Abstract
Here we report a novel two-dimensional LC-MS method that combines offline hydroxyapatite (HA) chromatography with online reversed-phase liquid chromatography mass spectrometry (HA/RP LC-MS). The 2D-LC-MS method was used to enrich and characterize histones and their posttranslational modifications. The 2D HA/RP LC-MS approach separates histones based on their relative binding affinity to DNA and relative hydrophobicity. HA/RP separations showed improvement in the number of histone isoforms detected as compared with 1D reversed-phase LC-MS of acid extracted histones. The improved histone fractionation resulted in better detection of lower abundant histone variants as well as their posttranslationally modified isoforms. Histones eluted from the HA/RP in the following order: H1, H2A/H2B heterodimers followed by H3/H4 heterotetramers, as predicted from their spatial organization in nucleosomes for binding affinity to DNA. Comparison between HA purified and acid-extracted histones revealed similar histone profiles with the exception that the HA fractions showed a greater number of H1 isoforms. Two elution conditions were also examined: batch elution and salt-gradient elution. While both elution techniques were able to sufficiently fractionate the histones, the salt-gradient approach has the most potential for purification of selected histone isoforms.
Keywords: Histone, chromatin, chromosomal proteins, hydroxyapatite chromatography, LC-MS
INTRODUCTION
The DNA in eukaryotic cells is packaged into an organized structure collectively called chromatin. The basic structural unit of chromatin, the nucleosome, is composed of a histone octamer wrapped by 145–147 base pairs of double stranded DNA in approximately two left-handed superhelical turns [1]. The histone octamer contains each of two molecules of H2A, H2B, H3 and H4. Higher-order chromatin structure is assembled and stabilized by linker histones binding to linker DNA between the nucleosomes [2]. Multiple levels of chromatin condensation result in the 1.8-meter long chromosomal DNA compacted to fit within a 6–10 μm nucleus.
Histones are the most abundant, highly conserved chromosomal proteins [3]. They have a globular structured domain containing alpha helical histone folds and the N- and C-terminal unstructured tails that protrude from the nucleosome [1] [4]. Histones receive a wide variety of posttranslational modifications (PTMs) such as acetylation, methylation, ubiquitination, ADP ribosylation and phosphorylation [5]. Histone modifications are involved in a variety of nuclear functions such as gene transcription, DNA replication and repair, silencing and chromosome segregation [5; 6; 7] [8]. These modifications modulate the chromatin structure as it changes the net charge in chromatin domains and also facilitates the binding of their cognate effector molecules, which are involved in chromatin functions [9; 10; 11; 12; 13]. Larger group modifications such as ubiquitination may affect chromatin architecture through steric hindrance and by targeting the modified proteins to the proteasome for degradation [14] [15].
Molecular studies of histones rely on efficient purification in their native form. The isolation and analysis of histones is complicated due to the presence of a variety of sequence variants and histone isoforms resulted from a large number of posttranslational modifications [5; 16]. Two commonly used approaches to purify histones are acid-extraction and hydroxyapatite chromatography (HA) [17]. The use of HA to fractionate histones, non-histone proteins, histone oligmers and nucleic acids dates back to the 1970s [18; 19; 20; 21; 22; 23]. For instance, the chromosomal proteins in chicken oviduct were fractionated using HA by Bloom and Anderson in 1978 [21], and Bluthmann et al. employed HA to isolate non-histone proteins of chromatin for the investigation of transcriptional specificity in vitro [18]. HA chromatography has been used to fractionate proteins according to their spatial arrangement within the histone octamer as well as their DNA binding properties [21]. Separation requires binding of native chromatin-DNA to HA and then eluting the proteins by increasing salt concentration in the mobile phase.
While HA is capable of rapid large-scale preparation of both chromosomal proteins and DNA [18; 21] the purifications are most often performed in batch to yield bulk histones[23]. Linear salt gradients may also be employed to further improve the fractionation of histones[24; 25]. These HA purified histones are then characterized by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and/or western blots. While these approaches are highly sensitive, they lack the resolution of current methods that employ reversed-phase liquid chromatography coupled with mass spectrometry. Reversed-phase liquid chromatography mass spectrometry (RP LC-MS) has been demonstrated to be a powerful tool to profile and characterize histone variants and their posttranslational modifications[26]. MS based methods are not subject to the limitations of Edmond sequencing and immunoblotting that require high sample amounts or highly specific antibodies. Here we describe the novel combination of gradient HA chromatography and RP LC-MS resulting in significantly improved profiling of histone isoforms. HA separations were performed using salt-step or salt gradient elution rather than batch purifications. Fractions were then collected and characterized by RP LC-MS. The combined use of HA and RP LC-MS (aka 2D HA/RP LC-MS) showed much greater performance than RP LC-MS alone of HA batch purified histones. The approach allows for a more comprehensive and unbiased evaluation of histone profiles.
EXPERIMENTAL
Preparation of Chromatin
HeLa cells were used as a source for chromatin and were maintained as monolayer cultures in DMEM supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin and 100 units/mL penicillin, in a humidified, 37 °C, 5% CO2 incubator. Cells were grown on 10 cm culture plates to near confluence, and 5–40 plates (1.5 × 107 – 12 × 107 cells) were used for each experiment. Cultured cells were washed with PBS, scraped and pelleted by centrifugation. Cell pellets were resuspended in 10 mL buffer A (20 mM KiPO4 pH 6.7, 5 mM KCl, 1 mM MgCl2, 0.1% NP40), incubated on ice for 20 min with disruption by titration and nuclei collected by centrifugation. Nuclei were lysed in 3 mL of buffer B (0.1 M KiPO4 pH 6.7, 0.65 M NaCl, 0.1% triton-X100) for experiments that utilized isocratic HA chromatography, or 3 mL of buffer C (0.1 M KiPO4 pH 6.7, 5 mM KCl, 1mM MgCl2, 0.25 or 0.65 M NaCl, 0.1% triton-X100) for HA chromatography using a NaCl gradient elution. For NaCl gradient elution, frozen nuclei were thawed on ice, sonicated for 3 seconds (Branson digital sonifier, 40% efficiency) and dialyzed against 0.1 M KiPO4 containing 0.65 M NaCl, followed by sonication for 3 seconds. Chromatin in (~1.7 mg/mL) was loaded on to ~8 mL HA column. For isocratic two-step elution, the chromatin was sonicated for 10 seconds, clarified by centrifugation and immediately subjected to two-step isocratic HA chromatography. All buffers contained 1.0 mM sodium orthovanadate as a phosphatase inhibitor as well as protease inhibitors (Sigma Complete, Sigma-Aldrich, St. Louis, MO)
Hydroxyapatite chromatography
A 8 mL (~10 cm × 1 cm) column was packed with Macroprep 20 μm ceramic hydroxyapatite type I (Biorad, Hercules, CA) in 0.1 M KiPO4 pH 6.7. Separation was performed by use of a Fast Performance Liquid Chromatography (FPLC) (Amersham Biosciences, New Jersey). Chromatin was loaded onto the column at a flow rate of 0.5 mL/min after equilibration in a minimum of 3 column volumes of 0.1 M KiPO4 pH 6.7, 0.65 M NaCl (isocratic elution) or 0.1 M KiPO4 pH 6.7, 0.25 M or 0.65 M NaCl (gradient elution). Histones were eluted from the column using a 0.65 M to 2 M NaCl gradient or directly using 2 M NaCl for step elution. Fractions were collected every 2 mL for both methods. Proteins were TCA (trichroloacetic acid) precipitated prior to analysis by SDS-PAGE and HA/RP LC-MS.
Reversed-phase liquid chromatography mass spectrometry (RP LC-MS)
HA separated chromatin associated protein fractions were separated by use of reversed-phase HPLC (Waters 2690, Waters; Milford, MA) and detected with an ESI-TOF mass spectrometer. RP HPLC separations were carried out at a flow rate 50 μL/min on a 1.0 mm × 150 mm C18 column (Discovery Bio wide pore C18, 5 μm, 300 Å, Supelco, USA). The gradient was composed of mobile phase A (0.1% TFA in water) and mobile phase B (0.1% TFA in acetonitrile), where B linearly increased from 30% to 45% for 2 min, then to 60% by 20 min and was held at 60% for 4 min. Between each run, the column was washed with 95% B for 2 min and equilibrated at 30% B for 30 min. A Micromass LCT (Micromass; Wythenshawe, UK) mass spectrometer with an orthogonal electrospray source (Z-spray) was coupled to the outlet of the HPLC. Histones were infused into the electrospray source at the flow rate of 50 μL/min without splitting. ESI was performed at an ESI capillary voltage = 3 kV, source temperature = 100 °C and cone voltage = 50 V. Data were acquired in continuum mode at the rate of 1 spectrum sec−1. All spectra were obtained in the positive ion mode. The mass spectrometer was externally calibrated with NaI over the m/z range 500–2500. The protein masses were determined by deconvolution of the ESI spectra using the MaxEnt (Maximum Entropy [27; 28]) algorithm included in the MassLynx 4.0 data analysis software. The spectra across each chromatographic peak were combined and smoothed prior to deconvolution. Deconvolution was performed for protein mass range of 8000–40000. The MaxEnt algorithm was allowed to run to convergence.
RESULTS
HA/RP LC-MS profiles of histones from two-step batch elution
We initially evaluated the HA/RP LC-MS approach with a traditional two-step batch salt elution. HA/RP LC-MS profiles for low and high salt fractions are shown in Figure 1. In order to keep the discussion succinct, isoforms are grouped into families with the exception of minor isoforms that are normally difficult to observe by RP LC-MS. The rational for assigning protein IDs [26] as well as possible posttranslational modifications for each histone isoform family have been provided in great detail in other reports [29; 30; 31].
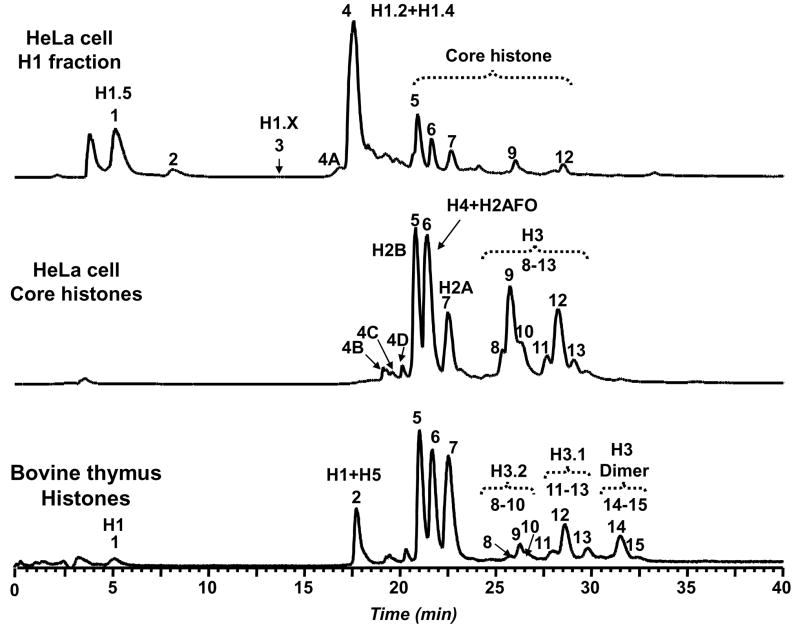
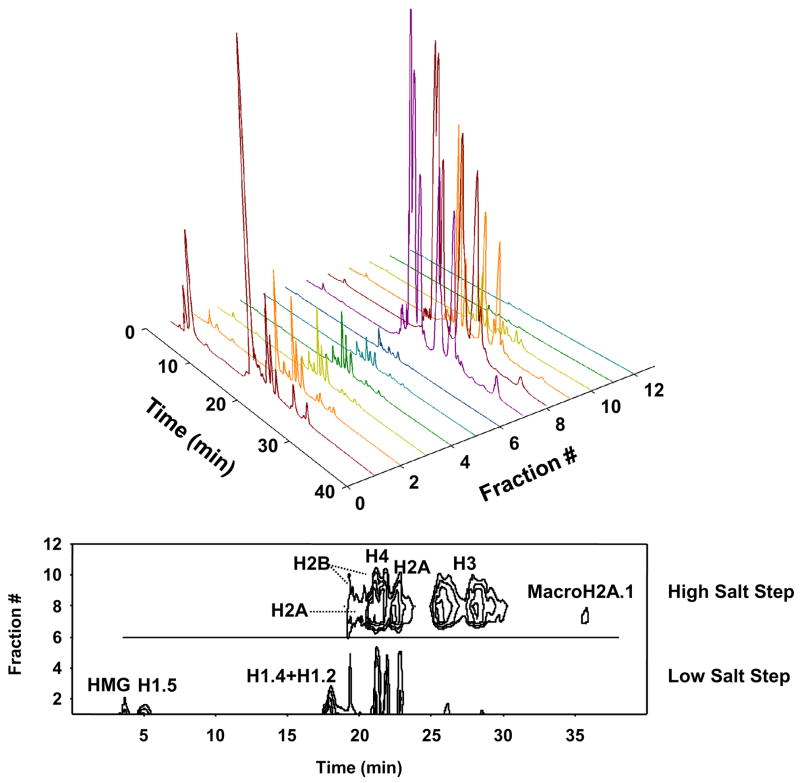
Figure 1.
Comparison of LC-MS profiles for linker and core histones in HeLa cells. Linker and core histone-containing fractions were collected using hydroxyapetite (HA) step-elution chromatography at 0.65 M and 2 M NaCl, respectively. The reference histone mixture from bovine thymus was obtained by acid extraction. The RPLC fraction annotations correspond to the following: 1 = H1.5, 2 = protein 22,770 Da, 3 = H1.X, 4A = protein 14,419 Da, 4B = H2B + 2MetOx, 4C = H2A + MetOx, 4D = H2B + MetOx, 4 = H1.2 + H1.4, 5 = H2B, 6 = H4 + H2AFO/R, 7 = 2A, 8–10 = H3.2, 11–13 = H3.1, 14–15 = H3 dimer.
The two-step elution effectively segregated linker and core histones as expected. The LC-MS profiles shows mixtures of histone isoforms consisting of histone variants and their posttranslational modifications. The low salt HA fraction predominantly contained the histone isoforms families H1.5 (RT = 5.18 min; RPLC fraction 1), H1.X (RT = 13.68 min; RPLC fraction 3) and H1.2 & H1.4 (RT = 17.52 min; RPLC fraction 4) as determined by molecular weight. Two unidentified proteins (RPLC fraction 2: MW = 22,770 Da and 4A: MW = 14,419 Da) were also observed in the low salt fraction. Core histones eluted from the high salt fraction in the following order: dioxidized H2B variants (RPLC fraction 4B), monooxidized H2A variants (RPLC fraction 4C), monooxidized H2B variants (RPLC fraction 4D), H2B variants (RPLC fraction 5), H4 and methionine-containing H2A variants (RPLC fraction 6), H2A variants without methionine (RPLC fraction 7), and finally the H3 variants (RPLC fractions 8–13). The observed oxidization was due to methionine conversion to methionine sulfoxide (MetOx). These observations were consistent with previously reported RP LC profiles[26]. SDS-PAGE analysis of the low and high salt fractions demonstrated selective displacement of linker histones and core histones from the HA column. Figure 2 shows the LC-MS traces performed on 12 equal column fractions (six each from low and high salt elutions) that contained the highest levels of histones as determined by SDS-PAGE
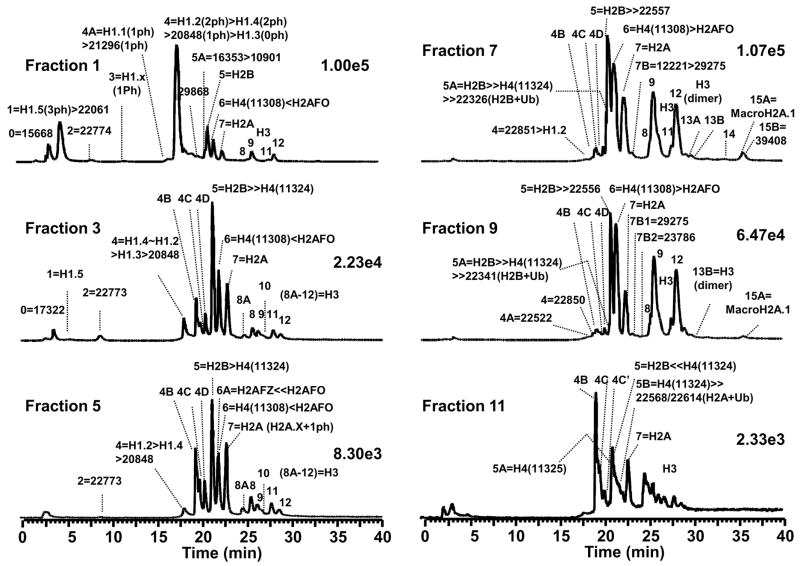
Figure 2.
LC-MS profiles of histone fractions from step elution. The number denoted by scientific notation refers to total ion current of the subsequent LC-MS. Fractions 1–6 were collected at 0.65 M NaCl and fractions 7–12 at 2 M NaCl.
Starting with the low-salt fraction, linker histones were most abundant in HA fraction 1, and diminished significantly in the subsequent fractions. Throughout the low salt elution, unidentified proteins with masses of 22,774 Da and 20,848 Da were observed. Core histones were first observed in HA fraction 2 and increased in abundance in subsequent fractions. The oxidized forms of H2B and H2A (RPLC fractions 4B–4D) increased in abundance from HA fraction 3 to 6. The dioxidized H2B (RPLC fraction 4B) were the predominant forms present in HA fraction 6. The level of oxidized H4 species also increased from HA fraction 4 to 6. Minor H2A variants H2AFZ (H2A family member Z) and H2AFX (H2A family member X) were present in the low salt fractions. The important H2AFZ (13,424 Da) variant was present in HA fractions 4–6 (RPLC fraction 6/6A), while H2AFX was observed in HA fraction 4 and its monophosphorylated isoform in HA fraction 5 (RPLC fraction 7/7A). All mass-based assignments as well as the identities of species mentioned here are shown in Figure 2 and listed in Table 1.
Table 1.
Protein identities (based on mass assignment) observed in the two-step salt elution
Histone Isoform Families (based on mass assignment) observed in the two-step salt elution
| Fraction | 0 | 1 | 2 | 3 | 4A | 4 |
|---|---|---|---|---|---|---|
| 1 | 15668 | H1.5>22061 | 22774 | H1.X | H1.0>21296 | H1.2>H1.4>20848>H1.3 |
| 2 | H1.0 | H1.5>22061 | 22774 | -- | H1.1>21298 | H1.4>H1.2>20847 |
| 3 | 17322 | H1.5 | 22773 | -- | -- | H1.4~H1.2>H1.3>20848 |
| 4 | 17322 | H1.5 | 22774 | -- | -- | H1.4~H1.2>20848 |
| 5 | -- | -- | 22773 | -- | -- | H1.2<H1.4<20848 |
| 6 | -- | -- | 22773 | -- | -- | H1.4~H1.2>H1.3 |
| 7 | -- | -- | -- | -- | -- | Small peak: 22851>H1.2 |
| 8 | -- | -- | -- | -- | -- | Small peak: 22850 |
| 9 | -- | -- | -- | -- | 22522 | Small peak: 22850 |
| 10 | -- | -- | -- | -- | -- | -- |
| 11 | -- | -- | -- | -- | -- | -- |
| 12 | -- | -- | -- | -- | -- | -- |
| Fraction | 4B | 4B′ | 4C | 4D | 5A | 5 |
| 1 | 13641 | 14441 | -- | 29868 | 16353>10901 | H2B(13777) |
| 2 | 13799>13673 | H2B(13809) | H2A(14025) | H2B(13793) | -- | H2B(13777) |
| 3 | H2B(13809) | H2B(13793) | H2A(14025) | H2B(13793) | -- | H2B(13777) |
| 4 | H2B(13809) | H2B(13809) | H2A(14025) | H2B(13793) | -- | H2B(13777)≫H4(11324) |
| 5 | H2B(13809) | -- | H2A(14025) | H2B(13793) | -- | H2B(13777)≫H4(11324) |
| 6 | H2B(13809) | H2B(13809) | H2A(14025) | H2B(13793) | -- | H2B(13777)~H4(11324) |
| 7 | H2B(13809) | H2B(13809) | H2A(14025) | H2B(13793) | H2B13778≫H4(11325)≫22326 (H2B+Ub) | H2B(13777)>22557 (H2A+Ub) |
| 8 | H2B(13809) | H2B(13809) | H2A(14024) | H2B(13793) | -- | H2B(13777) |
| 9 | H2B(13808) | H2B(13808) | H2A(14025) | H2B(13793) | H2B13779<H4(11324)≫22341 (H2B+Ub) | H2B(13776)≫22557 (H2A+Ub) |
| 10 | H2B(13809) | H2B(13809) | H2A(14024) | H2B(13793) | H4(11325) | H2B(13777)>H4(11324) |
| 11 | H2B(13809) | H2B(13825) | H2A(14025) | H2B(14041) | H4(11325) | H2B(13792)≪H4(11324) |
| 12 | H2B(13808) | H2B(13825) | H2A(14025) | H2B(14040) | -- | H4(11324) |
| Fraction | 5B | 6A | 6 | 7A | 7 | 7B |
| 1 | -- | -- | H4(11308)<H2AFO | -- | H2A | -- |
| 2 | -- | -- | H4(11308)<H2AFO | -- | H2A | -- |
| 3 | -- | -- | H4(11308)<H2AFO | -- | H2A | -- |
| 4 | -- | 22578 (H2A+Ub) ≪H2AFZ(13424)<H2AFO | H4(11308)<H2AFO | 7A(H2AFX)> 7(H2AFX) | H2A(H2AFX appear) | -- |
| 5 | -- | H2AFO | H4(11308)<H2AFO | -- | H2A (H2AFX+Ph) | -- |
| 6 | -- | -- | H4(11308)<H2AFZ(13424) <H2AFO | -- | H2A | -- |
| 7 | -- | -- | H4(11308)>H2AFO | -- | H2A | 12221>29275 |
| 8 | -- | -- | H4(11308)>H2AFO | -- | 11406<H2A | 11504 |
| 9 | -- | -- | H4(11308)>H2AFO | -- | H2A | 29275~23786 |
| 10 | -- | -- | H4(11308)>H2AFO | -- | H2A | -- |
| 11 | H4(11324)≫22568/22614 (H2A+Ub) | -- | No Peak 6 | -- | H2A | -- |
| 12 | H411340≫H2AFZ(13424) | -- | No Peak 6 | -- | H2A | -- |
| Fraction | 8A-12 | 13A | 13B | 14 | 15A | 15B |
| 1 | H3 | -- | -- | -- | -- | -- |
| 2 | H3 | -- | -- | -- | -- | -- |
| 3 | H3 | -- | -- | -- | -- | -- |
| 4 | H3 | -- | -- | -- | -- | -- |
| 5 | H3 | -- | -- | -- | -- | -- |
| 6 | H3 | -- | -- | -- | -- | -- |
| 7 | H3 | H3 Dimer | H3 Dimer | 39423 | MacroH2A.1 39406 | MacroH2A.1 39408 |
| 8 | H3 | H3 Dimer | -- | -- | MacroH2A.1 39406 | -- |
| 9 | H3 | -- | H3 Dimer | -- | MacroH2A.1 39406 | -- |
| 10 | H3 | -- | H3 Dimer | -- | -- | -- |
| 11 | H3 | -- | H3 Dimer | -- | -- | -- |
| 12 | H3 | -- | -- | -- | -- | -- |
The high-salt fraction contained many more isoforms than those typically observed when histones were purified by acid extraction. For example, the ubiquitinated H2B variants with molecular mass 22,326 Da (HA fraction 7/RPLC fraction 5A) and 22,341 Da (HA fraction 9/RPLC fraction 5A) were present. Ubiquitinated H2A forms were also observed with calculated mass of 22,557 Da (HA fraction 7/RPLC fraction 5), 22,556 Da (HA fraction 9/RPLC fraction 5) and 22,568 and 22,614 Da (HA fraction 11/RPLC fraction 5B). H3 dimers were present but at low abundance in HA fractions 7–11/RP LC fractions 13A and 13B. A species of 39,406 Da that corresponds to macroH2A.1 (MW = 39,400 Da) was observed in HA fractions 7–9/RPLC fractions 15A and 15B). Another species at 39,423 Da (HA fraction 7/RPLC fraction 14) was observed and is likely a modified isoform of macroH2A.1. In HA fractions 11 and 12, the dioxidized H2B variants (RPLC fraction 4B) became prominent, whereas the H2B variants in RPLC fraction 5 and H4 & Met-H2A variants in RPLC fraction 6 disappeared.
LC-MS profiles of histones from gradient salt elution
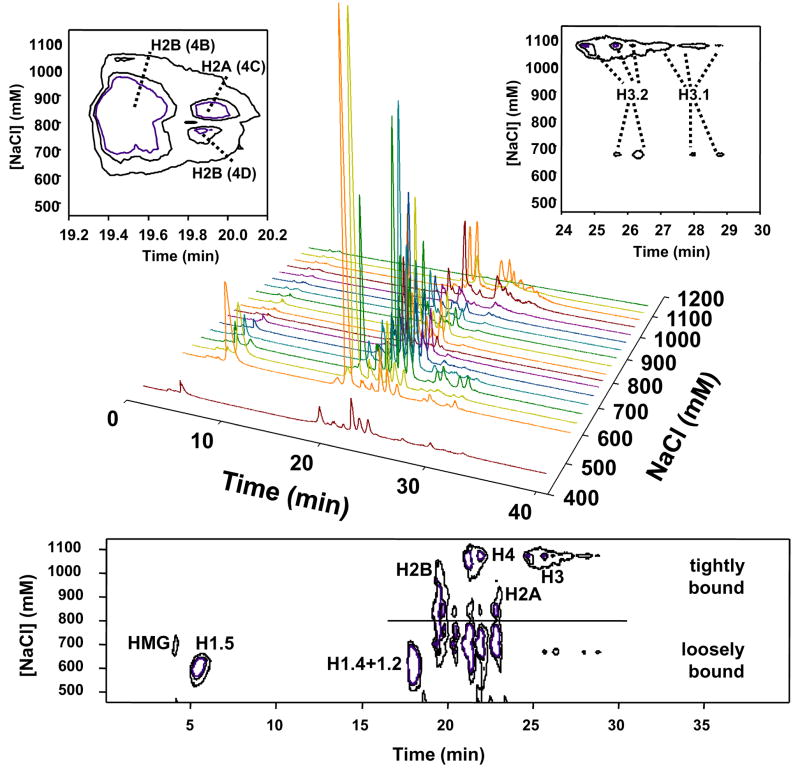
In an effort to improve upon the HA separation of histones we used a linear salt gradient to elute histones from the HA column. Fractions (2 mL each) were collected and analyzed by SDS-PAGE and RP LC-MS. Figure 3 shows the SDS-PAGE profile obtained from alternating fractions from the HA chromatographic separation utilizing the linear salt gradient. As expected, the chromatin-associated proteins eluted from the DNA are dominated by the highly abundant histones. HA fraction 1 (650 mM NaCl) showed the presence of linker histones and a small amount of core histones that were presumably loosely bound to the chromatin. A majority of linker histones dissociated from the HA column during the first 10 mL wash (with 650 mM NaCl), as evident from the LC-MS profile of HA fractions 2 and 3 in Figure 4 (corresponding to lanes 1–4 in Figure 3). A prolonged wash under similar conditions displaced ~20% of the H2A/H2B heteodimers from the column (HA fractions 8–12 in Figure 4, corresponding to lanes 6–14 in Figure 3). Further increase in salt concentration eluted the remainder of the H2A/H2B heterodimers along with the core H3 and H4 heterotetramers. The HA fractions 16–18 Figure 4, corresponding to lanes 19–21 in Figure 3 contain predominantly H3 and H4. The specific salt dependant elution of H2A/H2B and H3/H4 underscores the potential of HA chromatography to fractionate nucleosomal histones in their native associations.
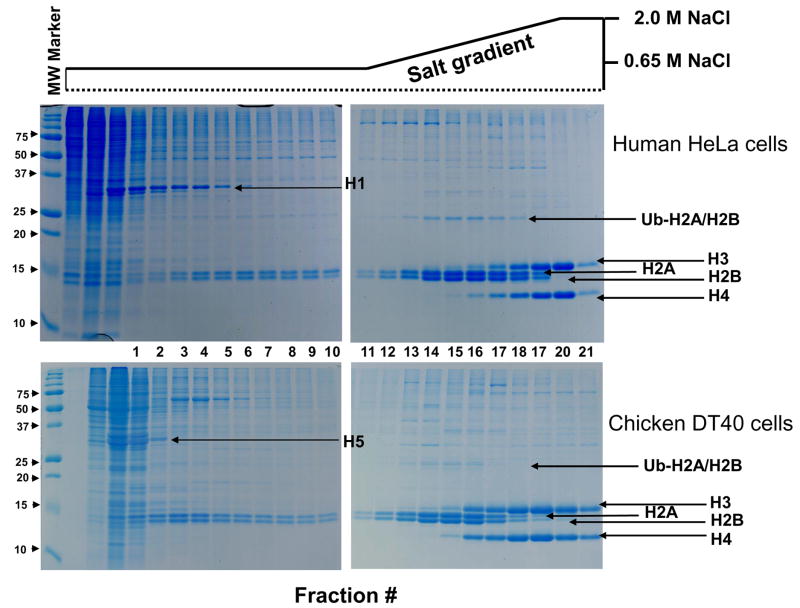
Figure 3.
Fractionation of nuclear proteins by hydroxyapatite (HA) chromatography. The gels show the elution profile of histones from a HA column using a linear NaCl gradient (0.65–2M). Data from HA separations of HeLa (top) and DT40 (bottom) chromatin illustrate the reproducibility of the approach. Nuclear proteins dissociated from the HA column was TCA precipitated and separated on 15% SDS-Polyacrylamide gel and stained with Coommassie G250. The schematic representation of the salt gradient is shown above.
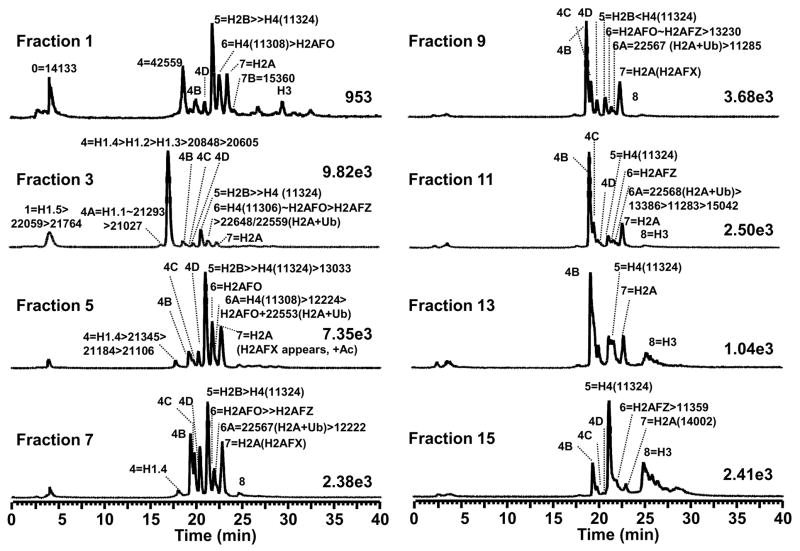
Figure 4.
LC-MS profiles of histone fractions from the gradient salt elution. The salt gradient was from 0.65–2 M NaCl. (The labels are the same as shown in Figure 3).
LC-MS analysis also revealed the enrichment of specific histone variants in selected fractions. For example, H2AFZ appeared in HA fraction 2/RPLC fraction 6 and it increased in abundance as the salt concentration increased from HA fraction 6 to 12. H2AFX and its monoacetylated form were observed in HA fraction 5/RPLC fraction 7 and reached its highest concentration in HA fraction 6. As the salt concentration increased, H2AFX species decreased in abundance from HA fraction 7 to 9 and disappeared in HA fraction 10. Ubiquitinated H2A was observed in HA fraction 5/RPLC fraction 6A and increased in abundance until HA fraction 11. Detailed annotations for RPLC fractions are shown in Figure 4 as well as listed in Table 2.
Table 2.
Protein identities (based on mass assignment) observed in the linear-salt gradient elution.
Histone Isoform Families (based on mass assignment) observed in the gradient salt elution
| Fraction | 0 | 1 | 2 | 3 | 4A | 4 |
|---|---|---|---|---|---|---|
| 1 | 14133 | -- | -- | -- | -- | 42559 |
| 2 | -- | H1.5>22058 | -- | -- | H1.1>21293>21028 | H1.2>H1.4>20847>H1.3>20605 |
| 3 | -- | H1.5>22059>21764 | -- | -- | H1.1~21293>21027 | H1.4>H1.2>H1.3>20848>20605 |
| 4 | 17371≫20763~20806 | H1.5 | -- | -- | H1.1~21293>22084 | H1.4>21345>21184>21106 |
| 5 | -- | -- | -- | -- | -- | H1.4>21343>21102 |
| 6 | -- | -- | -- | -- | -- | H1.4>H1.2 |
| 7 | -- | -- | -- | -- | -- | H1.4 |
| 8 | -- | -- | -- | -- | -- | -- |
| 9 | -- | -- | -- | -- | -- | -- |
| 10 | -- | -- | -- | -- | -- | -- |
| 11 | -- | -- | -- | -- | -- | -- |
| 12 | -- | -- | -- | -- | -- | -- |
| 13 | -- | -- | -- | -- | -- | -- |
| 14 | -- | -- | -- | -- | -- | -- |
| 15 | -- | -- | -- | -- | -- | -- |
| 16 | -- | -- | -- | -- | -- | -- |
| 17 | -- | -- | -- | -- | -- | -- |
| 18 | -- | -- | -- | -- | -- | -- |
| Fraction | 4B | 4C | 4D | 5 | 6 |
|---|---|---|---|---|---|
| 1 | H2B(13808) | -- | H2B(13792) | H2B(13777)≫H4(11324) | H4(11308)>H2AFO |
| 2 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13776)≫H4(11324) | H4(11308)>H2AFO>H2AFZ(13424) |
| 3 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13776)≫H4(11324) | H4(11308)~H2AFO>H2AFZ(13424) >22648/22559(H2A+Ub) |
| 4 | H2B(13808) | H2A(14024) | H2B(13792) | H2B 13776≫H4(11324) | H2AFZ(13424)<H4(11308)<H2AFO |
| 5 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13776)≫H4(11324)>13033 | H4(11308)<H2AFO |
| 6 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13776)≫H4(11324) | H2AFZ(13424)≪H2AFO |
| 7 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13777)>H4(11324) | H2AFZ(13424)≪H2AFO |
| 8 | H2B(13808) | H2A(14024) | H2B(13792) | H2B(13776)>H4(11324) | H2AFZ(13424)>H2AFO>10775 |
| 9 | H2B(13808) | H2A(14024) | H2B(13792) | 19843<22729<H2B(13776)<H11324 | H2AFZ(13424)~H2AFO>13230 |
| 10 | H2B(13809) | H2A(14024) | H2B(13793) | H2B(13792)<H4(11324) | H2AFZ(13424)>H2AFO>20134 |
| 11 | H2B(13809) | H2A(14024) | H2B(13793) | H2B(13823)≪H4(11324) | H2AFZ(13424) |
| 12 | H2B(13808) | H2A(14024) | -- | H4(11324) | H2AFZ(13423)>11340 |
| 13 | H2B(13808) | H2A(14024) | -- | H4(11324) | -- |
| 14 | H2B(13808) | -- | -- | H4(11324)>16651 | -- |
| 15 | H2B(13809) | H2A(14024) | H2B(13793) | H4(11324) | H2AFZ(13424)>11359 |
| 16 | H2B(13809) | -- | H2B(13793) | H4(11324) | H4(11308) |
| 17 | H2B(13809) | -- | -- | H4(11324) | 11340 |
| 18 | -- | -- | -- | H4(11325) | -- |
| Fraction | 6A | 7 | 7B | 8A-12 |
| 1 | -- | H2A | 15360 | H3 little |
| 2 | -- | H2A | -- | H3 little |
| 3 | -- | H2A | -- | H3 little |
| 4 | -- | H2A | -- | H3 little |
| 5 | H4(11308)>12224>H2AFO>22553 (H2A+Ub) | H2A (H2AFX appear, +Ac) | -- | H3 little |
| 6 | -- | H2A (H2AFX, no Ac) | -- | H3 little |
| 7 | 22567 (H2A+Ub)>12222 | H2A (H2AFX decreased) | 12221>29275 | H3 little |
| 8 | 22568 (H2A+Ub)>12222 | H2A (H2AFX decreased more) | 12220 | H3 little |
| 9 | 22568 (H2A+Ub)>11285 | H2A (little H2AFX) | H3 little | |
| 10 | 22568 (H2A+Ub) | H2A (No H2AFX) | -- | H3 little |
| 11 | 22569 (H2A+Ub)>13386>11283>15042 | H2A | -- | H3 little |
| 12 | -- | H2A | -- | H3 little |
| 13 | -- | H2A | -- | H3 |
| 14 | -- | H2A | -- | H3 |
| 15 | -- | H2A | -- | H3 |
| 16 | -- | -- | -- | H3 |
| 17 | -- | -- | -- | H3 |
| 18 | -- | -- | -- | H3 |
Fraction #s correspond to those shown in Figure 3.
Fraction 0 is the HA flow through fraction.
All the protein masses shown are the deconvoluted molecular weights reported as Daltons.
“~”: the relative abundances of two proteins are comparable.
“>” or “<”: the relative abundance of one protein is higher or lower than the other.
“≫” or “≪”: the relative abundance of two proteins was at least 5-fold difference.
The masses shown for H2A, H2B and H4 are the most predominant in the spectra.
“Ub”: ubiquitination
DISCUSSION
For better fractionation and characterization of histones, we have optimized a purification strategy by combining HA chromatography with RP LC-MS. Direct purification of histones from mammalian tissue/culture cells has considerable advantage over recombinant expressed histones or histones purified by acid extraction. Here, individual histones were dissociated from assembled nucleosomes under mild conditions at low temperatures. The purified histones retain their posttranslational modifications and can be further reconstituted to assemble nucleosomes in vitro for biophysical and biochemical studies. The inorganic matrix of hydroxyapatite immobilizes chromatin from which histones may elute as a function of increasing salt concentration and wash volume. Chromatin binds to the hydroxyapatite (HA) via a high affinity interaction between DNA and the calcium phosphate matrix [21]. Histone proteins are released from the DNA by virtue of their configuration in the nucleosome, hydrophobicity and other physical properties. Taken as a whole, the characteristic elution profile of histones from HA appears useful in the study of chromaitn dynamics in isoform populations.
Controlled sonication was used to shear the chromatin to a size range that reduced intrinsic viscosity to allow HA chromatography while maintaining the integrity of the nulceosomes. Overly aggressive sonication resulted in release of core histones from the nucleosome and thus poor recovery by HA. Inadequate sonication resulted in a highly viscous sample and development of high column pressures adversely affecting chromatographic performance. The entire isolation procedure was performed at 4°C in the constant presence of protease and phosphotase inhibitors, minimizing proteolysis and enzymatic activity that might affect histone isoform distributions. The optimized procedure is compatible with sample pretreatment using enzyme inhibitors such as vanadate and butyrate that appear to help maintain modified histones within chromatin.
The order of elution of histone from the HA column follows closely the spatial arrangement of the respective histone isoforms in the nucleosome. Linker histones facilitate the formation of the 30 nm chromatin fibers. They bind to DNA at the nucleosome entry site through a short linker DNA region and dissociate from the chromatin easily. Hence these isoforms eluted first at low salt concentrations. The distinct elution profile of various H1 isoforms as a function of time presumably depend on their net charge and differences in the size of the region of linker DNA to which they are bound. Charge-dependant dissociation of linker histones from nucleosomes have been reported from phosphorylation mutants by genetic studies [32] [33]. Upon complete release of H1, the chromatin is less constrained and the nucleosomes are more likely to be exposed to the high ionic environment. Consequently, core histones are displaced at higher salt concentration opposite to the order in which they are assembled within the nucleosome [34; 35]
We also evaluated recovery and reproducibility. Typically, we load 5 mg total chromatin onto the HA column. This quantity of chromatin can be obtained from 108 human cells (HeLa or 293T) or 109 chicken DT40 cells as quantified by nucleic acid concentration at OD 260. The total histones recovered from a typical preparation was estimated to be >1 mg based on BCA analysis. Histones from the HA fractions were also TCA precipitated (~5% of the proteins from each fraction) and analyzed by SDS-PAGE the order of histone elution and protein recovery. The binding of the nucleosomes to HA and salt dependant elution of core histones proved to be highly reproducible. There were essentially none or very little core histones present in the HA flow through fractions (Figure 3). Analysis of HA reproducibility for histones from human HeLa, 293T and chicken DT40 cells was performed. The HA gradient elution patterns for DT40 separations is shown in Figure 3 for comparison. Overall the HA separations proved to by highly reproducible.
For the LC-MS separations, we next evaluated the changes in relative abundance for histone isoforms purified from a bovine thymus histone standard using an identical methodology as for the HeLA cells. Bovine histone standards (N = 5) from the same preparation were analyzed over a 48-hour time period on the same LC-MS instrument. These samples yielded a coefficient of variance (CV) of 2–5% with regard to the relative abundance of histone variants that spanned the range of 103 relative abundance. The highest CV was associated with the lowest abundant isoform. In all cases, the relative standard deviation in histone abundance (RSD) was less than 5%.
In this application of 2D HA/RP LC-MS, seven H1 variants were observed in HeLa cells. The H1.2, H1.4 and H1.5 isoforms were the most predominant but lower abundant forms H1.X, H1.3, H1.1, and H1.0 were observed as well. Of note, these lower abundant forms were not observed when histones were purified using acid extraction [26]. The reversed-phase chromatography indicated that the hydrophobicity of the H1 forms increased in the following order: H1.0 < H1.5 < H1.X < H1.1 < H1.2/H1.3/H1.4. All the H1 variants observed were N-terminal acetylated and contained multiple phosphorylations [26]. Some of these proteins appear to be present as mono- and di-phosphorylated species. Chromatographic enrichment and analysis of these species is a focus of future studies.
Oxidized histones were more easily observed with HA purification as compared to acid extraction [26]. The improved detection capability suggests that oxidized H2B preferentially forms heterodimers with H2A variants that do not contain Met residues (fraction 5). In contrast, native H2B has a preference in its interaction with the Met-containing H2A variants (H2AFO and H2AFR, fraction 6). In addition, native H2A variants (RPLC fraction 7) remained at a consistent level as did the dioxidized H2B variants (RPLC fraction 4B). Moreover, the monooxidized H2A variants (RPLC fraction 4C) and monooxidized H2B variants (RPLC fraction 4D) maintained a consistent abundance across the HA fractions. These results are consistent with the hypothesis that it is the specificity between different variants that leads to heterodimer formation rather than a pure stochastic organization [36]. These heterodimers are released simultaneously from the DNA during HA chromatography and were detected by mass spectrometry. The results suggest that HA utilizing a salt gradient could be a powerful tool to study the specificity of histone interaction during global nucleosome disassembly/assembly.
The LC-MS profiles were also depicted as 3D contour plots for the two-step salt elution (Figure 5) and gradient salt elution (Figure 6). The two-step elution shows a clear demarcation between linker and core histones associated with the stepwise increase in salt concentration. With the 3D plot, the proteins can be classified and displayed based upon their affinity to DNA. For example, the concentration that separated strongly and weakly bound histones was 800 mM NaCl. These plots also demonstrate the potential for histone fractionation. With sufficient quantities of histones, selection of HA and RP conditions can be derived to greatly enhance the purification of selected histone isoforms. Such improvements will lead to better characterization of histone modifications on specific histone variants.
Figure 5.
3D plot for the two-step salt elution: Upper) 3D plot of time vs fraction # vs total ion current; Bottom) 3D contour plot.
Figure 6.
3D plots for the gradient elution: Upper) 3D plot of time vs fraction # vs total ion current; Bottom) 3D contour plot. The two upper highlight separation of H2A and H2B in RPLC fractions 4B–4D and H3 subtypes in RPLC fractions 8–13.
CONCLUSION
The integration of HA chromatography and reversed-phase liquid chromatography mass spectrometry (HA/RP LC-MS) is a powerful strategy for the purification and characterization of chromatin-associated proteins. The modified HA chromatography is useful for preparation of large amount of LC-MS-quality histones from cultured cells that are protected from degradation and other enzymatic activities. The qualitative analysis of salt elution profiles provided a relative assessment of DNA binding affinity of individual histones in the nucleosomes. Both SDS PAGE and RP LC-MS show that the H1 proteins are less tightly bound to DNA than the core histones, followed by the H2A–H2B heterodimers and the H3–H4 heterotetramers. These results agree with previous studies. Protein composition varied across the different salt elution fractions resulting in improved dynamic range of RP LC-MS as compared to acid-extracted and HA batch purified histones. Further optimization of the chromatographic conditions is expected to enrich specific histone variants and modified isoforms, which can be used for biochemical and biophysical studies.
Acknowledgments
This work was supported by NIH grants CA107106 and CA101956 to MAF, and CA67007, GM080176, and GM62556 to RAF. MAF and XS were also supported by Leukemica and Lymphoma Society (SCOR 7080-06) and the V Foundation/American Association for Cancer Research Translational Cancer Research Grant. The authors are grateful to Richard Sessler for initial technical support and Nanette Kleinholz at Campus Chemical Instrumentation Center of the Ohio State University for assistance with LC-MS.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Thoma F, Koller T. Influence of histone H1 on chromatin structure. Cell. 1977;12:101–7. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD. Structure of chromatin. Annu Rev Biochem. 1977;46:931–54. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 4.Khorasanizadeh S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–72. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- 5.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Li B, Carey M, Workman JL. The Role of Chromatin during Transcription. Cell. 2007;128:707–19. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Vidanes GM, Bonilla CY, Toczyski DP. Complicated tails: histone modifications and the DNA damage response. Cell. 2005;121:973–6. doi: 10.1016/j.cell.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Groth A, Rocha W, Verreault A, Almouzni G. Chromatin challenges during DNA replication and repair. Cell. 2007;128:721–33. doi: 10.1016/j.cell.2007.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–7. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 10.Kusch T, Workman JL. Histone variants and complexes involved in their exchange. Subcell Biochem. 2007;41:91–109. [PubMed] [Google Scholar]
- 11.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. DNA Damage-Dependent Acetylation and Ubiquitination of H2AX Enhances Chromatin Dynamics. Mol Cell Biol. 2007;27:7028–40. doi: 10.1128/MCB.00579-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dormann HL, Tseng BS, Allis CD, Funabiki H, Fischle W. Dynamic regulation of effector protein binding to histone modifications: the biology of HP1 switching. Cell Cycle. 2006;5:2842–51. doi: 10.4161/cc.5.24.3540. [DOI] [PubMed] [Google Scholar]
- 13.Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond.m. Nature. 2003;425:475–9. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- 14.Sun L, Chen ZJ. The novel functions of ubiquitination in signaling. Curr Opin Cell Biol. 2004;16:119–26. doi: 10.1016/j.ceb.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Jason LJ, Moore SC, Lewis JD, Lindsey G, Ausio J. Histone ubiquitination: a tagging tail unfolds? Bioessays. 2002;24:166–74. doi: 10.1002/bies.10038. [DOI] [PubMed] [Google Scholar]
- 16.Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–49. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- 17.Simon RH, Felsenfeld G. A new procedure for purifying histone pairs H2A + H2B and H3 + H4 from chromatin using hydroxylapatite. Nucleic Acids Res. 1979;6:689–96. doi: 10.1093/nar/6.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bluthmann H, Mrozek S, Gierer A. Non-histone chromosomal proteins. Their isolation and role in determining specificity of transcription in vitro. Eur J Biochem. 1975;58:315–26. doi: 10.1111/j.1432-1033.1975.tb02378.x. [DOI] [PubMed] [Google Scholar]
- 19.MacGillivray AJ, Rickwood D, Cameron A, Carroll D, Ingles CJ, Krauze RJ, Paul J. Methods for isolation and characterization of chromosomal nonhistone proteins. Fractionation of chromatin on hydroxyapatite and characterization of the nonhistone proteins by ion exchange chromatography and polyacrylamide gel electrophoresis. Methods Enzymol. 1975;40:160–171. doi: 10.1016/s0076-6879(75)40014-3. [DOI] [PubMed] [Google Scholar]
- 20.Rickwood D, MacGillivray AJ. Improved techniques for the fractionation of non-histone proteins of chromatin on hydroxyapatite. Eur J Biochem. 1975;51:593–601. doi: 10.1111/j.1432-1033.1975.tb03961.x. [DOI] [PubMed] [Google Scholar]
- 21.Bloom KS, Anderson JN. Fractionation and characterization of chromosomal proteins by the hydroxyapatite dissociation method. J Biol Chem. 1978;253:4446–50. [PubMed] [Google Scholar]
- 22.Fischman GJ, Lambert MW, Studzinski GP. Purification and properties of a nuclear DNA endonuclease from HeLa S3 cells. Biochim Biophys Acta. 1979;567:464–71. doi: 10.1016/0005-2744(79)90132-3. [DOI] [PubMed] [Google Scholar]
- 23.Zagariya A, Khrapunov S, Zacharias W. Rapid method for the fractionation of nuclear proteins and their complexes by batch elution from hydroxyapatite. J Chromatogr. 1993;648:275–8. doi: 10.1016/0021-9673(93)83311-f. [DOI] [PubMed] [Google Scholar]
- 24.Li W, Nagaraja S, Delcuve GP, Hendzel MJ, Davie JR. Effects of histone acetylation, ubiquitination and variants on nucleosome stability. Biochem J. 1993;296(Pt 3):737–44. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J Biol Chem. 2006;281:20036–44. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 26.Su X, Jacob NK, Amunugama R, Lucas DM, Knapp AR, Ren C, Davis ME, Marcucci G, Parthun MR, Byrd JC, Fishel R, Freitas MA. Liquid chromatography mass spectrometry profiling of histones. J Chromatogr B Analyt Technol Biomed Life Sci. 2007 doi: 10.1016/j.jchromb.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferrige AG, Seddon MJ, Green BN, Jarvis SA, Skilling J. Disentangling Electrospray Spectra with Maximum-Entropy. Rapid Communications in Mass Spectrometry. 1992;6:707–711. [Google Scholar]
- 28.Ferrige AG, Seddon MJ, Jarvis S. Maximum-Entropy Deconvolution in Electrospray Mass-Spectrometry. Rapid Communications in Mass Spectrometry. 1991;5:374–377. [Google Scholar]
- 29.Naldi M, Andrisano V, Fiori J, Calonghi N, Pagnotta E, Parolin C, Pieraccini G, Masotti L. Histone proteins determined in a human colon cancer by high-performance liquid chromatography and mass spectrometry. J Chromatogr A. 2006;1129:73–81. doi: 10.1016/j.chroma.2006.06.100. [DOI] [PubMed] [Google Scholar]
- 30.Su X, Jacob NK, Amunugama R, Lucas DM, Knapp AR, Ren C, Davis ME, Marcucci G, Parthun MR, Byrd JC, Fishel R, Freitas MA. Liquid chromatography mass spectrometry profiling of histones. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;850:440–54. doi: 10.1016/j.jchromb.2006.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang K, Tang H. Analysis of core histones by liquid chromatography-mass spectrometry and peptide mapping. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;783:173–9. doi: 10.1016/s1570-0232(02)00631-1. [DOI] [PubMed] [Google Scholar]
- 32.Dou Y, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol Cell. 2000;6:225–31. doi: 10.1016/s1097-2765(00)00024-1. [DOI] [PubMed] [Google Scholar]
- 33.Dou Y, Gorovsky MA. Regulation of transcription by H1 phosphorylation in Tetrahymena is position independent and requires clustered sites. Proc Natl Acad Sci U S A. 2002;99:6142–6. doi: 10.1073/pnas.092029599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hayes JJ, Lee KM. In vitro reconstitution and analysis of mononucleosomes containing defined DNAs and proteins. Methods. 1997;12:2–9. doi: 10.1006/meth.1997.0441. [DOI] [PubMed] [Google Scholar]
- 35.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 36.Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007;21:1519–29. doi: 10.1101/gad.1547707. [DOI] [PMC free article] [PubMed] [Google Scholar]