The cysteine thiolate ligand coordinated to the heme iron atom in cytochrome P450 enzymes is thought to be responsible for the unique spectroscopic and catalytic properties of these enzymes. In order to explore the role of the proximal ligand in these proteins, a variety of proximal ligands have been substituted for the cysteine by site-specific mutagenesis, including a histidine,[1a-c] methionine,[1d] and serine.[1e] None of these ligand substitutions yielded a protein with the spectroscopic or catalytic properties of a cytochrome P450 monooxygenase. However, recent computational studies suggested that a P450-like species might result from substitution of a selenolate for the thiolate ligand and, furthermore, that this substitution might accelerate the rate of formation and decelerate the rate of decay of the catalytic ferryl species, possibly making it observable.[2]
No selenolate-coordinated heme protein was known until our recent demonstration that such a protein is generated by the binding of PhSeH to a heme oxygenase cavity mutant in which the proximal histidine ligand had been replaced by an alanine.[3] Furthermore, binding of PhSeH to cytochrome P450cam yielded a hyperporphyrin spectrum analogous to that observed when a distal thiolate ligand is coordinated to the iron in addition to the proximal cysteine thiolate.[3] Nevertheless, only a brief meeting abstract exists describing a P450 enzyme in which the proximal cysteine has been replaced by a selenocysteine.[4]
Here we report the expression and characterization of a seleno cytochrome P450 in which the cysteine thiolate iron ligand is replaced by a selenocysteine. CYP119 was used for this substitution because the proximal ligand is the only cysteine in its sequence. The seleno protein was expressed in a cysteine auxotroph BL21(DE3)CysE strain of Escherichia coli that cannot synthesize cysteine due to a mutation in the CysE gene.[5] A pCWori vector containing the CYP119 gene encoding a 6-His tag at the C-terminus was transformed into the auxotrophic BL21(DE3)Cys cells and the seleno protein was expressed in minimal media containing L-selenocystine (see Supporting Information for a detailed protocol). The protein yield was 2.6 mg/L of culture after affinity purification, which is approximately 8-10 times lower than that of the normal thiolate-ligated protein. This approach results in over 70% replacement of the cysteine by a selenocysteine, as judged by the relative peak intensities of the Cys and SeCys proteins by LC/ESI-MS.
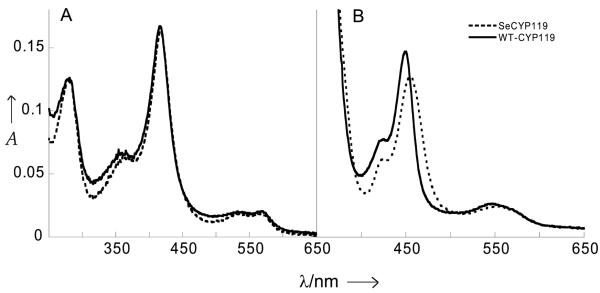
The UV-vis spectrum of ferric SeCYP119, the selenocysteine-substituted enzyme, has a Soret maximum at 417 nm very similar to the 416 nm maximum of WT CYP119.[6] Furthermore, the Q bands of the SeCYP119 protein correlate well with those of WT CYP119 (Figure 1A). The similarities of the UV-vis spectra of ferric SeCYP119 and WT CYP119 are consistent with the presence in both proteins of a hexacoordinated low-spin heme iron with a distal water ligand.
Figure 1.
Comparison of the UV-vis spectra of ~ 1.5 μM CYP119 and SeCYP119 proteins in 100 mM potassium phosphate buffer, pH 7.4. A) ferric resting state and B) ferrous-CO complex
The catalytic activity of the enzymes was examined using a hydrogen peroxide mediated shunt pathway in the presence of the substrate dodecanoic (lauric) acid. The specific activity of SeCYP119 was estimated to be 90 ± 20 pmoles min−1 nmole−1 of enzyme, which is approximately two fold less compared to 170 ± 18 pmoles min−1 nmole−1 for the WTCYP119.[7] Oxidation of lauric acid resulted in hydroxylation at the ω, ω-1 and ω-2 positions as determined by GC-MS. Interestingly, the regiospecificity of hydroxylation showed the same trend for both the WT and SeCYP119 proteins, with ω-1 being the predominant product followed by ω-2 and ω (see SI). Overall, our results suggest that the novel seleno mutant is indeed catalytically active.
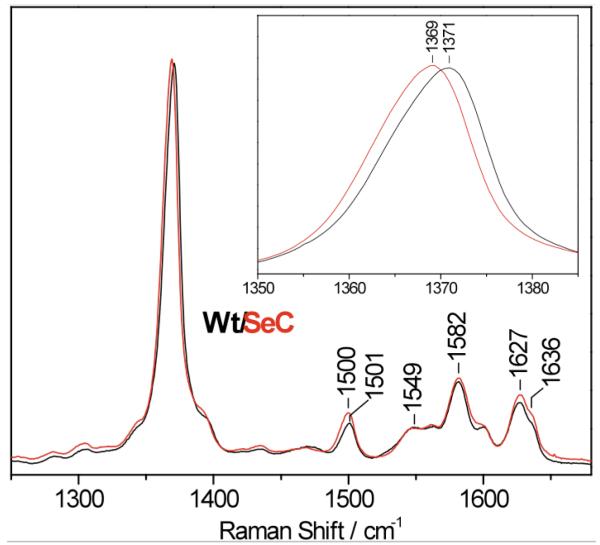
The EPR spectrum of ferric SeCYP119 is characteristic of a rhombic S = 1/2 low-spin heme iron (III) with g values that are distinct from those of the WT protein (Figure S4). Interpreting the changes in g values in electronic or structural terms is not straightforward since the Se substitution may affect tetragonal and rhombic splittings of the low-spin iron(III) d orbitals as well as the spin-orbit coupling constant.[8] The EPR spectra also support the presence of a WT CYP119 contaminant that varies from preparation to preparation but remains below 30%. The resonance Raman spectrum of ferric SeCYP119 also supports a hexacoordinate low-spin configuration[9] with porphyrin skeletal modes ν3, ν2, and ν10 at 1502, 1583, and 1636 cm−1, respectively (Figure 2). These frequencies are within 1-cm−1 of those observed in WT CYP119. The ν4 mode shows a 1-cm−1 downshift with the Se to S substitution which reflects an increased electron density on the antibonding porphyrin π* orbitals.[10]
Figure 2.
Resonance Raman spectra of ferric wild-type (WT) CYP119 (black) and SeCYP119 (red) at room temperature (excitation wavelength, 413 nm)
The impact of the Cys317SeCys substitution in SeCYP119 was investigated further by characterizing the ferric-nitrosyl complexes using resonance Raman spectroscopy and isotopic labeling of the nitrosyl group. In SeCYP119, the ν(Fe-NO) and ν(N-O) modes are observed at 526 (Δ15N = 3 cm−1) and 1838 cm-1 (Δ15N = 37 cm−1), respectively (Figure S5). These frequencies are 4 and 14 cm−1 lower than in the WT protein. These decreases in stretching frequencies can be interpreted in terms of a weakening of the Fe-NO and N-O bond strengths as the σ-trans effect of the coordinating selenolate ligand on the Fe-N-O unit increases compared to that of the thiolate in the WT protein. Recent DFT calculations have examined in detail the trans effect of thiolate ligation in ferric heme nitrosyl complexes.[11]
The UV-vis spectra of the ferrous-CO complexes show a pronounced shift in Soret maximum from 449 nm in WT CYP119 to 454 nm in SeCYP119 (Figure 1B), but the resonance Raman characterizations of these carbonyl complexes show nearly identical ν(Fe-CO) and ν(C-O) frequencies (Figure S6). Thus in the ferrous-CO complexes, and in contrast with the ferric-NO complexes, the trans effect of the selenolate and thiolate ligands on the Fe-C-O unit are equivalent.
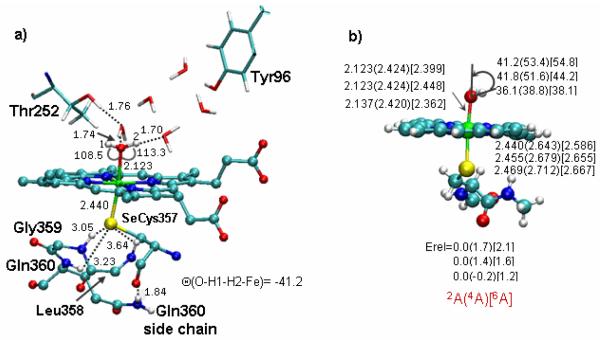
To model the three-dimensional structure of the Se-resting state in SeCYP119, we used theoretical DFT(B3LYP)/MM calculations of the type described before to probe an analogous SeCYP101 mutant.[2][12a] Three snapshots (60 ps ,120 ps and 200 ps) from the equilibrium trajectory of the WT enzyme were mutated by replacing cysteine by selenocysteine and fully optimized by DFT(B3LYP)/MM geometry optimization for the spin states with S = 1/2, 3/2, and 5/2.[12b] Details of the various calculations are provided in the Supporting Information (SI) document. All three snapshots display similar properties, and Figure 3 depicts representative geometric data, calculated at the highest basis set, of triple zeta quality augmented by polarization and diffuse functions (see SI). The Fe-Se distance is 2.44 Å, within the range found in iron-seleno complexes.[13] As seen from part b of the figure, the calculations predict small energetic gaps between the three spin states, which are packed within 2.1 kcal/mol or so. Given that B3LYP/MM slightly overestimates the stability of high spin states,[12b] along with the BLYP/MM data in the SI, which strongly prefer the doublet state, we can assign the ground state to have a doublet spin. This feature is similar to the computed situation for the WT and to the experimental finding that the ground state (doublet) is in spin-equilibrium with the higher spin states.[14] Comparisons to the WT geometries[12b] suggest that in both cases, the water ligand is tilted by 40-60° due to H-bond interactions within the protein pocket (Figure 3b). The Fe-O bond length is slightly shorter by ca 0.02 Å while the Fe-Se bond length is longer than Fe-S by ca 0.2 Å, as previously reported for the seleno and WT Cpd I species.[2] Thus, the DFT/MM calculations support the doublet spin-state assignment by EPR.
Figure 3.
B3LYP/MM calculated geometric features and spin-state energies of the SeCys mutant of CYP101: (a) Detailed structure of a representative snapshot (60 ps) in the doublet 2A state. (b) Relative QM/MM energies (kcal/mol) and key geometric parameters for different snapshots calculated with the largest basis set (BL//B1 in the SI). The geometric parameters correspond to snapshot 60 ps, snapshot 120 ps and snshot 200 ps (arranged vertically) in the 2A(4A)[6A] spin states
In conclusion, we report the expression and characterization of the first selenium incorporated P450 enzyme. Our preliminary results reveal that the spectral characteristics of this novel seleno CYP119 are comparable to those of the corresponding WT protein, indicating the presence of a hexacoordinated low-spin heme iron with water as a distal ligand. More importantly, the catalytic activity of the seleno mutant is comparable to that of the WT enzyme. Furthermore, computational calculations clearly support the experimentally assigned spin state. Future studies will focus on both examining how this substitution affects the stability of the putative Cpd I species and the development of novel catalysts.
Supplementary Material
Footnotes
This work is supported by NIH grants GM25515 (to PROM) and GM74785 (to PML) and by an ISF grant (to SS). Mass spectrometry was provided by the UCSF Mass Spectrometry Facility (A.L. Burlingame, Director) supported by the Biomedical Research Technology Program of the National Center for Research Resources, NIH NCRR BRTP 01614. We thank Dr. Hugues Ouellet for helpful discussions and suggestions.
Contributor Information
Yongying Jiang, Department of Pharmaceutical Chemistry University of California, 600 16th street, San Francisco, CA 94158 (USA).
Santhosh Sivaramakrishnan, Department of Pharmaceutical Chemistry University of California, 600 16th street, San Francisco, CA 94158 (USA).
Takahiro Hayashi, Department of Science and Engineering, Oregon Health & Science University, Beaverton, OR 97006 (USA).
Shimrit Cohen, Institute of Chemistry and the Lise Meitner-Minerva Center for Computational Quantum Chemistry, The Hebrew University, Jerusalem, 91904 (Israel).
Pierre Moënne-Loccoz, Department of Science and Engineering, Oregon Health & Science University, Beaverton, OR 97006 (USA).
Sason Shaik, Institute of Chemistry and the Lise Meitner-Minerva Center for Computational Quantum Chemistry, The Hebrew University, Jerusalem, 91904 (Israel).
Paul R. Ortiz de Montellano, Department of Pharmaceutical Chemistry University of California, 600 16th street, San Francisco, CA 94158 (USA).
References
- 1 a).Auclair K, Moënne-Loccoz P, de Montellano P. R. Ortiz. J. Am. Chem. Soc. 2001;123:4877. doi: 10.1021/ja0040262. [DOI] [PubMed] [Google Scholar]; b) Yoshioka S, Takahashi S, Hori H, Ishimori K, Morishima I. Eur. J. Biochem. 2001;268:252. doi: 10.1046/j.1432-1033.2001.01872.x. [DOI] [PubMed] [Google Scholar]; c) Vatsis KP, Peng H–M, Coon MJ. Arch. Biochem. Biophys. 2004;434:128. doi: 10.1016/j.abb.2004.10.015. [DOI] [PubMed] [Google Scholar]; d) Murugan R, Mazumdar S. ChemBioChem. 2005;6:1204. doi: 10.1002/cbic.200400399. [DOI] [PubMed] [Google Scholar]; e) Vatsis KP, Peng H–M, Coon MJ. J. Inorg. Biochem. 2002;91:542. doi: 10.1016/s0162-0134(02)00438-5. [DOI] [PubMed] [Google Scholar]
- 2.Cohen S, Kumar D, Shaik S. J. Am. Chem. Soc. 2006;128:2649. doi: 10.1021/ja056586c. [DOI] [PubMed] [Google Scholar]
- 3.Jiang Y, de Montellano P. R. Ortiz. Inorg. Chem. 2008;47:3480. doi: 10.1021/ic800148a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gromov I, Garcia-Rubio I, Aldag C, Schweiger A, Hilvert D. 6th European Federation EPR Groups Meeting; 2006. Abstract OT3. When this work was under review, Aldag et. al, published a paper on the expression and characterization of the seleno mutant of P450cam. See Aldag C, Gromov IA, Rubio IG, von Koenig K, Schlichting I, Jaun B, Hilvert D. Proc. Natl. Acad. Sci. U.S.A. 2009;106:5481. doi: 10.1073/pnas.0810503106.
- 5.Müller S, Senn H, Gsell B, Vetter W, Baron C, Böck A. Biochemistry. 1994;33:3404. doi: 10.1021/bi00177a034. [DOI] [PubMed] [Google Scholar]
- 6.Koo LS, Tschirret-Guth RA, Straub WE, Moë nne-Loccoz P, Loehr TM, de Montellano P. R. Ortiz. J. Biol. Chem. 2000;275:14112. doi: 10.1074/jbc.275.19.14112. [DOI] [PubMed] [Google Scholar]
- 7.The catalytic activity of SeP450cam is also reported to be 2 fold less than the activity of the corresponding WT control (see Ref 4).
- 8.Palmer G. In: Iron Porphyrins, Part II. Lever ABP, Gray HB, editors. Addison-Wesley; Reading MA: 1983. pp. 43–88. [Google Scholar]
- 9.Spiro TG, Li XY. Biological Applications of Raman Spectroscopy. Vol 3. John Wiley & Sons; New York: 1998. [Google Scholar]; Spiro TG. Resonance Raman spectra of hemes and metalloproteins. John Wiley & Sons; New York: 1998. pp. 1–37. [Google Scholar]
- 10.Spiro TG, Burke JM. J. Am. Chem. Soc. 1976;98:5482. doi: 10.1021/ja00434a013. [DOI] [PubMed] [Google Scholar]
- 11.Paulat F, Lehnert N. Inorg. Chem. 2007;46:1547. doi: 10.1021/ic070023f. [DOI] [PubMed] [Google Scholar]
- 12 a).Denisov, et al. J. Inorg. Biochem. 2001;87:215. doi: 10.1016/s0162-0134(01)00328-2. [DOI] [PubMed] [Google Scholar]; b) Schöneboom JC, Thiel W. J. Phys. Chem. B. 2004;108:7468. [Google Scholar]
- 13 a).Hauptmann R, Kliss R, Henkel G. Angew. Chem. Int. 1999;38:377. doi: 10.1002/(SICI)1521-3773(19990201)38:3<377::AID-ANIE377>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]; b) Song L–C, Sun Y, Hu Q–M, Liu YJ. Organomet. Chem. 2003;676:80. [Google Scholar]; c) Song L–C, Yang J, Hu Q–M, Wu Q–J. Organometallics. 2001;20:3293. [Google Scholar]; d) Song L–C, Fan H–T, Hu Q–M, Yang Z–Y, Yi S, Gong FH. Chem. Eur. J. 2003;9:170. doi: 10.1002/chem.200390010. [DOI] [PubMed] [Google Scholar]; e) El-khateeb M. Inorg. Chim. Acta. 2004;357:4341. [Google Scholar]
- 14 a).Thomann H, Bernardo M, Goldfarb D, Kroneck PMH, Ullrich V. J. Am. Chem.Soc. 1995;117:8243. [Google Scholar]; b) Sharrock M, Debrunner PG, Schulz C, Lipscomb JD, Marshall V, Gunsalus IC. Biochim. Biophys. Acta. 1976;420:8. doi: 10.1016/0005-2795(76)90340-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.