Abstract
Successful pregnancy depends on the precise regulation of extravillous trophoblast (EVT) invasion into the uterine decidua, primarily by decidua-derived factors. In humans, during early pregnancy interleukin 11 (IL11) is maximally expressed in the decidua, with its receptor, IL11 receptor alpha (IL11RA), also identified on invasive EVTs in vivo. Although a role for IL11 in EVT migration has been established, whether it also plays a role in regulating EVT invasion is unknown. We investigated whether IL11 influences human EVT invasion and the signaling pathways and underlying mechanisms that may be involved, using the HTR-8/SVneo immortalized EVT cell line and primary EVTs as models for EVTs. Interleukin 11 (100 ng/ml) significantly inhibited invasion of EVT cells by 40% to 60% (P < 0.001). This effect was abolished by inhibitors of signal transducer and activator of transcription 3 (STAT3) but not of mitogen-activated protein kinase (MAPK) pathways. Interleukin 11 (100 ng/ml) had no effect on matrix metallopeptidases 2 and 9 (MMP2 and MMP9), tissue inhibitors of MMP (TIMP1, TIMP2, and TIMP3), plasminogen activator urokinase (PLAU), plasminogen activator urokinase receptor (PLAUR), and serpin peptidase inhibitors 1 and 2 (SERPINE1 and SERPINE2) in EVT-conditioned media and/or cell lysates. Interleukin 11 (100 ng/ml) also did not regulate EVT cell adhesion or integrin expression. These data demonstrate that IL11 inhibits human EVT invasion via STAT3, indicating a likely role for IL11 in the decidual restraint of EVT invasion during normal pregnancy.
Keywords: cytokines, decidua, extravillous trophoblast, interleukin 11, placenta, pregnancy, STAT3, trophoblast
Interleukin 11 inhibits human trophoblast invasion via signal transducer and activator of transcription 3, indicating a likely important role in regulating the extent of trophoblast invasion during placentation.
INTRODUCTION
Extravillous trophoblast (EVT) invasion through the maternal decidua is critical to the formation of a functional placenta. Inadequate arterial remodeling associated with poor EVT invasion is a key pathological feature of preeclampsia and intrauterine growth restriction, whereas excessive EVT invasion occurs in placental pathologies, such as placental accreta or percreta [1]. Extravillous trophoblast invasion is tightly regulated by numerous growth and regulatory factors within the uterine endometrial microenvironment, primarily the decidua [2].
Interleukin 11 (IL11) belongs to the IL6-type family of cytokines (which includes leukemia inhibitory factor [LIF]) and signals via a heterodimeric receptor complex comprising the specific IL11 receptor alpha (IL11RA) chain and the associated signaling subunit, IL6 signal transducer (IL6ST; also known as gp130), primarily through the janus kinase/signal transducers and activators of transcription (JAK/STAT) signal transduction pathway. Interleukin 11 also promotes formation of active GTP-bound Ras and induces activation of mitogen-activated protein kinases (MAPKs [3]), with cross-talk between Ras-MAPK and JAK/STAT signaling pathways also occurring [3, 4]. Interleukin 11 influences both cellular proliferation and differentiation [5] and also importantly has roles in tumorigenesis, stimulating migration and/or invasion of gastric, colorectal, and breast carcinomas [6–12].
Interleukin 11 signaling is required unequivocally for blastocyst implantation in mice. Female mice with a null mutation in the IL11RA gene are infertile because of a defective decidualization response, resulting in dysregulated trophoblast invasion and necrotic loss of the fetus [13, 14]. In humans, during early pregnancy IL11 and IL11RA are maximally expressed in the decidua, where a role for IL11 in human stromal cell decidualization during pregnancy is well established [15]. IL11RA also is expressed by invasive EVTs in the nonhuman primate (cynomolgous monkey) [16] and in humans [17] in vivo, whereas in vitro, IL11 stimulates significant chemotactic migration of human EVTs [17].
Extravillous trophoblast invasion in vivo is a multistep process involving regulated proteolytic degradation of decidual/endothelial extracellular matrix (ECM; in the direction of invasion), then adhesion to ECM components, followed by active cell movement/migration through the degraded matrix [18]. For these processes, protease systems, including the matrix metallopeptidase (MMP) and plasminogen activator urokinase (PLAU; also known as urokinase plasminogen activator) systems, and cell adhesion molecules such as integrins, are all important [19]. Of the MMPs, MMP2 and MMP9 are particularly important during EVT invasion [20]. These are secreted as latent enzymes and are activated outside the cell or when bound to the cell membrane, where their activity is further regulated by the local concentration of tissue inhibitors of metallopeptidases (TIMP) 1, 2, and 3 [21]. Extravillous trophoblasts produce PLAU and express both plasminogen activator urokinase receptors (PLAUR; also known as urokinase plasminogen activator receptor) [22–24] required for PLAU activity and also PLAU inhibitors, serpine peptidase inhibitors (SERPINE) 1 and 2 (also known as plasminogen activator inhibitors 1 and 2), which regulate PLAU activity [22, 25]. Cell adhesion molecules, particularly the integrins, have important roles in the transformation of the EVT from a sessile to a motile phenotype [26]. Extravillous trophoblasts downregulate ITGA6B4 (a laminin receptor) but upregulate ITGA5B1 (a fibronectin receptor) and ITGA1B1 (a laminin/collagen receptor) as they invade into the maternal decidua, whereas EVTs within the maternal vasculature upregulate the vitronectin receptor ITGAvB3/B5 [27].
In the present study we hypothesized that IL11 produced by the decidua regulates human EVT invasion during early placentation. Because of the restricted availability of primary human EVT cells, extended studies were performed using an immortalized EVT cell line, the HTR-8/SVneo cells, followed by more limited studies using primary EVTs isolated from human chorionic villous explant cultures to validate the findings. We investigated the effect of IL11 on EVT invasion of Matrigel/ECM in vitro and the cellular signaling pathways (i.e., JAK/STAT or MAPK) involved via the use of specific inhibitors. We also determined its effects on the expression of specific members of the MMP and PLAU system and cellular adhesive properties that are important during EVT invasion in vivo.
MATERIALS AND METHODS
Tissue Collection
Informed consent was obtained from all participating patients. Ethics approval was granted by the Southern Health Human Research and Ethics committee. Placental samples were collected from healthy women undergoing first-trimester suction termination of pregnancy (6–10 wk) for psychosocial reasons. Tissues were washed in 0.9% saline before transfer to medium 199 (M199; Sigma, St. Louis, MO) containing an antibiotic-antimycotic mixture of 100 U/ml penicillin G sodium, 100 μg/ml erythromycin, 0.25 μg/ml amphotericin β-sulphate, and 20 μg/ml gentamicin sulphate (Invitrogen, Carlsbad, CA).
Placental Villous Explant Culture and EVT Cell Isolation
Primary EVT cells were prepared from the tips of placental terminal villi (n = 3 explant cultures) as described previously [17, 28, 29]. Small pieces of terminal villi were coated with Matrigel (BD Biosciences, Franklin Lakes, NJ) diluted 1:3, allowed to adhere to tissue culture flasks, and maintained in Dulbecco modified Eagle medium (DMEM)/F-12 (Invitrogen) culture media containing 10% fetal calf serum (FCS; JRH Biosciences, Lenexa, KS) and 1% antibiotics (penicillin, streptomycin, and fungizone; Invitrogen) to allow extensive outgrowth for 5–7 days. Outgrowing cells were detached by digestion with 0.03% trypsin-EDTA (Invitrogen) in PBS (pH 7.6) for 5 min at 37°C, followed by neutralization of trypsin activity with 10% FCS in M199. The dispersed cells were separated from tissue remnants using a 40-μm filter, placed in culture, and expanded. Cells then were incubated with mouse anti-human cluster of differentiation antigen-9 (CD9) antibody (DakoCytomation, Glostrup, Denmark), diluted 1:50, for 15 min at 4°C, washed, and incubated with mouse immunoglobulin G-coated magnetic microbeads (MACS kit; Miltenyi Biotech, Bergisch-Gladbach, Germany) for a further 15 min at 4°C. Trophoblast cells were separated from the mixed population of cells by negative selection via immunomagnetic column purification.
Immortalized EVT Cell Line (HTR-8/SVneo)
The EVT cell line HTR-8/SVneo was kindly provided by Dr. Charles Graham (Queen's University, Kingston, ON, Canada). The cell line was produced by immortalization of HTR-8 cells, an EVT cell line from primary explant cultures of human first-trimester placenta (8–10 wk gestation), with SV40 virus and selected on the basis of resistance to neomycin [30]. These cells exhibit markers of primary EVT cells in situ, including cytokeratins KRT7, KRT8, and KRT18, placental-type alkaline phosphatase, high-affinity PLAUR, human leukocyte antigen (HLA) framework antigen W6/32, insulinlike growth factor 2 (IGF2) mRNA and protein, a selective repertoire of integrins ITGA1, ITGA3, ITGA5, ITGAV, ITGB1, ITGAVB3/B5) [28], and HLA-G when cultured on Matrigel [31]. In the present study, HTR-8/SVneo cells were used between passages 70 and 75.
EVT Cell Culture
Extravillous trophoblast cells were seeded at subconfluence density in RPMI (Sigma; HTR-8/SVneo) or DMEM/F-12 (Invitrogen; primary EVTs) containing 10% FCS (JRH Biosciences) and 1% antibiotics (Invitrogen). Cells were washed with PBS without Ca2+ and Mg2+ (PBS−) and transferred to RPMI containing 1% charcoal-stripped (cs) FCS (HTR-8/SVneo) or DMEM/F-12 containing 0.2% csFCS (primary EVTs; experimental media) for 24 h. At the end of the experiment cell numbers were determined using a hemocytometer, conditioned media was collected, and RNA was extracted from the cells. Each experiment was performed two to three times.
Immunocytochemistry
Primary EVT cells were characterized and their identity and/or purity confirmed as described previously [17, 28, 29]. More than 95% of the freshly isolated cells were positive for cytokeratin 7 but negative for CD9 [17]. Further, these cells expressed HLA-G when grown on Matrigel [29]. Extravillous trophoblast cells were grown on chamber slides (BD Biosciences), then washed with cold PBS− and fixed in ice-cold ethanol for 10 min. To examine the expression of IL11RA in EVT cells, both primary EVT cells and HTR-8/SVneo cells were stained with mouse anti-human IL11RA (10 μg/ml; a kind gift from Dr. Lorraine Robb, Walter and Eliza Hall Institute, Melbourne, Australia). Primary human endometrial stromal cells were used as a positive control [32]. Briefly, rehydrated cells were treated with 3% hydrogen peroxide for 10 min, washed with Tris-buffered saline, and incubated with nonimmune block for 30 min. Primary antibody was applied to sections at 4°C for 16 h, followed by biotinylated horse anti-mouse IgG (1:200; Vector Laboratories, Burlingame, CA) and streptavidin-biotin-peroxidase complex ABC (DakoCytomation) for 30 min each. Peroxidase activity was visualized using diaminobenzidine substrate (DakoCytomation). Cells were counterstained with Harris hematoxylin (Sigma), air dried, and mounted.
RNA Extraction and Purification
Total cellular RNA was extracted with the RNeasy Minikit (Qiagen Sciences, Germantown, MD) according to the manufacturer's instructions. All samples were treated with RNase-free DNase (Ambion, Austin, TX) to remove any genomic DNA. RNA samples (1 μg) were reverse transcribed at 46°C for 1.5 h in a 20-μl reaction mixture using avian myeloblastosis mosaic virus reverse transcriptase (Promega, Madison, WI) and 100 ng random hexanucleotide primers (Amersham Biosciences, Piscataway, NJ). Negative controls were included where either RNA or the reverse transcriptase was omitted. The cDNA generated was subsequently amplified by PCR (Hybaid Express Block Cycler; Thermo Hybaid, Ashford, U.K.) using specific primers (Sigma) at 0.25 pmol/μl for IL11RA, IL6ST, and 18S as previously described, with term placenta and primary human endometrial stromal cells used as positive controls [32]. Reverse trascriptase-PCR was performed in a total volume of 40 μl containing 1 μl cDNA, single-strength PCR buffer, 20 μM dinucleotide triphosphates, 0.25 pmol/μl forward and reverse primers, and 2.5 units Taq DNA polymerase (Roche, Castle Hill, Australia). The PCR products were electrophoresed on a 2% agarose gel to establish the specificity of the PCR reaction and product size.
Invasion Assays
Extravillous trophoblast invasion was assessed using a commercially available in vitro invasion assay (Chemicon, Melbourne, Australia). Cells were transferred to experimental medium overnight, trypsinized, and cultured at a density of 10 × 104/250 μl (primary EVT) to 20 × 104/250 μl (HTR-8/SVneo cells) in Matrigel-coated polycarbonate membrane invasion chamber inserts, which were immersed in tissue culture wells (24-well plates) containing 1% csFCS/RPMI (HTR-8/SVneo) or 0.2% csFCS-DMEM/F-12 (primary EVTs). Cells were subject to different treatments: vehicle, recombinant human (rh) IL11 (1, 10, and 100 ng/ml; a gift from Dr. Lorraine Robb), and transforming growth factor beta 1 (TGFB1; 10 ng/ml; positive control; R&D Systems, Minneapolis, MN) in experimental medium (n ≥ 4 wells/treatment). In addition, HTR-8/SVneo cells were preincubated with inhibitors of STAT3 phosphorylation (STAT3i; 20 μM; Calbiochem, San Diego, CA) [33] or MAPK extracellular signal-regulated kinase and extracellular signal-regulated kinase activities (MAPKi; PD 98059; 30 μM; Calbiochem) for 45 min at 37°C prior to addition into invasion assays with exogenous IL11 (100 ng/ml). A dose-response was performed to determine the most effective dose of inhibitor to be used in these assays. After 48 h, media containing noninvaded cells were removed from the upper wells, spun to remove cells, aliquoted, and stored at −20°C until required for zymography or ELISA. Cells at the lower surface of the inserts were detached, lysed, and quantitated using CyQuant GR dye (according to the manufacturer's instructions; Molecular Probes, Eugene, OR). Each invasion assay was repeated three to four times.
Enzyme-Linked Immunosorbent Assays
Interleukin 11 secreted by EVT cells (24 h) was assayed in duplicate by commercially available ELISA kits (R&D Systems). Media from primary human endometrial stromal cells were included as quality control (to monitor precision) and positive control samples [15]. The lower detection limit of the assay was 15 pg/ml, and interassay and intraassay variabilities were 7.1% and 2.8%, respectively [15].
Total human free and MMP-bound TIMP1/2 in EVT-conditioned media was assayed in triplicate by commercially available ELISA kits (R&D Systems) according to manufacturer's instructions. The intraassay and interassay coefficients of variation for the TIMP1 ELISA were 5% and 4.9%, and for the TIMP2 ELISA were 6.5% and 7.8%, respectively. The lower detection limit of both assays was 0.156 ng/ml.
SERPINE1, SERPINE2, and PLAUR in EVT-conditioned media were assayed in triplicate using commercially available ELISA kits (American Diagnostica, Stamford, CT) according to the manufacturer's instructions. Preliminary experiments demonstrated that there was no difference between the levels of PLAUR detected in culture media and those in cell lysates (data not shown), as has also been published previously [34]. Therefore, only conditioned media were assayed. The intraassay and interassay coefficients of variation of the above ELISAs were all lower than 5%, whereas the lower detection limits of these assays were as follows: SERPINE1, 1 ng/ml; SERPINE2, 0.5 ng/ml; and PLAUR, 0.25 ng/ml.
Zymography
Proteinase activity in equal volumes of conditioned media was analyzed by zymography on 10% SDS-polyacrylamide gels containing 1 mg/ml gelatin (to detect MMP activity; Bio-Rad, Hercules, CA) or 2 mg/ml casein (Sigma) and 0.025 U/ml plasminogen (to detect PLAU activity; Sigma) under nonreducing conditions. Gels were washed five times for 5 min with Tris-HCl (pH 7.4) containing Triton X-100 to remove SDS and were incubated overnight at 37°C in 50 mM Tris-HCl (pH 7.4) containing 150 mM NaCl and 10 mM CaCl2 to allow enzymatic digestion of substrate. Gels then were stained with 30% methanol/10% acetic acid containing 0.2% Coomassie blue R250 for 30 min, destained for between 30 and 60 min, and dried between cellulose sheets. Gelatinase or plasminogen activity was visualized by negative staining. The MMP activity was identified by comparison with samples containing previously identified MMPs (media from baby hamster kidney cells stably transfected with human MMP9 provided by Dr. D. Edwards, University of East Anglia, Norwich, U.K.), whereas PLAU activity was identified by molecular weight, which was determined using molecular weight markers (Bio-Rad). The MMP identity of bands was confirmed by incubation of parallel gels in the presence of EDTA, whereas PLAU identity in samples was confirmed by resolving samples in gels containing casein but no plasminogen.
PLAU Activity Assay
Activity of PLAU also was determined by measuring plasminogen activation using commercially available kits (Chemicon) according to the manufacturer's instructions. Briefly, 100 μl standard/sample (i.e., conditioned medium, in duplicate) was added to sample buffer (100 mM Tris and 0.5% Triton X-100, pH 8.8) containing a chromogenic tripeptide PLAU substrate (supplied by the manufacturer) in a final volume of 200 μl. After incubation overnight at 37°C, samples were analyzed spectrophotometrically at 405 nm.
SDS-PAGE and Western Blotting
The EVT cells were grown to confluence, transferred to experimental medium for 24 h, and then treated with rhIL11 at 1, 10, or 100 ng/ml or vehicle (in experimental medium) for 10 min (for assessment of phosphorylated [p]-STAT3/STAT3) or for 48 h (for analysis of TIMP3). Conditioned media were collected, cells were washed with ice-cold PBS− and lysed in 250 μl/flask of ice-cold lysis buffer (50 mM Tris base, 150 mM NaCl, 2 mM EDTA, 2 mM EGTA, 25 mM NaF, and 25 mM β-glycerolphosphate; pH 7.5), and 2 μl/well protease inhibitor cocktail set (Calbiochem) was added. Cell extracts were centrifuged and supernatant protein quantified using the BCA protein assay kit (Pierce, Rockford, IL) according to the manufacturer's instructions. Equal amounts of protein (i.e., cell lysates: 15 μg for STAT3, 25 μg for pSTAT3, 30 μg for TIMP3) were resolved on 10% SDS-PAGE gels, transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk in TBS with 0.1% Tween-20 (Bio-Rad), and probed separately with monoclonal antibodies against pSTAT3, total STAT3 (Tyr705; 5.6 ng/ml; Cell Signaling Technology Inc., Beverly, MA), and TIMP3 (1 μg/ml; R&D Systems) overnight at 4°C. The membranes were washed in TBS with 0.1% Tween-20 (Bio-Rad) and incubated for 1 h with horseradish peroxidase-conjugated rabbit secondary antibody (1:1500; DakoCytomation). Horseradish peroxidase activity was detected using enhanced chemiluminescence reagent (Pierce). Membranes were exposed to autoradiographic film (Amersham Biosciences) with the exposure time adjusted to keep the integrated optical density within a linear and nonsaturated range. Membranes were stripped and incubated with anti-beta-tubulin (1:5000; Sigma) as loading control (data not shown).
Adhesion Assays
Adhesion of HTR-8/SVneo cells to ECM components was determined using commercially available cell adhesion assay kits (Cytomatrix Screening Kit; Chemicon) according to the manufacturer's instructions. These kits comprised 96-well plates coated with ECM substrates, fibronectin (FN), vitronectin (VN), laminin (LN), collagen type I (COL1), and collagen type IV (COL4). HTR-8/Svneo cells were grown to 50% confluence, transferred to experimental medium for 24 h, and then treated with rhIL11 (100 ng/ml) or vehicle (in experimental medium) for 48 h. Cells then were detached, washed twice with PBS containing Ca2+ and Mg2+ (PBS+), resuspended thoroughly in serum-free media (to avoid clumping of cells), and seeded in ECM-coated wells at a density of 5 × 104 to 7.5 × 104 cells/well in 100 μl serum-free media (four to six wells per ECM per treatment) for 1.5 h at 37°C in a CO2 incubator. Wells were washed with PBS+ to eliminate unadhered cells, stained with 0.2% crystal violet solution in 10% ethanol for 5 min at room temperature, and washed with PBS+ (to eliminate unincorporated crystal violet), and the cell-bound stain was solubilized with 100 μl solubilization buffer (a 50:50 mixture of 0.1 M of NaH2PO4, pH 4.5, and 50% ethanol) for 5 min at room temperature with shaking. Absorbance (representing the number of adhered cells) was measured at 540–570 nm. The BSA-coated wells were included for each treatment as negative controls. The experiment was repeated four times.
Integrin Assays
Specific integrin expression by HTR-8/SVneo cells was determined using commercially available integrin assay kits (Chemicon) similar to the ECM cell adhesion assay kits described above, according to the manufacturer's instructions. These kits comprised 96-well plates coated with monoclonal antibodies against various integrin subunits and/or dimers. The protocol was exactly as per the ECM cell adhesion assays, except that cells were incubated for 2 h at 37°C to facilitate adhesion to ITGA5B1, ITGAvB3, ITGAvB5, ITGA1, and ITGA5 integrin-coated wells (four to six wells per integrin per treatment). The experiment was repeated four times.
Statistical Analyses
Data are expressed as mean ± SEM for each treatment compared with control. Statistical analysis was performed on raw data by a Student t-test and/or using one-way ANOVA followed by Tukey posthoc test (P < 0.05 taken as significant) after testing for normal distribution using PRISM version 3.00 for Windows (GraphPad).
RESULTS
Expression of IL11RA, IL6ST, and IL11 in EVT Cells
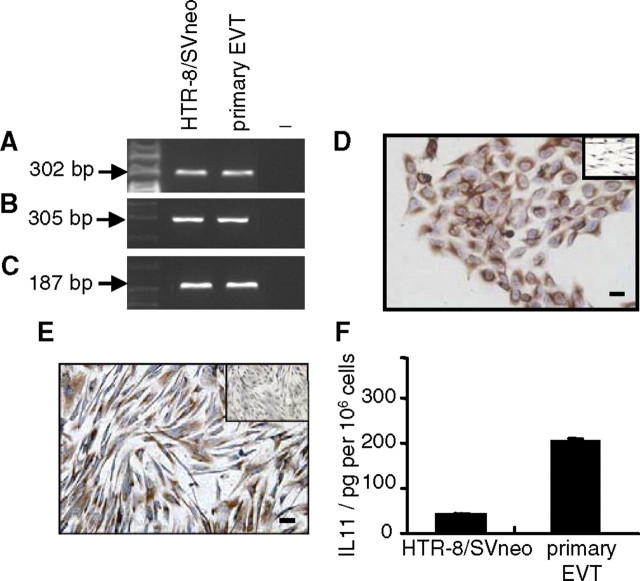
IL11RA (Fig. 1A) and IL6ST (Fig. 1B) mRNAs were expressed by HTR-8/SVneo cells and primary EVTs. 18S mRNA expression was used as the loading control (Fig. 1C). Immunoreactive IL11RA also was detected in both HTR-8/SVneo cells (Fig. 1D) and primary EVTs (Fig. 1E). Interleukin 11 was secreted by primary EVTs at greater levels (approximately 10-fold greater) than that secreted by HTR-8/SVneo cells per 106 cells (Fig. 1F).
FIG. 1.
Characterization of human EVT cells. Reverse transcriptase-PCR analysis of (A) IL11RA, (B) IL6ST mRNA, and (C) 18S mRNA expression (loading control). −, Negative controls. Immunoreactive IL11RA protein in (D) HTR-8/SVneo and (E) primary EVTs (insets, negative controls). F) Interleukin 11 secretion by EVT cells (mean ± SEM). Data are shown for representatives of four independent experiments in each case. Bars = 50 μm.
IL11 Inhibits EVT Invasion via STAT3 Activation and not MAPK
Treatment of HTR-8/SVneo cells with IL11 for 48 h inhibited cell invasion in a dose-dependent manner (Fig. 2A) compared with controls, with maximum inhibition (approximately 60%) observed at the highest dose of 100 ng/ml (n = 3 experiments; P < 0.001). Treatment of primary EVTs with IL11 (100 ng/ml) for 48 h also inhibited cell invasion by approximately 40% compared with controls (Fig. 2B). TGFB1 (10 ng/ml) was used as a positive control (Fig. 2), and it inhibited invasion of HTR-8/SVneo cells by approximately 40% and primary EVTs by approximately 90%. Incubation of HTR-8/SVneo cells with STAT3i but not MAPKi, abolished IL11-mediated inhibition of EVT invasion, restoring the number of invaded HTR-8/SVneo cells to control levels (n = 3 experiments; P > 0.05 versus controls). Incubation of HTR-8/SVneo cells with STAT3i (20 μM) or MAPKi (30 μM) alone, inhibitor diluent PBS− (STAT3i), or 0.001% dimethyl sulfoxide (MAPKi) had no significant effect on basal cell invasion (data not shown).
FIG. 2.
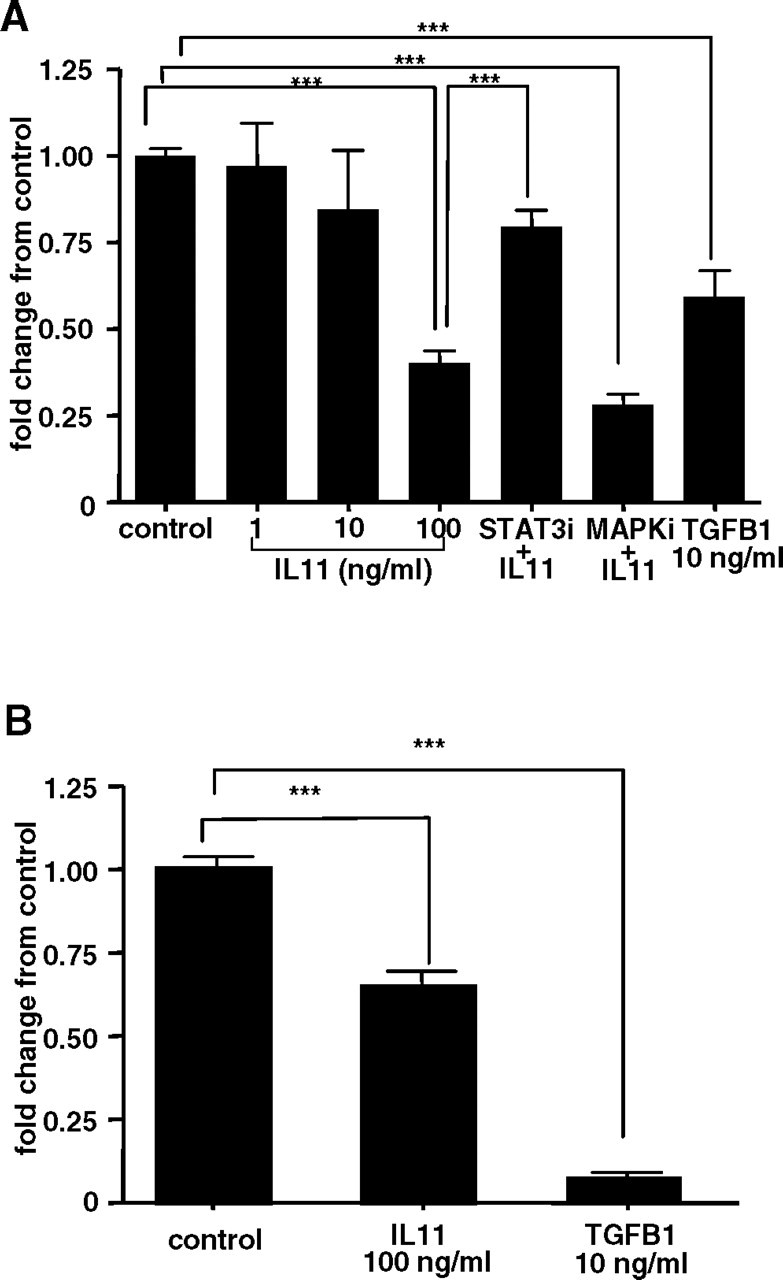
The effect of IL11 on invasion as assessed by Matrigel invasion assays of (A) HTR-8/SVneo and (B) primary EVTs during 48 h. Interleukin 11 (100 ng/ml) significantly inhibited invasion of HTR-8/SVneo cells during 48 h compared with vehicle-treated controls (***P < 0.001). Positive control, TGFB1 (10 ng/ml) versus control (***P < 0.001). Incubation of HTR-8/SVneo cells with STAT3i but not MAPKi prevented IL11-mediated inhibition of EVT invasion (P > 0.05 versus controls, **P < 0.001 versus IL11 [100 ng/ml]). Data are combined from four independent experiments (quadruplicate wells per treatment) and are expressed as fold-change in cell invasion (±SEM) compared with controls.
IL11 Phosphorylates STAT3 in EVT Cells
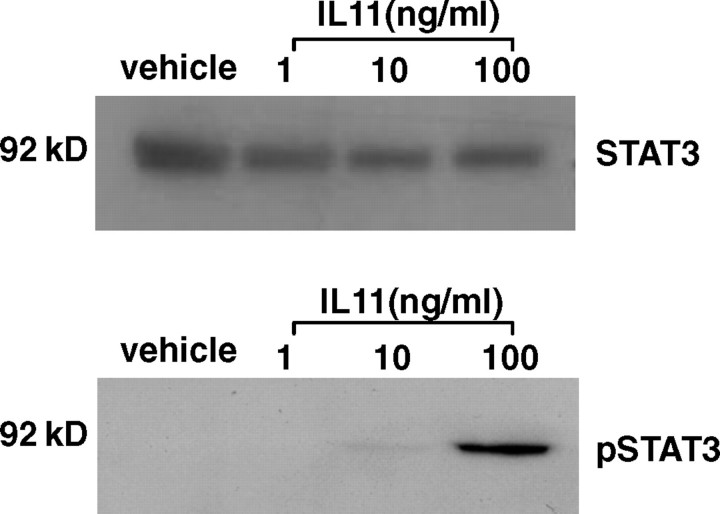
Interleukin 11 stimulated pSTAT3 in HTR-8/SVneo cells but had no effect on STAT3 abundance (n = 3 experiments; Fig. 3). Phosphorylated STAT3 was detectable at low levels following vehicle treatment but was upregulated following IL11 treatment (10 min), particularly at the 10- and 100-ng/ml doses (Fig. 3). Interleukin 11-mediated stimulation of pSTAT3 in primary EVTs has been demonstrated previously [17].
FIG. 3.
Interleukin 11-induced STAT3 phosphorylation in HTR-8/SVneo cells. Cells were cultured with vehicle or IL11 (1–100 ng/ml) for 10 min. Cell lysate proteins (15 μg for STAT3, 25 μg for p-STAT3) were electrophoresed by SDS-PAGE and immunoblotted with anti-phospho (Tyr705) STAT3 (bottom) or anti-STAT3 (top), followed by horseradish peroxidase-conjugated rabbit antiserum and visualized by chemiluminescence. Data shown are representative of four independent experiments. Kilodalton (kDa) labeled as kD.
IL11 Has No Effect on MMP2 and MMP9 Secretion/Activity
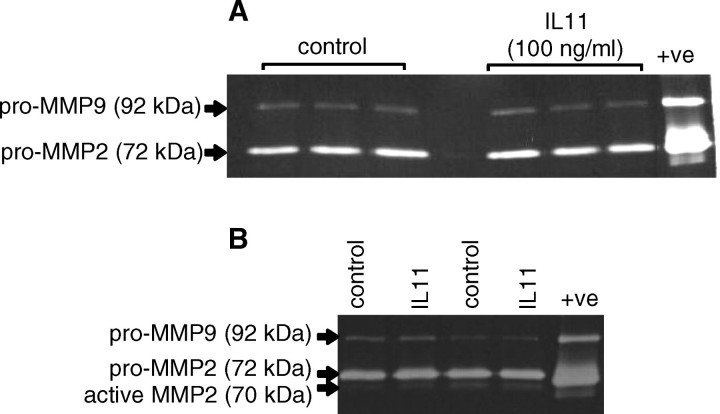
Gelatin zymography revealed a prominent band of 72 kDa (pro-MMP2) and a less intense band of 92 kDa (pro-MMP9) in the conditioned media from both vehicle and IL11-treated EVT cells during 48 h (Fig. 4A: HTR-8/SVneo; Fig. 4B: primary EVTs). In conditioned media from primary EVTs there was an additional band at approximately 70 kDa (active MMP2; Fig. 4B). There was no effect of IL11 (100 ng/ml) on secretion of latent/active forms of MMP2 or MMP9 compared with controls (Fig. 4). The MMP identity of the bands was proven because they were abolished when gels were incubated in the presence of EDTA (data not shown).
FIG. 4.
Gelatin zymography of conditioned media from EVT cells treated with IL11 (100 ng/ml) for 48 h. Interleukin 11 (100 ng/ml) had no significant effect on either pro or active forms of MMP2 or pro-MMP9 production by (A) HTR-8/SVNeo and (B) primary EVTs compared with controls. Data are shown from two independent experiments (primary EVTs) or three independent experiments (HTR8/SVneo). +ve represents positive control.
IL11 Has No Effect on TIMP Production by EVT
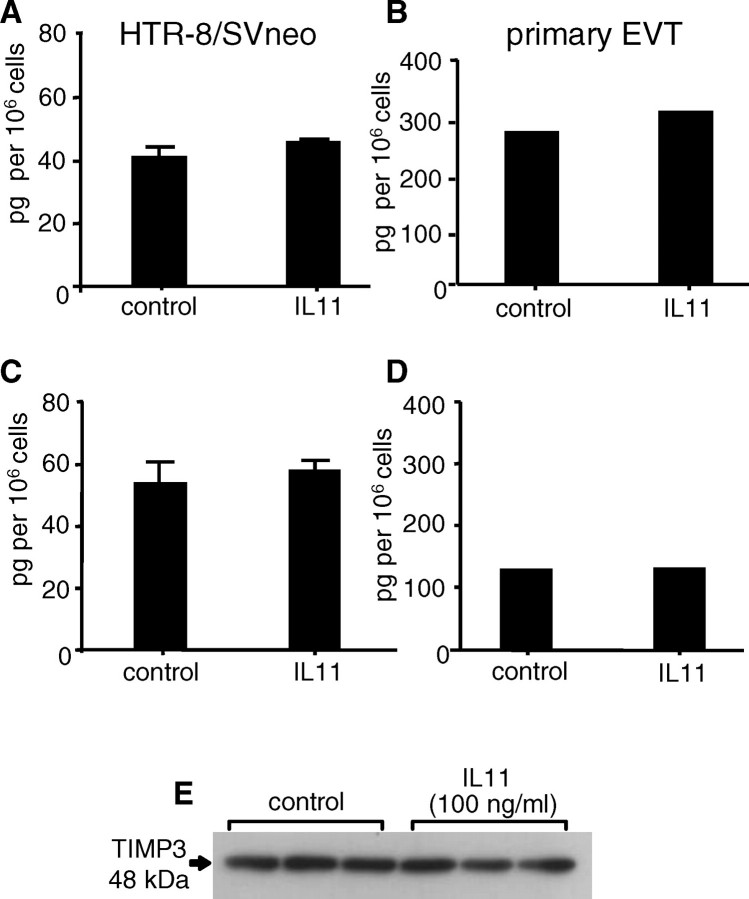
Treatment of HTR-8/SVneo cells (Fig. 5, A, C, and E) and primary EVTs (Fig. 5, B and D) with IL11 (100 ng/ml) for 48 h had no effect on secreted TIMP1 (Fig. 5, A and B), TIMP2 (Fig. 5, C and D), or cell-bound TIMP3 (Fig. 5E) compared with controls.
FIG. 5.
The effect of IL11 on TIMP1, TIMP2, and TIMP3 protein production by HTR-8/SVneo (A, C, E) and primary EVTs (B, D). Interleukin 11 (100 ng/ml) had no significant effect on soluble TIMP1 (A, B), TIMP2 (C, D), and cell-bound TIMP3 production (E) during 48 h compared with controls. Data are represented as mean concentrations of TIMPs 1 and 2 (±SEM) from two (primary EVTs) or three (HTR-8/SVneo) independent experiments. Data for TIMP3 are representative of three independent experiments.
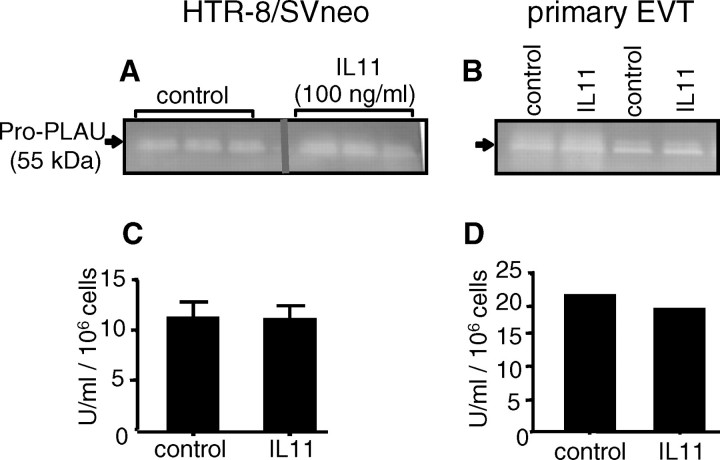
IL11 Has No Effect on PLAU Production/Activity
Plasminogen-casein zymography revealed the presence of two prominent bands in conditioned media from both vehicle and IL11-treated (100 ng/ml) EVT cells during 48 h (Fig. 6A: HTR-8/SVneo; Fig. 6B: primary EVTs). These bands were positively identified as PLAU because they were abolished in the absence of plasminogen in the gels (data not shown). Interleukin 11 had no effect on PLAU activity in these conditioned media compared with controls, as assessed by both plasminogen-casein zymography (Fig. 6, A and B) and spectrometric PLAU activity assay (Fig. 6, C and D).
FIG. 6.
The effect of IL11 on PLAU activity in conditioned media from HTR-8/SVneo (A, C) and primary EVTs (B, D). A, B) Plasminogen-casein zymography. C, D) PLAU activity assay. Interleukin 11 (100 ng/ml) had no significant effect on PLAU activity compared to controls during 48 h. Data are shown from two (primary EVTs) or three (HTR-8/SVneo) independent experiments (A, B) and are represented as mean PLAU activity (±SEM) from two (primary EVTs) or three (HTR-8/SVneo) independent experiments (C, D).
IL11 Has No Effect on SERPINE1, SERPINE2, and PLAUR Expression by EVTs
Treatment of HTR-8/SVneo cells (Fig. 7, A and D) and primary EVTs (Fig. 7, B, C, and E) with IL11 (100 ng/ml) for 48 h had no effect on secreted SERPINE1 (Fig. 7, A and B), SERPINE2 (Fig. 7C), or soluble PLAUR protein levels (Fig. 7, D and E) compared with controls. SERPINE2 was undetectable in conditioned media from HTR-8/SVneo cells (data not shown).
FIG. 7.
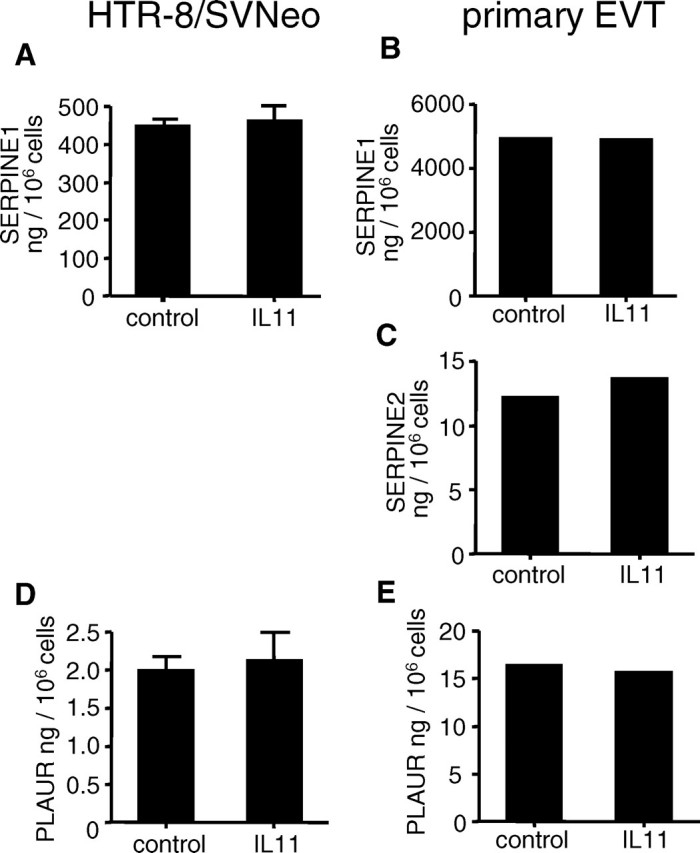
The effect of IL11 on SERPINE1, SERPINE2, and PLAUR protein production by HTR-8/SVneo (A, D) and primary EVTs (B, C, E). Interleukin 11 (100 ng/ml) had no significant effect on SERPINE1 (A, B), SERPINE2 (C), and PLAUR (D, E) secretion by EVT cells during 48 h compared with controls. Data are represented as mean concentration (±SEM) from two (primary EVTs) or three (HTR-8/SVneo) independent experiments.
IL11 Has No Effect on EVT-ECM Interactions
Treatment of HTR-8/SVneo cells with IL11 (100 ng/ml) for 48 h had no effect on cell binding to FN (Fig. 8A), VN (Fig. 8B), LN (Fig. 8C), COL1 (Fig. 8D), and COL4 (Fig. 8E) compared with controls.
FIG. 8.
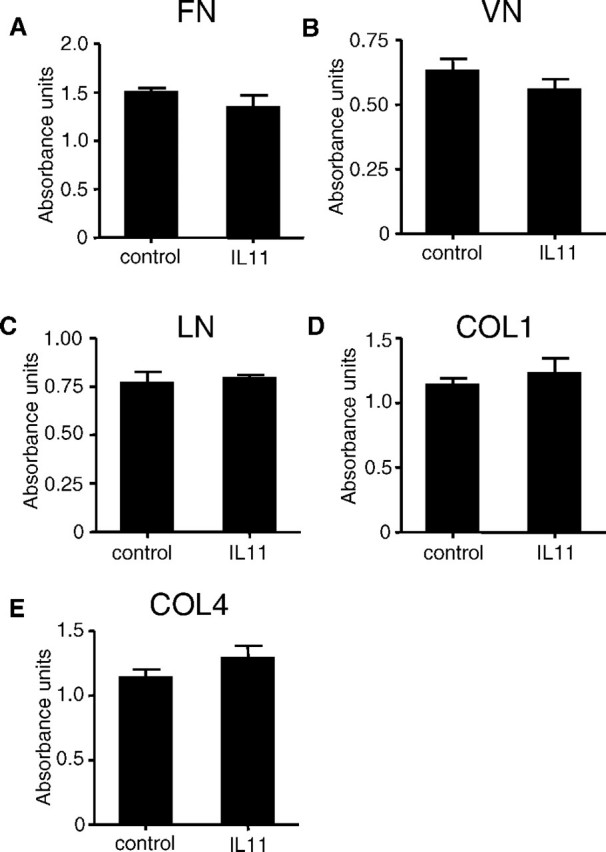
The effect of IL11 on cellular adhesion of HTR-8/SVneo cells to ECM components as assessed by colorimetric adhesion assays. Treatment of HTR-8/SVneo cells with IL11 (100 ng/ml) for 48 h had no effect on EVT binding to (A) FN, (B) VN, (C) LN, (D) COL1, and (E) COL4 compared with controls. Data are combined from four independent experiments.
IL11 Has No Effect on EVT Expression of Integrins
Treatment of HTR-8/SVneo cells with IL11 (100 ng/ml) for 48 h had no effect on cellular expression of integrin dimers ITGA5B1 (Fig. 9A), ITGAvB3 (Fig. 9B), and ITGAvB5 (Fig. 9C), or monomers ITGA1 (Fig. 9D) and ITGA5 (Fig. 9E) compared with controls.
FIG. 9.
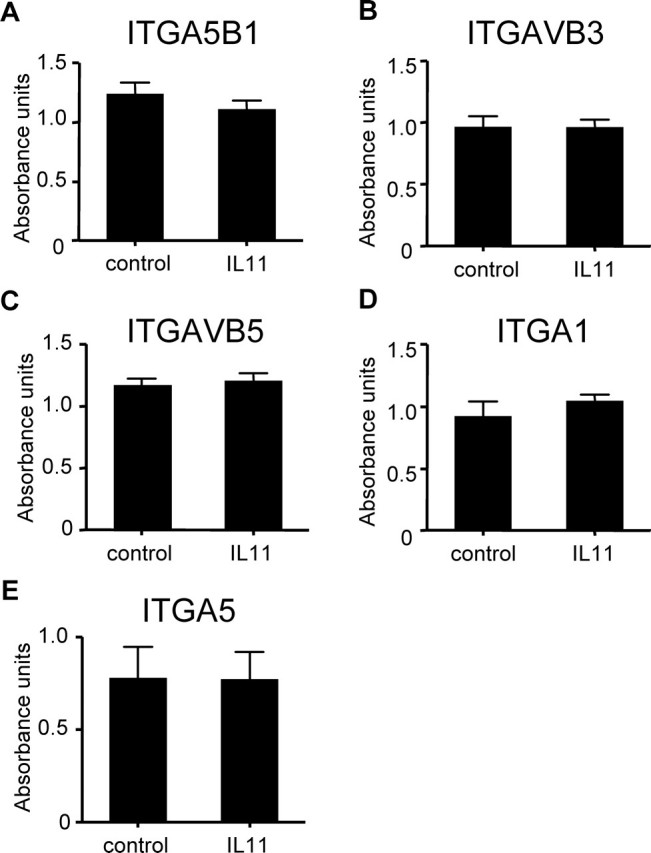
The effect of IL11 on the expression of selected integrins by HTR-8/SVneo cells as assessed by colorimetric integrin-binding assays. Treatment of HTR-8/SVneo cells with IL11 (100 ng/ml) for 48 h had no effect on EVT expression of (A) ITGA5B1, (B) ITGAVB3, (C) ITGAVB5, (D) ITGA1, and (E) ITGA5 compared with controls. Data are combined from four independent experiments.
DISCUSSION
This study has demonstrated for the first time that IL11 inhibits human EVT invasion, suggesting a likely role for IL11 in the decidual restraint of EVT invasion during early placental development. Such restraint is important in regulating the depth of EVT invasion to ensure that the needs of the fetus are met without compromising the survival/health of the mother. This finding was consistent using the immortalized EVT cell line HTR-8/SVneo and primary EVT cells. Importantly, the data presented here also indicate that IL11-mediated inhibition of EVT invasion is likely to be via STAT3 and not MAPK signaling pathways.
Interleukin 11 and IL11RA mRNA and protein are expressed in human endometrium throughout the menstrual cycle [32, 35, 36]. Interleukin 11 increases in luminal/glandular epithelium in the mid-late secretory phase (the receptive phase) but is maximally expressed in the decidua and chorionic villi during early pregnancy (first trimester) [16, 37]. Interestingly, abnormalities in decidual and villous trophoblast IL11 have been identified in anembryonic pregnancies that lead to early miscarriage [37], whereas IL11 is reduced in the pregnant sera of women with early spontaneous abortions [38]. These reports suggest a critical role for IL11 signaling during early pregnancy. In this regard, a role for IL11 in progressing human endometrial stromal cell decidualization in vitro has been demonstrated [15].
IL11RA has also been identified recently in EVTs of the cell column and invasive interstitial/endovascular EVTs of the nonhuman primate [16] and humans [17] in vivo, whereas in vitro, a novel functional role for IL11 in stimulating chemotactic migration of human EVT cells has also been demonstrated [17]. From the current work it appears that IL11 exerts independent effects on parameters of EVT function, stimulating EVT migration but inhibiting EVT invasion. Likewise, importantly, conditioned media from decidualized stromal cells, of which IL11 is an important product [15], similarly stimulate chemotactic migration of human EVT cells [39, 40] but inhibit their invasion in vitro [2]. Unlike EVT migration, EVT invasion of the maternal decidua and spiral arterioles involves considerable complexity that includes regulated proteolytic degradation of decidual/endothelial ECM (in the direction of invasion) and altered adhesion to ECM, followed by active cell movement/migration through the degraded matrix [18].
First-trimester human trophoblast cells share intrinsic invasive properties of malignant/tumor cells in vitro [41], suggesting that if dysregulated, invasion would proceed excessively into the myometrium, as observed in pathological conditions, such as placental accreta. In contrast with tumor invasion, EVT invasion during normal pregnancy is precisely regulated both spatially and temporally [1]. Such precision requires a balanced interplay between factors that promote and restrain EVT invasion. Existing evidence suggests that the control of EVT invasion is provided by the uterine microenvironment, primarily products of the decidua [2], such as the TIMPs [2], insulinlike growth factor-binding protein 1 (IGFBP1) [42], and TGFB1 [2], all of which inhibit EVT invasion in vitro [2, 43, 44]. That the decidua acts as a barrier to excessive EVT invasion is evidenced by a number of studies. In mice, transfer of blastocysts to ectopic nondecidualized soft tissues results in more extensive trophoblast invasion than observed within the uterus [45–48]. This phenomenon is also highlighted in the absence of adequately formed deciduas, such as in the Il11ra null mouse [14]. Likewise, human ectopic pregnancies also serve as a naturally occurring model of dysregulated EVT invasion [49]. Consequently, factors that promote EVT invasion are primarily thought to be trophoblast derived. Consistent with this paradigm, invading EVTs secrete MMPs [50] and also IGF2, which promote their invasion [51, 52]. From the results of this study, it is clear that the amount of IL11 produced by EVTs in vitro is negligible in comparison with that produced by the decidua (which is in the nanogram range) [15]. Therefore, it is likely that IL11 produced predominantly by the decidua [15, 16, 53] may be another such factor involved in the negative regulation of human EVT invasion.
In most cell types, IL11 mediates its effects through the JAK/STAT signaling pathway, primarily via STAT3 [54]. However, IL11 signaling may also involve the Ras-MAPK pathway [3], with cross-talk between the JAK/STAT and MAPK signaling pathways also occurring [3, 4]. This study demonstrates that IL11 exerts its effects on EVT invasion via STAT3 and not MAPK. STAT3 is of special interest in the context of malignant cellular properties because aberrant STAT3 activity is directly linked with oncogenesis [55]. Constitutive STAT3 activity is also detected in invasive human first-trimester trophoblast cells and choriocarcinoma cell lines, whereas it is absent in term placenta [56]. In contrast with IL11, LIF positively regulates invasion of JEG-3 choriocarcinoma cells and primary trophoblast cells via STAT3 [57]. This role for STAT3 in promoting invasion was demonstrated using RNAi [58]. Nevertheless, in addition to its proinvasive role, STAT3 may also upregulate factors that inhibit cellular invasion. In this regard, STAT3 upregulates TIMP1 during tumor invasion/metastasis [59]. Furthermore, during pregnancy in mice, IL11 via STAT3 upregulates the decidual protease inhibitor alpha-2-macroglobulin, which is crucial in restraining trophoblast invasion in this species [60]. In the same manner, IL11-mediated STAT3 may also upregulate trophoblast-derived factors that inhibit their invasiveness.
TGFB1 negatively regulates EVT invasion via multiple mechanisms that include downregulation of PLAU and upregulation of TIMP1, TIMP2 [2, 61, 62], SERPINE1 [63], integrins [64], and HGF [62]. Alternatively, IGFBP1 negatively regulates EVT invasion by stimulating TIMP1 [65]. In contrast, IL11 had no effect on the expression/activity of specific members of protease systems (i.e., MMP/TIMP system and PLAU/PLAUR/SERPINE systems) and integrins most commonly associated with EVT invasion. This is not entirely surprising, because several factors apart from those investigated in this study have been implicated in EVT invasion. Such examples include other MMPs, various other families of cell adhesion molecules, cytokines, and chemokines [66].
In conclusion, this is the first study to demonstrate a likely role for IL11 acting via STAT3 (but not MAPK) in the decidual restraint of human EVT invasion during normal pregnancy. Importantly, this is also the first study to demonstrate that a protein may inhibit (rather than stimulate) trophoblast invasion via STAT3. Trophoblast invasion can be considered a fine balance between the demands of the fetus to invade the mother and those of the mother to survive this invasion [1]. Failure of EVT invasion and arterial remodeling lead to spontaneous abortion, whereas defects in this process lead to preeclampsia and intrauterine growth restriction [67]. Conversely, excessive EVT invasion leads to placental accreta or percreta [67]. This study suggests that IL11 via its effects on EVT invasion may play a regulatory role in the establishment of a successful pregnancy.
Acknowledgments
We thank Dr. Lorraine Robb and the Walter and Eliza Hall Institute of Medical Research for provision of the IL11RA antibody; Ms. Claire Walker for excellent technical assistance; Ms. Lynsey Marshall for assistance with Western blot data; Ms. Sue Panckridge for assistance with preparation of the figures; and the patients who generously donated the tissues.
Footnotes
1Supported by a Monash Graduate Scholarship to P.P. and by National Health and Medical Research Council grants 388916 to E.D. and 388901 and 494802 to L.A.S.
REFERENCES
- Anin SA, Vince G, Quenby S. Trophoblast invasion. Hum Fertil (Camb) 2004; 7: 169 174. [DOI] [PubMed] [Google Scholar]
- Graham CH, Lala PK. Mechanism of control of trophoblast invasion in situ. J Cell Physiol 1991; 148: 228 234. [DOI] [PubMed] [Google Scholar]
- Wang XY, Fuhrer DK, Marshall MS, Yang YC. Interleukin-11 induces complex formation of Grb2, Fyn, and JAK2 in 3T3L1 cells. J Biol Chem 1995; 270: 27999 28002. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JS, Busch S, Wengenmayer T, Foerster K, de la Motte T, Poehlmann TG, Markert UR. Signal transduction in trophoblast invasion. Chem Immunol Allergy 2005; 88: 181 199. [DOI] [PubMed] [Google Scholar]
- Du X, Williams DA. Interleukin-11: review of molecular, cell biology, and clinical use. Blood 1997; 89: 3897 3908. [PubMed] [Google Scholar]
- Arihiro K, Oda H, Kaneko M, Inai K. Cytokines facilitate chemotactic motility of breast carcinoma cells. Breast Cancer 2000; 7: 221 230. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Lacroix M, Lespagnard L, Larsimont D, Paesmans M, Body JJ. Interleukins-6 and −11 expression in primary breast cancer and subsequent development of bone metastases. Cancer Lett 2001; 169: 87 95. [DOI] [PubMed] [Google Scholar]
- Hanavadi S, Martin TA, Watkins G, Mansel RE, Jiang WG. Expression of interleukin 11 and its receptor and their prognostic value in human breast cancer. Ann Surg Oncol 2006; 13: 802 808. [DOI] [PubMed] [Google Scholar]
- Yamazumi K, Nakayama T, Kusaba T, Wen CY, Yoshizaki A, Yakata Y, Nagayasu T, Sekine I. Expression of interleukin-11 and interleukin-11 receptor alpha in human colorectal adenocarcinoma; immunohistochemical analyses and correlation with clinicopathological factors. World J Gastroenterol 2006; 12: 317 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki A, Nakayama T, Yamazumi K, Yakata Y, Taba M, Sekine I. Expression of interleukin (IL)-11 and IL-11 receptor in human colorectal adenocarcinoma: IL-11 up-regulation of the invasive and proliferative activity of human colorectal carcinoma cells. Int J Oncol 2006; 29: 869 876. [PubMed] [Google Scholar]
- Nakayama T, Yoshizaki A, Izumida S, Suehiro T, Miura S, Uemura T, Yakata Y, Shichijo K, Yamashita S, Sekin I. Expression of interleukin-11 (IL-11) and IL-11 receptor alpha in human gastric carcinoma and IL-11 upregulates the invasive activity of human gastric carcinoma cells. Int J Oncol 2007; 30: 825 833. [PubMed] [Google Scholar]
- Ernst M, Najdovska M, Grail D, Lundgren-May T, Buchert M, Tye H, Matthews VB, Armes J, Bhathal PS, Hughes NR, Marcusson EG, Karras JG, et al. STAT3 and STAT1 mediate IL-11-dependent and inflammation-associated gastric tumorigenesis in gp130 receptor mutant mice. J Clin Invest 2008; 118: 1727 1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilinski P, Roopenian D, Gossler A. Maternal IL-11Ralpha function is required for normal decidua and fetoplacental development in mice. Genes Dev 1998; 12: 2234 2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robb L, Li R, Hartley L, Nandurkar HH, Koentgen F, Begley CG. Infertility in female mice lacking the receptor for interleukin 11 is due to a defective uterine response to implantation. Nat Med 1998; 4: 303 308. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E, Robb L, Salamonsen LA. Interleukin 11 advances progesterone-induced decidualization of human endometrial stromal cells. Mol Hum Reprod 2002; 8: 636 643. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E, Robb L, Liu YX, Enders AC, Martin H, Stoikos C, Wallace E, Salamonsen LA. IL-11 and IL-11Ralpha immunolocalisation at primate implantation sites supports a role for IL-11 in placentation and fetal development. Reprod Biol Endocrinol 2003; 1: 34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paiva P, Salamonsen LA, Manuelpillai U, Walker C, Tapia A, Wallace EM, Dimitriadis E. Interleukin-11 promotes migration, but not proliferation, of human trophoblast cells, implying a role in placentation. Endocrinology 2007; 148: 5566 5572. [DOI] [PubMed] [Google Scholar]
- Mareel M, Leroy A. Clinical, cellular, and molecular aspects of cancer invasion. Physiol Rev 2003; 83: 337 376. [DOI] [PubMed] [Google Scholar]
- Staun-Ram E, Shalev E. Human trophoblast function during the implantation process. Reprod Biol Endocrinol 2005; 3: 56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staun-Ram E, Goldman S, Gabarin D, Shalev E. Expression and importance of matrix metalloproteinase 2 and 9 (MMP-2 and -9) in human trophoblast invasion. Reprod Biol Endocrinol 2004; 2: 59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Meisser A, Bischof P. Metalloproteinases and human placental invasiveness. Placenta 2006; 27: 783 793. [DOI] [PubMed] [Google Scholar]
- Hofmann GE, Glatstein I, Schatz F, Heller D, Deligdisch L. Immunohistochemical localization of urokinase-type plasminogen activator and the plasminogen activator inhibitors 1 and 2 in early human implantation sites. Am J Obstet Gynecol 1994; 170: 671 676. [DOI] [PubMed] [Google Scholar]
- Multhaupt HA, Mazar A, Cines DB, Warhol MJ, McCrae KR. Expression of urokinase receptors by human trophoblast. A histochemical and ultrastructural analysis. Lab Invest 1994; 71: 392 400. [PubMed] [Google Scholar]
- Floridon C, Nielsen O, Holund B, Sunde L, Westergaard JG, Thomsen SG, Teisner B. Localization and significance of urokinase plasminogen activator and its receptor in placental tissue from intrauterine, ectopic and molar pregnancies. Placenta 1999; 20: 711 721. [DOI] [PubMed] [Google Scholar]
- Feinberg RF, Kao LC, Haimowitz JE, Queenan JT, Jr, Wun TC, Strauss JF, III, Kliman HJ. Plasminogen activator inhibitor types 1 and 2 in human trophoblasts. PAI-1 is an immunocytochemical marker of invading trophoblasts. Lab Invest 1989; 61: 20 26. [PubMed] [Google Scholar]
- Damsky CH, Librach C, Lim KH, Fitzgerald ML, McMaster MT, Janatpour M, Zhou Y, Logan SK, Fisher SJ. Integrin switching regulates normal trophoblast invasion. Development 1994; 120: 3657 3666. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, Damsky CH. Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest 1997; 99: 2139 2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Lysiak JJ, Graham CH, Hearn S, Han VK, Lala PK. Characteristics of trophoblast cells migrating from first trimester chorionic villus explants and propagated in culture. Placenta 1995; 16: 413 433. [DOI] [PubMed] [Google Scholar]
- Tapia A, Salamonsen LA, Manuelpillai U, Dimitriadis E. Leukemia inhibitory factor promotes human first trimester extravillous trophoblast adhesion to extracellular matrix and secretion of tissue inhibitor of metalloproteinases-1 and −2. Hum Reprod 2008; 23: 1724 1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham CH, Hawley TS, Hawley RG, MacDougall JR, Kerbel RS, Khoo N, Lala PK. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res 1993; 206: 204 211. [DOI] [PubMed] [Google Scholar]
- Kilburn BA, Wang J, Duniec-Dmuchowski ZM, Leach RE, Romero R, Armant DR. Extracellular matrix composition and hypoxia regulate the expression of HLA-G and integrins in a human trophoblast cell line. Biol Reprod 2000; 62: 739 747. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E, Salamonsen LA, Robb L. Expression of interleukin-11 during the human menstrual cycle: coincidence with stromal cell decidualization and relationship to leukaemia inhibitory factor and prolactin. Mol Hum Reprod 2000; 6: 907 914. [DOI] [PubMed] [Google Scholar]
- Catalano RD, Johnson MH, Campbell EA, Charnock-Jones DS, Smith SK, Sharkey AM. Inhibition of Stat3 activation in the endometrium prevents implantation: a nonsteroidal approach to contraception. Proc Natl Acad Sci U S A 2005; 102: 8585 8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash GE, Otun HA, Innes BA, Bulmer JN, Searle RF, Robson SC. Low oxygen concentrations inhibit trophoblast cell invasion from early gestation placental explants via alterations in levels of the urokinase plasminogen activator system. Biol Reprod 2006; 74: 403 409. [DOI] [PubMed] [Google Scholar]
- Cork BA, Li TC, Warren MA, Laird SM. Interleukin-11 (IL-11) in human endometrium: expression throughout the menstrual cycle and the effects of cytokines on endometrial IL-11 production in vitro. J Reprod Immunol 2001; 50: 3 17. [DOI] [PubMed] [Google Scholar]
- Cork BA, Tuckerman EM, Li TC, Laird SM. Expression of interleukin (IL)-11 receptor by the human endometrium in vivo and effects of IL-11, IL-6 and LIF on the production of MMP and cytokines by human endometrial cells in vitro. Mol Hum Reprod 2002; 8: 841 848. [DOI] [PubMed] [Google Scholar]
- Chen HF, Lin CY, Chao KH, Wu MY, Yang YS, Ho HN. Defective production of interleukin-11 by decidua and chorionic villi in human anembryonic pregnancy. J Clin Endocrinol Metab 2002; 87: 2320 2328. [DOI] [PubMed] [Google Scholar]
- Koumantaki Y, Matalliotakis I, Sifakis S, Kyriakou D, Neonaki M, Goymenou A, Koumantakis E. Detection of interleukin-6, interleukin-8, and interleukin-11 in plasma from women with spontaneous abortion. Eur J Obstet Gynecol Reprod Biol 2001; 98: 66 71. [DOI] [PubMed] [Google Scholar]
- Hanna J, Goldman-Wohl D, Hamani Y, Avraham I, Greenfield C, Natanson-Yaron S, Prus D, Cohen-Daniel L, Arnon TI, Manaster I, Gazit R, Yutkin V, et al. Decidual NK cells regulate key developmental processes at the human fetal-maternal interface. Nat Med 2006; 12: 1065 1074. [DOI] [PubMed] [Google Scholar]
- Wright JK, Dunk CE, Perkins JE, Winterhager E, Kingdom JC, Lye SJ. EGF modulates trophoblast migration through regulation of Connexin 40. Placenta 2006; 27 (suppl A): S114 S121. [DOI] [PubMed] [Google Scholar]
- Yagel S, Parhar RS, Jeffrey JJ, Lala PK. Normal nonmetastatic human trophoblast cells share in vitro invasive properties of malignant cells. J Cell Physiol 1988; 136: 455 462. [DOI] [PubMed] [Google Scholar]
- Han VK, Bassett N, Walton J, Challis JR. The expression of insulin-like growth factor (IGF) and IGF-binding protein (IGFBP) genes in the human placenta and membranes: evidence for IGF-IGFBP interactions at the feto-maternal interface. J Clin Endocrinol Metab 1996; 81: 2680 2693. [DOI] [PubMed] [Google Scholar]
- Fisher SJ, Damsky CH. Human cytotrophoblast invasion. Semin Cell Biol 1993; 4: 183 188. [DOI] [PubMed] [Google Scholar]
- Irwin JC, Giudice LC. Insulin-like growth factor binding protein-1 binds to placental cytotrophoblast alpha5beta1 integrin and inhibits cytotrophoblast invasion into decidualized endometrial stromal cultures. Growth Horm IGF Res 1998; 8: 21 31. [DOI] [PubMed] [Google Scholar]
- Kirby DR. Development of mouse eggs beneath the kidney capsule. Nature 1960; 187: 707 708. [DOI] [PubMed] [Google Scholar]
- Bevilacqua EM, Abrahamsohn PA. Growth of mouse embryos implanted in the subcutaneous tissue of recipient mice. J Exp Zool 1991; 257: 386 400. [DOI] [PubMed] [Google Scholar]
- Bevilacqua E, Abrahamsohn PA. Invasiveness of mouse trophoblastic cells in connective tissue. Acta Anat (Basel) 1994; 150: 246 252. [DOI] [PubMed] [Google Scholar]
- Aplin J. Maternal influences on placental development. Semin Cell Dev Biol 2000; 11: 115 125. [DOI] [PubMed] [Google Scholar]
- von Rango U, Alfer J, Kertschanska S, Kemp B, Muller-Newen G, Heinrich PC, Beier HM, Classen-Linke I. Interleukin-11 expression: its significance in eutopic and ectopic human implantation. Mol Hum Reprod 2004; 10: 783 792. [DOI] [PubMed] [Google Scholar]
- Aplin JD. Implantation, trophoblast differentiation and haemochorial placentation: mechanistic evidence in vivo and in vitro. J Cell Sci 1991; 99 (pt 4): 681 692. [DOI] [PubMed] [Google Scholar]
- Hamilton GS, Lysiak JJ, Han VK, Lala PK. Autocrine-paracrine regulation of human trophoblast invasiveness by insulin-like growth factor (IGF)-II and IGF-binding protein (IGFBP)-1. Exp Cell Res 1998; 244: 147 156. [DOI] [PubMed] [Google Scholar]
- McKinnon T, Chakraborty C, Gleeson LM, Chidiac P, Lala PK. Stimulation of human extravillous trophoblast migration by IGF-II is mediated by IGF type 2 receptor involving inhibitory G protein(s) and phosphorylation of MAPK. J Clin Endocrinol Metab 2001; 86: 3665 3674. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E, Stoikos C, Baca M, Fairlie WD, McCoubrie JE, Salamonsen LA. Relaxin and prostaglandin E(2) regulate interleukin 11 during human endometrial stromal cell decidualization. J Clin Endocrinol Metab 2005; 90: 3458 3465. [DOI] [PubMed] [Google Scholar]
- Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J 2003; 374: 1 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene 2000; 19: 2474 2488. [DOI] [PubMed] [Google Scholar]
- Corvinus FM, Fitzgerald JS, Friedrich K, Markert UR. Evidence for a correlation between trophoblast invasiveness and STAT3 activity. Am J Reprod Immunol 2003; 50: 316 321. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JS, Tsareva SA, Poehlmann TG, Berod L, Meissner A, Corvinus FM, Wiederanders B, Pfitzner E, Markert UR, Friedrich K. Leukemia inhibitory factor triggers activation of signal transducer and activator of transcription 3, proliferation, invasiveness, and altered protease expression in choriocarcinoma cells. Int J Biochem Cell Biol 2005; 37: 2284 2296. [DOI] [PubMed] [Google Scholar]
- Poehlmann TG, Fitzgerald JS, Meissner A, Wengenmayer T, Schleussner E, Friedrich K, Markert UR. Trophoblast invasion: tuning through LIF, signalling via Stat3. Placenta 2005; 26 (suppl A): S37 S41. [DOI] [PubMed] [Google Scholar]
- Dien J, Amin HM, Chiu N, Wong W, Frantz C, Chiu B, Mackey JR, Lai R. Signal transducers and activators of transcription-3 up-regulates tissue inhibitor of metalloproteinase-1 expression and decreases invasiveness of breast cancer. Am J Pathol 2006; 169: 633 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L, Devi S, Bowen-Shauver J, Ferguson-Gottschall S, Robb L, Gibori G. The role of interleukin-11 in pregnancy involves up-regulation of alpha2-macroglobulin gene through janus kinase 2-signal transducer and activator of transcription 3 pathway in the decidua. Mol Endocrinol 2006; 20: 3240 3250. [DOI] [PubMed] [Google Scholar]
- Karmakar S, Das C. Regulation of trophoblast invasion by IL-1beta and TGF-beta1. Am J Reprod Immunol 2002; 48: 210 219. [DOI] [PubMed] [Google Scholar]
- Tse WK, Whitley GS, Cartwright JE. Transforming growth factor-beta1 regulates hepatocyte growth factor-induced trophoblast motility and invasion. Placenta 2002; 23: 699 705. [DOI] [PubMed] [Google Scholar]
- Graham CH. Effect of transforming growth factor-beta on the plasminogen activator system in cultured first trimester human cytotrophoblasts. Placenta 1997; 18: 137 143. [DOI] [PubMed] [Google Scholar]
- Irving JA, Lala PK. Functional role of cell surface integrins on human trophoblast cell migration: regulation by TGF-beta, IGF-II, and IGFBP-1. Exp Cell Res 1995; 217: 419 427. [DOI] [PubMed] [Google Scholar]
- Bischof P, Meisser A, Campana A. Involvement of trophoblast in embryo implantation: regulation by paracrine factors. J Reprod Immunol 1998; 39: 167 177. [DOI] [PubMed] [Google Scholar]
- Lyall F. Mechanisms regulating cytotrophoblast invasion in normal pregnancy and pre-eclampsia. Aust N Z J Obstet Gynaecol 2006; 46: 266 273. [DOI] [PubMed] [Google Scholar]
- Norwitz ER. Defective implantation and placentation: laying the blueprint for pregnancy complications. Reprod Biomed Online 2006; 13: 591 599. [DOI] [PubMed] [Google Scholar]