Abstract
Most tailed bacteriophages with double-stranded DNA genomes code for a scaffolding protein, which is required for capsid assembly, but is removed during capsid maturation and DNA packaging. The gpO scaffolding protein of bacteriophage P2 also doubles as a maturation protease, while the scaffolding activity is confined to a 90 residue C-terminal “scaffolding” domain. Bacteriophage HK97 lacks a separate scaffolding protein; instead, an N-terminal “delta” domain in the capsid protein appears to serve an analogous role. We asked whether the C-terminal scaffolding domain of gpO could work as a delta domain when fused to the gpN capsid protein. Varying lengths of C-terminal sequences from gpO were fused to the N-terminus of gpN and expressed in E. coli. The presence of just the 41 C-terminal residues of gpO increased the fidelity of assembly and promoted the formation of closed shells, but the shells formed were predominantly small, 40 nm shells, compared to the normal, 55 nm P2 procapsid shells. Larger scaffolding domains fused to gpN caused the formation of shells of varying size and shape. The results suggest that while fusing the scaffolding protein to the capsid protein assists in shell closure, it also restricts the conformational variability of the capsid protein.
Keywords: Virus, Assembly, Procapsid, Size determination, Cryo-electron microscopy
Introduction
Capsid proteins of double-stranded (ds) DNA bacteriophages often have an intrinsic ability to assemble into shell-like structures in the absence of any other gene products. However, these structures are usually aberrant in nature. Regulation of capsid assembly usually requires the action of a scaffolding protein, which acts during capsid assembly, but is typically absent from the mature capsid [1, 2].
The dsDNA bacteriophage P2 forms an icosahedral capsid with T = 7 symmetry from 415 copies of the 40.2 kDa gpN capsid protein (five copies of gpN are replaced by the portal protein at the unique vertex) [3]. Correct assembly requires the 284-residue gpO protein, which serves a dual role as an internal scaffolding protein and a protease [4, 5]. The gpO protein is autoproteolytically cleaved to a 141 amino acid N-terminal fragment, O*, that remains inside the mature capsid after DNA packaging [6, 7]. Concurrently, gpN is N-terminally processed—presumably by gpO—to a 36.7 kDa cleavage product, N* [8–10].
In the presence of the genetically unrelated satellite phage P4, however, gpN is assembled into a smaller capsid with T = 4 symmetry, suitable for packaging the 11.6 kbp P4 genome, but unable to package the larger P2 genome (33.6 kbp) [11]. This size determination is dependent on the P4-encoded Sid protein [12], which forms an external scaffold around the P4 procapsids [13, 14]. Sid promotes the formation of P4-like T = 4 procapsids even in the absence of gpO [14, 15], although formation of viable P4 phage is dependent on gpO [8, 16].
We previously showed that the C-terminal 90 amino acids (residues 195–284) of gpO were required and sufficient to promote assembly of well-formed P2-like procapsids [7, 17]. This sequence interacts only transiently with gpN, and forms dimers or trimers in solution, suggesting that scaffolding activity involves gpO oligomerization [17]. The protease activity was found to require the three amino acids Asp19, His 48, and Ser107, which form a serine protease-like catalytic triad in the N-terminal half of the protein [17]. The C-terminal domain of gpO is predominantly α-helical, with a long helix predicted between residues 197 and 240 and a shorter helix from residue 246–257 (Fig. 1), suggesting a structural similarity with the helix-loop-helix motif of the scaffolding proteins of bacteriophages ϕ29 and P22 [18, 19].
Fig. 1.
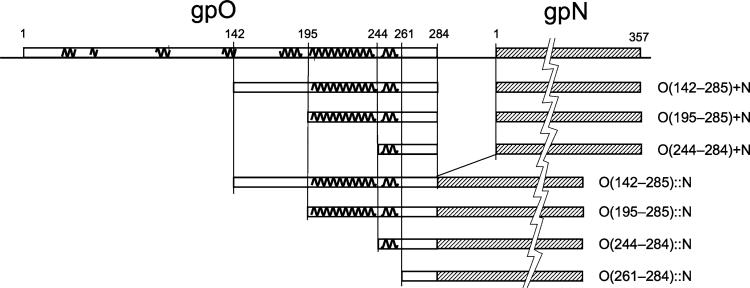
gpO and gpN co-expression and fusion protein constructs. Predicted α-helices are indicated as squiggly lines on top of the gpO protein. The 357 residue gpN protein is shown cross-hatched and is not drawn to scale, as indicated by the jagged line. The gpO truncations start at the indicated residues (142, 195, 244, or 261) and end at the gpO C-terminus (residue 284). All constructs include the full-length gpN protein
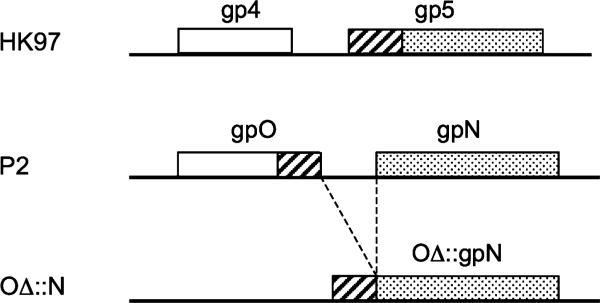
The lambdoid phage HK97 and its relatives are unusual among the dsDNA phages in that they do not employ a separate scaffolding protein. Instead, this role appears to be played by a 102 residue N-terminal “delta” (δ) domain of the capsid protein (gp5), which is subsequently cleaved off during capsid maturation [20, 21] (Fig. 2). Like most scaffolding proteins, the δ domain is also predominantly α-helical [20]. Other HK97-like phages have δ domains varying in length from 102 to 159 residues, which seems to be correlated with capsid size (James Conway and Robert Duda, pers. comm.) [22]. The gene for the capsid protein is preceded in the HK97 genome by a separate protease gene (gene 4).
Fig. 2.
Schematic diagram comparing the order of the protease (open box), scaffolding (cross hatched) and capsid (dotted) activities in the genomes of HK97 and P2 (drawn to scale). In HK97, the gene encoding the protease (gp4) precedes the gene for the capsid protein (gp5), which incorporates an N-terminal scaffolding domain. In P2, the protease and scaffolding activities are both contained within the gpO protein. The last line represents any of the various constructs that were made by fusing the C-terminal scaffolding domain of O with the N capsid gene
In light of the similarities between the δ domain of gp4 and the C-terminal “scaffolding” domain of gpO, we asked whether the C-terminal 90 residues (or less) of gpO could function as a δ domain, fused to the gpN capsid protein that follows it in the genome (Figs. 1, 2). Perhaps the gpO protein might represent an in-sequence fusion between protease and scaffolding activities found in a progenitor phage, while HK97 represents a similar fusion of the scaffolding activity to the capsid protein?
We show that the gpO scaffolding domain does indeed promote shell closure when it is fused to gpN, but the resulting shells are predominantly of small “P4-like” size, in addition to closed shells of aberrant shapes. Our results suggest that the presence of gpO fused to gpN restricts the conformational variability of gpN and thereby affects capsid size, but that assembly fidelity depends on transient gpO–gpN interactions.
Materials and methods
Cloning and expression of tandem and fusion constructs
The truncated O genes were made by PCR from genomic P2 DNA and inserted between the NcoI and NdeI sites of the vector pET16b (Novagen) under the control of the T7 promoter as previously described [5, 7]. Tandem clones of truncated O with full-length N were produced by inserting the N gene, including an artificial ribosome binding site, between the NdeI and XhoI sites in the appropriate O deletion construct. Clones expressing the truncated gpO::gpN fusion products were generated using either the QuikChange Lightning or QuikChange II Site-Directed Mutagenesis kits (Stratagene) by removing sequences between and including the NdeI and XhoI sites, thus fusing the 3′ end of O to the 5′ end of N. The co-expression clones O(244–484)N::N + N, O(195–284)194 N::N + N, and O(142–284)N::N + N were produced by the addition of full-length N between the XhoI and BamHI sites of the respective fusion clones.
For expression, all clones were grown at 37°C in LB with 100 μg/ml ampicillin until OD600 reached 0.4–0.6, induced by the addition of 0.5 mM IPTG, and harvested after 2 h. The cloning was done in the E. coli recA– strain DH5α (Invitrogen), while expression was performed in BL21 (DE3) (Novagen).
Protein purification
The harvested cells were resuspended in lysis buffer (10 mM Tris–HCl pH 7.4, 200 mM NaCl, 1 mM PMSF, 1% Triton X-100, and 0.5% deoxycholate) and frozen overnight at -20°C. The thawed and resuspended cells were lysed by sequential passages at 1,000; 5,000; 10,000, and 15,000 psi through an Emulsiflex EF-C3 high-pressure cell disruptor (Avestin Inc., Ottawa). The lysates were clarified by centrifugation at 11,800×g for 30 min. In order to collect the formed particles, the clarified supernatants were centrifuged at 40,000 rpm (114,000×g) for 1 h in a Beckman Type 60Ti rotor. The pellets were resuspended in procapsid buffer (50 mM Tris–HCl, pH 8.0, 100 mM NaCl, 10 mM MgCl2), loaded onto 10–40% sucrose gradients prepared in the same buffer and centrifuged for 2 h at 30,000 rpm (110,000×g) in a Beckman SW 41 rotor. One milliliter gradient fractions were collected manually. Total lysates, pellets, and sucrose gradient fractions were analyzed by SDS–PAGE. Fractions were pooled and then pelleted by centrifugation for 40 min at 50,000 rpm (178,000×g) in a Beckman Type 60Ti rotor. The resulting pellet was resuspended in procapsid buffer and prepared for EM.
Electron microscopy and image analysis
The virus samples were dialyzed against EM buffer (10 mM Tris–HCl, pH 8.0, 20 mM NaCl, 2 mM MgCl2)on a 0.025 lm filter for 30 min and diluted in EM buffer to a suitable concentration. Cryo-EM was done by standard methods [23]: 3 μl of sample was applied to C-flat holey film (Protochips, Inc.), blotted briefly before plunging into liquid ethane and transferred to a Gatan 626 cryo-sample holder. All samples were observed in an FEI Tecnai F20 electron microscope operated at 200 kV and images were captured on a 4 k × 4 k Gatan Ultrascan CCD camera at magnifications of 38,000× or 65,500×.
Image analysis and generation of radial plots was done using programs from the EMAN suite [24].
Results
Co-expression of gpN and truncated gpO proteins
In this and previous studies, we generated a range of clones expressing gpN alone in combination with various full-length and truncated forms of gpO [4, 5, 7, 15, 17]. The co-expression clones that express the truncated gpO proteins together with gpN are denoted as O(142–284) + N, O(195–284) + N, etc., where the numbers in parenthesis indicate the start and end of the included gpO sequence (Fig. 1).
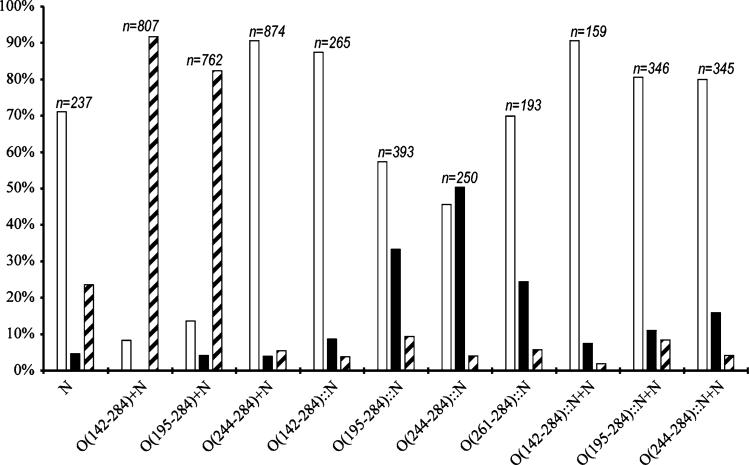
All clones expressing the gpN capsid protein produce capsid-related particles of various sizes and shapes. These particles were harvested by centrifugation at 114,000×g, followed by separation on 10–40% sucrose gradients and analysis by SDS–PAGE to assay for capsid formation and gpO incorporation. The capsid protein-containing fractions were pooled in order to include all capsid-related structures in the sample, concentrated by centrifugation and analyzed by negative stain electron microscopy (EM) and cryo-EM as previously described [23]. The particles in the micro-graphs were counted, classified according to whether they were aberrant or closed and isometric. Isometric shells generally fall into discrete size classes according to the rules of triangulation [25]. Most of the isometric shells produced by the gpO + gpN co-expressions fell into two size classes, namely T = 7 or “P2-like” shells of 55 nm diameter and T = 4 or “P4-like” shells of around 40 nm diameter. For the purpose of classification, shells of ≥46 nm in diameter were considered large or “P2-like” shells, while those <46 nm in diameter were classified as small or “P4-like” shells, and denoted as such in the following discussion. These data are represented graphically in Fig. 3.
Fig. 3.
Size distribution histogram of the particles formed by gpN alone and the various co-expression and fusion constructs described in the text. Open bars represent aberrant particles; solid bars, P4-like, small particles (<46 nm); and hatched bars, P2-like, large particles (≥46 nm). The number of particles counted for each construct (n) is shown above the bars
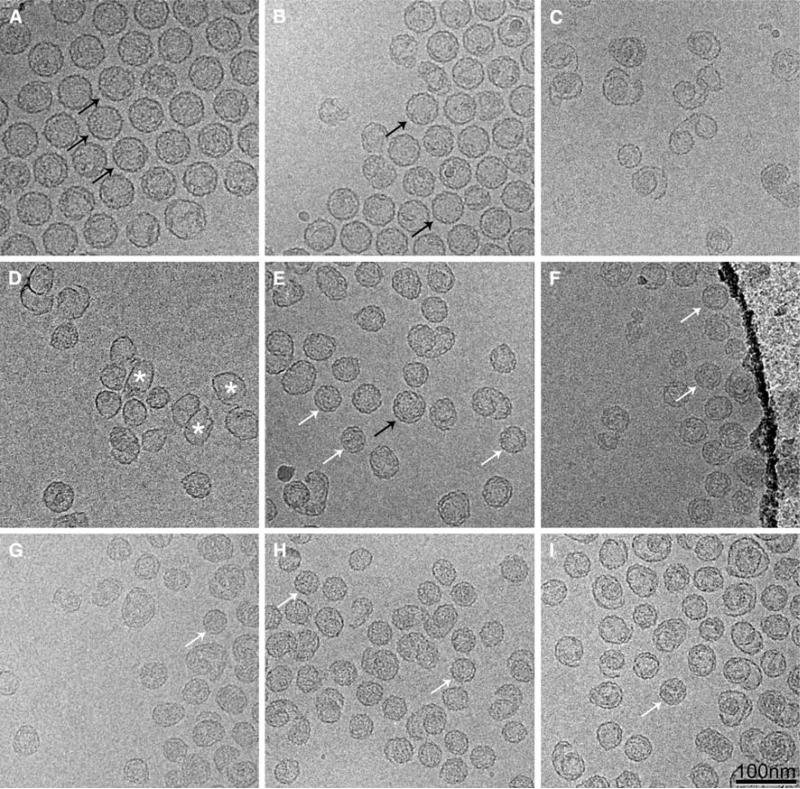
As previously described, expression of gpN alone leads to the formation of predominantly aberrant, unclosed shells [4, 15] (Fig. 3). We also showed that O(142–284), which represents the C-terminal half of gpO that is proteolytically removed during capsid maturation, and O(195–284), containing the last 90 amino acids of gpO, were both able to efficiently promote the formation of procapsids [7, 17]. Thus, co-expression clones O(142–284) + N and O(195–284) + N yielded 92 and 82% well-formed P2-size procapsids, respectively (Fig. 3). O(195–184) includes a segment of predicted α-helical structure from amino acid 197 to 257 that is involved in oligomerization and is important for scaffolding activity (Fig. 1)[7, 17]. Shorter gpO constructs, such as O(244–284), that did not include this helix, failed to promote capsid formation [17], resulting in 83% aberrant shells (Fig. 3). Examples of particles produced by the O(142–284) + N, O(195–284) + N, and O(244–284) + N co-expression clones are shown in Fig. 4a–c.
Fig. 4.
Cryo-EM of structures produced by expression of co-expression clones O(141–284) + N (A), O(195–284) + N (B) and O(244–284) + N (C); fusion protein clones O(141–284)::N (D), O(195– 284)::N (E) and O(244–284)::N (F); and fusion/co-expression clones O(141–284)::N + N (G) and O(195–284)::N + N (H) and O(244–284)::N + N (I). Examples of well-formed particles of P2 and P4 size are indicated in each panel (where available) by black (P2) and white (P4) arrows. Some of the many thin-walled shells produced in (D) are indicated with asterisks. Scale bar, 100 nm
Expression of gpO::gpN fusion protein
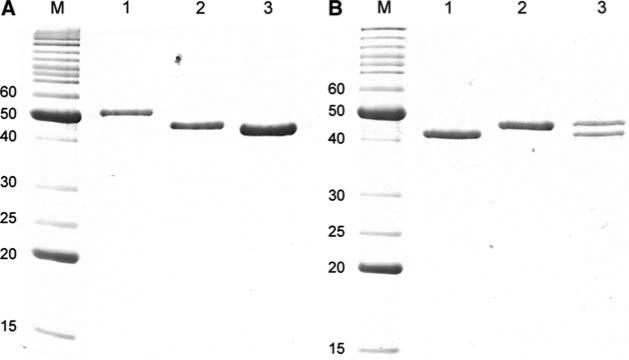
In order to test whether the α-helical scaffolding domain of gpO could function as a δ domain fused to gpN, several fusion protein constructs were generated by removing the interspersing non-coding sequences from the O(142–284) + N, O(195–284) + N, and O(244–284) + N co-expression clones (Fig. 1, 2). In one additional clone, only residues 261–284 of gpO were fused to gpN. The resulting fusion clones were denoted as O(142–284)::N, O(195– 284)::N, O(244–284)::N, and O(261–284)::N (Fig. 1) and produced fusion proteins of the expected size (Fig. 5a). In all cases, most of the protein was found in a rapidly sedimenting fraction that was pelleted at 114,000×g and purified on sucrose gradients, as described above. The size distribution of the resulting shells is shown in Fig. 3.
Fig. 5.
a Coomassie-stained SDS–PAGE of proteins expressed by fusion protein clones. Lane 1, O(142–284)::N; Lane 2, O(195–284)::N; lane 3, O(244–284)::N. b SDS–PAGE of gpN alone (lane 1), the O(195–284)::N fusion protein (lane 2), and the proteins expressed from the O(195–284)::N + N co-expression clone (lane 3). M marker, MW indicated (kDa)
Unlike the O(142–284) + N co-expression, expression of the O(142–284)::N fusion protein yielded shells that were predominantly aberrant (88%) in shape and size and mostly thin-walled (Fig. 4d). Expression of the O(194–284)::N fusion protein, on the other hand, yielded more isometric, closed shells (Fig. 4e). Only 57% of the shells were aberrant in this case. Many of these shells appeared to be closed shells of aberrant shapes rather than open “spiral” forms. Interestingly, the isometric shells were predominantly of small, P4-like size (33% of the total), a significant shift from the predominantly large, P2-like procapsids produced in the co-expressions (Fig. 3). Only 9% of the shells had the large, 55 nm size indicative of P2 procapsids.
Expression of the O(244–284)::N fusion protein resulted in an even greater proportion of closed, isometric shells (54%) than O(195–284)::N (Fig. 4e). The majority of these (50%) were small, P4-like shells. This was surprising, since O(244–284) does not include the important α-helical sequence and since co-expression of gpN and O(244–284) led to predominantly (83%) aberrant, mostly unclosed shells (Fig. 4c). As might be expected, expression of the fusion protein O(261–284)::N produced predominantly (70%) aberrant particles, about the same as that produced by gpN alone. Of those 30% shells that were closed and isometric, however, the majority were of small size (24%), unlike the case with expression of gpN alone, where most shells are of large size (24% of total) (Fig. 3).
Co-expression with combinations of O::N fusion protein with gpN
Having the gpO scaffolding protein fused to the gpN capsid protein naturally results in an equimolar ratio of scaffolding to capsid. In capsids produced by co-expression of gpO and gpN, however, the ratio of gpO:gpN is about 1:3 [5, 7]. In order to test the effect of this ratio on the size of the assembled capsids, three co-expression clones were generated, in which either the O(142–284)::N, O(195–284)::N, or O(244–284)::N fusion protein was expressed in tandem with gpN, resulting in clones O(142–284)::N + N, O(195–284)::N + N, and O(244–284)::N + N, respectively. When these clones were expressed, approximately equimolar amounts of unfused gpN and gpO::gpN fusion protein were produced (Fig. 5b). Expression of either of the three clones resulted in predominantly aberrant capsids (from 80% to 90%; Fig. 3; Fig. 4g–i). The 10–20% isometric capsids that formed were mostly of small P4 size (Fig. 4g–i). Few larger P2-size procapsids were produced, demonstrating that the ratio of scaffolding to capsid protein per se is not sufficient to enable the formation of large shells. We considered the possibility that the increased the presence of aberrant shells in these co-expressions was due to the formation of aberrant shells consisting only of gpN, with the fusion proteins segregating into separate particles.
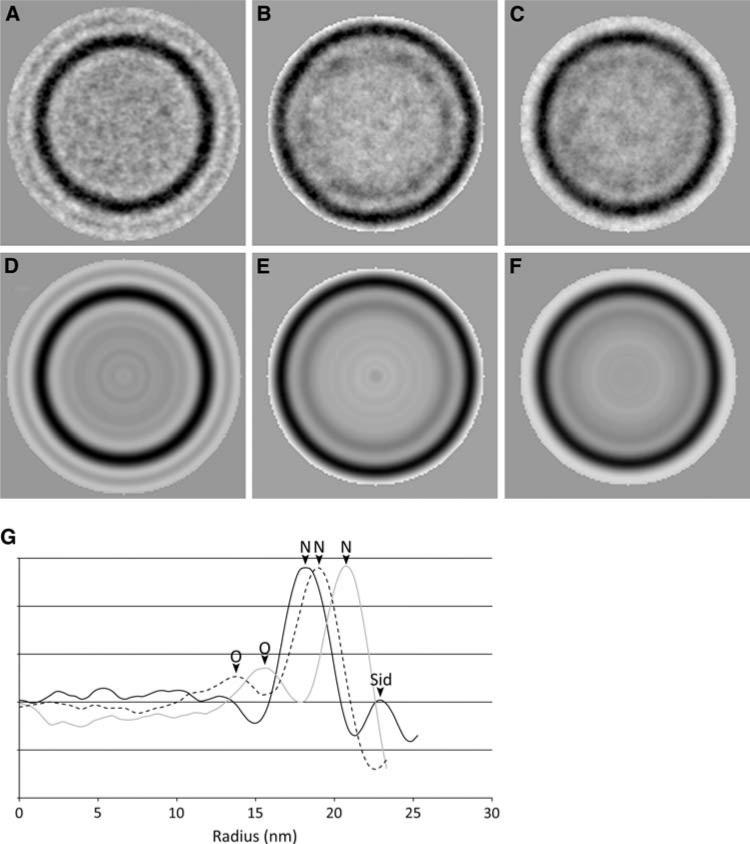
In order to analyze the scaffolding content of the expression constructs, isometric, P4-like shells were picked from the O(195–284)::N fusion and O(195–284)::N + N co-expressions (69 and 71 particles, respectively), aligned and averaged together rotationally (Fig. 6). These images were compared to a rotational average similarly generated from non-scaffolding containing P4 procapsids generated by co-expression of gpN and Sid [14]. The density was then plotted as a function of radius (Fig. 6g). These plots show clearly the presence of an additional inner shell in O(195–284)::N that is not seen in the gpN + Sid procapsids. The O(195–284)::N + N also have an internal shell, but in this case the density is lower and has a wider distribution, suggesting that these shells contain a mixture of unfused gpN and fusion protein and that the scaffolding domains are more flexible. (gpN + Sid procapsids contain an additional peak at higher radius, corresponding to the Sid external scaffold.) The O(195–284)::N shells are larger than the gpN + Sid procapsids, while O(195–284)::N + N capsids have an intermediate size, perhaps due to spatial constraints in the inner shell that force the capsid protein further out when the capsid is packed with scaffolding domains.
Fig. 6.
Radial profiles of fusion protein capsids. Top row, two-dimensional averages of a 71 P4 procapsids from a gpN + Sid coexpression, b the O(195–284)::N fusion protein (69 particles), and (c) the O(195–284)::N + N fusion co-expression clone (71 particles). Bottom row (d–f), radial averages of the same three sets of images. The graph (g) shows the radially averaged density as a function of radius (nm) for the gpN + Sid co-expression (solid black line), O(195–284)::N fusion (gray line) and O(195– 284)::N + N (dashed black line). The density maxima corresponding to gpN, gpO, and Sid are indicated on the graph
Discussion
An icosahedral structure has 60 symmetrically equivalent positions. In an icosahedral virus with more than 60 subunits, the capsid proteins, therefore, exist in more than one unique environment. The larger the icosahedral shell is, the greater the number of different environments exists for the capsid protein. These different environments are usually reflected in conformational differences in the capsid proteins [25, 26], requiring the capsid protein to have a certain amount of conformational flexibility. A priori, there are several ways in which a scaffolding protein can regulate capsid assembly: (1) by promoting the formation of interactions between capsid subunits that would otherwise be unable to interact; (2) by affecting the conformation of the capsid protein; or (3) by forming a template or core on which the capsid protein assembles [1, 26].
In the P2/P4 system, correct assembly of T = 7 shells is dependent on the gpO scaffolding protein, while formation of T = 4 capsids is directed by the P4-encoded external scaffolding protein, Sid, which appears to be a strong promoter of capsid assembly and causes the formation of small P4 capsids in the absence or presence of gpO [4, 5]. gpN alone assembles into mostly aberrant shells in the absence of either gpO or Sid. Nevertheless, a fair number of well-formed isometric shells of both P2 and P4 size are made (24 and 5%, respectively), indicating that gpN has the intrinsic potential for correct capsid formation, with a preference for P2-like, large shells. The role of gpO may thus be to restrict the conformational repertoire of gpN, rather than to promote any specific gpN–gpN interactions, much like a molecular chaperone acts to prevent improper protein associations.
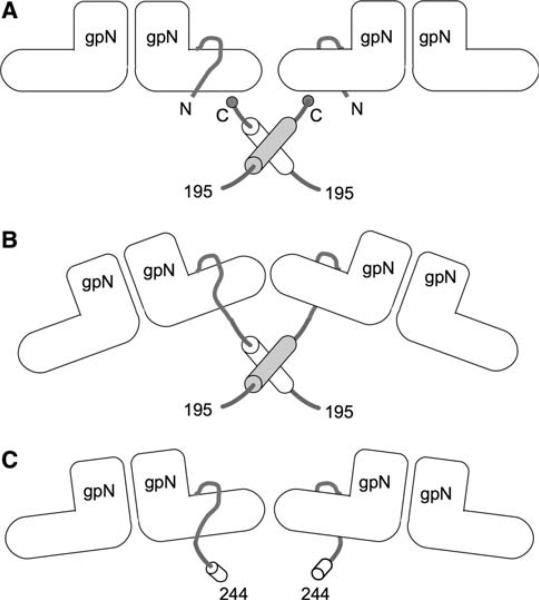
We previously demonstrated that the C-terminal 90 amino acids of gpO constitute a scaffolding domain that promotes the assembly of correctly formed P2-size procapsids through oligomerization and the formation of transient gpO–gpN interactions [7] (Fig. 7a). In the present study, we asked whether the scaffolding domain of gpO would promote procapsid formation when fused to the gpN capsid protein, similar to the δ domain of HK97.
Fig. 7.
Model for gpO–gpN interaction. The “L” shapes represent the gpN monomers. The N-terminal arm is indicated. a shows the interaction between O(195–284) and gpN in an O(195–284) + N co-expression experiment. The C-terminal Cys284 residues of gpO are thought to interact transiently with gpN, while the α-helical regions (residues 195–243, shown as cylinders) dimerize to promote shell assembly. b and c show the O(195–284) and O(244–284) fragments of gpO, respectively, covalently fused to the N-terminus of gpN in the O(195–284)::N and O(244–284)::N fusion clones
Indeed, the O(195–284)::N fusion protein did form more closed, isometric capsids (42% of the total) than gpN protein alone (29%). Most of these capsids (33%) were of the P4 size (Fig. 3). The smaller fusion, O(244–284)::N, produced a still more homogeneous population of P4 size particles (50%). In contrast, neither O(142–284)::N nor O(261–284)::N were efficient at forming proper procapsids of either size, yielding 88 and 70% aberrant capsids, respectively. None of the fusions gave appreciable amounts of P2-size capsids, demonstrating that there is no direct correlation between length of the fusion and capsid size and that assembly fidelity is independent of capsid size determination. These results suggest that the presence of gpO fused to gpN limits the conformational flexibility of gpN. The formation of T = 7 shells requires a greater range of conformational variation compared to that of T = 4 shells. In the absence of gpO, gpN has a preference for the formation of T = 7 capsids (Fig. 3).
The C-terminal α-helix (residues 195–243) of gpO is involved in gpO oligomerization and plays an important role in capsid assembly [17] (Fig. 7a). The relatively large proportion of aberrant capsids formed in the O(195–284)::N fusion compared to the O(195–284) + N co-expression may have been caused by improper positioning of gpO relative to gpN, which would be fixed by the location of the gpN N-terminus (Fig. 7b). Oligomerization of gpO in this position may have led to straining of the capsid lattice and consequently an increased proportion of aberrant shells. The even larger O(142–284)::N fusion might experience additional steric hindrance due to the large size of the gpO fragment. The fact that the O(142–284)::N shells tended to have a thin-walled, “expanded shell” phenotype walls suggests that this steric hindrance is incompatible with the small procapsid-like shell type.
The shorter O(244–284)::N fusion, on the other hand, lacks the α-helical oligomerization sequence. The remaining 40 amino acids are apparently sufficient to restrict the conformational variability of gpN so that P4-like, small shells are formed at relatively high fidelity, without the complications of gpO oligomerization (Fig. 7c). In contrast, the gpO fragment contained within the smallest fusion protein, O(261–284)::N, is apparently too small to restrict the gpN protein sufficiently and does not cause a significant increase in assembly fidelity over gpN alone (about 70% for both). This fusion does cause a shift in capsid size preference from P2-like (6%) to P4-like (24%), however (Fig. 3). These results show that oligomerization of the α-helical region is not absolutely required for promoting shell closure.
The ratio of gpN to gpO in vivo is about 3-to-1 [5]. If gpO binding restricts the conformational repertoire of gpN and thereby prevents the formation of larger shells, the addition of free gpN should increase the ratio of gpN-to-gpN bound to gpO and allow the formation of larger, P2-like shells. We tested this hypothesis by generating clones co-expressing gpN with O(142–284)::N, O(195–284)::N, and O(244–284)::N, resulting in an approximate 2:1 ratio of scaffolding domains to capsid protein (Fig. 5b). This would permit the unconstrained gpN protein to intercalate between the conformationally constrained fusion protein. Although the resulting shells did contain a mixture of fusion protein and free gpN (Fig. 6), the fidelity of assembly did not improve, nor did the shell size shift towards the larger, P2-like shells. Clearly, other mechanisms are at play in defining the assembly of larger shells from gpN protein. For example, it may be that correct assembly is dependent on only transient interactions between the scaffolding domain of gpO and gpN.
In summary, the α-helical region of gpO does have many of the properties of the HK97 gp5 δ domain, including the ability to promote the formation of closed, isometric shells. However, gpO is unable to provide the required conformational switching to form a large T = 7 shell when it is fused to gpN. This could be due to the fact that the interaction between gpO and gpN needs to be transient [7], or a result of the way the fused gpO protein is constrained by the N-terminus of gpN. It may be of interest to incorporate a linker between gpO and gpN that will allow more leeway in gpO position on the inner surface of gpN. It would also be interesting to see if the δ domain of HK97 gp5 would work as a scaffolding protein when fused to gpN.
Acknowledgment
We are grateful to Cindy Rodenburg for assistance with the molecular biology aspects of this work. This work was partly supported by National Institutes of Health (NIH) grant R21 AI071982 to T.D. while M.S.S. was partly supported by NIH training grant T32 AI007150.
Contributor Information
Jenny R. Chang, Department of Microbiology, University of Alabama at Birmingham, 845 19th St South, BBRB 311, Birmingham, AL 35294, USA Department of Biology, University of Alabama at Birmingham, Birmingham, AL, USA.
Michael S. Spilman, Department of Microbiology, University of Alabama at Birmingham, 845 19th St South, BBRB 311, Birmingham, AL 35294, USA Department of Biochemistry and Molecular Genetics, University of Alabama at Birmingham, Birmingham, AL, USA.
Terje Dokland, Department of Microbiology, University of Alabama at Birmingham, 845 19th St South, BBRB 311, Birmingham, AL 35294, USA dokland@uab.edu.
References
- 1.Dokland T. Scaffolding proteins and their role in viral assembly. Cell. Mol. Life Sci. 1999;56:580–603. doi: 10.1007/s000180050455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fane BA, Prevelige PE. Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem. 2003;64:259–299. doi: 10.1016/s0065-3233(03)01007-6. [DOI] [PubMed] [Google Scholar]
- 3.Dokland T, Lindqvist BH, Fuller SD. Image reconstruction from cryo-electron micrographs reveals the morphopoietic mechanism in the P2–P4 bacteriophage system. EMBO J. 1992;11:839–846. doi: 10.1002/j.1460-2075.1992.tb05121.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marvik OJ, Sharma P, Dokland T, Lindqvist BH. Bacteriophage P2 and P4 assembly: alternative scaffolding proteins regulate capsid size. Virology. 1994;200:702–714. doi: 10.1006/viro.1994.1234. [DOI] [PubMed] [Google Scholar]
- 5.Wang S, Chang JR, Dokland T. Assembly of bacteriophage P2 and P4 procapsids with internal scaffolding protein. Virology. 2006;348:133–140. doi: 10.1016/j.virol.2005.12.021. [DOI] [PubMed] [Google Scholar]
- 6.Rishovd S, Marvik OJ, Jacobsen E, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: identification and characterization of the portal protein. Virology. 1994;200:744–751. doi: 10.1006/viro.1994.1238. [DOI] [PubMed] [Google Scholar]
- 7.Chang JR, Poliakov A, Prevelige PE, Mobley JA, Dokland T. Incorporation of scaffolding protein gpO in bacteriophages P2 and P4. Virology. 2008;370:352–361. doi: 10.1016/j.virol.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lengyel JA, Goldstein RN, Marsh M, Sunshine MG, Calendar R. Bacteriophage P2 head morphogenesis: cleavage of the major capsid protein. Virology. 1973;53:1–23. doi: 10.1016/0042-6822(73)90461-3. [DOI] [PubMed] [Google Scholar]
- 9.Rishovd S, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: protein processing and capsid size determination. Virology. 1992;187:548–554. doi: 10.1016/0042-6822(92)90457-z. [DOI] [PubMed] [Google Scholar]
- 10.Marvik OJ, Jacobsen E, Dokland T, Lindqvist BH. Bacteriophage P2 and P4 morphogenesis: assembly precedes proteolytic processing of the capsid proteins. Virology. 1994;205:51–65. doi: 10.1006/viro.1994.1619. [DOI] [PubMed] [Google Scholar]
- 11.Lindqvist BH, Deho G, Calendar R. Mechanisms of genome propagation and helper exploitation by satellite phage P4. Microbiol. Rev. 1993;57:683–702. doi: 10.1128/mr.57.3.683-702.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shore D, Deho G, Tsipis J, Goldstein R. Determination of capsid size by satellite bacteriophage P4. Proc. Natl Acad. Sci. USA. 1978;75:400–404. doi: 10.1073/pnas.75.1.400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marvik OJ, Dokland T, Nøkling RH, Jacobsen E, Larsen T, Lindqvist BH. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J. Mol. Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 14.Dokland T, Wang S, Lindqvist BH. The structure of P4 procapsids produced by coexpression of capsid and external scaffolding proteins. Virology. 2002;298:224–231. doi: 10.1006/viro.2002.1485. [DOI] [PubMed] [Google Scholar]
- 15.Wang S, Palasingam P, Nøkling RH, Lindqvist BH, Dokland T. In vitro assembly of bacteriophage P4 procapsids from purified capsid and scaffolding proteins. Virology. 2000;275:133–144. doi: 10.1006/viro.2000.0521. [DOI] [PubMed] [Google Scholar]
- 16.Six EW. The helper dependence of satellite bacteriophage P4: which gene functions of bacteriophage P2 are needed by P4? Virology. 1975;67:249–263. doi: 10.1016/0042-6822(75)90422-5. [DOI] [PubMed] [Google Scholar]
- 17.Chang JR, Spilman MS, Rodenburg CM, Dokland T. Functional domains of the bacteriophage P2 scaffolding protein: identification of residues involved in assembly and protease activity. Virology. 2009;384:144–150. doi: 10.1016/j.virol.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morais MC, Kanamaru S, Badasso MO, Koti JS, Owen BAL, McMurray CT, Anderson DL, Rossmann MG. Bacteriophage phi29 scaffolding protein gp7 before and after prohead assembly. Nat. Struct. Biol. 2003;10:572–576. doi: 10.1038/nsb939. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Parker MH, Weigele P, Casjens S, Prevelige PE, Krishna NR. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000;297:1195–1202. doi: 10.1006/jmbi.2000.3620. [DOI] [PubMed] [Google Scholar]
- 20.Conway JF, Duda RL, Cheng N, Hendrix RW, Steven AC. Proteolytic and conformational control of virus capsid maturation: the bacteriophage HK97 system. J. Mol. Biol. 1995;253:86–99. doi: 10.1006/jmbi.1995.0538. [DOI] [PubMed] [Google Scholar]
- 21.Hendrix RW. Bacteriophage HK97: assembly of the capsid and evolutionary connections. Adv. Virus Res. 2005;64:1–14. doi: 10.1016/S0065-3527(05)64001-8. [DOI] [PubMed] [Google Scholar]
- 22.Effantin G, Boulanger P, Neumann E, Letellier L, Conway JF. Bacteriophage T5 structure reveals similarities with HK97 and T4 suggesting evolutionary relationships. J. Mol. Biol. 2006;361:993–1002. doi: 10.1016/j.jmb.2006.06.081. [DOI] [PubMed] [Google Scholar]
- 23.Dokland T, Ng ML. Transmission electron microscopy of biological specimens. In: Dokland T, et al., editors. Techniques in Microscopy for Biomedical Applications. World Scientific Press; Singapore: 2006. [Google Scholar]
- 24.Ludtke SJ, Baldwin PR, Chiu W. EMAN: semiautomated software for high-resolution single-partice reconstructions. J. Struct. Biol. 1999;128:82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 25.Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol. 1962;27:1–24. doi: 10.1101/sqb.1962.027.001.005. [DOI] [PubMed] [Google Scholar]
- 26.Dokland T. Freedom and restraint: themes in virus capsid assembly. Structure. 2000;8:R157–R167. doi: 10.1016/s0969-2126(00)00181-7. [DOI] [PubMed] [Google Scholar]