class analyses of alcohol abuse and other related phenotypes (Foroud et al., 1998), low response to alcohol (Wilhelmsen et al., 2003; Schuckit et al., 2005), and alcohol dependence with EEG correlates (Porjesz et al., 2002). For the purposes of the present set of analyses, we selected areas in the genome linked to any alcohol-related phenotypes where there was convergent evidence from at least two studies for QTL(s) within 50 cM with a relatively high level of statistical confidence in linkage (log of the odds of linkage [LOD] scores greater than 1.5). Initially, regions were identified using data from the following studies: COGA, SW Indian family study, Mission Indian family study, Pittsburgh family study, Irish Sib-Pair study, smoking behavior sample, and the Framingham study.
2.2 Identification of mouse phenotypes and QTLs
Most QTL mapping in mice has employed the B6 x D2-derived (BXD) RI strains. A recently curated alcohol-related data base (http://www.ohsu.edu/parc/data/qtl/by_phen.shtml) was used and the search for mouse QTLs was restricted to these data. Of the many phenotypic domains relevant for rodent models, two have been targeted extensively in genetic studies. The first domain is related to alcohol dependence. Because the acknowledgement of many DSM dependence symptoms cannot be reported by nonhuman animals, the existence of physical dependence is often inferred from the appearance of withdrawal signs when the drug is discontinued (Heilig et al., 2010). Convulsions, seizures, or “running fits” are withdrawal symptoms seen in all species that have been investigated thus far (Metten and Crabbe, 1996). In mice, the handling-induced convulsion (HIC) is an easily quantified sign of withdrawal, characterized initially by Dora Goldstein (Goldstein and Pal, 1971). HIC severity waxes and wanes during the hours of withdrawal, and shows dose- and duration-dependent effects of alcohol exposure. Many studies have explored the genetic bases of withdrawal severity in mice using the HIC as the index of severity, e.g. (Crabbe, 1996; Homanics et al., 1998; Metten and Crabbe, 2005), and thus this study focused on the existing QTLs mapped for withdrawal HIC. The severity of the HIC also increases in severity with repeated cycles of exposure, a phenomenon likened to the “kindling” of severity of successive withdrawal episodes in humans (Becker, 1996). Several other behavioral signs of withdrawal have been documented in rodents, but, as there were no available QTL data, they were not considered for the present set of analyses.
The other general behavioral domain for which there are QTL data in rodents pertains to ethanol's reinforcing effects. The most widely used phenotype in rodents is ethanol preference drinking. Genetic differences in relative preference for ethanol-containing solutions over water have proven to be stable across a variety of laboratories and test conditions for nearly 50 years (Wahlsten et al., 2006). Many lines of rats and mice have been bred for high vs. low preference in attempts to uncover the genetic determinants (Grahame et al., 1999; Bell et al., 2006). In mouse studies with targeted gene manipulations, roughly 2/3 either increase or decrease ethanol preference drinking (Crabbe et al., 2006). There are extensive data in populations derived from B6 and D2-derived mouse lines identifying QTLs for preference drinking (see references above). Despite its ubiquitous use, a limitation of the preference phenotype is uncertainty about how directly it reflects the rewarding value of alcohol's interoceptive effects (see Leeman et al, 2010). If one genotype drinks more alcohol by choice than another, is that because it is more sensitive to ethanol's presumed positive reinforcing value, or is it less sensitive, and therefore needs more alcohol to experience reward? Because animals with unlimited access to alcohol in preference drinking tests rarely achieve blood ethanol levels consistent with behavioral intoxication (Crabbe, 2008), this question is difficult to answer.
The genetics of alcohol reinforcement has been discussed in depth (see Cunningham and Phillips, 2003). Other tests of reinforcement (e.g., oral operant self-administration procedures), show that animals will work to obtain access to or avoid ethanol, but there are no QTL data available for these models. Another way to assess alcohol's reward value is to use classical conditioning. Conditioning can be used to associate the effects of an ethanol injection with the environment in which it was administered (place conditioning), or to associate alcohol's effects with ingestion of a novel flavor (taste conditioning). However, there are very limited QTL data available for these models. The reinforcing effect of ethanol has also been modeled in mice based upon the locomotor stimulant response, and with its tendency to sensitize, or grow more pronounced with repeated administrations of the drug. The putative link to reinforcement obtains from the participation of brain dopaminergic systems, particular corticolimbic pathways, in both reward and motor stimulation. For a discussion of genetic contributions to the stimulant effects, see Phillips (1997). There are numerous QTL reported for ethanol-stimulated activity, but they are quite complex (e.g., different time windows after injection yield different signals); thus, these were not included in the present analyses.
Other phenotypes for which BXD QTL data exists were not utilized in the present set of analyses for a number of reasons. Although there are robust human data for sites in the genome associated with a low level of response to alcohol (see Wilhelmsen et al., 2003; Schuckit et al., 2005), a mouse phenotype that was a plausible surrogate was not readily available. Mice (or rats) will lose the ability to right themselves after an anesthetic dose of ethanol, and the duration of this response has been the subject of selective breeding with mice (McClearn and Kakihana, 1981) and rats. Several QTLs for this trait have been identified, but it is not clear what the appropriate human phenotype that is analogous to this trait might be.
2.3 Syntenic mapping
For phenotypes and QTLs that met the criteria stated above, published LOD plots and established maximal QTL confidence intervals with translating markers were used in the QTL studies to compare current physical map builds of human (NCBI 36 assembly of the human genome, November 2005) and mouse (NCBI m37 mouse assembly, April 2007, B6 strain) using both NCBI comparative maps (www.ncbi.nlm.nih.gov/projects/homology/maps) and Ensembl synteny maps (www.ensembl.org). Using comparative maps at NCBI, syntenic regions between mouse and human anchored by one species were identified. Anchoring to a mouse QTL, the maximal mouse chromosomal region that showed association with phenotypic scores (based upon LOD results) was determined and the syntenic human chromosomal regions identified. Then overlap of these human syntenic regions with the published human QTL regions was scanned for the respective phenotypes. Similarly, we also anchored with human QTLs and looked at the mouse syntenic regions for overlaps with published mouse QTL regions.
3. Results
Table 1 presents loci in the human genome where there was suggestion of evidence for linkage to alcohol-related phenotypes in more than one study. Locations were identified within broad areas on chromosomes 1, 2, 4 (two QTLs), 5, 6, 15 and 16. Table 2 lists the mouse QTLs for preference and withdrawal. For mouse preference traits, there were 5 QTLs and for acute or chronic withdrawal, 7 QTLs. The boundaries of the region of probable linkage were established for each human QTL. Synteny maps then allowed for the identification of regions of mouse chromosomes that corresponded to those QTLs. Analogous searches were then performed starting with each mouse QTL region. As a result of these initial steps, two cases of apparent concurrence of QTL signal in both species were identified.
Table 1.
Approximate locations of QTLs for alcohol-related traits in humans
| Human Chromsome | Location | Trait | Sample Group | References |
|---|---|---|---|---|
| 1 | distal | Alcohol dependence/affective disorder/harm avoidance/ smoking | COGA | Reich et al., 1998; Nurnberger et al.,2001; Dick et al., 2002; Bierut et al., 2004 |
| Alcohol dependence | Pitts S-B | Hill et al., 2004 | ||
| Tobacco usage | MI | Ehlers et al., 2006 | ||
| 2 | medial | Alcohol dependence | COGA | Reich et al., 1998 |
| ASPD | MI | Ehlers et al., 2008 | ||
| Conduct symptoms | COGA | Dick et al., 2004 | ||
| 4 | medial | Alcohol dependence | SWI | Long et al., 1998 |
| Heavy drinking | Framingham | Wyszynski et al., 2003 | ||
| Alcohol dependence | COGA | Reich et al., 1998 | ||
| ß-EEG | COGA | Porjesz et al., 2002 | ||
| 4 | distal | Alcohol dependence drinking symptoms | MI | Ehlers et al., 2004 |
| Alcohol dependence drinking symptoms. | Irish S-P | Prescott et al., 2006 | ||
| Alcohol dependence | COGA | Reich et al., 1998; Corbett et al., 2005 | ||
| Max drinks | COGA | Saccone et al., 2000 | ||
| Having had 6 or more drinks | Smoking behavior | Wilhelmsen et al., 2005 | ||
| 5 | medial | Craving for alcohol | MI | Ehlers & Wilhelmsen, 2005 |
| Alcohol dependence | Pitts S-B | Hill et al., 2004 | ||
| 6 | distal | Any drug dependence | MI | Ehlers et al., 2006 |
| Alcohol dependence | Pitts S-B | Hill et al., 2004 | ||
| Alcohol dependence | COGA | Cantor and Lanning, 1999 | ||
| 15 | proximal | Alcohol dependence with withdrawal | MI | Ehlers et al., 2004 |
| Alcohol dependence / harm avoidance | COGA | Dick et al., 2002 | ||
| 16 | proximal | Alcohol dependence severity/latent class analysis | COGA | Foroud et al., 1998 |
| Alcohol dependence with withdrawal | MI | Ehlers et al., 2004 |
Table 2.
Approximate locations of QTLs for alcohol-related traits in mice
| Mouse Chromsome | Location | Trait | QTL | References |
|---|---|---|---|---|
| 1 | proximal | Chronic alcohol withdrawal | Caws1/Alcw5 | Bergeson et al., 2003 |
| 1 | medial-distal | Alcohol preference drinking | Ap1q | Tarantino et al., 1998 |
| 1 | distal | Acute Alcohol withdrawal | Alcw1 | Buck et al., 1997; Kozell et al., 2008 |
| Chronic alcohol withdrawal | Calw1/Alcdp1 | Buck et al., 2002 | ||
| 2 | proximal | Alcohol preference drinking | Alcp1 | Melo et al., 1996 |
| Alcohol preference drinking | Etp1/Etohc1 | Phillips et al., 1994 | ||
| Alcohol preference drinking | Etp2/Etohc2 | Phillips et al., 1994 | ||
| 3 | distal | Alcohol preference drinking | Etp3 | Phillips et al., 1994 |
| 4 | proximal | Acute Alcohol withdrawal | Alcw2 | Buck et al., 1997; Fehr et al., 2002; Shirley et al., 2004 |
| Chronic alcohol withdrawal | Caws2 | Bergeson et al., 2003 | ||
| 4 | distal | Alcohol preference drinking | Ap3q | Tarantino et al., 1998 |
| 8 | proximal | Chronic alcohol withdrawal | Caws3/Alcw6 | Bergeson et al., 2003 |
| 9 | proximal-medial | Alcohol preference drinking | Ap5q | Tarantino et al., 1998 |
| Alcohol preference drinking | Etp5/Etohc3 | Phillips et al., 1994, 1998 | ||
| 11 | proximal | Acute Alcohol withdrawal | Alcw3 | Buck et al., 1997 |
| Chronic alcohol withdrawal | Caws4/Alcw3 | Bergeson et al., 2003 | ||
| 14 | distal | Chronic alcohol withdrawal | Caws5/Alcw7 | Bergeson et al., 2003 |
| 19 | proximal-distal | Chronic alcohol withdrawal | Calw2/Alcdp2 | Buck et al., 2002 |
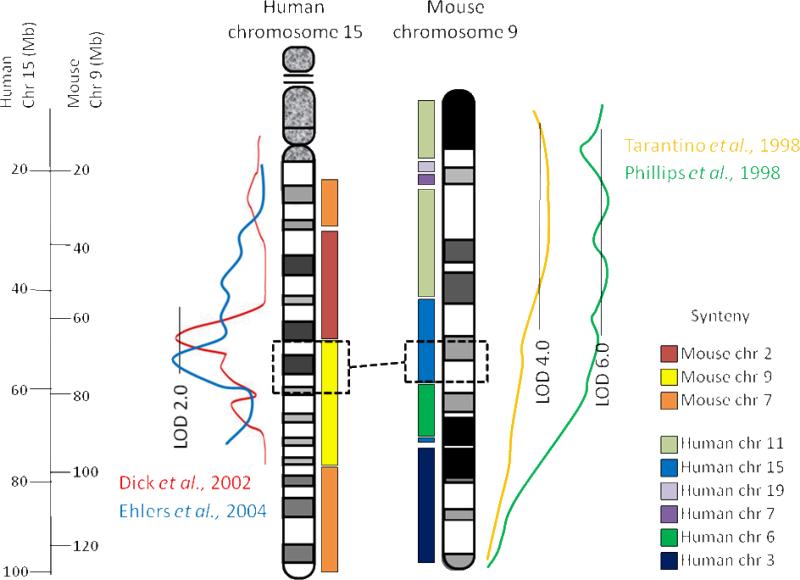
3.1 Human chromosome 15 syntenic to mouse chromosome 9
Two human studies have associated alcohol dependence with human chromosome 15q (Dick et al., 2002; Ehlers et al., 2004) with peak LOD scores >2 near 50 Mb on the current human physical map. Both of these QTLs on human chromosome 15 are syntenic to parts of mouse chromosomes 9 and 2 (Figure 1). Similarly, two groups have identified large alcohol preference QTLs (Etp5 and Ap5q), with LOD scores between 5 and 8, on mouse chromosome 9 (Phillips et al., 1994, 1998; Tarantino et al., 1998). Relying on the robustness of both the mouse and human QTLs, it was possible to narrow the QTL regions of interest in both species based on human/mouse synteny. If these QTLs are in the overlapping syntenic areas, then the focus can be restricted to the homologous candidate QTGs that lie within these narrower intervals (Figure 1). Additionally, since the human chromosome 15 QTLs have better defined regions (sharper peak LOD plots) than the mouse chromosome 9 QTLs, this information can be useful to further narrow the mouse QTL regions beyond just the syntenic regions (Figure 1).
Figure 1.
Synteny between human chromosome 15q and mouse chromosome 9 is illustrated with the colored bars. Proximally to distally, human chromosome 15 shares primary conserved regions with mouse chromosomes 7, 2, 9, and a different region of chromosome 7. Similarly, mouse chromosome 9 shares regions with human chromosomes 11, 19, 7, 15, 6, and 3. For both mouse and human there are smaller syntenic regions that we are not able to illustrate in detail here. Lines depicting LOD score plots are adapted from published QTLs to illustrate the statistical strength of linkage for human alcohol dependence QTLs (blue line from Ehlers et al. 2004; red line from Dick et al. 2002) shown to the left of human chromosome 15. The evident peaks were near 53 and 50 Mb, respectively. The analogous LOD plots for mouse alcohol preference QTLs are shown to the right of mouse chromosome 9 (orange line, Ap5q, from Tarantino et al. 1998; green line, Etp5, from Phillips et al. 1998). The dashed black boxes and line demonstrate the narrowed region of interest based upon mutual synteny of the mouse and human QTLs. The region of human chromosome 15 in the dashed black box is syntenic to the region of mouse chromosome 9 dashed black box, but in reverse orientation, so the most distal gene in human chromosome 15 region is the most proximal in mouse chromosome 9 region and vice versa. Comparison suggests that the human QTL is in the more proximal section of human 15 syntenic with mouse 9, and the more distal section of mouse 9 syntenic with human 15.
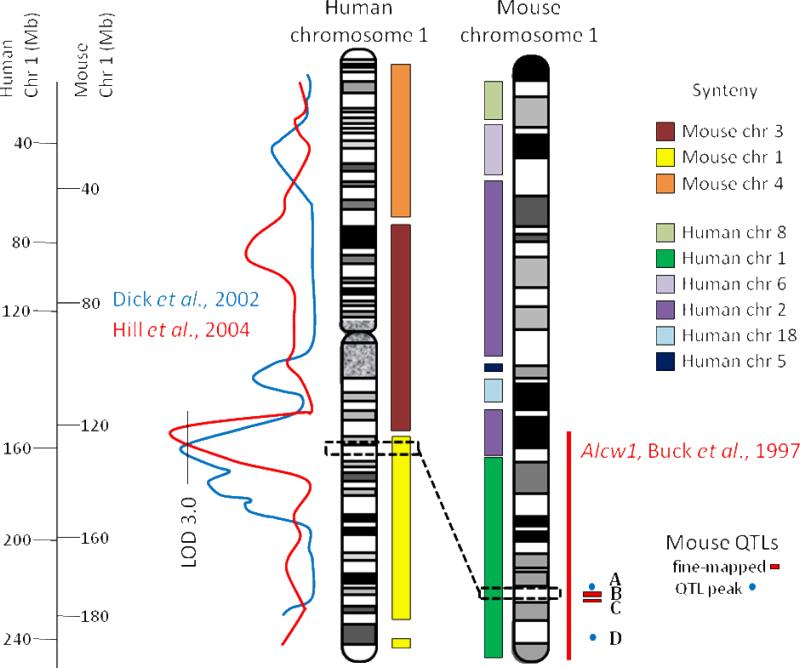
3.2 Human chromosome 1 syntenic to mouse chromosome 1
Two human studies have identified alcohol dependence QTLs (LODs >3) on the q-arm of human chromosome 1 (Dick et al., 2002; Hill et al., 2004). A third human QTL for tobacco usage has also been identified in this same region (Ehlers et al., 2006). This region of human chromosome 1 is primarily syntenic to mouse chromosomes 1 (distal) and 3 (Figure 2). Coincident reciprocal QTLs for ethanol consumption and acute withdrawal are identified on mouse chromosomes 1 and 3 (Hitzemann et al., 2009), but have not been fine-mapped. Buck et al. (1997, 2002) also identified robust QTLs for both acute and chronic ethanol withdrawal on distal chromosome 1 (Alcw1 and Alcdp1, respectively). Recently, the development and testing of a panel of interval-specific congenic strains for distal chromosome 1 delineated two QTLs within the starting Alcw1/Alcdp1 interval. The more proximal QTL affects ethanol withdrawal, but not pentobarbital or zolpidem withdrawal (Kozell et al., 2008), while the more distal QTL affects ethanol, pentobarbital, and zolpidem withdrawal (Kozell et al., 2009). Both of these QTLs are syntenic to human chromosome 1q23.2-1q23.3 (Figure 2). The narrowed mouse QTLs can inform the human QTLs and suggest prioritization of a much smaller number of QTG candidates. High priority candidates include Kcnj9, which encodes GIRK3 (Kir3.3), a subunit member of a family of G-protein-dependent inwardly-rectifying K+ (GIRK) channels that mediate inhibitory effects of Gi/o-coupled receptors (see Kozell et al., 2009), and a gene network crucially involved in oxidative/cellular stress (Denmark and Buck, 2008), indicating both established and novel aspects of the neurobiological response to ethanol.
Figure 2.
Synteny between human chromosome 1 and mouse chromosome 1 is illustrated with the colored bars. Human chromosome 1 shares primary conserved regions with mouse chromosomes 4, 3, and 1. Mouse chromosome 1 shares regions syntenic with human chromosomes 8, 6, 2, 5, 18, and 1. For both mouse and human there are smaller syntenic regions that we are not able to illustrate in detail here. Lines depicting LOD score plots are adapted from published QTLs to illustrate human alcohol dependence QTLs (blue line from Dick et al., 2002; red line from Hill et al., 2004) shown to the left of human chromosome 1. The red line to the right of mouse chromosome 1 shows the original 1-LOD confidence interval for the alcohol withdrawal QTL (Alcw1, Buck et al., 1997). Fine-mapping has delineated two QTLs (B and C with 1.7 Mb and 0.44 Mb QTL intervals, respectively) within the starting QTL region . The more proximal QTL (B) affects ethanol withdrawal, but not pentobarbital or zolpidem withdrawal (Kozell et al., 2008), while the more distal QTL (C) affects ethanol, pentobarbital, and zolpidem withdrawal (Kozell et al., 2009). Both QTLs are syntenic to human chromosome 1q23.2-1q23.3. Additionally, two reciprocal, coincident QTLs for ethanol consumption and ethanol withdrawal are located within this region (Hitzemann et al., 2009); the positions of the peak LOD values are indicated because these QTLs are not yet fine-mapped (A and D). The dashed black boxes and line indicate the QTL intervals on mouse (A, B, and also C) and human based on the fine mapping of the mouse QTLs and synteny with human distal chromosome 1. The region of mouse chromosome 1 in the dashed black box is syntenic to the region of human chromosome 1 dashed black box, but in reverse orientation, so the most distal gene in mouse chromosome 1 region is the most proximal in human chromosome 1 region and vice versa. Additionally, the most distal mouse QTL peak (D) is syntenic to very distal human chromosome 1 and is not included in the synteny block shown. Comparison of the narrowed mouse QTL on chromosome 1 with the human QTL suggests that the human QTL can also be narrowed to the small region on chromosome 1.
4. Discussion
The results of these preliminary analyses of potential synteny between human and mouse QTLs, while limited in scope, do provide evidence that this approach is promising. Mapping animal QTLs onto human linkage findings has the potential to narrow down a broad region of linkage and provides further confidence that there are gene(s) in the region involved in alcohol-related phenotypes. This in turn may help prioritize regions for follow-up in both human and animal studies. Our finding of the utility of this approach is consistent with a previous study that compared genome-wide association studies of SNPs from individuals with dependence on a variety of addictive substances to QTL results from studies of addiction-related phenotypes in mice that focused on alcohol, methamphetamine and barbiturates (see Uhl et al., 2008). The present study differs from previous attempts as it focused on human QTL data from linkage analyses and on genomic locations where more than one study had found suggestive evidence for linkage. It also intentionally restricted the search to two mouse phenotypes.
To date, mouse alcohol response QTL analyses have not detected consilience with the site on human chromosome 4 near the ADH gene cluster, which has been replicated repeatedly in human studies. While it is not known for certain what genes are responsible for the QTL on chromosome 4 there is some evidence from human studies that ADH1B may contribute to the association (Ehlers et al., 2004; MacGregor et al., 2008). Numerous candidate gene studies have also provided evidence for association with ADH gene polymorphisms and alcohol dependence in human studies (see Edenberg et al., 2006). If ADH polymorphisms do account for most of the QTL on human chromosome 4 then the present data may suggest that variations in ADH are less functionally linked to alcohol-related phenotypes in mice than in humans. This may also reflect that fact that one advantage of animal studies is that ethanol metabolism and resulting differences in blood/brain alcohol concentrations can be and frequently are controlled for statistically, which would serve to mitigate detection of ADH or other ethanol metabolism genes as QTLs in many animal studies. That said, even when blood alcohol levels are allowed to vary freely in animal studies, they do not seem to correlate with behavioral sensitivity, suggesting that metabolic differences are not the most important determinants of genetic differences in sensitivity to ethanol (Crabbe et al, 2005). Finally, it must be kept in mind that, to date, most QTL mapping of alcohol-related phenotypes in mice has employed crosses derived from two strains. As a result, substantial existing genetic variation is not represented, leading to “blind spots” (Roberts et al., 2007) that limit comparison to human data. It remains to be determined whether or not this limitation contributes to the current lack of evidence for mouse alcohol response QTLs syntenic with the human chromosome 4 QTL. This issue will mitigate as future studies increasingly use the collaborative cross that includes wild-derived mouse strains to perform genome-wide QTL analyses, including chromosomal regions with little or no interrogation in BXD analyses.
Two examples of positive findings of synteny were observed in the present study. Alcohol withdrawal QTLs on distal mouse chromosome 1 affecting withdrawal after acute and chronic ethanol exposure (Alcw1and Alcdp1, respectively, Buck et al., 1997, 2002) were found to be syntenic to human chromosome 1q23.2-23.3 (Kozell et al., 2008). Two human studies have identified alcohol dependence QTLs (LODs >3) on 1q (Dick et al., 2002; Hill et al., 2004), with additional studies providing supporting evidence for association of 1q markers with alcoholism (Aragaki et al., 1999; Turecki et al., 1999; Guerrini et al., 2005). A third human QTL for tobacco usage has also been identified in this same region (Ehlers et al., 2006). In the second example, a QTL on human chromosome 15 for alcohol dependence severity/withdrawal (Dick et al., 2002; Ehlers et al., 2004) was found to map to a region on mouse chromosome 9 where multiple studies have found QTLs for alcohol preference (Phillips et al., 1994, 1998; Tarantino et al., 1998). It is of interest in both of these cases that while the QTLs were found to be syntenic that the exact phenotypes between humans and mice did not necessarily overlap. These findings suggest that trying to match exact phenotypes in humans and animals may not be necessary or even optimal for determining whether similar genes influence alcohol-related behaviors between the two species. While these two examples offer additional support for these QTLs and are promising for narrowing these QTLs, specific QTGs have not yet been confirmed for these phenotypes.
The present report is part of a larger effort to develop greater consilience between phenotypes for alcohol dependence used in human studies and those used in animal models. The aim of the current set of analyses was to identify a limited set of alcohol-related phenotypes in humans and in mouse by which to compare QTL data between the species using syntenic mapping. The intent of this effort was not meant to be exhaustive but rather to demonstrate how the technique might be applied to a few phenotypes and their QTLs and to identify specific phenotypes in both mice and humans where future studies might improve both data collection and analyses in order to optimize the eventual success of this endeavor. This will contribute significantly to progress in understanding the genetic determination of alcohol behaviors, but there are some limitations. First, the chromosomal regions showing linkage in human and animals studies are often broad and imprecise (at least initially) in terms of their localization of the underlying susceptibility variant(s). Simulation studies have demonstrated considerable variation in the linkage peaks observed for complex phenotypes (Roberts et al., 1999). This has also been borne out in ongoing gene identification projects; for example, the COGA detected linkage on chromosome 4 (Reich et al., 1998; Williams et al., 1999; Saccone et al., 2000) and has subsequently identified several genes in the region as associated with alcohol dependence (Edenberg et al., 2004, 2006, 2008). However, these genes are located throughout the QTL region, with the strongest associations (Edenberg et al., 2004) not localized directly under the strongest linkage peak with alcohol dependence (Williams et al., 1999).
Second, in some cases the behavioral QTGs identified will be the same in mouse and man (Mogil et al., 2003), and in other cases QTL research will identify networks of genes relevant to ethanol response in both mouse and man. For example, there is considerable evidence in the animal literature for a relationship between the Gabra1-Garba6-Gabrg2-Gabrb2 cluster of GABAA receptor genes and alcohol-related phenotypes, but evidence for their involvement in human alcohol dependence has been mixed (Dick et al., 2005, 2006b). However, other GABA-A receptor genes, such as GABRA2, have been robustly associated with alcohol dependence in human studies across multiple independent projects. (Covault et al., 2004, 2008; Edenberg et al., 2004; Lappalainen et al., 2005; Dick et al., 2006c; Fehr et al., 2006; Matthews et al., 2007). In another example, alcohol consumption is associated with the alpha-synuclein gene, SNCA, in alcohol preferring and alcohol non-preferring rats (Liang et al., 2003) and craving for alcohol in humans (Foroud et al., 2007), despite there being no association with clinical diagnoses in the sample. This underscores the importance of phenotype choice in gene identification efforts and the need to examine alcohol-related phenomena in addition to DSM alcohol dependence diagnoses.
The task of identifying the correct phenotypes to be used in humans and animals is not straightforward, as alcohol-related concepts are often operationalized in a different manner in animal and human studies. Most obviously, clinical studies often times rely on diagnoses or symptom counts as primary phenotypes whereas animal models have adopted a more quantitative assessment of behavioral traits. For example, in mice, the severity of behavioral convulsions is commonly used as the primary measure of the alcohol withdrawal syndrome (Buck et al., 1997, 2002; Crabbe, 1998). Other withdrawal signs that are more common in humans (e.g., tremors, tachycardia, diaphoreses) are less studied in animals (Kosobud and Crabbe, 1986; Belknap et al., 1987, Philibin et al, 2008). Similarly, while level of response to alcohol in animals is measured using a variety of behaviors including the duration of the loss of righting reflex, in humans a self-report measure of the number of drinks needed to achieve a subjective “effect” the first five times an individual drank alcohol is often used (Schuckit et al., 2001). Such differences in phenotype definitions may contribute to discrepancies between the human and animal literatures. With this in mind a concerted effort for both animal and human researchers to adopt alcohol related measures that may be more reliably collected in both species may provide more usable data for meaningful comparisons.
Finally, we return to the original goal of this analysis. Here, we entertained the possibility that the path to greater phenotypic consilience might be illuminated by searching for consilience in QTL mapping of chromosomal regions. The strength of the correspondence seen in the two cases that are elucidated here suggests that syntenic mapping will continue to be a valuable exercise. This has the potential to be one of the fastest paths to the underlying genes currently available, and that additional examples of conserved genetic signal will be discovered as the fidelity of both human and mouse maps continues to improve. This task may be enhanced by the use of similar maps between studies and by author's providing accurate confidence intervals. . Human linkage studies are can suffer from lack of power, which can make replication of a linkage finding very challenging (see Sham et al., 2002). Another option would be to consider cases where an initial linkage signal has been followed up with fine-mapping that resulted in the identification of associated genetic variants, which have been confidently confirmed in subsequent studies. Because of technical limitations to the existing data, the lack of widespread concurrence across species cannot be taken as evidence of lack of actual correspondence. However, the limited success of this search for correspondences suggests that this is not the most fruitful path towards suggesting better phenotypes to either the human or animal modeling communities.
Acknowledgements
Supported in part by the National Institute on Alcohol Abuse and Alcoholism Grants AA 006059, AA010201, the National Center on Minority Health and Health Disparities (NCMHD), and U54 RR025204 to CLE, AA010760, and AA12714 to JCC, AA011114 and DA005228 to KJB, and VA Merit Review awards to KJB and JCC. DMD acknowledges support from a NARSAD young investigator award. The authors thank Shirley Sanchez for editing the manuscript.
References
- Agrawal A, Pergadia ML, Saccone SF, Lynskey MT, Wang JC, Martin NG, Statham D, Henders A, Campbell M, Garcia R, Broms U, Todd RD, Goate AM, Rice J, Kaprio J, Heath AC, Montgomery GW, Madden PA. An autosomal linkage scan for cannabis use disorders in the nicotine addiction genetics project. Arch Gen Psychiatry. 2008;65:713–721. doi: 10.1001/archpsyc.65.6.713. [DOI] [PubMed] [Google Scholar]
- Aragaki C, Quiaoit F, Hsu L, Zhao LP. Mapping alcoholism genes using linkage/linkage disequilibrium analysis. Genet Epidemiol. 1999;17(Suppl 1):S43–S48. doi: 10.1002/gepi.1370170708. [DOI] [PubMed] [Google Scholar]
- Barr CS, Goldman D. Non-human primate models of inheritance vulnerability to alcohol use disorders. Addict Biol. 2006;11:374–385. doi: 10.1111/j.1369-1600.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- Becker HC. The alcohol withdrawal “kindling” phenomenon: clinical and experimental findings. Alcohol Clin Exp Res. 1996;20:121A–124A. doi: 10.1111/j.1530-0277.1996.tb01760.x. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Laursen SE, Crabbe JC. Ethanol and nitrous oxide produce withdrawal-induced convulsions by similar mechanisms in mice. Life Sci. 1987;41:2033–2040. doi: 10.1016/0024-3205(87)90477-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Atkins AL. The replicability of QTLs for murine alcohol preference drinking behavior across eight independent studies. Mamm Genome. 2001;12:893–899. doi: 10.1007/s00335-001-2074-2. [DOI] [PubMed] [Google Scholar]
- Bell RL, Rodd ZA, Lumeng L, Murphy JM, McBride WJ. The alcohol-preferring P rat and animal models of excessive alcohol drinking. Addict Biol. 2006;11:270–288. doi: 10.1111/j.1369-1600.2005.00029.x. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Zahniser NR, Johnson TE. Confirmation and fine mapping of ethanol sensitivity quantitative trait loci, and candidate gene testing in the LXS recombinant inbred mice. J Pharmacol Exp Ther. 2006;319:299–307. doi: 10.1124/jpet.106.103572. [DOI] [PubMed] [Google Scholar]
- Bennett B, Carosone-Link P, Beeson M, Gordon L, Phares-Zook N, Johnson TE. Genetic dissection of quantitative trait locus for ethanol sensitivity in long- and short-sleep mice. Genes Brain Behav. 2008;7:659–668. doi: 10.1111/j.1601-183X.2008.00403.x. [DOI] [PubMed] [Google Scholar]
- Bergeson SE, Kyle WR, Crabbe JC, Metten P, Gene E,V, Belknap JK. Chromosomal loci influencing chronic alcohol withdrawal severity. Mamm Genome. 2003;14:454–463. doi: 10.1007/s00335-002-2254-4. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, Edenberg HJ, Cloninger CR, Begleiter H, Conneally PM, Crowe RR, Hesselbrock V, Li TK, Nurnberger JI, Jr., Porjesz B, Schuckit MA, Reich T. A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet A. 2004;124:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- Browman KE, Crabbe JC. Quantitative trait loci affecting ethanol sensitivity in BXD recombinant inbred mice. Alcohol Clin Exp Res. 2000;24:17–23. [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck KJ, Rademacher BS, Metten P, Crabbe JC. Mapping murine loci for physical dependence on ethanol. Psychopharmacology (Berl) 2002;160:398–407. doi: 10.1007/s00213-001-0988-8. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Lanning CD. Comparison of evidence supporting a chromosome 6 alcoholism gene. Genet Epidemiol. 1999;17(Suppl 1):S91–S96. doi: 10.1002/gepi.1370170716. [DOI] [PubMed] [Google Scholar]
- Corbett J, Saccone NL, Foroud T, Goate A, Edenberg H, Nurnberger J, Porjesz B, Begleiter H, Reich T, Rice JP. A sex-adjusted and age-adjusted genome screen for nested alcohol dependence diagnoses. Psychiatr Genet. 2005;15:25–30. doi: 10.1097/00041444-200503000-00005. [DOI] [PubMed] [Google Scholar]
- Cotton NS. The familial incidence of alcoholism: a review. J Stud Alcohol. 1979;40:89–116. doi: 10.15288/jsa.1979.40.89. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Hesselbrock V, Nellissery M, Kranzler HR. Allelic and haplotypic association of GABRA2 with alcohol dependence. Am J Med Genet B (Neuropsychiatr Genet ) 2004;129B:104–109. doi: 10.1002/ajmg.b.30091. [DOI] [PubMed] [Google Scholar]
- Covault J, Gelernter J, Jensen K, Anton R, Kranzler HR. Markers in the 5'-region of GABRG1 associate to alcohol dependence and are in linkage disequilibrium with markers in the adjacent GABRA2 gene. Neuropsychopharmacology. 2008;33:837–848. doi: 10.1038/sj.npp.1301456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabbe JC, Kosobud A, Young ER, Janowsky JS. Polygenic and single-gene determination of responses to ethanol in BXD/Ty recombinant inbred mouse strains. Neurobehav Toxicol Teratol. 1983;5:181–187. [PubMed] [Google Scholar]
- Crabbe JC. A genetic animal model of alcohol withdrawal. Alcohol Clin Exp Res. 1996;20:96A–100A. doi: 10.1111/j.1530-0277.1996.tb01754.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Provisional mapping of quantitative trait loci for chronic ethanol withdrawal severity in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1998;286:263–271. [PubMed] [Google Scholar]
- Crabbe JC, Metten P, Cameron AJ, Wahlsten D. An analysis of the genetics of alcohol intoxication in inbred mice. Neurosci Biobehav Rev. 2005;28:785–802. doi: 10.1016/j.neubiorev.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Phillips TJ, Harris RA, Arends MA, Koob GF. Alcohol-related genes: contributions from studies with genetically engineered mice. Addict Biol. 2006;11:195–269. doi: 10.1111/j.1369-1600.2006.00038.x. [DOI] [PubMed] [Google Scholar]
- Crabbe JC. Neurogenetic studies of alcohol addiction. Philos Trans R Soc Lond B Biol Sci. 2008;363:3201–3211. doi: 10.1098/rstb.2008.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder CM. Ethanol targets: a BK channel cocktail in C. elegans. Trends Neurosci. 2004;27:579–582. doi: 10.1016/j.tins.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Cunningham CL. Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology (Berl) 1995;120:28–41. doi: 10.1007/BF02246142. [DOI] [PubMed] [Google Scholar]
- Cunningham CL, Phillips TJ. Genetic basis of ethanol reward. In: Maldonado R, editor. Molecular biology of drug addiction. Humana Press; Totowa, N.J.: 2003. pp. 263–294. [Google Scholar]
- Demarest K, McCaughran J, Jr., Mahjubi E, Cipp L, Hitzemann R. Identification of an acute ethanol response quantitative trait locus on mouse chromosome 2. J Neurosci. 1999;19:549–561. doi: 10.1523/JNEUROSCI.19-02-00549.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denmark DL, Buck KJ. Molecular analyses and identification of promising candidate genes for loci on mouse chromosome 1 affecting alcohol physical dependence and associated withdrawal. Genes Brain Behav. 2008;7:599–608. doi: 10.1111/j.1601-183X.2008.00396.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Nurnberger J, Jr., Edenberg HJ, Goate A, Crowe R, Rice J, Bucholz KK, Kramer J, Schuckit MA, Smith TL, Porjesz B, Begleiter H, Hesselbrock V, Foroud T. Suggestive linkage on chromosome 1 for a quantitative alcohol-related phenotype. Alcohol Clin Exp Res. 2002;26:1453–1460. doi: 10.1097/01.ALC.0000034037.10333.FD. [DOI] [PubMed] [Google Scholar]
- Dick DM, Li TK, Edenberg HJ, Hesselbrock V, Kramer J, Kuperman S, Porjesz B, Bucholz K, Goate A, Nurnberger J, Foroud T. A genome-wide screen for genes influencing conduct disorder. Mol Psychiatry. 2004;9:81–86. doi: 10.1038/sj.mp.4001368. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Foroud T. No association of the GABAA receptor genes on chromosome 5 with alcoholism in the collaborative study on the genetics of alcoholism sample. Am J Med Genet B Neuropsychiatr Genet. 2005;132B:24–28. doi: 10.1002/ajmg.b.30058. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151–157. doi: 10.1007/s11920-006-0015-1. [DOI] [PubMed] [Google Scholar]
- Dick DM, Rose RJ, Kaprio J. The next challenge for psychiatric genetics: characterizing the risk associated with identified genes. Ann Clin Psychiatry. 2006a;18:223–231. doi: 10.1080/10401230600948407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Wetherill LF, Xuei X, Goate A, Hesselbrock V, Schuckit M, Crowe R, Edenberg HJ, Foroud T. Association between GABRA1 and drinking behaviors in the collaborative study on the genetics of alcoholism sample. Alcohol Clin Exp Res. 2006b;30:1101–1110. doi: 10.1111/j.1530-0277.2006.00136.x. [DOI] [PubMed] [Google Scholar]
- Dick DM, Bierut L, Hinrichs A, Fox L, Bucholz KK, Kramer J, Kuperman S, Hesselbrock V, Schuckit M, Almasy L, Tischfield J, Porjesz B, Begleiter H, Nurnberger J, Jr., Xuei X, Edenberg HJ, Foroud T. The role of GABRA2 in risk for conduct disorder and alcohol and drug dependence across developmental stages. Behav Genet. 2006c;36:577–590. doi: 10.1007/s10519-005-9041-8. [DOI] [PubMed] [Google Scholar]
- Dick DM, Aliev F, Wang JC, Saccone S, Hinrichs A, Bertelsen S, Budde J, Saccone N, Foroud T, Nurnberger J, Jr., Xuei X, Conneally PM, Schuckit M, Almasy L, Crowe R, Kuperman S, Kramer J, Tischfield JA, Hesselbrock V, Edenberg HJ, Porjesz B, Rice JP, Bierut L, Goate A. A Systematic single nucleotide polymorphism screen to fine-map alcohol dependence genes on chromosome 7 identifies association with a novel susceptibility gene ACN9. Biol Psychiatry. 2008;63:1047–1053. doi: 10.1016/j.biopsych.2007.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgon T, D'Addario C, Uhl GR. Linkage disequilibrium, haplotype and association studies of a chromosome 4 GABA receptor gene cluster: candidate gene variants for addictions. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:854–860. doi: 10.1002/ajmg.b.30349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggirala R, Almasy L, Blangero J. Smoking behavior is under the influence of a major quantitative trait locus on human chromosome 5q. Genet Epidemiol. 1999;17(Suppl 1):S139–S144. doi: 10.1002/gepi.1370170724. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr., O'Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74:705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T. The genetics of alcoholism: identifying specific genes through family studies. Addict Biol. 2006;11:386–396. doi: 10.1111/j.1369-1600.2006.00035.x. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Chen HJ, Tian H, Wetherill LF, Dick DM, Almasy L, Bierut L, Bucholz KK, Goate A, Hesselbrock V, Kuperman S, Nurnberger J, Porjesz B, Rice J, Schuckit M, Tischfield J, Begleiter H, Foroud T. Association of alcohol dehydrogenase genes with alcohol dependence: a comprehensive analysis. Hum Mol Genet. 2006;15:1539–1549. doi: 10.1093/hmg/ddl073. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Xuei X, Wetherill LF, Bierut L, Bucholz K, Dick DM, Hesselbrock V, Kuperman S, Porjesz B, Schuckit MA, Tischfield JA, Almasy LA, Nurnberger JI, Jr., Foroud T. Association of NFKB1, which encodes a subunit of the transcription factor NF-kappaB, with alcohol dependence. Hum Mol Genet. 2008;17:963–970. doi: 10.1093/hmg/ddm368. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Slutske WS, Gilder DA, Lau P, Wilhelmsen KC. Age at first intoxication and alcohol use disorders in Southwest California Indians. Alcohol Clin Exp Res. 2006;30:1856–1865. doi: 10.1111/j.1530-0277.2006.00222.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic screen for loci associated with tobacco usage in Mission Indians. BMC Med Genet. 2006 Feb 10;7:9. doi: 10.1186/1471-2350-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Wilhelmsen KC. Genomic screen for substance dependence and body mass index in Southwest California Indians. Genes Brain Behav. 2007;6:184–191. doi: 10.1111/j.1601-183X.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- Ehlers CL, Gilder DA, Slutske WS, Lind PA, Wilhelmsen KC. Externalizing disorders in American Indians: Comorbidity and a genome wide linkage analysis. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:690–698. doi: 10.1002/ajmg.b.30666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers CL, Chester JA. Alcoholism. In: Squire LR, editor. Encyclopedia of Neuroscience. Academic Press; Oxford: 2009. pp. 231–236. [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr C, Sander T, Tadic A, Lenzen KP, Anghelescu I, Klawe C, Dahmen N, Schmidt LG, Szegedi A. Confirmation of association of the GABRA2 gene with alcohol dependence by subtype-specific analysis. Psychiatr Genet. 2006;16:9–17. doi: 10.1097/01.ypg.0000185027.89816.d9. [DOI] [PubMed] [Google Scholar]
- Fernandez JR, Vogler GP, Tarantino LM, Vignetti S, Plomin R, McClearn GE. Sex-exclusive quantitative trait loci influences in alcohol-related phenotypes. Am J Med Genet. 1999;88:647–652. doi: 10.1002/(sici)1096-8628(19991215)88:6<647::aid-ajmg13>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Foroud T, Bucholz KK, Edenberg HJ, Goate A, Neuman RJ, Porjesz B, Koller DL, Rice J, Reich T, Bierut LJ, Cloninger CR, Nurnberger JI, Jr., Li TK, Conneally PM, Tischfield JA, Crowe R, Hesselbrock V, Schuckit M, Begleiter H. Linkage of an alcoholism-related severity phenotype to chromosome 16. Alcohol Clin Exp Res. 1998;22:2035–2042. [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- Foroud T, Wetherill LF, Liang T, Dick DM, Hesselbrock V, Kramer J, Nurnberger J, Schuckit M, Carr L, Porjesz B, Xuei X, Edenberg HJ. Association of alcohol craving with alpha-synuclein (SNCA). Alcohol Clin Exp Res. 2007;31:537–545. doi: 10.1111/j.1530-0277.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Gehle VM, Erwin VG. Common quantitative trait loci for alcohol-related behaviors and CNS neurotensin measures: voluntary ethanol consumption. Alcohol Clin Exp Res. 1998;22:401–408. [PubMed] [Google Scholar]
- Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, Kranzler HR. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Pal N. Alcohol dependence produced in mice by inhalation of ethanol: grading the withdrawal reaction. Science. 1971;172:288–290. doi: 10.1126/science.172.3980.288. [DOI] [PubMed] [Google Scholar]
- Grahame NJ, Li TK, Lumeng L. Selective breeding for high and low alcohol preference in mice. Behav Genet. 1999;29:47–57. doi: 10.1023/a:1021489922751. [DOI] [PubMed] [Google Scholar]
- Grant KA, Bennett AJ. Advances in nonhuman primate alcohol abuse and alcoholism research. Pharmacol Ther. 2003;100:235–255. doi: 10.1016/j.pharmthera.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Green AS, Grahame NJ. Ethanol drinking in rodents: is free-choice drinking related to the reinforcing effects of ethanol? Alcohol. 2008;42:1–11. doi: 10.1016/j.alcohol.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisel JE, Metten P, Wenger CD, Merrill CM, Crabbe JC. Mapping of quantitative trait loci underlying ethanol metabolism in BXD recombinant inbred mouse strains. Alcohol Clin Exp Res. 2002;26:610–616. [PubMed] [Google Scholar]
- Guerrini I, Cook CC, Kest W, Devitgh A, McQuillin A, Curtis D, Gurling HM. Genetic linkage analysis supports the presence of two susceptibility loci for alcoholism and heavy drinking on chromosome 1p22.1-11.2 and 1q21.3-24.2. BMC Genet. 2005;6:11. doi: 10.1186/1471-2156-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Bucholz KK, Madden PA, Dinwiddie SH, Slutske WS, Bierut LJ, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic and environmental contributions to alcohol dependence risk in a national twin sample: consistency of findings in women and men. Psychol Med. 1997;27:1381–1396. doi: 10.1017/s0033291797005643. [DOI] [PubMed] [Google Scholar]
- Heberlein U. Genetics of alcohol-induced behaviors in Drosophila. Alcohol Res Health. 2000;24:185–188. [PMC free article] [PubMed] [Google Scholar]
- Heilig M, Egli M, Crabbe JC, Becker HC. Acute withdrawal, protracted abstinence and negative affect in alcoholism: are they linked? Addict Biol. 2010;15:169–184. doi: 10.1111/j.1369-1600.2009.00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Hum Genet. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitzemann R, Edmunds S, Wu W, Malmanger B, Walter N, Belknap J, Darakjian P, McWeeney S. Detection of reciprocal quantitative trait loci for acute ethanol withdrawal and ethanol consumption in heterogeneous stock mice. Psychopharmacology (Berl) 2009;203:713–722. doi: 10.1007/s00213-008-1418-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homanics GE, Le NQ, Kist F, Mihalek R, Hart AR, Quinlan JJ. Ethanol tolerance and withdrawal responses in GABA(A) receptor alpha 6 subunit null allele mice and in inbred C57BL/6J and strain 129/SvJ mice. Alcohol Clin Exp Res. 1998;22:259–265. [PubMed] [Google Scholar]
- Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- Johnson FW, Gruenewald PJ, Treno AJ, Taff GA. Drinking over the life course within gender and ethnic groups: a hyperparametric analysis. J Stud Alcohol. 1998;59:568–580. doi: 10.15288/jsa.1998.59.568. [DOI] [PubMed] [Google Scholar]
- Karpyak VM, Kim JH, Biernacka JM, Wieben ED, Mrazek DA, Black JL, Choi DS. Sequence variations of the human MPDZ gene and association with alcoholism in subjects with European ancestry. Alcohol Clin Exp Res. 2009;33:712–721. doi: 10.1111/j.1530-0277.2008.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kuo PH, Todd WB, Kalsi G, Neale MC, Sullivan PF, Walsh D, Patterson DG, Riley B, Prescott CA. A joint genomewide linkage analysis of symptoms of alcohol dependence and conduct disorder. Alcohol Clin Exp Res. 2006;30:1972–1977. doi: 10.1111/j.1530-0277.2006.00243.x. [DOI] [PubMed] [Google Scholar]
- Kirstein SL, Davidson KL, Ehringer MA, Sikela JM, Erwin VG, Tabakoff B. Quantitative trait loci affecting initial sensitivity and acute functional tolerance to ethanol-induced ataxia and brain cAMP signaling in BXD recombinant inbred mice. J Pharmacol Exp Ther. 2002;302:1238–1245. doi: 10.1124/jpet.302.3.1238. [DOI] [PubMed] [Google Scholar]
- Kohnke MD. Approach to the genetics of alcoholism: a review based on pathophysiology. Biochem Pharmacol. 2008;75:160–177. doi: 10.1016/j.bcp.2007.06.021. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Crabbe JC. Ethanol withdrawal in mice bred to be genetically prone or resistant to ethanol withdrawal seizures. J Pharmacol Exp Ther. 1986;238:170–177. [PubMed] [Google Scholar]
- Kozell L, Belknap JK, Hofstetter JR, Mayeda A, Buck KJ. Mapping a locus for alcohol physical dependence and associated withdrawal to a 1.1 Mb interval of mouse chromosome 1 syntenic with human chromosome 1q23.2-23.3. Genes Brain Behav. 2008;7:560–567. doi: 10.1111/j.1601-183X.2008.00391.x. [DOI] [PubMed] [Google Scholar]
- Kozell LB, Walter NA, Milner LC, Wickman K, Buck KJ. Mapping a barbiturate withdrawal locus to a 0.44 Mb interval and analysis of a novel null mutant identify a role for Kcnj9 (GIRK3) in withdrawal from pentobarbital, zolpidem, and ethanol. J Neurosci. 2009;29:11662–11673. doi: 10.1523/JNEUROSCI.1413-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen J, Kranzler HR, Petrakis I, Somberg LK, Page G, Krystal JH, Gelernter J. Confirmation and fine mapping of the chromosome 1 alcohol dependence risk locus. Mol Psychiatry. 2004;9:312–319. doi: 10.1038/sj.mp.4001429. [DOI] [PubMed] [Google Scholar]
- Lappalainen J, Krupitsky E, Remizov M, Pchelina S, Taraskina A, Zvartau E, Somberg LK, Covault J, Kranzler HR, Krystal JH, Gelernter J. Association between alcoholism and gamma-amino butyric acid alpha2 receptor subtype in a Russian population. Alcohol Clin Exp Res. 2005;29:493–498. doi: 10.1097/01.alc.0000158938.97464.90. [DOI] [PubMed] [Google Scholar]
- Leeman RF, Heilig M, Cunningham CL, Stephens DN, Duka T, O'Malley SS. Ethanol consumption: how should we measure it? Achieving consilience between human and animal phenotypes. Addict Biol. 2010;15:109–124. doi: 10.1111/j.1369-1600.2009.00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Spence J, Liu L, Strother WN, Chang HW, Ellison JA, Lumeng L, Li TK, Foroud T, Carr LG. alpha-Synuclein maps to a quantitative trait locus for alcohol preference and is differentially expressed in alcohol-preferring and -nonpreferring rats. Proc Natl Acad Sci U S A. 2003;100:4690–4695. doi: 10.1073/pnas.0737182100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Lovinger DM, Crabbe JC. Laboratory models of alcoholism: treatment target identification and insight into mechanisms. Nat Neurosci. 2005;8:1471–1480. doi: 10.1038/nn1581. [DOI] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PA, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Hum Mol Genet. 2008;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AG, Hoffman EK, Zezza N, Stiffler S, Hill SY. The role of the GABRA2 polymorphism in multiplex alcohol dependence families with minimal comorbidity: within-family association and linkage analyses. J Stud Alcohol Drugs. 2007;68:625–633. doi: 10.15288/jsad.2007.68.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride WJ, Li TK. Animal models of alcoholism: neurobiology of high alcohol-drinking behavior in rodents. Crit Rev Neurobiol. 1998;12:339–369. doi: 10.1615/critrevneurobiol.v12.i4.40. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Kakihana R. Selective breeding for ethanol sensitivity: short-sleep and long-sleep mice. In: McClearn GE, Deitrich RA, Erwin VG, editors. Development of animal models as pharmacogenetic tools. NIAAA, USDHHS, PHS, Alcohol, Drug Abuse, and Mental Health Administration; Rockville, MD: 1981. pp. 147–159. [Google Scholar]
- McGue M. Genes, environment, and the etiology of alcoholism. In: Zucker R, Boyd G, Howard J, editors. The Development of Alcohol Problems: Exploring the Biopsychosocial Matrix of Risk. NIAAA, Research Monograph No. 26, NIH Pub No. 94-3495, U.S. Dept. of Health and Human Services; Washington, DC: 1994. pp. 1–40. [Google Scholar]
- McGue M. The behavioral genetics of alcoholism. Curr Directions Psychol Sci. 1999;8:109–115. [Google Scholar]
- Melo JA, Shendure J, Pociask K, Silver LM. Identification of sex-specific quantitative trait loci controlling alcohol preference in C57BL/ 6 mice. Nat Genet. 1996;13:147–153. doi: 10.1038/ng0696-147. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Dependence and withdrawal. In: Deitrich RA, Erwin VG, editors. Pharmacological effects of ethanol on the nervous system. CRC Press; Boca Raton, FL: 1996. pp. 269–290. [Google Scholar]
- Metten P, Phillips TJ, Crabbe JC, Tarantino LM, McClearn GE, Plomin R, Erwin VG, Belknap JK. High genetic susceptibility to ethanol withdrawal predicts low ethanol consumption. Mamm Genome. 1998;9:983–990. doi: 10.1007/s003359900911. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Alcohol withdrawal severity in inbred mouse (Mus musculus) strains. Behav Neurosci. 2005;119:911–925. doi: 10.1037/0735-7044.119.4.911. [DOI] [PubMed] [Google Scholar]
- Mogil JS, Wilson SG, Chesler EJ, Rankin AL, Nemmani KV, Lariviere WR, Groce MK, Wallace MR, Kaplan L, Staud R, Ness TJ, Glover TL, Stankova M, Mayorov A, Hruby VJ, Grisel JE, Fillingim RB. The melanocortin-1 receptor gene mediates female-specific mechanisms of analgesia in mice and humans. Proc Natl Acad Sci U S A. 2003;100:4867–4872. doi: 10.1073/pnas.0730053100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurnberger JI, Jr., Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Philibin SD, Cameron AJ, Metten P, Crabbe JC. Motor impairment: a new ethanol withdrawal phenotype in mice. Behav Pharmacol. 2008;19:604–614. doi: 10.1097/FBP.0b013e32830ded27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Crabbe JC, Metten P, Belknap JK. Localization of genes affecting alcohol drinking in mice. Alcohol Clin Exp Res. 1994;18:931–941. doi: 10.1111/j.1530-0277.1994.tb00062.x. [DOI] [PubMed] [Google Scholar]
- Phillips TJ. Behavior genetics of drug sensitization. Crit Rev Neurobiol. 1997;11:21–33. doi: 10.1615/critrevneurobiol.v11.i1.20. [DOI] [PubMed] [Google Scholar]
- Phillips TJ, Belknap JK, Buck KJ, Cunningham CL. Genes on mouse chromosomes 2 and 9 determine variation in ethanol consumption. Mamm Genome. 1998;9:936–941. doi: 10.1007/s003359900903. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Goate A, Rice JP, O'Connor SJ, Rohrbaugh J, Kuperman S, Bauer LO, Crowe RR, Schuckit MA, Hesselbrock V, Conneally PM, Tischfield JA, Li TK, Reich T, Begleiter H. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci U S A. 2002;99:3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr., Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Hum Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- Risinger FO, Cunningham CL. Ethanol-induced conditioned taste aversion in BXD recombinant inbred mice. Alcohol Clin Exp Res. 1998;22:1234–1244. [PubMed] [Google Scholar]
- Roberts A, Pardo-Manuel d, V, Wang W, McMillan L, Threadgill DW. The polymorphism architecture of mouse genetic resources elucidated using genome-wide resequencing data: implications for QTL discovery and systems genetics. Mamm Genome. 2007;18:473–481. doi: 10.1007/s00335-007-9045-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65:876–884. doi: 10.1086/302528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodd ZA, Bell RL, Sable HJ, Murphy JM, McBride WJ. Recent advances in animal models of alcohol craving and relapse. Pharmacol Biochem Behav. 2004;79:439–450. doi: 10.1016/j.pbb.2004.08.018. [DOI] [PubMed] [Google Scholar]
- Rodriguez LA, Plomin R, Blizard DA, Jones BC, McClearn GE. Alcohol acceptance, preference, and sensitivity in mice. II. Quantitative trait loci mapping analysis using BXD recombinant inbred strains. Alcohol Clin Exp Res. 1995;19:367–373. doi: 10.1111/j.1530-0277.1995.tb01517.x. [DOI] [PubMed] [Google Scholar]
- Ruf C, Carosone-Link P, Springett J, Bennett B. Confirmation and genetic dissection of a major quantitative trait locus for alcohol preference drinking. Alcohol Clin Exp Res. 2004;28:1613–1621. doi: 10.1097/01.alc.0000145693.58448.95. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Kwon JM, Corbett J, Goate A, Rochberg N, Edenberg HJ, Foroud T, Li TK, Begleiter H, Reich T, Rice JP. A genome screen of maximum number of drinks as an alcoholism phenotype. Am J Med Genet. 2000;96:632–637. doi: 10.1002/1096-8628(20001009)96:5<632::aid-ajmg8>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- Saccone NL, Rochberg N, Neuman RJ, Rice JP. Covariates in linkage analysis using sibling and cousin pairs. Genet Epidemiol. 2001;21(Suppl 1):S540–S545. doi: 10.1002/gepi.2001.21.s1.s540. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Saccone NL, Neuman RJ, Rice JP. Genetic analysis of the maximum drinks phenotype. BMC Genet. 2005;6(Suppl 1):S124. doi: 10.1186/1471-2156-6-S1-S124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchis-Segura C, Spanagel R. Behavioural assessment of drug reinforcement and addictive features in rodents: an overview. Addict Biol. 2006;11:2–38. doi: 10.1111/j.1369-1600.2006.00012.x. [DOI] [PubMed] [Google Scholar]
- Schuckit MA. Drug and alcohol abuse: a clinical guide to diagnosis and treatment. 4th ed Plenum Medical Book Co.; New York: 1995. [Google Scholar]
- Schuckit MA, Edenberg HJ, Kalmijn J, Flury L, Smith TL, Reich T, Bierut L, Goate A, Foroud T. A genome-wide search for genes that relate to a low level of response to alcohol. Alcohol Clin Exp Res. 2001;25:323–329. [PubMed] [Google Scholar]
- Schuckit MA, Wilhelmsen K, Smith TL, Feiler HS, Lind P, Lange LA, Kalmijn J. Autosomal linkage analysis for the level of response to alcohol. Alcohol Clin Exp Res. 2005;29:1976–1982. doi: 10.1097/01.alc.0000187598.82921.27. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell S, Cherny SS, Abecasis GR. Powerful regression-based quantitative-trait linkage analysis of general pedigrees. Am J Hum Genet. 2002;71:238–253. doi: 10.1086/341560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, Kadambi B, Sadek H, Silverman MA, Webb BT, Neale MC, Bulik CM, Joyce PR, Kendler KS. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4:129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- Swan GE, Hops H, Wilhelmsen KC, Lessov-Schlaggar CN, Cheng LS, Hudmon KS, Amos CI, Feiler HS, Ring HZ, Andrews JA, Tildesley E, Benowitz N. A genome-wide screen for nicotine dependence susceptibility loci. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:354–360. doi: 10.1002/ajmg.b.30315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Bhave SV, Hoffman PL. Selective breeding, quantitative trait locus analysis, and gene arrays identify candidate genes for complex drug-related behaviors. J Neurosci. 2003;23:4491–4498. doi: 10.1523/JNEUROSCI.23-11-04491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabakoff B, Saba L, Printz M, Flodman P, Hodgkinson C, Goldman D, Koob G, Richardson HN, Kechris K, Bell RL, Hubner N, Heinig M, Pravenec M, Mangion J, Legault L, Dongier M, Conigrave KM, Whitfield JB, Saunders J, Grant B, Hoffman PL, On S, Trait Markers Of Alcoholism WI Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 2009;7:70. doi: 10.1186/1741-7007-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res. 1998;22:1099–1105. [PubMed] [Google Scholar]
- Treadwell JA. Integrative strategies to identify candidate genes in rodent models of human alcoholism. Genome. 2006;49:1–7. doi: 10.1139/g05-083. [DOI] [PubMed] [Google Scholar]
- Turecki G, Rouleau GA, Alda M. Family density of alcoholism and linkage information in the analysis of the COGA data. Genet Epidemiol. 1999;17(Suppl 1):S361–S366. doi: 10.1002/gepi.1370170761. [DOI] [PubMed] [Google Scholar]
- Uhl GR, Drgon T, Johnson C, Fatusin OO, Liu QR, Contoreggi C, Li CY, Buck K, Crabbe J. “Higher order” addiction molecular genetics: convergent data from genome-wide association in humans and mice. Biochem Pharmacol. 2008;75:98–111. doi: 10.1016/j.bcp.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieten C, Seaton KL, Feiler HS, Wilhelmsen KC. The University of California, San Francisco Family Alcoholism Study. I. Design, methods, and demographics. Alcohol Clin Exp Res. 2004;28:1509–1516. doi: 10.1097/01.alc.0000142261.32980.64. [DOI] [PubMed] [Google Scholar]
- Wahlsten D, Bachmanov A, Finn DA, Crabbe JC. Stability of inbred mouse strain differences in behavior and brain size between laboratories and across decades. Proc Natl Acad Sci U S A. 2006;103:16364–16369. doi: 10.1073/pnas.0605342103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NA, Bottomly D, Laderas T, Mooney MA, Darakjian P, Searles RP, Harrington CA, McWeeney SK, Hitzemann R, Buck KJ. High throughput sequencing in mice: a platform comparison identifies a preponderance of cryptic SNPs. BMC Genomics. 2009;10:379. doi: 10.1186/1471-2164-10-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, Antonarakis SE, Attwood J, Baertsch R, Bailey J, Barlow K, Beck S, Berry E, Birren B, Bloom T, Bork P, Botcherby M, Bray N, Brent MR, Brown DG, Brown SD, Bult C, Burton J, Butler J, Campbell RD, Carninci P, Cawley S, Chiaromonte F, Chinwalla AT, Church DM, Clamp M, Clee C, Collins FS, Cook LL, Copley RR, Coulson A, Couronne O, Cuff J, Curwen V, Cutts T, Daly M, David R, Davies J, Delehaunty KD, Deri J, Dermitzakis ET, Dewey C, Dickens NJ, Diekhans M, Dodge S, Dubchak I, Dunn DM, Eddy SR, Elnitski L, Emes RD, Eswara P, Eyras E, Felsenfeld A, Fewell GA, Flicek P, Foley K, Frankel WN, Fulton LA, Fulton RS, Furey TS, Gage D, Gibbs RA, Glusman G, Gnerre S, Goldman N, Goodstadt L, Grafham D, Graves TA, Green ED, Gregory S, Guigo R, Guyer M, Hardison RC, Haussler D, Hayashizaki Y, Hillier LW, Hinrichs A, Hlavina W, Holzer T, Hsu F, Hua A, Hubbard T, Hunt A, Jackson I, Jaffe DB, Johnson LS, Jones M, Jones TA, Joy A, Kamal M, Karlsson EK, Karolchik D, Kasprzyk A, Kawai J, Keibler E, Kells C, Kent WJ, Kirby A, Kolbe DL, Korf I, Kucherlapati RS, Kulbokas EJ, Kulp D, Landers T, Leger JP, Leonard S, Letunic I, Levine R, Li J, Li M, Lloyd C, Lucas S, Ma B, Maglott DR, Mardis ER, Matthews L, Mauceli E, Mayer JH, McCarthy M, McCombie WR, McLaren S, McLay K, McPherson JD, Meldrim J, Meredith B, Mesirov JP, Miller W, Miner TL, Mongin E, Montgomery KT, Morgan M, Mott R, Mullikin JC, Muzny DM, Nash WE, Nelson JO, Nhan MN, Nicol R, Ning Z, Nusbaum C, O'Connor MJ, Okazaki Y, Oliver K, Overton-Larty E, Pachter L, Parra G, Pepin KH, Peterson J, Pevzner P, Plumb R, Pohl CS, Poliakov A, Ponce TC, Ponting CP, Potter S, Quail M, Reymond A, Roe BA, Roskin KM, Rubin EM, Rust AG, Santos R, Sapojnikov V, Schultz B, Schultz J, Schwartz MS, Schwartz S, Scott C, Seaman S, Searle S, Sharpe T, Sheridan A, Shownkeen R, Sims S, Singer JB, Slater G, Smit A, Smith DR, Spencer B, Stabenau A, Stange-Thomann N, Sugnet C, Suyama M, Tesler G, Thompson J, Torrents D, Trevaskis E, Tromp J, Ucla C, Ureta-Vidal A, Vinson JP, Von Niederhausern AC, Wade CM, Wall M, Weber RJ, Weiss RB, Wendl MC, West AP, Wetterstrand K, Wheeler R, Whelan S, Wierzbowski J, Willey D, Williams S, Wilson RK, Winter E, Worley KC, Wyman D, Yang S, Yang SP, Zdobnov EM, Zody MC, Lander ES. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Whatley VJ, Johnson TE, Erwin VG. Identification and confirmation of quantitative trait loci regulating alcohol consumption in congenic strains of mice. Alcohol Clin Exp Res. 1999;23:1262–1271. doi: 10.1111/j.1530-0277.1999.tb04287.x. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Schuckit M, Smith TL, Lee JV, Segall SK, Feiler HS, Kalmijn J. The search for genes related to a low-level response to alcohol determined by alcohol challenges. Alcohol Clin Exp Res. 2003;27:1041–1047. doi: 10.1097/01.ALC.0000075551.02714.63. [DOI] [PubMed] [Google Scholar]
- Wilhelmsen KC, Swan GE, Cheng LS, Lessov-Schlaggar CN, Amos CI, Feiler HS, Hudmon KS, Ring HZ, Andrews JA, Tildesley E, Benowitz NL, Hops H. Support for previously identified alcoholism susceptibility Loci in a cohort selected for smoking behavior. Alcohol Clin Exp Res. 2005;29:2108–2115. doi: 10.1097/01.alc.0000191773.68675.71. [DOI] [PubMed] [Google Scholar]
- Williams JT, Begleiter H, Porjesz B, Edenberg HJ, Foroud T, Reich T, Goate A, Van EP, Almasy L, Blangero J. Joint multipoint linkage analysis of multivariate qualitative and quantitative traits. II. Alcoholism and event-related potentials. Am J Hum Genet. 1999;65:1148–1160. doi: 10.1086/302571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynski DF, Panhuysen CI, Ma Q, Yip AG, Wilcox M, Erlich P, Farrer LA. Genome-wide screen for heavy alcohol consumption. BMC Genet. 2003;4(Suppl 1):S106. doi: 10.1186/1471-2156-4-S1-S106. [DOI] [PMC free article] [PubMed] [Google Scholar]