Abstract
Defining mechanisms by which differentiated, contractile smooth muscle cells become proliferative and secretory in response to mechanical and environmental stress is crucial for determining the contribution of airway smooth muscle (ASM) to inflammatory responses that result in airway disease. Regulation by microRNAs (miRNAs) has emerged as an important post-transcriptional mechanism regulating gene expression that may modulate ASM phenotype, but little is known about the expression and functions of miRNA in smooth muscle. In the present study we used microarrays to determine whether miRNAs in human ASM cells are altered by a proinflammatory stimulus. In ASM cells exposed to IL-1β, TNF-α, and IFN-γ, we found 11 miRNAs to be significantly down-regulated. We verified decreased expression of miR-25, miR-140*, mir-188, and miR-320 by quantitative PCR. Analysis of miR-25 expression indicates that it has a broad role in regulating ASM phenotype by modulating expression of inflammatory mediators such as RANTES, eotaxin, and TNF-α; genes involved in extracellular matrix turnover; and contractile proteins, most notably myosin heavy chain. miRNA binding algorithms predict that miR-25 targets Krüppel-like factor 4 (KLF4), a potent inhibitor of smooth muscle–specific gene expression and mediator of inflammation. Our study demonstrates that inhibition of miR-25 in cytokine-stimulated ASM cells up-regulates KLF4 expression via a post-transcriptional mechanism. This provides novel evidence that miR-25 targets KLF4 in ASM cells and proposes that miR-25 may be an important mediator of ASM phenotype.
Keywords: smooth muscle, gene expression, miRNA, inflammation, remodeling
CLINICAL RELEVANCE.
The study of miRNA in airway smooth muscle is an emerging area of interest in lung biology. Our studies have identified a novel miRNA target that may profoundly affect airway smooth muscle phenotype and contribute significantly to the knowledge of phenotypic plasticity in smooth muscle. This may lead to new therapeutics for lung disease such as asthma.
MicroRNAs (miRNAs) are 21- to 23-nucleotide noncoding RNA molecules that have emerged as important post-transcriptional regulators of cell growth, differentiation, and apoptosis (1, 2). Both cloning and bioinformatics approaches have been used to identify miRNA individually or as polycistronic clusters in introns, exons, and regions between protein-coding genes (3). In addition, it has recently been reported that miRNAs can exist as elements in promoter regions of select genes (4). Currently, 721 human miRNAs are listed in the miRNA registry (miRBase 14.0, September 2009) (5), but it has been estimated that there may be 1,000 or more human miRNAs that target approximately 30% of the genome (3).
In the lung, miRNA expression has been examined in tumors and under inflammatory conditions. Members of the let-7 family have been identified as suppressors of lung tumor growth in vivo and in cultured cells, which may lead to novel molecular therapies for lung cancer (6, 7). Expression profiling in patients with lung cancer has also identified an miRNA signature that predicts survival and relapse (8). During inflammatory stimulation of lung alveolar epithelial cells, rapid changes in miR-146a expression occur and negatively regulate the release of IL-1β–induced IL-8 and regulated upon activation, normal T cell expressed and secreted (RANTES) (9). Exposure to lipopolysaccharide (LPS) in vivo causes rapid changes in mouse lung expression of multiple miRNAs that correlate with reduction of inflammatory mediators such as tumor necrosis factor (TNF)-α and macrophage inflammatory protein (MIP)-2 (10). In addition, miRNA binding sites have been identified in the asthma-susceptibility marker HLA-G where the 3′-untranslated region (UTR) contains sites for miR-148a, miR-148b, and miR-152 (11).
Little is known of the role of miRNA in airway smooth muscle (ASM), although other smooth muscle tissues have been studied. For example, miR-143 is highly expressed in leiomyosarcoma (12) and the regulation of miRNA by ovarian steroids has been demonstrated in the myometrium and leiomyoma (13). In the vasculature, a polymorphism in the 3′-UTR of the angiotensin II type I receptor (AT1R) has been associated with cardiovascular disease and has recently been identified as a target for miR-155 (14). Inhibition of miR-155 in vascular smooth muscle cells (VSMC) represses AT1R expression and decreases receptor-mediated activation. Microarray analysis has also recently demonstrated aberrant miRNA expression after vascular injury (15). This study identified miR-21 as a mediator of neointimal lesion formation that increases proliferation and inhibits apoptosis in VSMC.
It is recognized that chronic inflammatory events in the lung mediate processes by which ASM cells undergo reversible switching between a contractile phenotype and a more immature proliferative, synthetic, or secretory phenotype (16, 17) that produces Th1/Th2 cytokines, CC and C-X-C chemokines, chemotactic proteins, and peptide growth factors (18, 19). The molecular mechanisms regulating this phenotypic plasticity are not well understood. We undertook the present studies to determine whether regulation of miRNA expression may play a role in these processes. We examined miRNA expression in cultured ASM cells exposed to a proinflammatory stimulus of IL-1β, TNF-α, and IFN-γ. We present data identifying miR-25 as an miRNA down-regulated with inflammatory stimulation that affects expression of inflammatory mediators, extracellular matrix (ECM), and contractile proteins. We further investigate a predicted interaction between miR-25 and Krüppel-like factor 4 (KLF4), a transcription factor that participates in the regulation of serum-response factor (SRF)-dependent smooth muscle–specific gene expression and is a mediator of the inflammatory response in macrophages (20). These data support a role for miR-25 in post-transcriptional regulation of KLF4 expression and illustrate the potential of miR-25 as a novel modulator of smooth muscle phenotype.
MATERIALS AND METHODS
Human ASM RNA
Total RNA from human tracheal smooth muscle was obtained from Ambion (Austin, TX). The sex, age, race, and cause of death were noted for each donor as follows. Male, 20 years old, African American, suicide; male, 59 years old, white, lung cancer; male, 68 years old, unknown race, lung cancer; female, 87 years old, myocardial infarction.
Cell Culture
Primary cultured human airway myocytes were obtained from second- to fourth-generation mainstem bronchi of patients undergoing lung resection surgery at the University of Manitoba as previously described (21). Alternatively, human bronchial smooth muscle cells were obtained from Lonza (Walkersville, MD). Cells from passages four through seven were used and grown in a humidified 5% CO2 atmosphere at 37°C in M199 supplemented with 10% new born calf serum (Invitrogen, Carlsbad, CA), 0.5 μg/L epidermal growth factor, and 2 μg/L fibroblast growth factor. Cultures were grown to confluence and growth-arrested for 48 hours in serum-free media consisting of a 50:50 mix of F12/DMEM supplemented with 10 ml/L ITS+Premix containing 5 μg/ml insulin, 5 μg/ml transferrin, and 5 ng/ml selenium (BD Bioscience, Bedford, MA). Cells were then treated with 10 ng/ml IL-1β, TNF-α, and IFN-γ for 24 hours or left untreated. IL-1β, TNF-α, and other growth factors were purchased from Sigma (St. Louis, MO). IFN-γ was purchased from R&D Systems, Inc. (Minneapolis, MN).
miRNA Arrays
Expression of miRNAs in intact human ASM and in ASM cells was evaluated using mirVana miRNA bioarrays V2 (Ambion). The probes available on the arrays were spotted in quadruplicate with positive controls for labeling and orientation. Two arrays were spotted on each slide. For experiments using human ASM tissue, miRNA was purified from 20 μg of total RNA fractionated using the flashPAGE system as directed by the manufacturer (Ambion). This system uses capillary gel electrophoresis to isolate small RNA less than 50 nucleotides. This was experimentally verified by following fractionation of a small nucleotide RNA marker in control experiments. The filter-based flashPAGE clean-up kit was used to concentrate the purified miRNA. miRNA was then polyadenylated and labeled with Cy5 using the mirVana miRNA labeling kit and arrays hybridized and washed as directed. For miRNA from ASM cells, total RNA was extracted using TRIzol reagent and purified using the PureLink miRNA isolation kit (Invitrogen). Purified miRNA was quantified using RiboGreen and the quality of RNA estimated by absorbance ratios of 260/280 nm > 1.8. An aliquot of 2 μg miRNA was lyophilized and resuspended in 15.5 μl of RNAse-free water for labeling. The NCode miRNA labeling system (Invitrogen) was used for labeling and polyadenylation of miRNA followed by ligation of a capture sequence before hybridization overnight. Hybridized arrays were washed and incubated with AlexaFluor capture reagents to amplify signal intensity in these samples.
miRNA Array Analysis
Hybridized slides were scanned using a Perkin Elmer ScanArray 4000 Microarray Scanner (Waltham, MA) at the Nevada Genomics Center. GenePix 4.1 software (22) was used to quantify raw signal intensities. The remaining analysis and normalization was completed in collaboration with the Nevada Bioinformatics Center at the University of Nevada. For the ASM tissue arrays, the local background as computed by GenePix was subtracted from each signal value as a first quality control step. A noise threshold was computed using the empty wells included on the arrays. The noise threshold for each array was computed independently as the sum of the mean and 1/2 the standard deviation of the background-corrected positive signal levels of the 194 empty wells on each array (23). All spots with expression greater than this threshold were retained, and the remaining set of per-array replicates of each miRNA was examined for outliers based on the coefficient of variation. Only one replicate of one spot was deleted due to an abnormally large coefficient of variation > 0.08, which is approximately 1% of all sets of detectable replicates. Remaining replicated spots were then averaged, and averaged spot intensities were normalized per array by division of the median array signal. Normalized intensities were then averaged across sample replicates, except for the female patient, which was only analyzed on one array. Heat maps of normalized expression levels were generated using Java TreeView v.1.1.3.
miRNA arrays for cytokine-stimulated ASM cells were completed in duplicate from two different cultures. In this experiment, the local background was not available. Again, the same noise threshold was used to filter miRNA signals that were deemed detectable. Remaining spot replicates were averaged after exclusion of those replicates that caused abnormally large coefficients of variations. Only 22 signal intensities were excluded in sets of replicates with coefficients of variation greater than 0.8. Of all 2,504 sets of detectable replicates, 1% was associated with coefficients of variation greater than this chosen threshold of 0.8. This yielded 642 remaining features to be considered for further analysis, including controls. The ratios of cytokine-stimulated to nontreated, control miRNA expression were computed for each of the four arrays, and then normalized per array via a simple division by the median ratio of each array. Normalized ratios were then averaged for each of the two different cultures assayed and log2-transformed.
In Silico Analysis of Predicted miRNA Targets
Publicly available algorithms (PicTar, pictar.mdc-berlin.de; TargetScan 4.1, www.targetscan.org; miRBase, microrna.sanger.ac.uk; miRNA.org, www.microrna.org; Patrocles, www.patrocles.org) were used to determine potential binding sites of miRNA to mRNA of target proteins. All algorithms gave more weight to “seed” region bases 2–9 of miRNA (24–28).
Quantitative PCR
RNA was extracted using TRIzol reagent and enriched for the miRNA fraction using the PureLink miRNA isolation kit (Invitrogen) or enriched for the total RNA fraction containing miRNA using miRVana isolation protocols (Ambion). Purified miRNA was quantified using RiboGreen and the quality of RNA estimated by absorbance ratios of 260/280 nm greater than 1.8. TaqMan miRNA assays (Applied Biosystems, Foster City, CA), were used to prepare cDNA from 250 ng RNA and Ct values from standard curves used to quantify relative expression of specific miRNA normalized to U6. Expression of contractile proteins or KLF4 was evaluated using TaqMan gene expression assays from first-strand cDNA prepared from 2 μg total RNA diluted 1:5 as previously described (29) and normalized relative to 18S rRNA. The specific TaqMan gene expression assays were as follows: myosin heavy chain (MYH11), Hs00224610; calponin (CNN1), Hs00154543; TAGLN (SM22), Hs00162558; KLF4, Hs00358836.
PCR Arrays
PCR arrays (SABiosciences, Frederick, MD) were used examine expression levels of extracellular matrix proteins (ECM) and Th1-Th2-Th3 inflammatory mediators affected by miR-25. First-strand cDNA was synthesized from 2 μg total RNA diluted 1:5 (29). The reactions contained 1× SYBR PCR master mix and 2 μl diluted cDNA in a final volume of 25 μl and were amplified at 95°C, 15 seconds; 60°C, 1 minute for 40 cycles. Cytokine-stimulated ASM cells transfected with the negative anti-miR control or anti–miR-25 were compared and analyzed across multiple plates using the ΔΔCt method and normalized to β-actin.
Western Blot
Whole cell lysates were prepared as previously described (29, 30). Protein concentrations were determined by the bicinchoninic acid method using bovine serum albumin as the standard. Total protein (10 μg) was separated by 4 to 20% SDS-PAGE and transferred to nitrocellulose. Blots were blocked in Odyssey blocking buffer diluted 1:1 in PBS for 1 hour (Licor Biosciences, Lincoln, NE) before incubation with primary antibody. Anti-KLF4 was obtained from Millipore (Billerica, MA), anti-GAPDH from Santa Cruz Biotechnologies (Santa Cruz, CA) and anti-myosin heavy chain from Biomedical Technologies, Inc. (Stoughton, MA). Secondary antibodies were conjugated to AlexaFluor 680 (Molecular Probes, Eugene, OR) or IRDye800 (Rockland Immunochemicals, Gilbertsville, PA) for fluorometric detection using an Odyssey infrared imaging system. All densitometric analyses took place within the linear range of the immunoreactive signal using Odyssey system software.
ELISA
Cell culture media was assayed for eotaxin (R&D Systems), RANTES, or TNF-α (Invitrogen) by ELISA according to the manufacturer's protocols.
Anti-miR Transfections
RNA oligonucleotides that inhibit miR-25 or a nonbinding negative control were designed and synthesized by Ambion. ASM cells were grown to approximately 80% confluence before transfection with 90 nM anti-miR miRNA inhibitors using siPORTNeoFX (Ambion) diluted in OptiMEM. After 48 hours, cultures were treated with 10 ng/ml IL-1β, TNF-α, and IFN-γ for 24 hours and total RNA, miRNA, or protein isolated as described. Using FAM-labeled negative control anti-miR oligonucleotides, we estimate 40% transfection efficiency under these conditions (unpublished observation).
Statistics
Paired t tests were conducted using GraphPad software (La Jolla, CA).
RESULTS
miRNA Expression in Human Airway Smooth Muscle
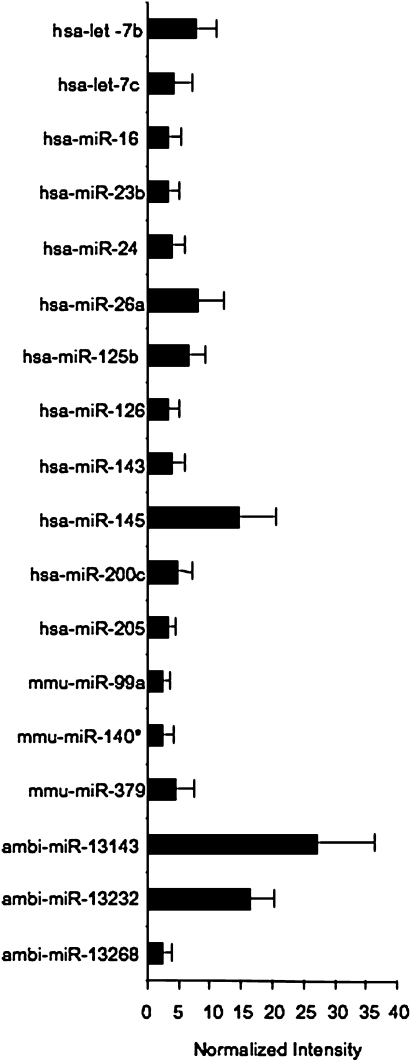
We initially wanted to survey miRNA expression in intact ASM tissue, since miRNA expression has not been described in detail in ASM tissue samples. Total RNA was obtained from tracheal smooth muscle of four donors ranging in age from 20 to 87 years old. The donors included one female, were of varying races and care was taken to ensure that the cause of death was not due to clinical signs of asthma. These arrays contained a panel of 328 human, 114 mouse, and 52 rat miRNAs from the Sanger miRBase sequence database version 8.0 along with 152 novel human miRNAs with the possibility of overlap between species for some probes. We found 60 miRNAs expressed at a detectable level in all four donors and another 118 miRNAs expressed in three of the four donors (see Table E1 in the online supplement). Normalized median signal intensities were used to generate a heat map of normal miRNA expression in ASM tissue from miRNA expressed in all four donors (Figure E1). There were 18 miRNAs with expression levels ≥ 2-fold above normalized expression values (Figure 1).
Figure 1.
miRNA expression in human airway smooth muscle (ASM). Total RNA from human airway smooth muscle was obtained commercially. The miRNA fraction was purified by capillary gel electrophoresis and quantified for labeling as described. Each sample was hybridized in duplicate, except for that from donor 4, which was hybridized to one array only. Arrays were scanned with a Perkin Elmer ScanArray 4000 Microarray Scanner and analyzed with Gene Pix 4.1 software. Data were normalized to the median signal intensity across all four samples, and normalized intensity of the 18 miRNA expressed ≥ 2-fold over the median expression levels for each array is shown (n = 4 ± SEM).
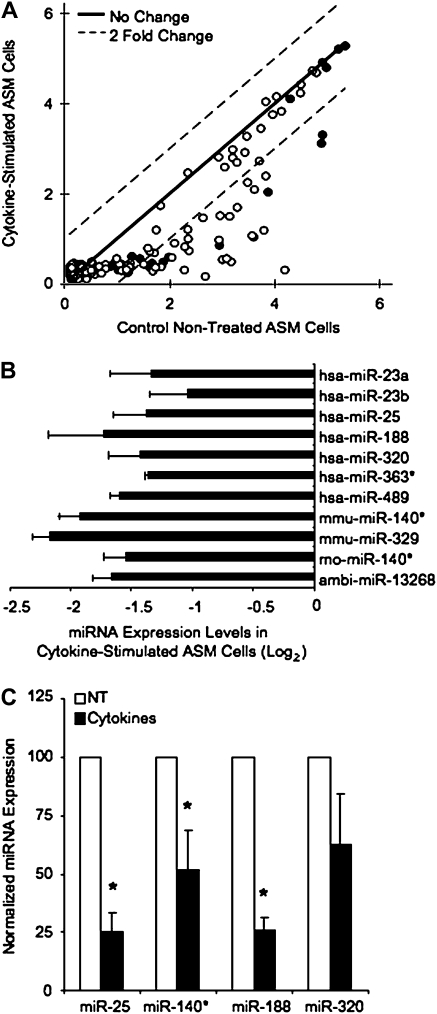
Cytokine-Stimulated miRNA Expression in ASM Cells
The effects of cytokine treatment on miRNA expression were examined in ASM cultures stimulated with IL-1β, TNF-α, and IFN-γ, which has been previously demonstrated to induce expression of inflammatory mediators in these cells (30, 31). A comparison of miRNA expression under cytokine-stimulated and nontreated conditions from hybridized miRNA arrays is shown in Figure 2A. We also used this data to compare the miRNAs expressed in cultured ASM cells with those miRNAs expressed in intact tissue (Table E3). Of the 640 miRNAs spotted on the array, 134 miRNAs were expressed in all replicates. A majority of the miRNAs on the array exhibited very low signal intensity, and none of the miRNAs expressed were significantly up-regulated with cytokine treatment (Table E2). We did observe 11 miRNA down-regulated ≥ 2-fold in both cultures with cytokine treatment (Figure 2B). These miRNAs include described human miRNAs miR-23a, -23b, -25, -188, -320, -363, and -489, as well as mouse and rat miRNAs homologous to miR-140*, mouse miR-329, and a novel miRNA, abi-13268. We verified the expression of select miRNA down-regulated with cytokine stimulation using human-specific TaqMan expression assays (Figure 2C). As seen on the array, miR-25, miR-140*, and miR-188 were significantly down-regulated with cytokine stimulation, while expression of miR-329 was also markedly decreased.
Figure 2.
Cytokine stimulation affects miRNA expression in human ASM cells. Human ASM cultures were grown to confluence and serum-starved for 48 hours before cytokine treatment with 10 ng/ml IL-1β, TNF-α, and IFN-γ for 24 hours. RNA was isolated and miRNA arrays hybridized and analyzed as described. (A) A scatter plot summarizes the mean signal intensity on a log2 scale from the 134 miRNA expressed in two different ASM cultures (shown in solid and open circles) assayed in duplicate. Dotted lines indicate a 2-fold expression change between cytokine-stimulated and nontreated cultures; the solid line indicates no changes. (B) Ratios of miRNA expression in cytokine-stimulated versus nontreated cultures were generated from miRNA array analysis. Normalized ratios of the 11 miRNA down-regulated ≥ 2-fold with cytokine treatment are shown. (C) TaqMan miRNA expression assays were used to verify changes in miRNA expression seen on the arrays. Human ASM cultures were treated as above, RNA extracted with TRIzol, and miRNA further purified for cDNA synthesis. RT-PCR were performed with TaqMan miRNA assays using 250 ng RNA enriched for miRNA. miRNA expression was normalized to U6 expression in each sample. Data is expressed as the % change in miRNA expression from nontreated cultures (100%) (n = 3–6 ± SEM). *Statistically significant difference from control, nontreated cultures (P < 0.05).
Functions of miR-25 on ASM Cell Phenotype
To begin to understand the functions of miRNA in cytokine-stimulated ASM cells, we searched multiple miRNA target-predicting algorithms to generate possible candidates of miR-25 binding. We chose miR-25 as our initial candidate because it was consistently down-regulated with cytokine stimulation in miRNA array studies and by quantitative PCR analysis. Targets that were predicted in more than one database include the vitronectin receptor integrin αV (ITGAV), myosin 1B (MYO1B), myocyte enhancer factor 2D (MEF2D), Krüppel like factor 4 (KLF4), and suppressor of cytokine signaling 5 (SOCS5). These results suggested that miR-25 may have a broader role in regulating ASM plasticity by affecting inflammatory responses, extracellular matrix (ECM) turnover, and possibly contractile phenotype.
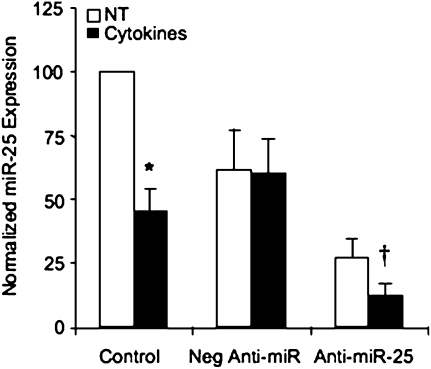
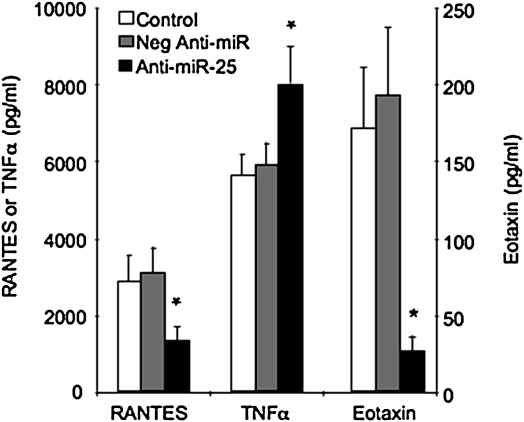
To begin to study functions of miR-25, we optimized a system to manipulate endogenous miRNA expression using RNA oligonucleotides designed to inhibit miRNA, known as anti-miRs. As shown in Figure 3, transfection of the miR-25–specific anti-miR down-regulated miR-25 in both nontreated and cytokine-stimulated cultures. This validates the use of anti–miR-25 in subsequent experiments and further supports our data demonstrating that this cytokine stimulus down-regulates miR-25 expression. After this, we used samples of cytokine-stimulated ASM cells transfected with the anti-miRs to survey expression of a wide variety of inflammatory mediators or extracellular matrix (ECM) genes using PCR arrays. The inflammatory mediator PCR array contained primers for a variety of Th1 and Th2 mediators and receptors, transcription factors, and signaling molecules associated with immune responses (Table 1; Table E4). Inhibition of miR-25 in cytokine-stimulated cells down-regulates expression of RANTES, IL-18, and eotaxin greater than 2-fold while up-regulating expression of TNF-α as well as receptors for RANTES, IL-6, and IL-12. Effects of miR-25 on secretion of RANTES, eotaxin, and TNF-α were verified by ELISA (Figure 4). Secretion of these mediators was not detectable in nontreated cultures (data not shown), but was induced by treatment with 10 ng/ml IL-1β, TNF-α, and IFN-γ for 24 hours in control, nontransfected cultures and in cultures transfected with the negative anti-miR. Transfection with anti–miR-25 dramatically reduces cytokine-stimulated secretion of RANTES and eotaxin and stimulates secretion of TNF-α. These results demonstrate that expression of miR-25 does affect the inflammatory response of ASM cells by modulating secretion of both proinflammatory mediators and chemokines associated with inflammatory airway remodeling (18).
Figure 3.
Anti-miR oligonucleotides down-regulate miR-25 expression. Cultures were transfected with 90 nM anti-miR miRNA oligonucleotide inhibitors specific for miR-25 (Anti–miR-25) or a negative nonbinding control (Neg Anti-miR) using siPORTNeoFX for 48 hours. Control cultures received mock transfection conditions. Cultures were then treated with 10 ng/ml IL-1β, TNF-α, and IFN-γ (Cytokines) or not treated (NT) for 24 hours. miRNA expression assays used to analyze expression of miR-25 normalized to U6 in the same samples. Data were expressed as the % control from control, NT cells (n = 5 ± SEM). *Statistically significant difference from miRNA expression levels seen nontreated cultures; †statistically significant difference from control, cytokine-stimulated cultures (P < 0.05).
TABLE 1.
INHIBITION OF miR-25 REGULATES EXPRESSION OF INFLAMMATORY MEDIATORS IN CYTOKINE-STIMULATED AIRWAY SMOOTH MUSCLE CELLS
| Gene Name | Fold Regulation by Anti–miR-25 |
|---|---|
| Chemokine (C-C motif) ligand 5; CCL5 | −44.76 |
| Interleukin 18; IL18 | −2.53 |
| Chemokine (C-C motif) ligand 11; CCL11 | −2.05 |
| Interleukin 23, alpha subunit p19; IL23A | −1.98 |
| Chemokine (C-C motif) receptor 2; CCR2 | −1.83 |
| CD86 molecule; CD86 | −1.77 |
| Interleukin 18 receptor 1; IL18R1 | −1.69 |
| Fas ligand; FASLG | −1.60 |
| CD40 ligand; CD40LG | −1.56 |
| Inhibin, alpha; INHA | −1.55 |
| Suppressor of cytokine signaling 2; SOCS2 | 1.64 |
| NFAT, cytoplasmic, calcineurin-dependent 2; NFATC2 | 1.74 |
| Growth factor independent 1 transcription repressor; GFI1 | 1.88 |
| CD27 molecule; CD27 | 1.95 |
| Interleukin 6 receptor; IL6R | 2.21 |
| Hepatitis A virus cellular receptor 2; HAVCR2 | 2.23 |
| Interleukin 12 receptor, beta 2; IL12RB2 | 2.30 |
| Chemokine (C-C motif) receptor 5; CCR5 | 2.32 |
| Tumor necrosis factor; TNF | 2.57 |
| Linker for activation of T cells; LAT | 2.72 |
Figure 4.
miR-25 regulates expression of inflammatory mediators. ASM cells were transfected with the negative anti-miR control or anti–miR-25 as described and stimulated with 10 ng/ml IL-1β, TNF-α, and IFN-γ, for 24 hours. Control cultures received mock transfection conditions. RANTES, TNF-α, or eotaxin secretion was measured in media samples by ELISA, n = 6 ± SEM. *Statistically significant difference from control, cytokine-stimulated cultures (P < 0.05).
Accumulation of extracellular matrix (ECM) proteins is another feature of airway remodeling, and bronchoalveloar lavage fluid from individuals with asthma contains increased levels of fibronectin, laminin, and hyaluronate, reflecting increased ECM turnover (32, 33). We again chose to use PCR arrays to survey whether miR-25 had effects on expression of genes encoding ECM and basement membrane proteins, their receptors, or other structural proteins associated with cell ECM interactions (Table 2; Table E5). Inhibition of miR-25 in cytokine-stimulated ASM cells down-regulates expression of a wide variety of these proteins, with greater than 2-fold down-regulation of delta catenin, type XI collagen, thrombospondin, and ADAMTS8 metallopeptidases. To a lesser extent, E-cadherin, types V and XV collagen, integrins αM and β2, matrix metalloproteinases (MMP) 9, ADAMTS1, and fibronectin are also down-regulated. In addition, inhibition of miR-25 also up-regulated expression of tenascin C, sarcoglycan ɛ, and MMP 15 greater than 2-fold, and to a lesser extent up-regulated expression of tissue inhibitor of metalloproteinase (TIMP2), laminin α2, selectin P, selectin L, MMP3, and MMP14. These results demonstrate that expression of miR-25 does affect expression of a wide variety of ECM proteins that could affect ECM turnover and airway remodeling.
TABLE 2.
INHIBITION OF miR-25 REGULATES EXPRESSION OF EXTRACELLULAR MATRIX PROTEINS IN CYTOKINE-STIMULATED AIRWAY SMOOTH MUSCLE CELLS
| Gene Name | Fold Regulation by Anti–miR-25 |
|---|---|
| Catenin, delta 1; CTNND1 | −5.74 |
| Collagen, type XI, alpha 1; COL11A1 | −3.31 |
| Thrombospondin 2; THBS2 | −3.10 |
| ADAM metallopeptidase with thrombospondin motif, 8; ADAMTS8 | −2.22 |
| Cadherin 1, type 1, E-cadherin; CDH1 | −1.88 |
| Collagen, type XV, alpha 1; COL15A1 | −1.81 |
| Secreted phosphoprotein 1; SPP1 | −1.69 |
| Integrin, beta 2; ITGB2 | −1.60 |
| ADAM metallopeptidase with thrombospondin motif, 1; ADAMTS1 | −1.60 |
| Integrin, alpha M; ITGAM | −1.60 |
| Fibronectin 1; FN1 | −1.57 |
| Matrix metallopeptidase 9; MMP9 | −1.55 |
| Collagen, type V, alpha 1; COL5A1 | −1.51 |
| Laminin, gamma 1; LAMC1 | −1.49 |
| TIMP metallopeptidase inhibitor 2; TIMP2 | 1.57 |
| Matrix metallopeptidase 14; MMP14 | 1.59 |
| Laminin, alpha 2; LAMA2 | 1.65 |
| Collagen, type XIV, alpha 1; COL14A1 | 1.66 |
| Matrix metallopeptidase 3; MMP3 | 1.70 |
| Selectin P (CD62); SELP | 1.74 |
| Selectin L; SELL | 1.78 |
| Spastic paraplegia 7; SPG7 | 1.90 |
| Tenascin C; TNC | 2.14 |
| Sarcoglycan, epsilon; SGCE | 2.19 |
| Matrix metallopeptidase 15; MMP15 | 2.58 |
miR-25 Regulates Expression of Selected Contractile Proteins
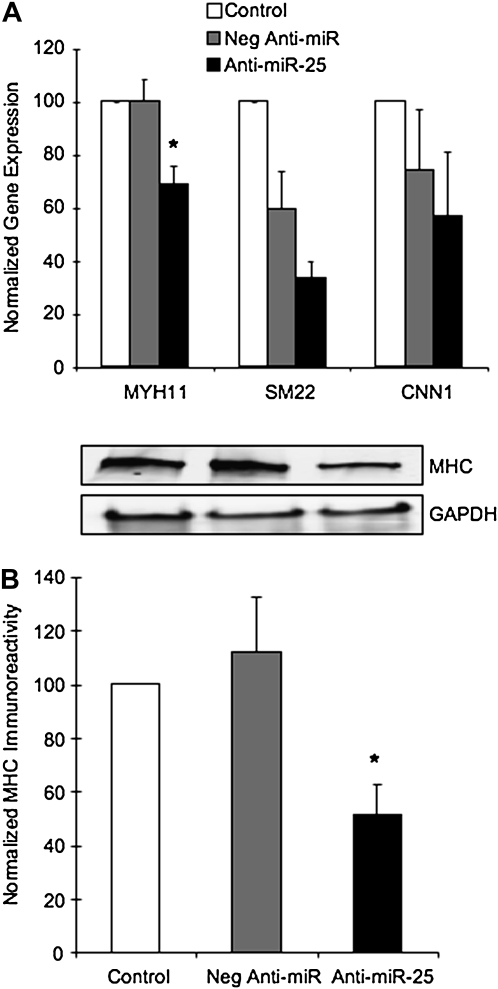
To determine whether miR-25 targeted any proteins important in regulating contractile ASM phenotype, we analyzed gene expression of selected contractile proteins after anti–miR-25 transfection. Figure 5A summarizes quantitative RT-PCR analysis of mRNA expression for myosin heavy chain (MHC, MYH11) SM22 or calponin (CNN1) after cytokine treatment with 10 ng/ml IL-1β, TNF-α, and IFN-γ. Treatment with inflammatory cytokines increased gene expression of MHC and SM22, but not calponin (data not shown). Transfection with anti–miR-25 decreased cytokine-stimulated myosin heavy chain expression by 25%. This result was verified by immunoblotting analysis (Figure 5B), in which anti–miR-25 decreased MHC by 50%. SM22 expression was also decreased by anti–miR-25, although the negative nonbinding anti-miR control also had an effect on expression levels. Calponin expression was not affected by anti–miR-25. These results indicate the miR-25 plays a role in modulating mRNA expression of these contractile proteins, particularly MHC. There are no predicted binding sites for miR-25 in the 3′UTR of MHC. Thus, any regulation of MHC by this miRNA is likely to be indirect, possibly through a transcription factor that modulates ASM phenotype.
Figure 5.
miR-25 affects expression of contractile proteins. ASM cells were transfected with the negative anti-miR control or anti–miR-25 as described and stimulated with 10 ng/ml IL-1β, TNF-α, and IFN-γ, for 24 hours. (A) mRNA expression was analyzed using TaqMan gene expression assays for myosin heavy chain (MHC, MYH11), SM22, or calponin (CNN1) and normalized to 18S rRNA. (B) MHC immunoreactivity was quantified after SDS-PAGE and normalized to GAPDH, as shown in representative immunoblots. Data are expressed relative to expression levels in cytokine-stimulated mock-transfected cells (Control, 100%) (n = 5 ± SEM). *Statistically significant difference from cytokine-stimulated cultures transfected with the negative anti-miR (P < 0.05).
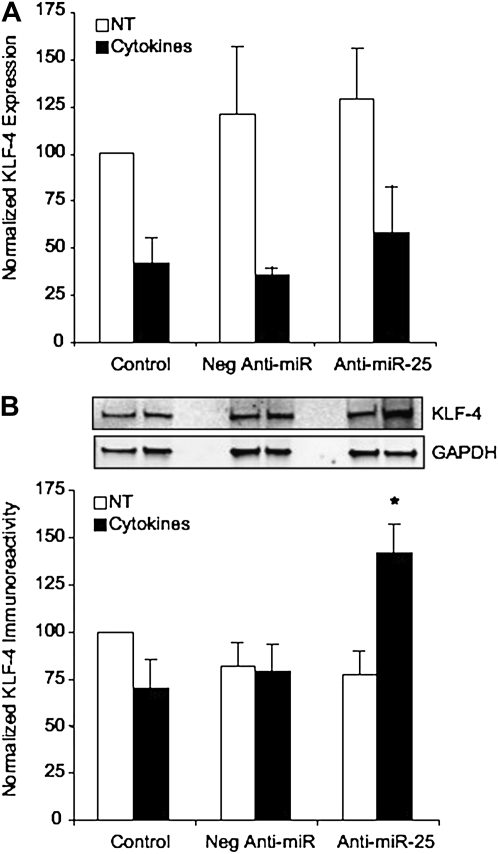
miR-25 Regulates KLF4 Expression
Bioinformatics analysis using multiple miRNA-binding site algorithms identified two possible miR-25–binding regions in the 3′UTR of human KLF4 at base pairs 362–368 and 674–681. As KLF4 has potential inhibitory effects of smooth muscle contractile phenotype gene expression, we investigated whether inhibition of miR-25 affected KLF4 expression. In these experiments, cytokine stimulation of ASM cells resulted in down-regulation of KLF4 mRNA expression (Figure 6). However, transfection with anti–miR-25 did not further affect KLF4 mRNA expression (Figure 6A). Analysis of protein expression levels determined that KLF4 immunoreactivity decreased with cytokine stimulation, although not as dramatically as that seen with mRNA expression levels. In nontreated cultures, anti–miR-25 did not have any significant effect on KLF4 expression. However, in cytokine-stimulated cultures, transfection with anti–miR-25 up-regulates KLF4 protein abundance considerably (Figure 6B). Consistent with current understanding of the mechanism of action of miRs, miR-25 plays a post-transcriptional role in silencing KLF4 expression in ASM cells; thus, inhibition of miR-25 represses these silencing mechanisms, allowing for KLF4 protein accumulation. This provides novel evidence that miR-25 targets KLF4 in ASM cells and suggests that this may be one mechanism by which miR-25 modulates ASM phenotype.
Figure 6.
Effects of miR-25 on KLF4 expression. ASM cells were transfected with the negative anti-miR control or anti–miR-25 as described and stimulated with 10 ng/ml IL-1β, TNF-α, and IFN-γ (Cytokines), or not treated (NT), for 24 hours. (A) KLF4 gene expression was analyzed using TaqMan assays and normalized to 18S rRNA. (B) KLF4 immunoreactivity was quantified after SDS-PAGE and normalized to GAPDH, as shown in representative immunoblots. All data were expressed as the % control from nontreated, mock-transfected cells (Control, 100%) (n = 6 ± SEM). *Statistically significant difference from control cultures or cultures transfected with the negative anti-miR (P < 0.05).
DISCUSSION
miRNAs have emerged as a class of small, noncoding RNAs that post-transcriptionally regulate gene expression by binding to the 3′ UTR of target mRNA to either repress translation or induce cleavage (3). We undertook studies to begin elucidating the role of miRNA in ASM by profiling the miRNA expressed in ASM and determining whether inflammatory stimulation modulated miRNA expression. We assessed a wide variety of known and novel human miRNAs, along with homologous rat and mouse targets, using commercially available microarrays. Initially, we chose to examine miRNA expression in intact ASM to identify those miRNAs expressed in ASM, which has not been reported to date. This data set is representative of a small number of patients and expression levels varied, which could be accounted for by differences in age, race, sex, or cause of death. We identified 60 miRNAs as having consistent expression levels above background. It is not surprising that only a small fraction of the known miRNAs were expressed in intact ASM, since miRNA expression has been shown to be tissue and stimulus specific (34). In this study, when miRNA expression was compared between organs in the mouse, only two miRNAs (miR-195 and miR-200c) were found to be specific for the lung, while nine miRNAs were expressed in at least one other organ. In addition, a comparison of the miRNA expressed in intact tissue compared with cultured cells demonstrates that there are differences in miRNA expression. While some of these differences may be attributed to changes in ASM cells in culture, it is difficult to study the molecular biology and biochemistry of miRNA functions in vivo without a working knowledge obtained in vitro.
In examining the regulation of miRNA expression after an inflammatory stimulus, we treated cultures with a cocktail of IL-1β, TNF-α, and IFN-γ. This stimulus was chosen to more closely approximate the environment in vivo since it has been found in bronchoalveolar lavage fluid from symptomatic patients with asthma (35, 36). This inflammatory cocktail has also been previously used by our laboratory and others to elicit the production of inflammatory mediators in ASM cells, including IL-1β, IL-6, IL-8, cyclooxygenase-2, and monocyte chemotactic proteins (29, 31). Several studies in the lung have reported rapid up-regulation of miRNA, including miR-25, in response to IL-1β and LPS within 3 hours (10). Surprisingly, under the conditions reported here, only down-regulation of miRNA expression was observed with 24 hours of exposure to an inflammatory stimulus. This suggests that profiles of miRNA expression are likely time-dependent, and the 11 miRNAs down-regulated in our study may represent a subset of miRNAs affected by chronic environmental changes.
To further understand the role of miRNA in ASM, we searched publicly available miRNA targeting algorithms for possible targets relevant to ASM functions. These algorithms find complimentary targets in the 3′ UTR of mRNA and puts more weight on the complementarity of the “seed” region (bases 2–9) of the miRNA with 3′ UTR. We singled out miR-25 for further functional analysis, since it is predicted to target a variety of genes that may regulate ASM plasticity. One of the approaches we used was to survey gene expression of a wide variety of inflammatory mediators, extracellular matrix (ECM) genes and selected contractile proteins involved in maintaining and altering ASM phenotype in cultures transfected with miR-25 inhibitors. Since miRNAs generally function as translational repressors, this approach was not designed to directly examine targets of miR-25 binding, but rather to determine whether miR-25 has any widespread effects on gene expression. The results indicate that inhibition of miR-25 does affect expression of inflammatory mediators by decreasing expression and secretion of RANTES and eotaxin, while increasing levels of TNF-α. Inhibition of miR-25 also affected expression of numerous ECM genes with a number of targets, laminin α2 and sarcoglycan ɛ for example, linked to the regulation of, or identified as, markers for phenotypic plasticity of ASM (37–39). Moreover, alterations in miR-25 expression affected expression of contractile proteins, most notably myosin heavy chain, which may have profound implications on development of a mature, contractile phenotype during airway remodeling.
Another approach we took to examining functions of miR-25 was to study the effects of miR-25 on KLF4, a target predicted by multiple binding algorithms. KLF4 was of interest since it has been proposed to play an important role in inflammation, proliferation, and differentiation of VSMC (40, 41) and participates in phenotypic switching as a potent inhibitor of smooth muscle–specific gene expression (42). KLF4 also participates in the inflammatory response, where it is a key regulator of monocyte differentiation (20) and cooperates in the transactivation of proinflammatory genes in response to IFN-γ, LPS, or TNF-α. One would expect KLF4 expression to increase with cytokine stimulation of ASM cells, since one hypothesis would be that KLF4 represses smooth muscle–specific gene expression in ASM cells to modulate a secretory phenotype under these conditions. This was not observed in our experiments, since both mRNA and to a lesser extent protein expression of KLF4 was decreased. It may be that serum deprivation of the cells alters KLF4 expression or that we are missing any rapid changes in KLF4 expression with 24 hours of cytokine stimulation. Endogenous KLF4 expression and its regulation have not been reported in ASM cells, and these will be questions for further study. We have determined that under the conditions in our study, inhibition of miR-25 up-regulates KLF4 immunoreactivity but not mRNA expression. Thus, miR-25 appears to regulate KLF4 expression post-transcriptionally, a hallmark of miRNA activity.
The data presented here demonstrate for the first time that proinflammatory cytokines regulate miRNA in ASM. This work also illustrates that expression of miR-25 affects a broad range of genes involved in inflammation and ECM turnover as well as myosin heavy chain to likely affect ASM phenotype. Furthermore, this work has identified an effect of miR-25 on the transcription factor KLF4, which is important in the multifunctional behavior of smooth muscle cells. These results demonstrate that while the study of miRNA in smooth muscle homeostasis and pathology is in its infancy, characterizing miRNA expression and identifying protein targets will provide valuable insight about regulation of smooth muscle cell functions, which may lead to the development of novel therapeutics.
Supplementary Material
Acknowledgments
The authors recognize Shanti Rawat, Vani Kilari and Margaret Elorza and the staff at the Nevada Genomics Center for excellent technical assistance.
These studies were supported by the National Institutes of Health HL080960 to C.A.S., HL077726 to W.T.G., and RR016464 from the Nevada INBRE Program at the National Center for Research Resources. A.J.H. is supported through the Canada Research Chairs Program and Canadian Institutes for Health Research.
This article contains an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1165/rcmb.2009-0123OC on June 18, 2009
Conflict of Interest Statement: K.S. received a grant from the National Institutes of Health (NIH) for $5,001 to $10,000. A.J.H. received a research grant from Merk Frost for $50,001 to $100,000, GlaxoSmithKline for more than $100,001, and a support grant for meeting proceedings publication from Merk Frost for less than $1,000. W.T.G. received an RO1 research grant from NIH for more than $100,001.
References
- 1.Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 2006;94:776–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett 2005;579:5911–5922. [DOI] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281–297. [DOI] [PubMed] [Google Scholar]
- 4.Younger ST, Pertsemlidis A, Corey DR. Predicting potential miRNA target sites within gene promoters. Bioorg Med Chem Lett 2009;19:3791–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res 2004;32:D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, Weidhaas JB, Brown D, Bader AG, Slack FJ. The Let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle 2008;7:759–764. [DOI] [PubMed] [Google Scholar]
- 7.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the Let-7 microRNA family. Proc Natl Acad Sci USA 2008;105:3903–3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu SL, Chen HY, Chang GC, Chen CY, Chen HW, Singh S, Cheng CL, Yu CJ, Lee YC, Chen HS, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell 2008;13:48–57. [DOI] [PubMed] [Google Scholar]
- 9.Perry MM, Moschos SA, Williams AE, Shepherd NJ, Larner-Svensson HM, Lindsay MA. Rapid changes in microRNA-146a expression negatively regulate the IL-1beta-induced inflammatory response in human lung alveolar epithelial cells. J Immunol 2008;180:5689–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moschos SA, Williams AE, Perry MM, Birrell MA, Belvisi MG, Lindsay MA. Expression profiling in vivo demonstrates rapid changes in lung microRNA levels following lipopolysaccharide-induced inflammation but not in the anti-inflammatory action of glucocorticoids. BMC Genomics 2007;8:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan Z, Randall G, Fan J, Camoretti-Mercado B, Brockman-Schneider R, Pan L, Solway J, Gern JE, Lemanske RF, Nicolae D, et al. Allele-specific targeting of microRNAs to HLA-G and risk of asthma. Am J Hum Genet 2007;81:829–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian S, Lui WO, Lee CH, Espinosa I, Nielsen TO, Heinrich MC, Corless CL, Fire AZ, van de Rijn, M. MicroRNA expression signature of human sarcomas. Oncogene 2008;27:2015–2026. [DOI] [PubMed] [Google Scholar]
- 13.Pan Q, Luo X, Chegini N. Differential expression of microRNAs in myometrium and leiomyomas and regulation by ovarian steroids. J Cell Mol Med 2008;12:227–240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 Receptor +1166 A/C polymorphism attenuates microrna-155 binding. J Biol Chem 2007;282:24262–24269. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res 2007;100:1579–1588. [DOI] [PubMed] [Google Scholar]
- 16.Halayko AJ, Camoretti-Mercado B, Forsythe SM, Vieira JE, Mitchell RW, Wylam ME, Hershenson MB, Solway J. Divergent differentiation paths in airway smooth muscle culture: induction of functionally contractile myocytes. Am J Physiol 1999;276:L197–L206. [DOI] [PubMed] [Google Scholar]
- 17.Hirst SJ, Walker TR, Chilvers ER. Phenotypic diversity and molecular mechanisms of airway smooth muscle proliferation in asthma. Eur Respir J 2000;16:159–177. [DOI] [PubMed] [Google Scholar]
- 18.Singer CA, Salinthone S, Baker KJ, Gerthoffer WT. Synthesis of immune modulators by smooth muscles. Bioessays 2004;26:646–655. [DOI] [PubMed] [Google Scholar]
- 19.Halayko AJ, Solway J. Molecular mechanisms of phenotypic plasticity in smooth muscle cells. J Appl Physiol 2001;90:358–368. [DOI] [PubMed] [Google Scholar]
- 20.Alder JK, Georgantas RW III, Hildreth RL, Kaplan IM, Morisot S, Yu X, McDevitt M, Civin CI. Kruppel-like factor 4 is essential for inflammatory monocyte differentiation in vivo. J Immunol 2008;180:5645–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naureckas ET, Ndukwu IM, Halayko AJ, Maxwell C, Hershenson MB, Solway J. Bronchoalveolar lavage fluid from asthmatic subjects is mitogenic for human airway smooth muscle. Am J Respir Crit Care Med 1999;160:2062–2066. [DOI] [PubMed] [Google Scholar]
- 22.Fielden MR, Halgren RG, Dere E, Zacharewski TR. GP3: GenePix post-processing program for automated analysis of raw microarray data. Bioinformatics 2002;18:771–773. [DOI] [PubMed] [Google Scholar]
- 23.Younossi ZM, Baranova A, Ziegler K, Del Giacco L, Schlauch K, Born TL, Elariny H, Gorreta F, VanMeter A, Younoszai A, et al. A genomic and proteomic study of the spectrum of nonalcoholic fatty liver disease. Hepatology 2005;42:665–674. [DOI] [PubMed] [Google Scholar]
- 24.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat Genet 2005;37:495–500. [DOI] [PubMed] [Google Scholar]
- 25.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005;120:15–20. [DOI] [PubMed] [Google Scholar]
- 26.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. MiRBase: tools for microRNA genomics. Nucleic Acids Res 2008;36:D154–D158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 2008;36:D149–D153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5:396–400. [DOI] [PubMed] [Google Scholar]
- 29.Salinthone S, Singer CA, Gerthoffer WT. Inflammatory gene expression by human colonic smooth muscle cells. Am J Physiol Gastrointest Liver Physiol 2004;287:G627–G637. [DOI] [PubMed] [Google Scholar]
- 30.Singer CA, Baker KJ, McCaffrey A, AuCoin DP, Dechert MA, Gerthoffer WT. p38 MAPK and NF-KappaB mediate COX-2 expression in human airway myocytes. Am J Physiol Lung Cell Mol Physiol 2003;285:L1087–L1098. [DOI] [PubMed] [Google Scholar]
- 31.Hedges JC, Singer CA, Gerthoffer WT. Mitogen-activated protein kinases regulate cytokine gene expression in human airway myocytes. Am J Respir Cell Mol Biol 2000;23:86–94. [DOI] [PubMed] [Google Scholar]
- 32.Altraja A, Laitinen A, Virtanen I, Kampe M, Simonsson BG, Karlsson SE, Hakansson L, Venge P, Sillastu H, Laitinen LA. Expression of laminins in the airways in various types of asthmatic patients: a morphometric study. Am J Respir Cell Mol Biol 1996;15:482–488. [DOI] [PubMed] [Google Scholar]
- 33.Bousquet J, Chanez P, Lacoste JY, Enander I, Venge P, Peterson C, Ahlstedt S, Michel FB, Godard P. Indirect evidence of bronchial inflammation assessed by titration of inflammatory mediators in BAL fluid of patients with asthma. J Allergy Clin Immunol 1991;88:649–660. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Weng T, Gou D, Chen Z, Chintagari NR, Liu L. Identification of rat lung-specific microRNAs by micoRNA microarray: valuable discoveries for the facilitation of lung research. BMC Genomics 2007;8:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattoli S, Mattoso VL, Soloperto M, Allegra L, Fasoli A. Cellular and biochemical characteristics of bronchoalveolar lavage fluid in symptomatic nonallergic asthma. J Allergy Clin Immunol 1991;87:794–802. [DOI] [PubMed] [Google Scholar]
- 36.Broide DH, Lotz M, Cuomo AJ, Coburn DA, Federman EC, Wasserman SI. Cytokines in symptomatic asthma airways. J Allergy Clin Immunol 1992;89:958–967. [DOI] [PubMed] [Google Scholar]
- 37.Tran T, McNeill KD, Gerthoffer WT, Unruh H, Halayko AJ. Endogenous laminin is required for human airway smooth muscle cell maturation. Respir Res 2006;7:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran T, Ens-Blackie K, Rector ES, Stelmack GL, McNeill KD, Tarone G, Gerthoffer WT, Unruh H, Halayko AJ. Laminin-binding integrin alpha7 is required for contractile phenotype expression by human airway myocytes. Am J Respir Cell Mol Biol 2007;37:668–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma P, Tran T, Stelmack GL, McNeill K, Gosens R, Mutawe MM, Unruh H, Gerthoffer WT, Halayko AJ. Expression of the dystrophin-glycoprotein complex is a marker for human airway smooth muscle phenotype maturation. Am J Physiol Lung Cell Mol Physiol 2008;294:L57–L68. [DOI] [PubMed] [Google Scholar]
- 40.Adam PJ, Regan CP, Hautmann MB, Owens GK. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22alpha in vivo. J Biol Chem 2000;275:37798–37806. [DOI] [PubMed] [Google Scholar]
- 41.Autieri MV. Kruppel-like factor 4: transcriptional regulator of proliferation, or inflammation, or differentiation, or all three? Circ Res 2008;102:1455–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem 2005;280:9719–9727. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.