Abstract
The p53 tumor suppressor inhibits the proliferation of cells which undergo prolonged activation of the mitotic checkpoint. However, the function of this antiproliferative response is not well defined. Here we report that p53 suppresses structural chromosome instability following mitotic arrest in human cells. In both HCT116 colon cancer cells and normal human fibroblasts, DNA breaks occurred during mitotic arrest in a p53-independent manner, but p53 was required to suppress the proliferation and structural chromosome instability of the resulting polyploid cells. In contrast, cells made polyploid without mitotic arrest exhibited neither significant structural chromosome instability nor p53-dependent cell cycle arrest. We also observed that p53 suppressed both the frequency and structural chromosome instability of spontaneous polyploids in HCT116 cells. Furthermore, time-lapse videomicroscopy revealed that polyploidization of p53−/− HCT116 cells is frequently accompanied by mitotic arrest. These data suggest that a function of the p53-dependent postmitotic response is the prevention of structural chromosome instability following prolonged activation of the mitotic checkpoint. Accordingly, our study suggests a novel mechanism of tumor suppression for p53, as well as a potential role for p53 in the outcome of antimitotic chemotherapy.
Keywords: p53, cell cycle arrest, chromosomal instability, DNA damage, mitotic checkpoint, polypoidization
Introduction
The p53 tumor suppressor represents a central defense against human cancer (Vousden and Lane, 2007). Its inactivation is one of the most common alterations in human tumors, and numerous studies have established the tumor suppressing properties of p53 (Toledo and Wahl, 2006). A principal mechanism of this tumor suppression is the induction of growth arrest and/or apoptosis in cells which suffer DNA damage (Vousden and Lu, 2002). In this way, p53 inhibits the propagation of cells which harbor potentially oncogenic DNA alterations. In addition, other forms of stress have been shown to activate p53-dependent responses (Vousden and Lane, 2007). One example is prolonged activation of the mitotic checkpoint, which elicits a p53-dependent cell cycle arrest (Ganem and Pellman, 2007). This “postmitotic” response, so named because growth arrest is actually imposed on cells which have exited from prolonged mitosis, has been observed in numerous cell systems (Andreassen et al., 2001; Chan et al., 2008; Cross et al., 1995; Di Leonardo et al., 1997; Lanni and Jacks, 1998; Minn et al., 1996; Rajagopalan et al., 2004). Despite the ubiquity of the postmitotic response, its function is not well defined (Ganem and Pellman, 2007; Stukenberg, 2004).
One clue to the function of the postmitotic response may be that prolonged activation of the mitotic checkpoint has been causally implicated in tumorigenesis (Dalton and Yang, 2009). Indeed, mitosis is frequently prolonged in cancer cells, and several genetic and epigenetic defects which cause mitotic arrest can contribute to cancer (Dalton and Yang, 2009). For some of these defects, such as inactivation of Rb and hCDC4, oncogenic activation of c-Myc, and the presence of supernumerary chromosomes and/or centrosomes, prolonged mitosis is one of many cellular effects which may or may not be oncogenic (Fujiwara et al., 2005; Hernando et al., 2004; Rajagopalan et al., 2004; Yang et al., 2008). However, mitotic arrest and cancer also develop in mice overexpressing Mad2, a protein principally involved in mitotic checkpoint signaling, providing strong evidence that prolonged mitotic checkpoint activation can directly promote tumorigenesis (Sotillo et al., 2007). Accordingly, the p53-dependent postmitotic response may serve to inhibit the propagation of cells which acquire oncogenic properties during prolonged activation of the mitotic checkpoint.
What aspects of mitotic arrest might be oncogenic? Certainly, one candidate is aneuploidy and/or tetraploidy resulting from the chromosome missegregation and/or cytokinesis failure which can follow prolonged activation of the mitotic checkpoint (Ganem and Pellman, 2007). Indeed, in some contexts, aneuploidy and tetraploidy have themselves been causally implicated in tumorigenesis (Fujiwara et al., 2005; Weaver et al., 2007). At the same time, we and others recently found that mitotic arrest can induce structural chromosome changes resulting from double-stranded DNA breaks (Dalton et al., 2007; Quignon et al., 2007; Stevens et al., 2007). Given the role of structural chromosome aberrations in tumorigenesis, these observations suggest that one way prolonged mitosis could promote cancer is through introduction of DNA breaks. By extension, one function of the p53-dependent postmitotic response may be to prevent this structural chromosome instability. To investigate this possibility, we have measured structural chromosome instability resulting from mitotic arrest in human colon cancer cells and normal fibroblasts which differ only in their p53 status. Our results demonstrate that, by imposing growth arrest and/or apoptosis in cells whose DNA is damaged during mitotic arrest, p53 suppresses structural chromosome instability following prolonged mitotic checkpoint activation in human cells.
Materials and Methods
Cell lines and treatments
IMR90 HDFs were obtained from the ATCC. p53+/+ and p53−/− HCT116 cells were kindly provided by B. Vogelstein (Johns Hopkins Medical Institution, Baltimore, MD). HCT116 cells were cultured in McCoy’s and seeded at a density of 3 × 104 cells/cm2 onto fibronectin-coated dishes or slides 24h prior to experiments. IMR90 cells were cultured in DMEM and also seeded at a density of 3 × 104 cells/cm2 24h prior to experiments. Nocodazole and blebbistatin (Sigma) were used at 200nM and 150μM, respectively, the minimum concentrations which completely inhibited cell division in HCT116 cells (data not shown). γ-irradiation was performed with a Cs-137 Gammacell. Stealth™ Select siRNAs targeted to Eg5, control siRNA, and Lipofectamine RNAiMax™ were obtained from, and used according to the instructions of, Invitrogen. miRNAs targeting p53, or a nonspecific control, were also obtained from Invitrogen, initially as DNA oligos. These oligos were then cloned into the pLenti6-GW/EmGFP-miR lentiviral expression vector (Invitrogen), and these vectors were transfected into 293FT cells along with the pLP/VSVG, pLP1, and pLP2 plasmids (Invitrogen) to produce miRNA-containing lentivirus. HCT116 and IMR90 were transduced with high-titer virus, and pools of hundreds of blasticidin-resistant colonies were expanded to produce stable knockdown cell lines.
Immunodetection
For immunocytochemistry, cells were fixed with 2% formaldehyde/PBS and permeabilized with 0.2% Triton-X 100. Antibody incubations were 1h at room temperature, and DNA was counterstained with DAPI. Images were acquired with a Zeiss Axioskop 2 Plus microscope. For MPM-2 flow cytometry, cells were harvested by trypsinization and fixed overnight at −20°C in 70% ethanol. Antibody incubations were 1h at room temperature, and DNA was counterstained with propidium iodide. For γ-H2AX/CC-3 flow cytometry, cells were harvested by trypsinization, fixed with 2% formaldehyde/PBS, permeabilized with methanol, and incubated overnight at 4°C with primary antibodies, followed the next day by 1h incubations with secondary antibodies. Like MPM-2, CC-3 is an antibody which specifically stains mitotic cells, but is an IgG2a allotype, and can thus be used simultaneously with IgG1 antibodies, such as γ-H2AX (Thibodeau and Vincent, 1991). Data were acquired using a FACSCalibur (Becton-Dickinson) and analyzed with Flowjo. Immunoblotting was performed as previously described (Yoon et al., 2005).
The following antibodies were used: mouse anti-γ-H2AX (Upstate), rat anti-α-tubulin (Chemicon), mouse MPM-2 (Upstate), mouse CC-3 (a gift from M. Vincent, Université Laval, Québec, Qc, Canada), rabbit anti-Eg5 (Abcam), goat anti-p53 (Santa Cruz), rabbit anti-cleaved-PARP (Cell Signaling), and mouse anti-actin (Sigma). All fluorescent secondary antibodies were Alexa-conjugates (Molecular Probes).
Cytogenetic analyses
Chromosome spreads were prepared using standard cytogenetic techniques, DNA was stained with DAPI, and images were obtained using a Zeiss Axioskop 2 Plus microscope. Scoring of chromosome aberrations was performed according to the classification system of Savage (Savage, 1976). Furthermore, all scoring was performed, where possible, in a blinded fashion. For color karyotyping of spontaneous polyploid p53−/− cells, analysis was performed by Chrombios GmbH (Munich, Germany).
Colony survival
Following 48h nocodazole treatment, HCT116 cells were harvested by trypsinization and seeded into T75 flasks. After 9 days, colonies were stained with methylene blue, photographs of the flasks were taken, and colony number was quantified using Metamorph imaging software. For quantification of untreated control colonies, 1/100 the number of cells used in nocodazole-treated samples were seeded into T75 flasks and colony number was normalized accordingly, as use of the same number of cells as nocodazole-treated samples produced colonies which were too dense to be quantified (Figure 2F).
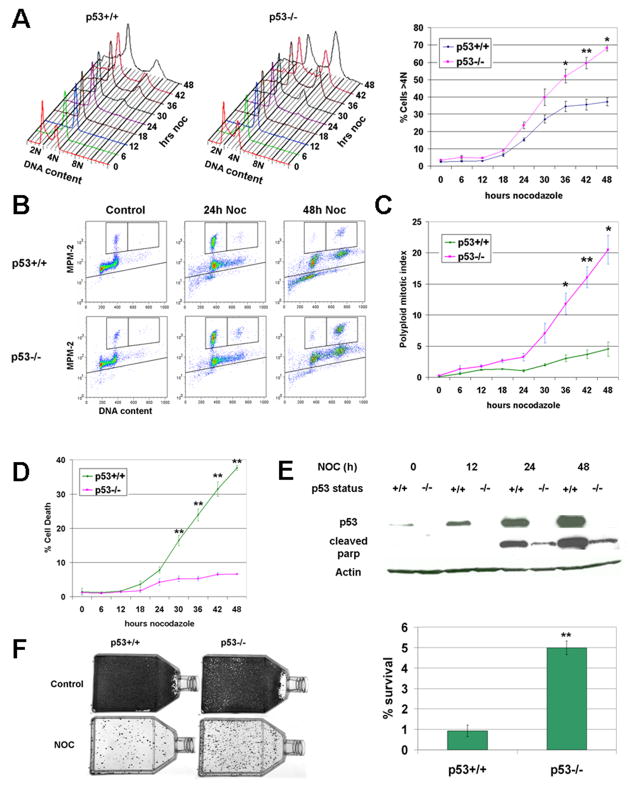
Figure 2. p53 inhibits the polyploidization and survival of HCT116 cells following prolonged mitotic arrest.
A. Example of cell ploidy (left panel) and quantification of polyploidy (right panel) in nocodazole-treated cells. Means and SEMs are from 2–3 independent experiments. * = p<0.05, ** = p<0.01 for t-tests. B. Dot plots of cells stained for MPM-2 and DNA content after 24h and 48h nocodazole treatment. C. Quantification of polyploid (> 4N) mitotic index (upper right gate in Figure 2B) after 48h nocodazole. Means and SEMs are from 2–3 independent experiments. * = p<0.05, ** = p<0.01 for t-tests. D. Quantification of cell death (lower gate in Figure 2B) during 48h nocodazole treatment. Cells that were entirely negative for the MPM-2 phosphoepitope were found, through fluorescent microscopy, to be apoptotic (Supporting Figure S1). Means and SEMs are from 2–3 independent experiments. * = p<0.05, ** = p<0.01 for t-tests. E. Western blot analysis of p53 and cleaved PARP levels during 48h nocodazole. Actin was used as a loading control. F. Example of colony survival after 48h nocodazole (left panel) and its quantification (right panel). % survival is the number of nocodazole-treated colonies divided by the number of control colonies, although more sparse control flasks were used for quantification (see Materials and methods). Means and SEMs are from three independent experiments. ** = p<0.01 for t-test.
Time-lapse imaging
Phase-contrast images of p53−/− HCT116 cells grown inside a 37°C, 5% CO2 chamber were automatically obtained at 6 min intervals in multiple locations over 48h using an Olympus IX81 microscope. All images were analyzed with Slidebook.
Results
Both p53+/+ and p53−/− cells acquire γ-H2AX foci during mitotic arrest
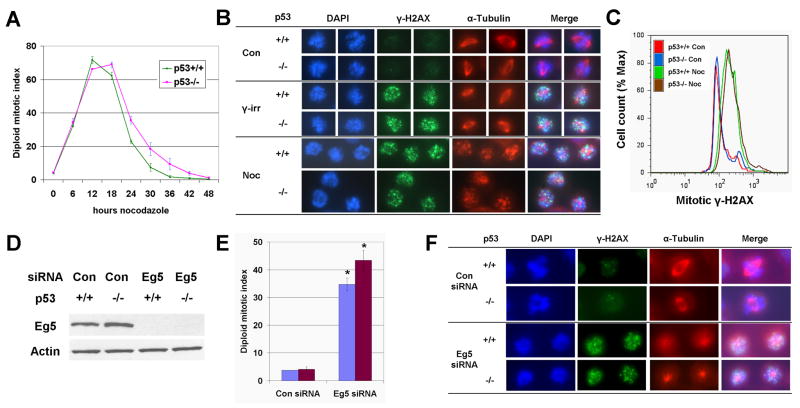
To examine the role of p53 in structural chromosome instability following prolonged mitosis, we first asked whether p53 influences the acquisition of DNA damage during pharmacologic induction of mitotic arrest in isogenic p53+/+ and p53−/− HCT116 cells (Bunz et al., 1998). Upon treatment with the microtubule-depolymerizing agent nocodazole, both p53+/+ and p53−/− cells exhibited a transient rise in mitotic index which peaked at 12–18h (Figure 1A). This was followed by “mitotic slippage,” a process whereby mitotically-arrested cells return to interphase without undergoing cell division (Figure 1A) (Rieder and Maiato, 2004). To determine the extent of DNA damage acquired during this arrest, we measured the formation of γ-H2AX, the phosphorylated form of histone H2AX which forms around sites of DNA breaks (Rogakou et al., 1999). Similar to our previous findings (Dalton et al., 2007), nocodazole-arrested prometaphase p53+/+ cells showed an increase in γ-H2AX foci, as compared to prometaphase controls (Figure 1B). Increased γ-H2AX foci were also observed in p53−/− cells (Figure 1B), and flow cytometric analysis demonstrated that the magnitude of this increase was comparable to p53+/+ cells (Figure 1C). To determine whether increased γ-H2AX was indeed the result of mitotic arrest, and not some other effects of nocodazole, we performed the assay after siRNA-mediated knockdown of the Eg5 mitotic kinesin protein, whose inactivation prevents centrosome separation, produces a monopolar spindle, and thereby induces mitotic arrest (Koller et al., 2006). Indeed, knockdown of Eg5 produced an elevated mitotic index, monopolar spindles, and increased γ-H2AX foci in mitotic p53+/+ and p53−/− cells (Figure 1D–F). Thus, both pharmacologic and genetic manipulations which provoke mitotic arrest through distinct mechanisms produce evidence of DNA breaks in p53+/+ and p53−/− HCT116 cells.
Figure 1. Both p53+/+ and p53−/− HCT116 cells acquire DNA damage during mitotic arrest.
A. Diploid mitotic index of HCT116 cells during 48h nocodazole, as determined by MPM-2 flow cytometry. Means and SEMs are from 2–3 independent experiments. B. Images of prometaphase cells stained for γ-H2AX and α-tubulin. Nuclei were counterstained with DAPI. Noc = 18h nocodazole. γ-irr = 30 min after 2 Gy γ-irradiation. C. Flow cytometric analysis of mitotic γ-H2AX. Cells were treated with or without 18h nocodazole and stained for CC-3 and γ-H2AX. CC-3-positive cells, which are mitotic (Thibodeau and Vincent, 1991), were gated, and γ-H2AX signals of the gated cells are shown. Data are representative of two independent experiments. D. Western blot analysis of Eg5 in HCT116 cells 24h after transfection with Eg5-specific, or control, siRNA. Actin was used as a loading control. E. Diploid mitotic index of HCT116 cells 24h after transfection with Eg5-specific, or control, siRNA, as determined by MPM-2 flow cytometry. Means and SEMs are from two independent experiments. * = p<0.05, for t-tests, as compared to control. F. Images of prometaphase cells stained for γ-H2AX and α-tubulin 24h after transfection with Eg5-specific, or control, siRNA.
p53 inhibits the polyploidization and survival of postmitotic cells
Having observed that p53 does not influence the acquisition of DNA damage during mitotic arrest, we next investigated whether the cellular consequences of this damage are dependent on p53. We first determined the fates of p53+/+ and p53−/− cells after nocodazole treatment. Similar to results from previous studies (Castedo et al., 2006; Kim et al., 2004; Vogel et al., 2004), p53−/− cells exhibited greater polyploidization and reduced apoptosis when compared to p53+/+ cells during 48h nocodazole (Figure 2A–E and Supporting Figure S1). Because the specificity of gene targeting in cancer cells is not always certain (Matoba et al., 2006; Pfleghaar et al., 2005), we independently confirmed the p53-dependence of these phenotypes using miRNA (Supporting Figure S2). Moreover, because short-term rates of apoptosis do not always correspond to long-term rates of survival (Bunz et al., 1999), we also determined the clonogenicity of these nocodazole-treated cells. This analysis revealed that colony survival of p53−/− cells exposed to 48h nocodazole, while low overall, was 5-fold higher than that of p53+/+ cells (Figure 2F). Collectively, these data indicate that p53 inhibits polyploidization, promotes apoptosis, and suppresses clonogenicitiy following pharmacologic induction of mitotic arrest in HCT116 cells.
p53 inhibits structural chromosome instability in postmitotic cells
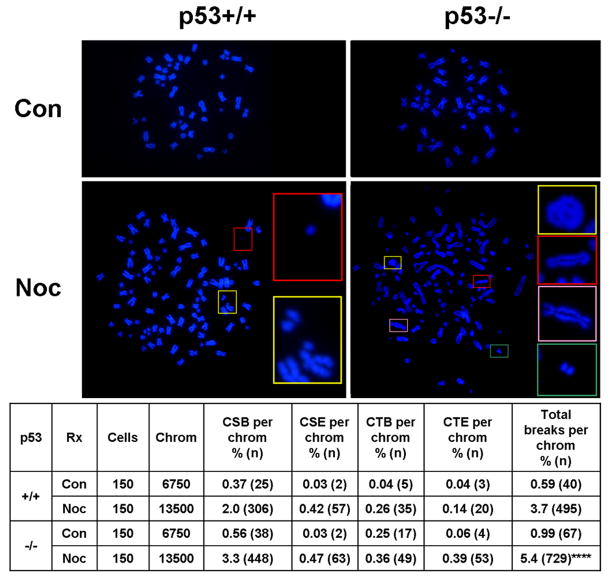
By inducing growth arrest and apoptosis in cells which have undergone prolonged mitotic arrest, p53 could act to suppress the structural chromosome changes which result from DNA damage acquired during prolonged mitosis. If true, p53−/− cells made polyploid following prolonged mitosis might be expected to exhibit not only greater survival but also more structural chromosome aberrations than their p53+/+ counterparts. To test this hypothesis, we examined p53+/+ and p53−/− HCT116 cells for the presence of chromosome aberrations after 48h of nocodazole. Consistent with our previous findings (Dalton et al., 2007), nocodazole-induced polyploid HCT116 cells exhibited multiple types of structural chromosome aberrations (Figure 3). Notably, nocodazole-treated p53−/− cells exhibited a 50% higher burden of aberrations than p53+/+ cells (p < 0.0001) (Figure 3). This result suggests that p53 preferentially inhibits the polyploidization and survival of those cells which suffer the greatest DNA damage during mitotic arrest. In this way, p53 suppresses structural chromosome instability following mitotic arrest in HCT116 cells.
Figure 3. p53 suppresses structural chromosome instability following mitotic arrest in HCT116 cells.
Examples of chromosome spreads in cells treated with or without nocodazole (Noc) for 48h are shown in upper panels. Insets show chromosome fragments (upper left, lower left, lower right), a chromatid break (lower left), a ring chromosome (upper right), and dicentric chromosomes (upper-middle right, lower-middle right). Quantification of chromosome aberrations is shown in table. Con = control cells. Noc = 48h nocodazole. Cells = number of cells analyzed from three independent experiments. Chrom = number of chromosomes, obtained by multiplying cell number by either 45 chromosomes/cell for control diploids (4N), or 90 chromosome/cell for nocodazole-induced polyploids (8N). CSB = chromosome break. CSE = chromosome exchange. CTB = chromatid break. CTE = chromatid exchange. Total breaks = CSB + CTB + 2(CSE + CTE), as two breaks produce exchanges. **** = p<0.0001, for chi square test, as compared to nocodazole-treated p53+/+ cells.
Polyploidy is not responsible for structural chromosome instability following mitotic arrest
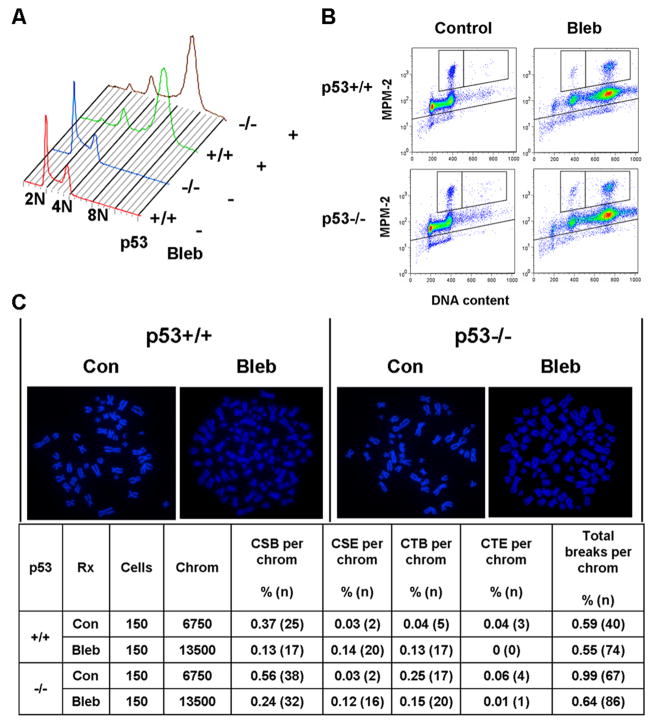
To address the possibility that the polyploid state itself might elicit structural chromosome changes (Ganem et al., 2007), we determined the cell fate and genomic stability of p53+/+ and p53−/− HCT116 cells treated with the myosin inhibitor blebbistatin, which creates polyploid cells through inhibition of cytokinesis without provoking mitotic arrest (Wong and Stearns, 2005). In contrast to nocodazole, blebbistatin was capable of producing polyploidy without significant cell death, cell cycle arrest, or increased chromosome aberrations (Figure 4A–C). Moreover, no difference in these parameters was observed between p53+/+ and p53−/− cells (Figure 4A–C). This result indicates that it is prolonged mitotic arrest, and not polyploidy per se, that elicits significant DNA damage and p53-dependent suppression of structural chromosome instability in nocodazole-treated HCT116 cells. Interestingly, although the total number of breaks per chromosome was not increased by blebbistatin treatment, the frequencies of certain aberrations did change (Figure 4C). This pattern was also observed following cytokinesis inhibition using dihydrocytochalasin B (unpublished observations). Thus, while polyploidization itself does not account for the observed structural chromosome instability following prolonged mitotic arrest, it may be that polyploidization, or pharmacologic inhibition of cytokinesis, may induce some degree of DNA damage.
Figure 4. Polyploidization through inhibition of cytokinesis does not elicit significant DNA damage, postmitotic arrest, or cell death in HCT116 cells.
A. Cell ploidy after 24h blebbistatin. B. Dot plot of cells stained for MPM-2 and DNA content after 24h blebbistatin. C. Examples of chromosome spreads (images) and quantification of chromosome aberrations (table) after 24h blebbistatin. Con = control cells. Bleb = 24h blebbistatin. Cells = number of cells analyzed. Chrom = number of cells multiplied by either 45 chromosomes/cell for control diploids (4N), or 90 chromosomes/cell for blebbistatin-induced polyploids (8N). CSB = chromosome break. CSE = chromosome exchange. CTB = chromatid break. CTE = chromatid exchange. Total breaks = CSB + CTB + 2(CSE + CTE), as two breaks produce exchanges.
p53 suppresses structural chromosome instability following mitotic arrest in normal human fibroblasts
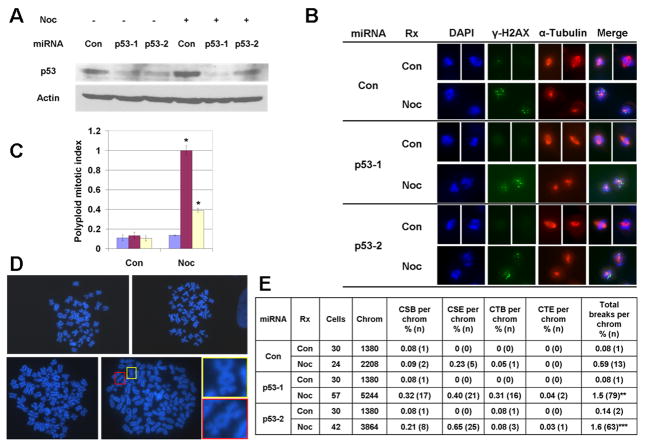
To test whether p53 suppresses structural chromosome instability following mitotic arrest in untransformed human cells, we examined the response to nocodazole of human diploid fibroblasts (HDFs) in which p53 was silenced through lentivirus-mediated expression of miRNAs (Figure 5A). In HDFs expressing either p53 or control miRNAs, nocodazole produced an increase in γ-H2AX foci during mitotic arrest (Figure 5B). Thus, as in HCT116 cells, p53 does not influence the acquisition of DNA damage during mitotic arrest in normal human cells. Also similar to HCT116 cells, HDFs expressing p53 miRNA exhibited an increase in polyploidization (3.5– to 7.5-fold, p<0.05) and structural chromosome instability (2.6- to 2.8-fold, p<0.01) following nocodazole treatment, as compared to control miRNA (Figure 5C–E). In addition to harboring chromosome aberrations like fragments and dicentrics, a minority of HDFs also exhibited evidence of chromosome pulverization, which can occur when micronuclei enter mitosis prematurely (Supporting Figure S3) (Ikeuchi et al., 1972; Kato and Sandberg, 1967). Nocodazole-induced cell death, however, did not appear to be influenced by p53 knockdown in HDFs (Supporting Figure S4), demonstrating that the role of p53 on apoptosis following prolonged mitotic arrest is cell type-dependent, as previously discussed. These data therefore demonstrate that, by inducing postmitotic cell cycle arrest, p53 suppresses structural chromosome instability following mitotic arrest in untransformed, as well as transformed, human cells.
Figure 5. p53 suppresses structural chromosome instability following mitotic arrest in IMR90 human diploid fibroblasts (HDFs).
A. Western blot analysis of p53 after 72h nocodazole in cells stably expressing either control or two independent p53 miRNAs. Actin was used as a loading control. B. Images of prometaphase cells stained for γ-H2AX and α-tubulin. Con = control. Noc = 18h nocodazole. C. Quantification of polyploid mitotic index in miRNA-expressing cells after 72h nocodazole, as determined by MPM-2 flow cytometry. Means and SEMs are from two independent experiments. * = p<0.05 for t-tests, as compared to miRNA-Con cells treated with 48h nocodazole. D. Examples of chromosome spreads in cells treated with or without 72h nocodazole. Untreated (upper) or nocodazole-treated (lower) cells expressing either control (left) or p53-1 (right) miRNAs. Insets show dicentric chromosomes. E. Quantification of chromosome aberrations. Con = control cells. Noc = 72h nocodazole. Cells = number of cells analyzed from two independent experiments. Chrom = number of chromosomes, obtained by multiplying cell number by either 46 chromosomes/cell for control diploids (2N), or 92 chromosomes/cell for nocodazole-induced polyploids (8N). CSB = chromosome break. CSE = chromosome exchange. CTB = chromatid break. CTE = chromatid exchange. Total breaks = CSB + CTB + 2(CSE + CTE), as two breaks produce exchanges. ** = p<0.01, *** = p<0.001, for chi square tests, as compared to nocodazole-treated cells expressing control miRNA.
p53 suppresses structural chromosome instability in spontaneous polyploids
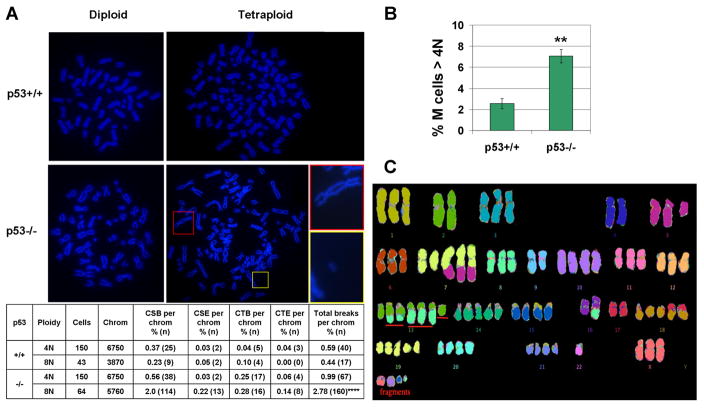
Interestingly, while performing cytogenetic analysis of HCT116 cells, we noticed that many spontaneous polyploid p53−/− cells contained chromosome aberrations, including chromosome fragments, dicentrics, and chromatid breaks (Figure 6A). Indeed, quantification revealed a 2.8-fold increase in the number of breaks per chromosome in polyploid (8N) vs. diploid (4N) p53−/− cells (p<0.0001) (Figure 6A). Color karyotyping of these spontaneous p53−/− polyploids (>4N) confirmed the presence of cells with chromosome fragments and chromosome rearrangements (Figure 6C). In contrast, spontaneous p53+/+ polyploids did not exhibit an elevated frequency of chromosome aberrations, when compared to p53+/+ diploid cells (Figure 6A). Moreover, we found that, while of low overall abundance, spontaneous polyploid mitotic cells were 3-fold more frequent in p53−/−, as compared to p53+/+, cells (Figure 6B), consistent with previous studies (Bunz et al., 2002, Pantic et al., 2006). These findings thus indicate that, in HCT116 cells, p53 suppresses both the frequency and the structural chromosome instability of spontaneous polyploids.
Figure 6. p53 suppresses the frequency and structural chromosome instability in spontaneous polyploid HCT116 cells.
A. Examples of chromosome spreads in spontaneous diploid (4N) or polyploid (8N) cells are shown in upper panels. Insets show a dicentric chromosome (red rectangle) and a chromosome fragment (yellow rectangle). Quantification of chromosome aberrations is shown in table. Con = control cells. Cells = number of cells analyzed from three independent experiments. Chrom = number of chromosomes, obtained by multiplying cell number by either 45 chromosomes/cell for diploid (4N), or 90 chromosomes/cell, for polyploid (8N), chromosomes/cell. CSB = chromosome break. CSE = chromosome exchange. CTB = chromatid break. CTE = chromatid exchange. Total breaks = CSB + CTB + 2(CSE + CTE), as two breaks produce exchanges. **** = p<0.0001, for chi square tests, as compared to diploid p53−/−, diploid p53+/+, or polyploid p53+/+ cells. B. Quantification of mitotic cells with polyploid DNA content, as determined by scoring of chromosome spreads. Means and SEMs are from 3 independent experiments. ** = p<0.01 for t-test. C. Example of chromosome aberrations in a polyploid p53−/− cell, as determined by color karyotyping. The red underlining indicates an aberration not present in the diploid p53+/+ and p53−/− HCT116 karyotype (Bunz et al., 2002). Chromosome fragments are also underlined in the lower left.
Mitotic slippage following prolonged mitosis occurs spontaneously in p53−/− HCT116 cells
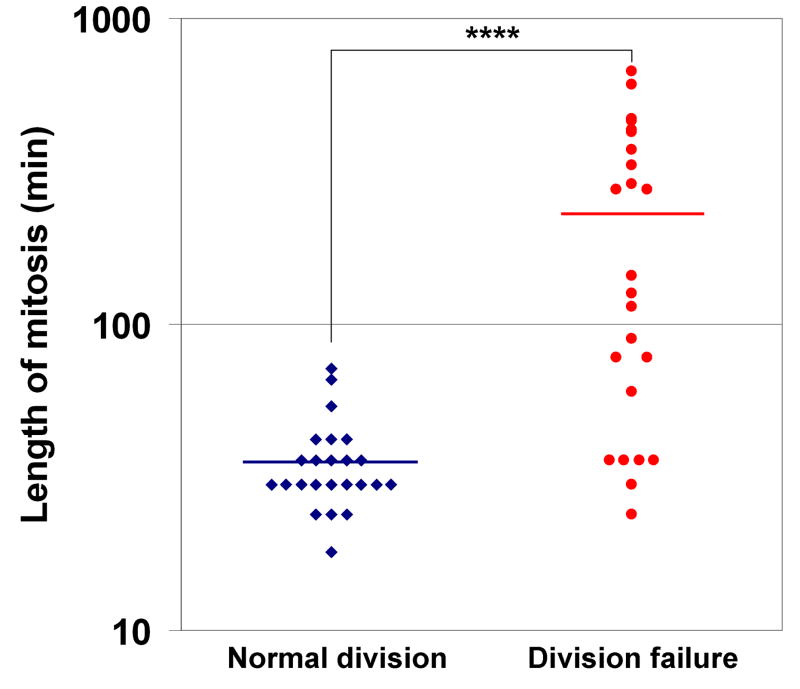
While the origin of spontaneously damaged p53−/− polyploids is unknown, one possibility is that they arise from cells which undergo mitotic slippage following mitotic arrest, for example due to the appearance of spontaneous spindle defects, which we previously observed at low frequency in HCT116 cells (Dalton et al., 2007). To examine this possibility, we examined mitotic progression using time-lapse videomicroscopy. This analysis revealed that 4% (24/592) of mitoses resulted in spontaneous cell division failure in p53−/− HCT116 cells. Moreover, the average length of mitosis in these cells was significantly longer than that of cells which completed a normal, bipolar mitosis (229 vs. 36 minutes, p<0.0001) (Figure 7). Indeed, some cells spent up to 10h in spontaneous mitotic arrest, before undergoing mitotic slippage (Supporting Videos S1–S3). These data thus indicate that mitotic slippage following prolonged mitosis can occur spontaneously, and are consistent with the possibility that this mechanism of polyploidization may be responsible for the increased structural chromosome instability in polyploid p53−/− HCT116 cells.
Figure 7. Mitosis is spontaneously prolonged in p53−/− HCT116 cells which fail to divide.
Dot plot of mitotic length, defined as the interval between the start of mitotic rounding up and anaphase (or mitotic slippage), in cells which underwent normal, bipolar cell division or which failed to divide. 24 cells in each category were scored, and the mean lengths of mitosis are represented by horizontal lines. **** = p<0.0001 for ttest.
Discussion
Our study demonstrates that in both human colon cancer cells and normal human fibroblasts, p53 suppresses structural chromosome instability following prolonged activation of the mitotic checkpoint. This finding has several important implications. Because (1) certain genetic and epigenetic alterations which elicit prolonged mitotic checkpoint activation are causally implicated in tumorigenesis (Dalton and Yang, 2009), (2) prolonged mitosis can provoke DNA breaks (Dalton et al., 2007; Quignon et al., 2007; Stevens et al., 2007), (3) DNA breaks can promote tumorigenesis (van Gent et al., 2001) and (4) inactivation of p53 is one of the most common oncogenic events in human cancer (Vousden and Lane, 2007), our findings suggest that suppression of structural chromosome instability following prolonged mitosis may represent a novel mechanism of tumor suppression for p53. Thus, similar to other sources of DNA damage such as replication stress and radiation, prolonged mitotic checkpoint activation may produce potentially oncogenic DNA damage which necessitates the antitumor activities of p53.
By extension, our data suggest that one function of the p53-dependent postmitotic response may be the suppression of structural chromosome instability. Accordingly, if polyploid cells generated through mitotic slippage evade this p53-dependent response, they may bear an increased risk of promoting cancer, due to genetic alterations acquired during their formation. What is more, polyploid cells may experience further DNA damage during subsequent mitosis, as the presence of supernumerary chromosomes and/or centrosomes itself prolongs activation of the mitotic checkpoint (Yang et al., 2008). Consistent with this idea, we previously observed evidence that spontaneous multipolar HCT116 cells, many of which are likely to be polyploid (Stewenius et al., 2005), may acquire DNA damage during a spontaneously prolonged mitosis (Dalton et al., 2007). Indeed, inactivation of p53 increased the frequency of these damaged multipolar cells (data not shown). Thus, p53-dependent postmitotic arrest may suppress a “vicious cycle” of structural chromosome instability occurring during the formation—and propagation—of polyploid cells. This may help explain why polyploid cells created through cytokinesis inhibition—and thus without prior mitotic arrest—have increased tumorigenicity in a p53-deficient background (Fujiwara et al., 2005).
Previous studies have shown that the role of p53 in the regulation of spontaneous structural chromosome integrity is context-dependent. For example, increased structural aberrations have been observed in bone marrow cells of p53−/− mice (Bouffler et al., 1995) and fibroblasts from patients with germline p53 mutations (Bischoff et al., 1990), but not in lymphoblastoid cells from patients with germline p53 mutations (Lalle et al., 1995) or isogenic human cancer cell lines (Bunz et al., 2002) (see Supporting Info for a discussion of the last example). Similar context-dependence has been observed in cytogenetic and comparative genomic hybridization studies of human tumor tissue, as p53 mutations correlate with structural chromosome instability in several case series (De Angelis et al., 1999; Jong et al., 2004; Kleivi et al., 2005), but not in others (Curtis et al., 2000; Eshleman et al., 1998; Westra et al., 2005). Interestingly, some of this context-dependence may be due to differences in the types of p53 mutations which occur, as there is evidence that gain-of-function p53 mutations are more likely than null p53 mutations to promote structural chromosome instability (Song et al., 2007). Conversely, much of the complexity may result from the different genetic backgrounds of tissue types and individual tumors in which p53 is mutated. Indeed, several studies have shown that while p53 mutation alone may have minimal effects on structural chromosome instability in certain mouse models, its mutation can greatly exacerbate structural chromosome instability produced by mutations in other genes, such as H2AX, 53BP1, telomerase, and others (Artandi et al., 2000; Bassing et al., 2003; Ward et al., 2005). In this way, p53 appears to facilitate the underlying tendencies to structural chromosome instability initiated by other defects. From this view, our data suggest that prolonged activation of the mitotic checkpoint may be one such instigator of structural chromosome instability that is facilitated by p53 mutation. Of note, this situation resembles the complexity recently discovered in numerical chromosome instability, where errors in chromosome missegregation require additional “aneuploidy-tolerating” defects in order to produce stable numerical changes (Thompson and Compton, 2008).
Our data also have implications for understanding the cellular responses to antimitotic chemotherapeutics. The p53-regulated survival of nocodazole-treated HCT116 cells suggests that p53 can be a determinant of antimitotic chemosensitivity. Indeed, we have also observed that p53 knockdown partially attenuated nocodazole-induced apoptosis in RKO colon cancer cells (unpublished observations). These findings are consistent with previous reports in HCT116 and MCF-7 cells (Castedo et al., 2006; Galmarini et al., 2001; Kienitz et al., 2005; Yamaguchi et al., 2004; Zhang et al., 2002). At the same time, studies in other cell systems have found that p53 inactivation confers no change—or even an increase—in antimitotic sensitivity, demonstrating the cell type-specificity of this effect (Minn et al., 1996; Tao et al., 2007; Wahl et al., 1996; Woods et al., 1995). Our own observation that p53 knockdown did not significantly affect nocodazole-induced apoptosis in HDFs is consistent with this idea. This context-dependence is likely influenced by the same factors enumerated above for the role of p53 in structural chromosome instability, as tissue of origin and p53 mutation type have previously been implicated in p53-dependent sensitivity to other chemotherapeutics (Lu and El-Deiry, 2009). Thus, a conservative interpretation of these findings is that p53 may be a determinant of antimitotic sensitivity in a subset of human tumors which also share other modulators of drug sensitivity. Identification of such modulators will be an interesting area for future work, and might include investigations into DNA repair pathways (Swanton et al., 2009), as well as other p53 family members, such as p63 and p73, both of which are known to influence chemosensitivity (Muller et al., 2006). Finally, with the ongoing development of pharmaceuticals which partially restore p53 activity in cancer cells (Lu and El-Deiry, 2009), it may be worthwhile to determine whether such agents might work in synergy with antimitotics by restoring p53-dependent apoptotic and growth arrest signaling following prolonged mitotic arrest.
An important but unanswered question is what triggers p53 during this process (Chan et al., 2008; Ganem and Pellman, 2007; Stukenberg, 2004). Although an initial proposal was that p53 is activated by a mechanism which senses, or counts, the presence of a tetraploid genome (Andreassen et al., 2001), subsequent studies demonstrated that tetraploid cells produced in the absence of prolonged mitosis do not necessarily undergo p53-dependent growth arrest (Uetake and Sluder, 2004; Uetake and Sluder, 2007; Wong and Stearns, 2005). Indeed, the results of our blebbistatin experiment (Figure 5) support the conclusions of these later studies. Another proposal has been that the transcriptional repression which occurs during mitosis leads to inhibition of p53 degradation, which in turn leads to progressive accumulation of p53 protein during prolonged mitotic arrest (Blagosklonny, 2006). However, this “mitotic timer” model for p53 is not supported by studies showing that conditions which dramatically shorten mitotic arrest do not diminish the accumulation of p53 (Chan et al., 2008; Vogel et al., 2004), nor by data showing that p53 accumulation occurs after, and not during, mitotic arrest (Minn et al., 1996).
Because DNA damage is a well-established activator of p53 (Vousden and Lu, 2002), it is tempting to speculate that postmitotic activation of p53 is induced, or influenced, by DNA damage acquired during mitotic arrest. Indeed, our finding that loss of p53 increases the burden of chromosome aberrations in nocodazole-induced polyploids—but does not affect the initial acquisition of DNA damage during mitotic arrest—suggests that the p53-dependent postmitotic response preferentially inhibits the most damaged cells. This, in turn, suggests that the extent of DNA damage acquired during prolonged mitosis influences cell fate. In this way, the postmitotic activation of p53 by DNA damage would be analogous to p53 activation following blockage of nucleotide biosynthesis, which while initially believed to be a non-genotoxic inducer of p53 (Linke et al., 1996), has recently been shown, with more sensitive assays, to cause DNA damage (Hastak et al., 2008).
At the same time, it remains possible that DNA damage is one, but not the only, determinant of p53 activation following prolonged mitotic arrest (Ganem and Pellman, 2007). Indeed, we previously observed significant cell-type variation in nocodazole-induced γ-H2AX foci (Dalton et al., 2007), which raises the question of whether DNA damage could account for p53 activation following mitotic arrest in all cell types. Along these lines, a lack of increased γ-H2AX in nocodazole-treated U20S cells has been reported (Aylon et al., 2006; Chan et al., 2008). However, multiple studies have now reported evidence that DNA damage accompanies treatments which induce mitotic arrest in a variety of cell lines (Dalton et al., 2007; Quignon et al., 2007; Shi et al., 2008; Stevens et al., 2007; Tighe et al., 2004; Wong and Stearns, 2005). Furthermore, it is important to note that a lack of increased γ-H2AX does not rule out the presence of DNA damage, as there are DNA lesions, such as single-stranded breaks and base alterations, which do not induce γ-H2AX (Rogakou et al., 1998). In fact, some of the chromatid-type aberrations we observed after prolonged mitotic arrest could result from lesions other than double-stranded breaks (Zhuanzi et al., 2007). Clearly, future studies into the mechanisms responsible for DNA damage during mitotic arrest will be needed to determine its role in postmitotic p53 activation. Nonetheless, we believe our data demonstrate that p53 functions to inhibit the potentially dangerous consequences of DNA damage acquired during mitotic arrest.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the National Institutes of Health to V.W.Y. (DK52230, DK64399, and CA84197) and to W.B.D. (5T32GM008367).
We thank D. Pallas, P. Doetsch, D. Jones, A. Corbett, G. Davis, and M. Wiltenburg for discussion and support, and B. Vogelstein for providing cell lines. This work was supported in part by grants from the National Institutes of Health to V.W.Y. (DK52230, DK64399, and CA84197) and to W.B.D. (5T32GM008367-18).
Footnotes
Conflict of Interest
None of the authors has any competing financial interests in relation to this work.
References
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–28. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–5. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Aylon Y, Michael D, Shmueli A, Yabuta N, Nojima H, Oren M. A positive feedback loop between the p53 and Lats2 tumor suppressors prevents tetraploidization. Genes Dev. 2006;20:2687–700. doi: 10.1101/gad.1447006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, et al. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell. 2003;114:359–70. doi: 10.1016/s0092-8674(03)00566-x. [DOI] [PubMed] [Google Scholar]
- Bischoff FZ, Yim SO, Pathak S, Grant G, Siciliano MJ, Giovanella BC, et al. Spontaneous abnormalities in normal fibroblasts from patients with Li-Fraumeni cancer syndrome: aneuploidy and immortalization. Cancer Res. 1990;50:7979–84. [PubMed] [Google Scholar]
- Blagosklonny MV. Prolonged mitosis versus tetraploid checkpoint: how p53 measures the duration of mitosis. Cell Cycle. 2006;5:971–5. doi: 10.4161/cc.5.9.2711. [DOI] [PubMed] [Google Scholar]
- Bouffler SD, Kemp CJ, Balmain A, Cox R. Spontaneous and ionizing radiation-induced chromosomal abnormalities in p53-deficient mice. Cancer Res. 1995;55:3883–9. [PubMed] [Google Scholar]
- Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- Bunz F, Fauth C, Speicher MR, Dutriaux A, Sedivy JM, Kinzler KW, et al. Targeted inactivation of p53 in human cells does not result in aneuploidy. Cancer Res. 2002;62:1129–33. [PubMed] [Google Scholar]
- Bunz F, Hwang PM, Torrance C, Waldman T, Zhang Y, Dillehay L, et al. Disruption of p53 in human cancer cells alters the responses to therapeutic agents. J Clin Invest. 1999;104:263–9. doi: 10.1172/JCI6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, et al. Apoptosis regulation in tetraploid cancer cells. EMBO J. 2006;25:2584–95. doi: 10.1038/sj.emboj.7601127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YW, On KF, Chan WM, Wong W, Siu HO, Hau PM, et al. The kinetics of p53 activation versus cyclin E accumulation underlies the relationship between the spindle-assembly checkpoint and the postmitotic checkpoint. J Biol Chem. 2008;283:15716–23. doi: 10.1074/jbc.M800629200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, et al. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–6. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- Curtis LJ, Georgiades IB, White S, Bird CC, Harrison DJ, Wyllie AH. Specific patterns of chromosomal abnormalities are associated with RER status in sporadic colorectal cancer. J Pathol. 2000;192:440–5. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH761>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Dalton WB, Nandan MO, Moore RT, Yang VW. Human cancer cells commonly acquire DNA damage during mitotic arrest. Cancer Res. 2007;67:11487–92. doi: 10.1158/0008-5472.CAN-07-5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton WB, Yang VW. The role of prolonged mitotic checkpoint activation in the formation and treatment of cancer. Future Oncology. 2009 doi: 10.2217/fon.09.118. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis PM, Clausen OP, Schjolberg A, Stokke T. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer. 1999;80:526–35. doi: 10.1038/sj.bjc.6690388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leonardo A, Khan SH, Linke SP, Greco V, Seidita G, Wahl GM. DNA rereplication in the presence of mitotic spindle inhibitors in human and mouse fibroblasts lacking either p53 or pRb function. Cancer Res. 1997;57:1013–9. [PubMed] [Google Scholar]
- Eshleman JR, Casey G, Kochera ME, Sedwick WD, Swinler SE, Veigl ML, et al. Chromosome number and structure both are markedly stable in RER colorectal cancers and are not destabilized by mutation of p53. Oncogene. 1998;17:719–25. doi: 10.1038/sj.onc.1201986. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–7. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Galmarini CM, Falette N, Tabone E, Levrat C, Britten R, Voorzanger-Rousselot N, et al. Inactivation of wild-type p53 by a dominant negative mutant renders MCF-7 cells resistant to tubulin-binding agent cytotoxicity. Br J Cancer. 2001;85:902–8. doi: 10.1054/bjoc.2001.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–40. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–62. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Hastak K, Paul RK, Agarwal MK, Thakur VS, Amin AR, Agrawal S, et al. DNA synthesis from unbalanced nucleotide pools causes limited DNA damage that triggers ATR-CHK1-dependent p53 activation. Proc Natl Acad Sci U S A. 2008;105:6314–9. doi: 10.1073/pnas.0802080105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430:797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- Ikeuchi T, Weinfeld H, Sandberg AA. Chromosome pulverization in micronuclei induced by tritiated thymidine. J Cell Biol. 1972;52:97–104. doi: 10.1083/jcb.52.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jong YJ, Li LH, Tsou MH, Chen YJ, Cheng SH, Wang-Wuu S, et al. Chromosomal comparative genomic hybridization abnormalities in early- and late-onset human breast cancers: correlation with disease progression and TP53 mutations. Cancer Genet Cytogenet. 2004;148:55–65. doi: 10.1016/s0165-4608(03)00205-x. [DOI] [PubMed] [Google Scholar]
- Kato H, Sandberg AA. Chromosome pulverization in human binucleate cells following colcemid treatment. J Cell Biol. 1967;34:35–45. doi: 10.1083/jcb.34.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienitz A, Vogel C, Morales I, Muller R, Bastians H. Partial downregulation of MAD1 causes spindle checkpoint inactivation and aneuploidy, but does not confer resistance towards taxol. Oncogene. 2005;24:4301–10. doi: 10.1038/sj.onc.1208589. [DOI] [PubMed] [Google Scholar]
- Kim KT, Ongusaha PP, Hong YK, Kurdistani SK, Nakamura M, Lu KP, et al. Function of Drg1/Rit42 in p53-dependent mitotic spindle checkpoint. J Biol Chem. 2004;279:38597–602. doi: 10.1074/jbc.M400781200. [DOI] [PubMed] [Google Scholar]
- Kleivi K, Diep CB, Pandis N, Heim S, Teixeira MR, Lothe RA. TP53 mutations are associated with a particular pattern of genomic imbalances in breast carcinomas. J Pathol. 2005;207:14–9. doi: 10.1002/path.1812. [DOI] [PubMed] [Google Scholar]
- Koller E, Propp S, Zhang H, Zhao C, Xiao X, Chang M, et al. Use of a chemically modified antisense oligonucleotide library to identify and validate Eg5 (kinesin-like 1) as a target for antineoplastic drug development. Cancer Res. 2006;66:2059–66. doi: 10.1158/0008-5472.CAN-05-1531. [DOI] [PubMed] [Google Scholar]
- Lalle P, Moyret-Lalle C, Wang Q, Vialle JM, Navarro C, Bressac-de Paillerets B, et al. Genomic stability and wild-type p53 function of lymphoblastoid cells with germ-line p53 mutation. Oncogene. 1995;10:2447–54. [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–64. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, Di Leonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes Dev. 1996;10:934–47. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- Lu C, El-Deiry WS. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 2009;14:597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, et al. p53 regulates mitochondrial respiration. Science. 2006;312:1650–3. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- Minn AJ, Boise LH, Thompson CB. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–31. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- Muller M, Schleithoff ES, Stremmel W, Melino G, Krammer PH, Schilling T. One, two, three--p53, p63, p73 and chemosensitivity. Drug Resist Updat. 2006;9:288–306. doi: 10.1016/j.drup.2007.01.001. [DOI] [PubMed] [Google Scholar]
- Pfleghaar K, Heubes S, Cox J, Stemmann O, Speicher MR. Securin is not required for chromosomal stability in human cells. PLoS Biol. 2005;3:e416. doi: 10.1371/journal.pbio.0030416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quignon F, Rozier L, Lachages AM, Bieth A, Simili M, Debatisse M. Sustained mitotic block elicits DNA breaks: one-step alteration of ploidy and chromosome integrity in mammalian cells. Oncogene. 2007;26:165–72. doi: 10.1038/sj.onc.1209787. [DOI] [PubMed] [Google Scholar]
- Rajagopalan H, Jallepalli PV, Rago C, Velculescu VE, Kinzler KW, Vogelstein B, et al. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell. 2004;7:637–51. doi: 10.1016/j.devcel.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–16. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Savage JR. Classification and relationships of induced chromosomal structual changes. J Med Genet. 1976;13:103–22. doi: 10.1136/jmg.13.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Orth JD, Mitchison T. Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res. 2008;68:3269–76. doi: 10.1158/0008-5472.CAN-07-6699. [DOI] [PubMed] [Google Scholar]
- Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol. 2007;9:573–80. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, et al. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JB, Liu G, Bremer SW, Ye KJ, Xu W, Xu J, et al. Mitotic cell death by chromosome fragmentation. Cancer Res. 2007;67:7686–94. doi: 10.1158/0008-5472.CAN-07-0472. [DOI] [PubMed] [Google Scholar]
- Stewenius Y, Gorunova L, Jonson T, Larsson N, Hoglund M, Mandahl N, et al. Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc Natl Acad Sci U S A. 2005;102:5541–6. doi: 10.1073/pnas.0408454102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukenberg PT. Triggering p53 after cytokinesis failure. J Cell Biol. 2004;165:607–8. doi: 10.1083/jcb.200405089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanton C, Nicke B, Schuett M, Eklund AC, Ng C, Li Q, et al. Chromosomal instability determines taxane response. Proc Natl Acad Sci U S A. 2009;106:8671–6. doi: 10.1073/pnas.0811835106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W, South VJ, Diehl RE, Davide JP, Sepp-Lorenzino L, Fraley ME, et al. An inhibitor of the kinesin spindle protein activates the intrinsic apoptotic pathway independently of p53 and de novo protein synthesis. Mol Cell Biol. 2007;27:689–98. doi: 10.1128/MCB.01505-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibodeau A, Vincent M. Monoclonal antibody CC-3 recognizes phosphoproteins in interphase and mitotic cells. Exp Cell Res. 1991;195:145–53. doi: 10.1016/0014-4827(91)90510-2. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol. 2008;180:665–72. doi: 10.1083/jcb.200712029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Taylor SS. Truncating APC mutations have dominant effects on proliferation, spindle checkpoint control, survival and chromosome stability. J Cell Sci. 2004;117:6339–53. doi: 10.1242/jcs.01556. [DOI] [PubMed] [Google Scholar]
- Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165:609–15. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell-cycle progression without an intact microtuble cytoskeleton. Curr Biol. 2007;17:2081–6. doi: 10.1016/j.cub.2007.10.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet. 2001;2:196–206. doi: 10.1038/35056049. [DOI] [PubMed] [Google Scholar]
- Vogel C, Kienitz A, Hofmann I, Muller R, Bastians H. Crosstalk of the mitotic spindle assembly checkpoint with p53 to prevent polyploidy. Oncogene. 2004;23:6845–53. doi: 10.1038/sj.onc.1207860. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–83. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- Wahl AF, Donaldson KL, Fairchild C, Lee FY, Foster SA, Demers GW, et al. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat Med. 1996;2:72–9. doi: 10.1038/nm0196-72. [DOI] [PubMed] [Google Scholar]
- Ward IM, Difilippantonio S, Minn K, Mueller MD, Molina JR, Yu X, et al. 53BP1 cooperates with p53 and functions as a haploinsufficient tumor suppressor in mice. Mol Cell Biol. 2005;25:10079–86. doi: 10.1128/MCB.25.22.10079-10086.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Westra JL, Boven LG, van der Vlies P, Faber H, Sikkema B, Schaapveld M, et al. A substantial proportion of microsatellite-unstable colon tumors carry TP53 mutations while not showing chromosomal instability. Genes Chromosomes Cancer. 2005;43:194–201. doi: 10.1002/gcc.20148. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CM, Zhu J, McQueney PA, Bollag D, Lazarides E. Taxol-induced mitotic block triggers rapid onset of a p53-independent apoptotic pathway. Mol Med. 1995;1:506–26. [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Chen J, Bhalla K, Wang HG. Regulation of Bax activation and apoptotic response to microtubule-damaging agents by p53 transcription-dependent and -independent pathways. J Biol Chem. 2004;279:39431–7. doi: 10.1074/jbc.M401530200. [DOI] [PubMed] [Google Scholar]
- Yang Z, Loncarek J, Khodjakov A, Rieder CL. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat Cell Biol. 2008;10:748–51. doi: 10.1038/ncb1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon HS, Ghaleb AM, Nandan MO, Hisamuddin IM, Dalton WB, Yang VW. Kruppel-like factor 4 prevents centrosome amplification following gamma-irradiation-induced DNA damage. Oncogene. 2005;24:4017–25. doi: 10.1038/sj.onc.1208576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Shi X, Zhang QJ, Hampong M, Paddon H, Wahyuningsih D, et al. Nocodazole-induced p53-dependent c-Jun N-terminal kinase activation reduces apoptosis in human colon carcinoma HCT116 cells. J Biol Chem. 2002;277:43648–58. doi: 10.1074/jbc.M203214200. [DOI] [PubMed] [Google Scholar]
- Zhuanzi W, Wenjian L, Dejuan Z, Wei W, Xigang J, Qingxiang G. Chromatid-type aberrations following irradiation in G0 lymphocytes with heavy ions. Mutat Res. 2007;617:98–103. doi: 10.1016/j.mrfmmm.2007.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.