Abstract
DNA-damaging agents usually produce a vast collection of lesions within the genome. Analysis of these lesions from the structural and biological viewpoints is often complicated by the reality that some of the lesions are chemically fragile, leading to an even larger set of secondary and tertiary products. In an effort to deconvolute complex DNA-damage spectra, a strategy is presented whereby an oligonucleotide containing a specific target for chemical reaction is allowed to react with a DNA-damaging agent. A large collection of HPLC-resolvable modified oligonucleotides is generated, and chromatographically distinct members of the set are then individually characterized using chemical, spectroscopic, biochemical, and genetic probes. The biological component of this “chemical–biological fingerprinting” tool is the use of polymerase bypass in vivo in cells having defined replication status and quantitative and qualitative patterns of lesion-directed mutagenesis, as key properties that complement physical analysis of modified DNA. This approach was applied to the complex product spectrum generated by peroxynitrite in the presence of CO2; peroxynitrite is a powerful oxidizing and nitrating agent generated as part of immune response. An oligonucleotide containing the primary oxidation product, 7,8-dihydro-8-oxoguanine (8-oxoGua), which is highly susceptible to further oxidation and/or nitration, was treated with peroxynitrite. Using mass spectrometry, coelution with authentic standards, sensitivity to piperidine, recognition and strand cleavage by the DNA repair enzyme MutM, and mutagenicity and genotoxicity in vivo, a matrix was created that defined the properties of the secondary DNA lesions formed when 3-morpholinosydnonimine (SIN-1) delivered a low, constant flux of peroxynitrite to an oligonucleotide containing 8-oxoGua. Two lesions were identified as the diastereomers of spiroiminodihydantoin (Sp), which had been observed previously in nucleoside-based experiments employing SIN-1. A third lesion, triazine, was tentatively identified. However, in addition to these lesions, a number of secondary lesions were generated that had chemical–biological fingerprints inconsistent with that of any known 8-oxoGua-derived lesion described to date. In vitro experiments showed that while some of these newly characterized secondary lesions were removed from DNA by MutM, others were in fact very poor substrates for this repair enzyme. These 8-oxoGua-derived lesions also showed varying degrees of sensitivity to piperidine. Furthermore, all of the secondary lesions observed in this work were potently mutagenic and genotoxic in Escherichia coli. Therefore, while 8-oxoGua itself is nontoxic and only mildly mutagenic in repair-proficient cells, peroxynitrite reveals the promutagenic potential and triggers the covert nature of this DNA lesion.
Introduction
Certain types of white blood cells produce reactive radicals as part of immune response. For instance, macrophages secrete •NO and O2•− to help the body defend against invading pathogens. These two reactive species combine in a diffusion-limited process to generate peroxynitrite (1), a powerful oxidizing and nitrating agent capable of reacting with many cellular components, including lipids, proteins, and DNA (2–9). In a state of chronic inflammation, peroxynitrite can be overproduced for prolonged periods of time. In fact, chronic inflammation has been linked to an increased risk of colon cancer in people with inflammatory bowel syndrome and also to an increased risk of gastric cancer in people infected with Helicobacter pylori (10).
With regard to DNA, guanine is the nucleobase most readily modified by peroxynitrite, with 5-guanidino-4-nitroimidazole (NI),1 8-nitroguanine (8-NO2-Gua), and 7,8-dihydro-8-oxoguanine (8-oxoGua) as the major products (5, 6, 11). NI is a chemically stable lesion, but 8-NO2-Gua is prone to depurination (12, 13). Furthermore, although 8-oxoGua does not readily depurinate, this lesion is chemically labile toward further oxidation and/or nitration, and several secondary lesions have been identified (Chart 1) (14, 15). These include guanidinohydantoin (Gh) and its isomer iminoallantoin (Ia) (16), the two diastereomers of spiroiminodihydantoin (Sp) (17, 18), oxaluric acid (Oa) (19, 20), urea (Ur) (21), N-nitrodehydroguanidinohydantoin (NO2-DGh) (22), dehydroguanidinohydantoin (DGh) (23, 24), 2,4,6-trioxo[1,3,5]triazinane-1-carboxamidine (CAC) (25), 4-hydroxy-2,5-dioxoimidazolidine-4-carboxylic acid (HICA) (26, 27), cyanuric acid (Ca) (25, 28), oxazolone (Oz) (29), imidazolone (Iz) (23), and 2-amino-4-hydroxy-s-triazine-6-carboxylic acid (triazine) (30, 31).
Chart 1.
Chemical Structures of DNA Lesions Discussed in This Work
The distribution of 8-oxoGua secondary products varies with both the flux (dose per unit time) of peroxynitrite and the nature of the 8-oxoGua substrate (i.e., whether it is present as the nucleoside monomer or within DNA). At the nucleoside level, addition of a large, instantaneous dose of peroxynitrite yielded qualitatively and quantitatively different products than did use of 3-morpholinosydnonimine (SIN-1) (18, 22, 26, 27, 32). SIN-1 decomposes slowly in solution to generate peroxynitrite at a continuous rate, and this method of delivery more closely approximates that of activated macrophages. To extend to the DNA level the aforementioned studies on monomers containing 8-oxoGua, Tretyakova and co-workers (20, 33) exposed oligonucleotides containing 8-oxoGua to an instantaneous dose of peroxynitrite, which models an acute bolus dose of oxidant, and identified the product lesions using mass spectrometry. Indeed, some of the 8-oxoGua-derived lesions identified in nucleoside experiments were also observed when the lesion was present within the context of DNA. However, novel products also emerged with the transition from nucleoside to oligonucleotide.
In the work presented here, we identified the chemical–biological fingerprints of the lesions formed when an oligonucleotide containing 8-oxoGua was exposed to a flux of peroxynitrite that modeled pathophysiological conditions associated with chronic inflammation, and we compared this work with the model studies using bolus administration described above. Our experimental strategy is outlined in Figure 1. After treatment with SIN-1, the oligonucleotide products were resolved by HPLC, characterized by mass spectrometry, probed for piperidine lability, and assayed for recognition and strand cleavage by the base-excision repair enzyme MutM. To complement the physical and biochemical characterizations of the oligonucleotide products, each product was incorporated into M13 bacteriophage genomes for analysis of lesion mutagenicity and genotoxicity in vivo.
Figure 1.
Overall experimental strategy. A single-stranded oligonucleotide containing 8-oxoGua was exposed to SIN-1, and the reaction products were fractionated by HPLC. The isolated oligonucleotide products were then either analyzed directly by mass spectrometry or gel electrophoresis or inserted into viral genomes for replication in E. coli.
As seen previously in the nucleoside models, we found that the flux of peroxynitrite modulated the 8-oxoGua-derived lesions formed within DNA. Additionally, when SIN-1 was used to deliver peroxynitrite to the system, some 8-oxoGua-derived products were retained in the transition from nucleoside to oligonucleotide, whereas others were not. Furthermore, novel 8-oxoGua-derived products were also observed within oligonucleotides. Importantly, in contrast to the mildy mutagenic and nontoxic 8-oxoGua from which they arose, all of the secondary lesions observed here were potently mutagenic and significantly genotoxic when replicated in Escherichia coli.
Experimental Procedures
Oligonucleotides
The single-stranded 16-mer sequence 5′-GAA GACCTXGGCGTCC-3′ (X = 8-oxoGua) was purchased from Midland Certified Reagent Co. In place of 8-oxoGua, the authentic standards contained either Gh, Sp1, Sp2 (the Sp diastereomers were named for their relative retention times on an anion-exchange HPLC column), Oa, Ur, or a THF abasic site. Oligonucleotides containing Gh (34), Sp1 (34), Sp2 (34), Oa (33), or Ur (35) were synthesized as previously described. Standard phosphoramidite chemistry was used to synthesize an oligonucleotide containing a THF abasic site. All oligonucleotides were purified by anion-exchange HPLC on a Dionex NucleoPac PA-100 column (4 × 250 mm) using 10% acetonitrile (solvent A) and 1.5 M ammonium acetate (pH 8) (solvent B) (1.0 mL/min; 10 to 50% B over 5 min, then 50 to 75% B over 25 min). The purified oligonucleotides were twice desalted with NAP-10 columns (Amersham). The molecular weight of all oligonucleotides was verified with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF-MS). DNA scaffolds (5′-GGTCTTCCACTGAATCATGGTCATAGC-3′ and 5′-AAAACGACGGCCAGTGAATTGGACGC-3′) were purchased from Integrated DNA Technologies.
Reaction of 8-oxoGua Oligonucleotide with SIN-1
The oligonucleotide containing 8-oxoGua was suspended in reaction buffer (10 mM sodium phosphate, 10 mM NaCl, and 25 mM NaHCO3,pH 7.5). SIN-1 (1.5 mM) was dissolved in reaction buffer immediately prior to use and added to the oligonucleotide solution to yield a final concentration of 75 μM oligonucleotide and 250 μM SIN-1 in a total volume of 100 μL. The reaction mixture was incubated for 5 h at 37 °C.
Isolation of Products by HPLC
The reaction mixture (100 μL) was immediately analyzed by HPLC on a Dionex NucleoPac PA-100 column using the gradient for oligonucleotide purification described above. The area under each peak was integrated and used to calculate the product distribution. Fractions were collected, and in some cases, multiple rounds of purification were used to remove traces of other products. Samples were reanalyzed by HPLC to ensure that only one peak was present in each sample. Prior to analysis by mass spectrometry, fractions (~0.5–1 mL each) were twice dialyzed against 2 mL of 30 mM ammonium acetate (pH 7) using YM-3 centricon filter tubes (3000 MWCO, Millipore). Following dialysis, samples were dried in vacuo, resuspended in water, and filtered using a C18-ZipTip (Millipore). Importantly, when reanalyzed by HPLC following this sample preparation, all the oligonucleotide products except the one labeled 4 in Figure 2 migrated with retention times identical to those observed in the initial analysis. This result revealed that 4 was not stable under the dialysis conditions and/or the drying procedure and therefore could not be analyzed further.
Figure 2.
HPLC chromatogram following the reaction of 8-oxoGua oligonucleotide (75 μM) with SIN-1 (250 μM) for 5 h at 37 °C in 10 mM sodium phosphate, 10 mM NaCl, and 25 mM NaHCO3 (pH 7.5) (solid line) overlaid with chromatograms (dashed lines) for (A) a standard representing the 8-oxoGua starting material and (B) oligo-nucleotide standards containing authentic 8-oxoGua-derived lesions.
Electrospray Ionization Mass Spectrometry
Electrospray ionization mass spectrometry (ESI-MS) was performed as described previously (36) on an Agilent 1100 MSD ESI-TOF, using approximately 20 pmol of each oligonucleotide. The mass spectrometer was operated in negative-ion mode.
Piperidine Lability and MutM Repair Studies
Oligonucleotides containing lesions were 5′-end labeled using T4 polynucleotide kinase (NEB) and γ-32P-ATP (PerkinElmer). DNA duplexes (0.5 μM) were formed by mixing equal concentrations of complementary strands in buffer (10 mM HEPES-KOH, 100 mM KCl, 10 mM EDTA, and 0.1 mg/mL BSA, pH 7.4) and heating to 70 °C followed by slow cooling to 20 °C. Analysis by MALDI-TOF-MS confirmed that the lesion-containing oligonucleotides were stable under these conditions. Duplex samples (10 μL) were treated with 10% (v/v) piperidine for 30 min at 90 °C or 5 units of MutM (Trevigen) for 1 h at 37 °C. All samples were then dried in vacuo, resuspended in denaturing formamide dye, and subjected to electrophoresis through a 20% denaturing polyacrylamide gel. The levels of strand cleavage were quantified using phosphorimagery (Imagequant).
Construction of Genomes Containing 8-oxoGua-Derived Lesions
Single-stranded M13 genomes containing a site-specific lesion were synthesized as previously described (37). Briefly, M13mp7L2 bacteriophage genome was linearized by EcoRI at a hairpin containing a unique restriction site. Two scaffolds, complementary to the 5′- or 3′-end of the 16-mer oligonucleotide insert and the linearized viral genome, were annealed to the linearized M13 DNA. The genome was recircularized by the addition of 5′-phosphorylated 16-mer insert and incubation with T4 DNA ligase. The 16-mer insert either contained 8-oxoGua or was an oligonucleotide product isolated from the reaction between the 8-oxoGua oligonucleotide and SIN-1. After ligation, the scaffolds were digested with T4 DNA polymerase. The genome constructs were phenol extracted, desalted with Sephadex G50 fine resin (Amersham), and stored at −20 °C. The stability of the lesion-containing DNAs was verified by MALDI-TOF-MS after the oligonucleotides were subjected to genome construction conditions.
Preparation of Electrocompetent Cells
LB medium (150 mL) was inoculated with 1 mL of saturated AB1157 E. coli and grown on a shaker at 37 °C to an optical density of 0.5 at 600 nm. The cells were pelleted, resuspended in 25 mL of 1 mM MgSO4, and transferred to a 150 × 15 mm Petri dish. When applicable, the SOS system was induced by irradiating the cells with 254 nm light (45 J/m2). The cells were then immediately transferred to 125 mL of 2×YT medium and grown for 40 min at 37 °C on a shaker. The cell cultures were twice pelleted and resuspended in 175 mL of water. After these two washes, the cells were once again pelleted and resuspended in 1.5 mL of 10% glycerol. These electrocompetent cells were stored at 4 °C and used within 24 h.
Analysis of the Mutational Spectrum
Electrocompetent cells (100 μL) and lesion-containing genome (5 μL) were mixed and chilled on ice for 45 min prior to transformation by electroporation (2.5 kV, 129 Ω, 2 mm gap cuvette). The number of independently transformed cells was 104–105 as determined by plating on a lawn of NR9050 E. coli (R. M. Schaaper, National Institute of Environmental Health Sciences). After electroporation, the sample was transferred to 10 mL of LB medium and incubated at room temperature for 30 min; the cultures were then grown for 4 h at 37 °C. Successful replication of the genome by E. coli generated progeny phage that were isolated by pelleting the cells and retaining the supernatant. To eliminate PCR amplification of nontransformed genomes, 100 μL of this supernatant, containing ~109 progeny phage, was added to 10 mL of LB medium and regrown in SCS110 E. coli (Stratagene). Single-stranded phage DNA was isolated using the QIAPrep Spin M13 kit (Qiagen), and the sequence area of interest was amplified by PCR as described previously (37). The REAP assay (as outlined in the Supporting Information) was then used to determine the identity of the base at the site originally occupied by the lesion from which the mutational profile is obtained (37).
Determination of Replication Efficiencies for 8-oxoGua-Derived Lesions
The CRAB assay (as outlined in the Supporting Information) was used to determine replication efficiencies for lesion-containing genomes (37). The procedure was similar to the REAP assay mentioned above, but used a primer set and PCR conditions different from those of the REAP assay and employed electroporations using a mixture of the lesion-containing genome and an internal standard. The internal standard contained no lesion and was three nucleotides larger than the lesion-containing genome. Following BbsI digestion of the PCR product, 5′-end labeling, and HaeIII restriction, the resultant 18-mer was quantified with respect to the internal standard (observed as a 21-mer). Replication efficiency was then determined by comparing this ratio to that obtained for a genome containing Gua.
Results
Separation of Oligonucleotide Products following Exposure of 8-oxoGua DNA to SIN-1
A single-stranded oligonucleotide containing a centrally located 8-oxoGua lesion was exposed to SIN-1 at an oligonucleotide/SIN-1 ratio of 1:3.3. After a defined period of reaction time, the oligonucleotide products were separated by HPLC, and individual peaks were collected. As shown in Figure 2A, the 8-oxoGua starting material was converted to no fewer than 11 products. After 5 h at 37 °C, half of the DNA starting material remained, and half had been converted to oligonucleotide products (Table 1). Eight products, those having the highest absorbance intensities at 260 nm, were isolated for further analysis and designated 1–8; each represented 2–13% of the overall composition. In some cases, multiple rounds of HPLC purification were used to resolve a single peak and remove traces of other products. All of the isolated products possessed a UV maximum at ~260 nm consistent with DNA; additionally, 3 had a second maximum centered at 338 nm (Figure 3). Using reaction times longer than 5 h did not change the overall yield or product distribution. Interestingly, under identical conditions, no reaction was observed in double-stranded DNA when the 8-oxoGua oligonucleotide was annealed to the complementary strand and C was placed opposite the lesion.
Table 1.
Composition (%) of the Mixture of Oligonucleotide Products
| Product | percent of mixture |
|---|---|
| 8-oxoGua oligonucleotidea | 51 |
| 1 | 2 |
| 2 | 2 |
| 3 | 7 |
| 4 | 10 |
| 5 | 3 |
| 6 | 13 |
| 7 | 6 |
| 8 | 4 |
5′-GAAGACCT(8-oxoGua)GGCGTCC-3′.
Figure 3.
UV-visible spectrum of 3.
Other doses of SIN-1 were utilized in addition to the 1:3.3 ratio described above. When equimolar amounts of 8-oxoGua oligonucleotide and SIN-1 were used, the same products were formed but in smaller amounts overall, as determined by HPLC. When the amount of SIN-1 was increased 10-fold to yield an oligonucleotide/SIN-1 ratio of 1:33.3, the 8-oxoGua starting material was nearly completely consumed, and although 7 and 8 were formed, their HPLC peaks were small compared to the one predominant peak observed (Supporting Information). This predominant peak had a retention time different from that of any of the products observed at lower SIN-1 doses. Furthermore, MALDI-TOF-MS analysis of this peak revealed five products having masses of [M + 16], [M + 56], [M + 73], [M + 85], and [M + 193] relative to the 8-oxoGua oligonucleotide. Because of the heterogeneity of this peak, subsequent studies focused on characterizing the products formed using a 1:3.3 ratio of oligonucleotide to SIN-1, where multiple stable products could be resolved and examined individually.
HPLC Analysis Using Authentic Standards
As a means of identifying 1–8, authentic standards containing known, previously identified lesions derived from 8-oxoGua were synthesized and analyzed by HPLC. Oligonucleotides containing either Gh, Sp1, Sp2, Oa, Ur, or a THF abasic site analogue in place of 8-oxoGua were examined for coelution with the reaction products (Figure 2B). Indeed, standards containing the Sp diastereomers Sp1 and Sp2 coeluted with 7 and 8, respectively. The Gh standard, which was present as a mixture of Gh and Ia isomers in solution (16), eluted as two peaks that appeared to coelute with two very minor reaction products that were formed in quantities (⪡1%) insufficient for further analysis. The standard containing Oa migrated only slightly slower than 8-oxoGua DNA under these chromatographic conditions, suggesting that a distinct peak representing Oa would not be observed because it would be obscured by the unreacted starting material. The standards containing Ur and the THF abasic site coelute, so for clarity only the Ur standard is shown in Figure 2B; these two standards had retention times very similar, yet not identical, to that of 4.
Isolated Oligonucleotide Products Characterized by Mass Spectrometry
Following dialysis to remove the salt used in anion-exchange HPLC, 1–8 were analyzed by ESI-MS (Table 2). Except for 3, which was shown by ESI-MS to represent multiple products, each peak isolated by HPLC was characterized by one mass and corresponded to a single oligonucleotide product. A mass of [M − 11] relative to the 8-oxoGua starting material was observed for both 1 and 2. Four products, with masses of [M − 10], [M + 16], [M + 72], and [M + 100], were obtained when 3 was analyzed by ESI-MS. During the course of sample preparation, 4 degraded and was not analyzed further. For 5 and 6, masses of [M − 9] and [M + 15], respectively, relative to the 8-oxoGua oligonucleotide were observed. Products 7 and 8, which coeluted with the Sp1 and Sp2 standards, respectively, both had masses of [M + 16] relative to the 8-oxoGua oligonucleotide.
Table 2.
ESI-MS Analysis of the Oligonucleotide Products
| Product | Ma | M relative to 8-oxoGua oligonucleotideb | observed ions (m/z) | charge state (z) |
|---|---|---|---|---|
| 8-oxoGua oligonucleotideb | 4920.88 | - | 1229.21 | −4 |
| 1639.28 | −3 | |||
| 1 | 4909.92 | [M−11] | 1226.47 | −4 |
| 1635.63 | −3 | |||
| 2 | 4909.92 | [M−11] | 1226.47 | −4 |
| 1635.62 | −3 | |||
| 3 | 4910.88 | [M−10] | 1226.71 | −4 |
| 4936.88 | [M + 16] | 1233.21 | −4 | |
| 4992.88 | [M + 72] | 1247.21 | −4 | |
| 5020.88 | [M + 100] | 1254.21 | −4 | |
| 5 | 4935.88 | [M−9] | 1226.96 | −4 |
| 6 | 4935.88 | [M + 15] | 1232.96 | −4 |
| 1644.27 | −3 | |||
| 7 | 4936.88 | [M + 16] | 1233.21 | −4 |
| 1644.61 | −3 | |||
| 8 | 4936.88 | [M + 16] | 1233.21 | −4 |
| 1644.61 | −3 |
Monoisotopic molecular weight calculated using the −4 charge state.
5′-GAAGACCT(8-oxoGua)GGCGTCC-3′.
Stability of Lesion-Containing Oligonucleotides toward Piperidine Treatment and Cleavage by MutM
In addition to characterization by mass spectrometry, the reaction products were probed for their sensitivities to piperidine and for the extents to which they are substrates for the base excision repair enzyme MutM (also known as formamidopyrimidine DNA glycosylase, or Fpg). Treatment with hot piperidine is commonly used to reveal DNA damage, and MutM is a glycosylase known to excise 8-oxoGua and some other lesions from DNA (38, 39). Double-stranded substrates were obtained by annealing the lesion-containing oligonucleotides to their complements, with either A, G, C, or T paired opposite the lesion.
After treatment with hot piperidine or MutM and analysis by PAGE (Supporting Information), percent strand cleavage at 8-oxoGua or the 8-oxoGua-derived lesion was quantified (Figure 4). Given its multicomponent nature, 3 was not studied. Importantly, in all cases, strand cleavage was observed only at 8-oxoGua or the 8-oxoGua-derived lesion. No strand breakage was seen at other sites within the oligonucleotide, nor were smaller fragments indicative of multiple cleavage sites observed. Furthermore, when the double-stranded substrate was heated to 90 °C for 30 min, no decomposition of the lesion within the substrate was observed by PAGE (Supporting Information).
Figure 4.
Percent strand cleavage of DNA containing 8-oxoGua, 1, 2, 5, 6, 7, or 8 following treatment with (A) MutM or (B) piperidine when the lesion is base-paired with either C (black), A (horizontal lines), G (gray), or T (diagonal lines).
Consistent with literature reports, the oligonucleotides containing 8-oxoGua were refractory to cleavage by piperidine (40). In contrast, the 8-oxoGua-derived products 1, 2, and 5 were sensitive to piperidine, showing 35–55% strand cleavage. Similar levels of strand cleavage were observed whether the lesion was paired with A or G, and slightly lower levels were observed when the lesion was paired with C or T. Oligonucleotides 6, 7, and 8 showed significantly less sensitivity to piperidine, with <10% of the substrate being cleaved in all cases.
Whereas 8-oxoGua was refractory to cleavage by piperidine, DNA containing this lesion was recognized by MutM (38). Enzymatic cleavage of the glycosidic bond followed by β- and δ-elimination generated a single-strand break, which was visualized by PAGE. When 8-oxoGua was paired with C, 90% strand cleavage was observed. However, when 8-oxoGua was paired with G or T, 38% or 70% strand cleavage was observed, respectively. The reduced level of activity when 8-oxoGua was paired with T or G as opposed to C is consistent with literature reports (41). Furthermore, as is also consistent with literature reports, no MutM activity was observed at 8-oxoGua when it was paired with A. In fact, the 8-oxoGua:A pair is the substrate for MutY, another glycosylase involved in minimizing the mutagenicity of 8-oxoGua in cells, which excises A misincorporated opposite 8-oxoGua (42).
In general, the trend of MutM having preferential activity on a lesion paired with C as opposed to the same lesion paired with T, G, or A was upheld for the 8-oxoGua-derived lesions studied here, except in the case of 8, for which the greatest amount of strand cleavage was observed when the lesion in 8 was paired with G. Interestingly, levels of MutM activity comparable to that for 8-oxoGua (~40%) were seen for the lesions in 7 and 8 paired with G. However, relative to that for the 8-oxoGua:C substrate, the amount of strand cleavage was significantly less for substrates containing the 8-oxoGua-derived lesions. Interestingly, although no activity was observed for the 8-oxoGua:A duplex, some of the 8-oxoGua-derived lesions were modest substrates for MutM when paired with A; 1–13% strand cleavage was observed for these duplexes.
Construction of Bacteriophage Genomes Containing 8-oxoGua-Derived Lesions
In order to explore the genetic properties of the 8-oxoGua-derived lesions replicated in vivo, the 16-mer reaction products were inserted into single-stranded M13 genomes. Genomes containing 1, 2, 3, 5, 6, 7, and 8 were assembled; the instability of 4 precluded further study of this product. When the lesion-containing genome was introduced into E. coli, the bacterial polymerases replicated it and produced viral progeny. Analysis of the DNA contained within this viral progeny using the REAP and CRAB methodologies developed previously in our laboratory (37) yielded information on both the mutagenicities and genotoxicities of the 8-oxoGua-derived lesions.
Efficiency of Lesion Replication in Wild-Type and SOS-Induced E. coli
The extent to which a lesion-containing bacteriophage genome is replicated within E. coli provides a measure of the genotoxicity of the lesion. Lesions that are poorly replicated are considered genotoxic, whereas readily replicated lesions are nontoxic. Relative to 8-oxoGua, which is efficiently replicated in wild-type and SOS-induced E. coli, the 8-oxoGua-derived lesions studied here were markedly genotoxic (Figure 5). In wild-type E. coli, genomes containing 1, 2, 3, 5, 6, 7, and 8 were replicated 4–19% as efficiently as the genome containing the 8-oxoGua oligonucleotide. Induction of the SOS response caused very little, if any, increase in replication efficiency for 1, 2, 3, 5, and 6. However, a dramatic increase in replication efficiency, from ~20% to ~60%, was observed for both 7 and 8 upon SOS induction.
Figure 5.
Lesion replication efficiencies, as compared with that of G, for 8-oxoGua and the 8-oxoGua-derived lesions in 1, 2, 3, 5, 6, 7, and 8 in wild-type (black) and SOS-induced (hatched) E. coli. Transformations were performed in triplicate, and error bars represent one standard deviation from the mean.
Mutational Profile of 8-oxoGua-Derived Lesions in Vivo
Mutation types and frequencies of the 8-oxoGua-derived lesions were determined and are shown in Figure 6. While the sensitivity of the REAP assay allows for observation of frameshift mutations that do not disturb the BbsI binding site, only point mutations were observed. Consistent with literature reports, 8-oxoGua was mildly mutagenic, yielding 5% G → T mutations when replicated in wild-type E. coli (43–45). Induction of the SOS response caused no change in the mutational profile. In contrast to 8-oxoGua, all of the 8-oxoGua-derived lesions were potently mutagenic, having mutation frequencies of >90% in both wild-type and SOS-induced E. coli. Very similar mutational profiles were observed for 1 and 2. Both yielded ~65% G → C, ~30% G → T, and ~5% G → A mutations in wild-type cells. Upon SOS induction, the distribution of mutations shifted such that G → C and G → T transversions were observed at 40% and 50%, respectively. In both wild-type and SOS-induced E. coli, the mixture of products represented by 3 gave ~30% G → C, ~55% G → T, and ~5% G → A mutations. Product 5 yielded 20% G → C, 39% G → T, and 28% G → A mutations in wild-type cells. A decrease in G → A transitions from 28% to 8% was observed upon SOS induction, along with a concomitant increase in both G → C and G → T transversions. Product 6 had a similar mutational profile in wild-type and SOS-induced cells: ~45% G → C, ~20% G → T, and ~25% G → A. A mixture of G → C and G → T mutations was observed for 7 and 8: 7 yielded 19% G → C and 80% G → T mutations, whereas 48% G → C and 49% G → T mutations were observed for 8. A small amount of G → A transitions (<2%) was also observed for 7 and 8.
Figure 6.
Mutation types and frequencies following replication of genomes containing the 8-oxoGua oligonucleotide, 1, 2, 3, 5, 6, 7, or 8 in wild-type (black) or SOS-induced (hatched) E. coli. Transformations were performed in triplicate, and error bars represent one standard deviation from the mean.
Discussion
Many previous studies have examined the reaction of peroxynitrite with 8-oxoGua. Indeed, in model studies using the O-acetyl-protected 8-oxoGua nucleoside, a multitude of secondary lesions have been identified and characterized (14, 15). Interestingly, the qualitative nature of the 8-oxoGua-derived lesions formed varied with the flux of peroxynitrite. When exposed to a large dose of peroxynitrite delivered to the system instantaneously, the 8-oxoGua nucleoside was converted primarily to DGh, NO2-DGh, and CAC (Chart 1) (23). Furthermore, DGh and NO2-DGh are hydrolytically unstable and can decompose to Oa and subsequently to Ur (23). In contrast, when peroxynitrite was delivered at a lower flux using SIN-1, the two Sp diasteromers, Gh, and HICA predominated (26, 27). These two classes of 8-oxoGua-derived products, seen with high and low fluxes of peroxynitrite, respectively, can be rationalized mechanistically (15). At high fluxes, peroxynitrite itself acts as the attacking nucleophile after the initial oxidation of 8-oxoGua, whereas at lower fluxes, water can also compete and act as a nucleophile.
A dependence on peroxynitrite flux was also observed in studies of the mutation profile in the supF gene of the pSP189 shuttle vector (46). Addition of a large, instantaneous dose of peroxynitrite caused the greatest amount of DNA damage; infusion of peroxynitrite using a syringe pump caused the least amount of damage, and exposure to SIN-1 generated an intermediate amount of DNA damage. Under all conditions, G → T transversions were predominant, but levels of G → C mutations were elevated following the instantaneous dose of peroxynitrite. The fact that the mutation distribution differed with the rate of peroxynitrite delivery raised the possibility that in vivo, the flux of peroxynitrite generated by cells of the immune system may modulate the 8-oxoGua-derived lesions formed. Intriguingly, although the pSP189 shuttle vector represents a double-stranded target and mutations were observed in the supF gene upon SIN-1 exposure, using mass spectrometry we saw no modification of a double-stranded oligonucleotide containing 8-oxoGua. However, the ratio of SIN-1 to base pairs was ~5-fold greater in the supF-targeted mutational studies, and additionally, the pSP189 vector does not intrinsically contain 8-oxoGua, thus complicating a direct comparison of our results with those of the supF studies.
In addition to studies using the 8-oxoGua nucleoside, experiments exploring the reactivity of this lesion within DNA have also been performed. Using single-stranded DNA containing 8-oxoGua and an instantaneous dose of peroxynitrite, Tretyakova and co-workers (33) identified 3a-hydroxy-5-imino-3,3a,4,5-tetrahydro-1H-imidazo[4,5d]imidazol-2-one, Ca, and CAC as the major products when peroxynitrite was present in a 5-fold excess. When this sample was left at room temperature overnight, Oa was also observed. In a second study, where peroxynitrite was also present in a 5-fold excess over the 8-oxoGua oligonucleotide, Oz and Oa were the major products, along with smaller amounts of Iz and Ca (20). The length of time between completion of the reaction and analysis of the reaction products, during which product hydrolysis and/or degradation can occur, may account for the different results in these two studies. When the dose of peroxynitrite was increased to 50-fold, Oa and Ca were the major products, with small amounts of Oz observed (20). Clearly, while some of the products identified using the 8-oxoGua nucleoside, namely, CAC and Oa, were also formed when 8-oxoGua was present within DNA, others were not. Additionally, when present within single-stranded DNA, 8-oxoGua was converted to Ca and Oz, which were not observed in nucleoside studies with peroxynitrite. It is noteworthy, however, that Ca has been identified as the major 1O2-mediated degradation product of the 8-oxoGua nucleoside (25, 28) and that Oz was observed following exposure of the 8-methoxyGua nucleoside to 1O2 (47).
Here we have carried out the first exploration of the lesions formed when SIN-1 is used to deliver a low flux of peroxynitrite to an oligonucleotide containing 8-oxoGua. At a concentration of 1 mM in bacterial culture media, SIN-1 decomposes to form peroxynitrite at a rate of 10 μM/min, as measured by the oxidation of dihydrorhodamine 123 (48, 49). We observed a comparable rate of peroxynitrite production (9 μM/min) in our phosphate buffer (data not shown). Since the chemistry of peroxynitrite is modulated by CO2 (50, 51), all experiments were conducted in the presence of 25 mM NaHCO3 to maintain the concentration of CO2 within the buffer at ~1 mM, which is comparable to physiological CO2 levels. Importantly, the molar excess of SIN-1 to 8-oxoGua employed here was the same as that utilized in nucleoside experiments.
Within the oligonucleotide used in these studies, 8-oxoGua represented the most chemically reactive site. Indeed, our PAGE experiments confirmed that the only piperidine- or MutM-sensitive site was 8-oxoGua or the 8-oxoGua-derived lesion, as strand cleavage was observed only at this particular location. Additionally, when loaded directly onto the gel without treatment all the lesion-containing DNAs migrated as a single band corresponding to a 16-mer, therefore indicating that no direct strand breaks occurred at the lesion or any other site within the oligonucleotide. The homogeneity of each oligonucleotide isolated by HPLC was further supported by ESI-MS data. Except for 3, each peak was characterized by a single molecular weight, indicating that one oligonucleotide, containing one particular lesion, was being analyzed. Furthermore, the lesion-containing oligonucleotides studied here were the final, stable products of the reaction between the 8-oxoGua DNA and SIN-1. No hydrolysis or decomposition was observed when the samples were incubated for an additional 24 h at 37 °C or for 30 min at 90 °C.
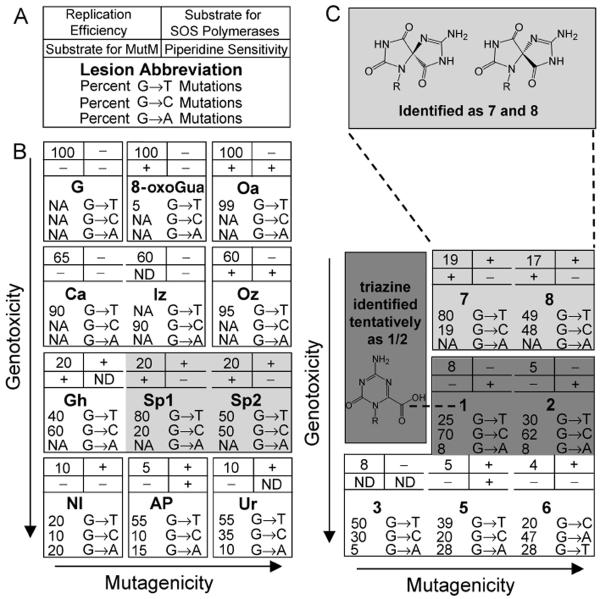
Interestingly, yet not surprisingly, the oligonucleotide products generated with SIN-1 differ from those seen with a larger flux of peroxynitrite. Furthermore, as seen when using a larger flux of peroxynitrite, the transition from nucleoside to oligonucleotide resulted in retention of some products but also the emergence of novel lesions. We characterized these 8-oxoGua-derived lesions using a combination of HPLC retention time, molecular weight, sensitivity to strand cleavage by piperidine, MutM activity, genotoxicity, and mutational profile. Collectively, these data were then compared to results from previous studies of 8-oxoGua-derived products, which provide an extensive array of lesion-specific biochemical and genetic properties for reference. In Figure 7, we present a synthesis of this collection of data, modeled after the periodic table. The genetic properties of peroxynitrite-induced lesions, following replication in E. coli, are used to group lesions on the basis of genotoxicity and mutagenicity, and a key is provided in Figure 7A. Data for previously identified peroxynitrite-induced lesions are summarized in Figure 7B. Certainly, one could imagine including additional lesion-specific information. Nonetheless, in much the same way as elements can be grouped and described on the basis of a collection of atomic properties, a unique signature or chemical–biological fingerprint emerges that is distinctive for each individual DNA lesion. In Figure 7C we include data for the 8-oxoGua-derived products induced by SIN-1 exposure that have been characterized in this work. All of the products observed in this work are potently mutagenic and toxic; if they were included in Figure 7B, they would be clustered at the bottom right of the figure.
Figure 7.
Chemical–biological fingerprints of G and the peroxynitrite-induced lesions. (A) Key. For an individual lesion, in the four boxes above the lesion abbreviation, genotoxicity is shown in the upper left, activity as a substrate for MutM in the lower left, activity as a substrate for SOS bypass polymerases in the upper right, and sensitivity to piperidine in the lower right. The profile of point mutations for the lesion is provided below the lesion abbreviation. Genotoxicity is represented by replication efficiency relative to G, and activity/sensitivity is indicated by + if the lesion is a substrate/is sensitive or by − if it is not. A lesion that is a substrate for MutM or is sensitive to piperidine is defined as one that yields ≥20% strand cleavage upon treatment when paired with C, A, G, or T. This particular percentage was selected because it defines two distinct groups of lesions in Figure 4. ND indicates that MutM activity and/or piperidine sensitivity of a lesion has not been explored. A substrate for the SOS bypass polymerases is defined as a lesion whose replication efficiency in SOS-induced cells is statistically higher than in noninduced cells. (B) Chemical–biological fingerprints of G, 8-oxoGua, and the previously identified peroxynitrite-induced lesions. Data are taken from the following references: 8-oxoGua (21, 35, 43, 60, 61); Oa (20, 21, 60, 62); Ca (20, 60, 63); Iz (64–67); Oz (33, 60); Gh (35, 52, 68); Sp1 (35, 61); Sp2 (35, 61); NI (64, 69); and Ur (21). The abbreviation AP represents an authentic abasic site (70). Although strand cleavage was observed following treatment of an AP-containing oligonucleotide with MutM, this cleavage does not require the glycosylase activity of the enzyme, and therefore, AP is not listed as a substrate for MutM. For genotoxicity and mutagenicity, the lesions 8-oxoGua, Iz, Gh, Sp1, Sp2, NI, and AP were present in a 5′-TXG-3′ (X = lesion) sequence context, as used in this work, whereas Oa, Ca, Oz, and Ur were present in a 5′-GXA-3′ sequence context. NA indicates a mutation frequency of <2%. Dividing lines are used to indicate a difference in genotoxicity or mutagenicity. If no line is present, the lesions are not distinguishable on the basis of that characteristic. For instance, Gh, Sp1, and Sp2 all are 100% mutagenic and therefore are not separated from each other. (C) Chemical–biological fingerprints of the 8-oxoGua-derived lesions characterized in this work.
By comparison to authentic standards, we have identified the lesions in 7 and 8 as having chemical–biological fingerprints that match those of the two diastereomers of Sp. The two reaction products, 7 and 8, coeluted with chemically synthesized oligonucleotide standards containing the Sp diastereomers. Furthermore, the mass increase of 16 Da was consistent with conversion of 8-oxoGua to Sp. Also, in accordance with previous work, 7 and 8 were substrates for MutM (52), and within this sequence context, the greatest amount of strand cleavage was observed when the lesion was paired with C or G. We have previously shown that although MutY provides protection from the miscoding potential of 8-oxoGua in vivo, it is unable to minimize the potent mutagenicity of Sp (35). Additionally, we report here the relative insensitivity of the Sp diastereomers to strand cleavage by piperidine. This organic base is known to cleave sufficiently weakened glycosydic bonds, and the lack of reactivity observed here can be added to the repertoire of properties that identify the Sp lesions. We have also demonstrated here the utility of using biological properties to identify a particular DNA lesion. Products 7 and 8 possess replication efficiencies and mutational profiles nearly identical to those observed previously for the Sp diastereomers present within a 5′-T(Sp)G-3′ sequence context when replicated in E. coli (35, 53). Recently, two groups (54, 55) have reported the absolute stereochemistry of the two Sp diastereomers; however, the conclusions of these reports differ, prohibiting us from confidently assigning the absolute stereochemistry of Sp1 and Sp2 in our system. Nonetheless, the two lesions, which were first identified in nucleoside studies, were also observed here in these DNA experiments. Furthermore, Sp has been observed in the genomic DNA of bacteria exposed to chromate, indicating the biological significance of 8-oxoGua oxidation (56).
In the case of Sp, use of authentic standards and comparison to literature reports allowed for positive identification of a lesion formed within DNA. These same methods can be used to exclude the formation of previously characterized 8-oxoGua-derived products. On the basis of HPLC retention times, besides the Sp1 and Sp2 standards, only the Gh/Ia standard appeared to coelute with any of the products observed when the 8-oxoGua oligonucleotide was exposed to SIN-1. However, if indeed Gh and Ia were generated, they were very minor products. The major products (1–6) did not coelute with any of the authentic standards utilized. Of course, many other lesions derived from 8-oxoGua have been identified in previous studies but were not used here as standards. In these cases, molecular weight can be a practical and useful identifier.
On the basis of mass, products 1 and 2 are consistent with triazine, which was identified previously as the oxidation product of the 8-oxoGua nucleobase exposed to K2S2O8 or KMnO4 (30) and also when 8-oxoGua, present as the nucleoside or within a single-stranded oligonucleotide, was treated with ammonium hydroxide (31). Furthermore, both 1 and 2 tested positive in a bromocresol green colorimetric indicator assay, which is specific for carboxylic acids; as expected, the products containing Sp1 and Sp2 tested negative. Resolution of this oligonucleotide product into two distinguishable peaks, 1 and 2, by HPLC may have been due to the existence of rotational isomers, which have been identified for derivatized triazines present in single-stranded DNA (57). However, in previous work, resolution was not observed when a reversed-phase C18 column (as opposed to the anion-exchange column used here) was employed; in that case, a single-stranded oligonucleotide containing triazine migrated as a single peak (57). Interestingly, in addition to having the same mass, 1 and 2 have nearly identical sensitivities to piperidine and, in fact, of all the oligonucleotides studied, display the highest sensitivity to piperidine. Additionally, 1 and 2 were equally modest substrates for MutM and showed similar replication efficiencies and mutational profiles in E. coli. When replicated in vivo, 1 and 2 also showed a change in mutational profile upon SOS induction, indicating that they are substrates for at least one of the SOS bypass polymerases.
In an effort to confirm the identities and obtain the exact masses of lesions 1 and 2, the oligonucleotide products containing these lesions were digested to nucleosides using nuclease P1 and shrimp alkaline phosphatase. Analysis using reversed-phase HPLC, however, revealed that the nucleoside products were not stable and/or eluted with the solvent front because of their high polarity. Both scenarios prohibited ESIMS analysis. In order to avoid the instability of 1 and 2 when they are present as nucleosides but still have products small enough to obtain exact masses, we next performed the experiment using a 3-mer containing 8-oxoGua. The addition of two other DNA bases was also expected to aid in increasing retention on the HPLC column. The 8-oxoGua was present within the same sequence context as in the 16-mer [5′-T(8-oxoGua)G-3′] and was allowed to react with the same molar ratio of SIN-1. The 3-mer starting material was indeed converted to multiple products, as revealed by LC–ESI-MS analysis; however, none of the products corresponded to an [M − 11] product. The observed products included 5′-TSpG-3′ and 5′-TUrG-3′. Therefore, we were unable to obtain exact masses of 1 and 2, and their assignment as triazine rotamers remains tentative.
The identification of 3 was complicated by the fact that, according to ESI-MS, this peak represented multiple products. Therefore, all data represented a compilation of characteristics from several lesions. Although two of the products had masses consistent with previously identified products, [M − 10] and [M + 16] corresponding to Gh and Sp, respectively, authentic standards containing these lesions did not coelute with 3. Taken together, the lesions represented by 3 were highly genotoxic, being replicated only 10% as well as 8-oxoGua and showing no statistically significant increase in replication efficiency upon induction of the SOS polymerases. When successfully replicated, these products were highly mutagenic, giving a mixture of mutations in which G → T > G → C > G → A. A particularly interesting and distinguishing feature of 3 is its UV absorbance at 338 nm. This lower-energy absorbance led us to suspect formation of a nitration product. When the 8-oxoGua nucleoside was exposed to peroxynitrite, the nitration products identified were NI, which has an absorbance at ~380 nm both as the nucleoside (58) and when present within DNA (59), and NO2-DGh, which absorbs only at 272 nm (22). We also considered the nitration of Gua, as opposed to 8-oxoGua, within the oligonucleotide. However, like NI, 8-NO2-Gua absorbs at a lower energy (~400 nm) (5) than 3. Furthermore, ESI-MS analysis of 3 did not reveal a product corresponding to NI, NO2-DGh, or 8-NO2-Gua. Therefore, one of the lesions represented by 3 may be a novel 8-oxoGua-derived nitration product.
Products 5 and 6 also contained novel 8-oxoGua-derived lesions. These lesions, having masses of [M − 9] and [M + 15], respectively, relative to 8-oxoGua, are not consistent with any previously identified 8-oxoGua-derived lesions. Of all the products observed here, 6 was present in the greatest amount, representing 13% of the overall yield. Similar to the other 8-oxoGua-derived products, the lesions in 5 and 6 were also highly genotoxic and potently mutagenic. Interestingly, in addition to causing both G → C and G → T mutations, the lesions in 5 and 6 caused the greatest amounts of G → A transitions (~30%) observed for any of the lesions studied here. Changes in the mutational profile with SOS induction indicate that 5 is a substrate for at least one of the bypass polymerases. Furthermore, a statistically significant increase in replication efficiency upon SOS induction indicated that 6 is also a substrate for at least one of the bypass polymerases.
On the basis of this work and previous nucleoside and DNA-based experiments, the ability of peroxynitrite to react with 8-oxoGua is apparent. However, we now also appreciate the complexity of this interaction. The lesions generated are highly dependent not only on the flux of peroxynitrite but also on the nature of the substrate. Here we have demonstrated that when SIN-1 is used to deliver peroxynitrite to an 8-oxoGua-containing single-stranded oligonucleotide, multiple 8-oxoGua-derived lesions are formed. While two of these 8-oxoGua-derived products, the Sp diastereomers, were also observed previously in nucleoside experiments, multiple novel products are seen when 8-oxoGua is present within DNA. Using a collection of spectroscopic, chemical, biochemical, and genetic methods, we have assigned the chemical–biological fingerprints of these lesions and demonstrated, by comparison to literature reports or authentic standards, that they are not consistent with any previously identified 8-oxoGua-derived lesion. Further studies, such as investigations utilizing NMR, would be useful for assigning the chemical structures of these lesions; in the present study, however, the quantities of products obtained were insufficient for such experiments. Nevertheless, we have shown that in contrast to 8-oxoGua, all of these 8-oxoGua-derived lesions are potently mutagenic, yielding G → T, G → C, and/or G → A mutations. Furthermore, these lesions are significantly genotoxic, serving as blocks to replication by E. coli DNA polymerases. Taken together, these results indicate that 8-oxoGua, which is itself <10% mutagenic in repair-proficient cells, represents a covertly miscoding nucleobase within DNA. Exposure to peroxynitrite activates its mutagenic potential by generating multiple DNA lesions that are >90% mutagenic in repair-proficient E. coli. Clearly, given the potent mutagenicities and genotoxicities of these lesions in a host that possesses all of its natural defenses, the cell has no efficient post-replicative pathway to deal with the mis-coding abilities of these lesions. Therefore, as we look to unravel the contributions of peroxynitrite overproduction to cancer and to understand spontaneous and induced mutagenesis, the covert nature of 8-oxoGua must be considered.
Supplementary Material
Acknowledgment
The authors extend their best wishes to Larry Marnett on the occasion of his 60th birthday and thank him for the leadership he has provided to this Journal and to our field over the years. We also thank Prof. Steven Tannenbaum for useful discussions and Dr. Peter Slade for insight and assistance with mass spectrometry. We acknowledge the NIH for funding (CA26731 and CA80024). S.D. is a Damon Runyon Postdoctoral Fellow supported by the Damon Runyon Cancer Research Foundation (DRG: 1848-04).
Footnotes
Abbreviations: NI, 5-guanidino-4-nitroimidazole; 8-NO2-Gua, 8-nitroguanine; 8-oxoGua, 7,8-dihydro-8-oxoguanine; Gh, guanidinohydantoin; Ia, iminoallantoin; Sp, spiroiminodihydantoin; Oa, oxaluric acid; Ur, urea; NO2-DGh, N-nitrodehydroguanidinohydantoin; DGh, dehydroguanidinohydantoin; CAC, 2,4,6-trioxo[1,3,5]triazinane-1-carboxamidine; HICA, 4-hydroxy-2,5-dioxoimidazolidine-4-carboxylic acid; Ca, cyanuric acid; Oz, oxazolone; Iz, imidazolone; triazine, 2-amino-4-hydroxy-s-triazine-6-carboxylic acid; SIN-1, 3-morpholinosydnonimine; MALDI-TOF-MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; ESI-MS, electrospray ionization mass spectrometry.
Supporting Information Available: HPLC chromatogram for the reaction between the 8-oxoGua oligonucleotide and SIN-1 (33.3-fold excess of SIN-1), PAGE results following piperidine and MutM treatment, and an outline of the CRAB and REAP assays used to determine replication efficiency and lesion mutagenicity, respectively. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Huie RE, Padmaja S. The reaction of NO with superoxide. Free Radical Res. Commun. 1993;18:195–199. doi: 10.3109/10715769309145868. [DOI] [PubMed] [Google Scholar]
- (2).Lymar SV, Jiang Q, Hurst JK. Mechanism of carbon dioxide-catalyzed oxidation of tyrosine by peroxynitrite. Biochemistry. 1996;35:7855–7861. doi: 10.1021/bi960331h. [DOI] [PubMed] [Google Scholar]
- (3).Salgo MG, Bermudez E, Squadrito GL, Pryor WA. DNA damage and oxidation of thiols peroxynitrite causes in rat thymocytes. Arch. Biochem. Biophys. 1995;322:500–505. doi: 10.1006/abbi.1995.1493. [DOI] [PubMed] [Google Scholar]
- (4).Salgo MG, Stone K, Squadrito GL, Battista JR, Pryor WA. Peroxynitrite causes DNA nicks in plasmid pBR322. Biochem. Biophys. Res. Commun. 1995;210:1025–1030. doi: 10.1006/bbrc.1995.1759. [DOI] [PubMed] [Google Scholar]
- (5).Yermilov V, Rubio J, Becchi M, Friesen MD, Pignatelli B, Ohshima H. Formation of 8-nitroguanine by the reaction of guanine with peroxynitrite in Vitro. Carcinogenesis. 1995;16:2045–2050. doi: 10.1093/carcin/16.9.2045. [DOI] [PubMed] [Google Scholar]
- (6).Kennedy LJ, Moore K, Jr., Caulfield JL, Tannenbaum SR, Dedon PC. Quantitation of 8-oxoguanine and strand breaks produced by four oxidizing agents. Chem. Res. Toxicol. 1997;10:386–392. doi: 10.1021/tx960102w. [DOI] [PubMed] [Google Scholar]
- (7).Uppu RM, Cueto R, Squadrito GL, Salgo MG, Pryor WA. Competitive reactions of peroxynitrite with 2′-deoxyguanosine and 7,8-dihydro-8-oxo-2′-deoxyguanosine (8-oxodG): Relevance to the formation of 8-oxodG in DNA exposed to peroxynitrite. Free Radical Biol. Med. 1996;21:407–411. doi: 10.1016/0891-5849(96)00220-1. [DOI] [PubMed] [Google Scholar]
- (8).Yermilov V, Yoshie Y, Rubio J, Ohshima H. Effects of carbon dioxide/bicarbonate on induction of DNA single-strand breaks and formation of 8-nitroguanine, 8-oxoguanine and base-propenal mediated by peroxynitrite. FEBS Lett. 1996;399:67–70. doi: 10.1016/s0014-5793(96)01288-4. [DOI] [PubMed] [Google Scholar]
- (9).Douki T, Cadet J. Peroxynitrite mediated oxidation of purine bases of nucleosides and isolated DNA. Free Radical Res. 1996;24:369–380. doi: 10.3109/10715769609088035. [DOI] [PubMed] [Google Scholar]
- (10).Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat. ReV. Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- (11).Joffe A, Mock S, Yun BH, Kolbanovskiy A, Geacintov NE, Shafirovich V. Oxidative generation of guanine radicals by carbonate radicals and their reactions with nitrogen dioxide to form site specific 5-guanidino-4-nitroimidazole lesions in oligodeoxynucleotides. Chem. Res. Toxicol. 2003;16:966–973. doi: 10.1021/tx025578w. [DOI] [PubMed] [Google Scholar]
- (12).Tretyakova NY, Burney S, Pamir B, Wishnok JS, Dedon PC, Wogan GN, Tannenbaum SR. Peroxynitrite-induced DNA damage in the supF gene: Correlation with the mutational spectrum. Mutat. Res. 2000;447:287–303. doi: 10.1016/s0027-5107(99)00221-3. [DOI] [PubMed] [Google Scholar]
- (13).Shafirovich V, Mock S, Kolbanovskiy A, Geacintov NE. Photochemically catalyzed generation of site-specific 8-nitroguanine adducts in DNA by the reaction of long-lived neutral guanine radicals with nitrogen dioxide. Chem. Res. Toxicol. 2002;15:591–597. doi: 10.1021/tx015593l. [DOI] [PubMed] [Google Scholar]
- (14).Neeley WL, Essigmann JM. Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- (15).Niles JC, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced oxidation and nitration products of guanine and 8-oxoguanine: Structures and mechanisms of product formation. Nitric Oxide. 2006;14:109–121. doi: 10.1016/j.niox.2005.11.001. [DOI] [PubMed] [Google Scholar]
- (16).Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of hydantoin products from one-electron oxidation of 8-oxo-7,8-dihydroguanosine in a nucleoside model. Chem. Res. Toxicol. 2001;14:927–938. doi: 10.1021/tx010072j. [DOI] [PubMed] [Google Scholar]
- (17).Luo W, Muller JG, Rachlin EM, Burrows CJ. Characterization of spiroiminodihydantoin as a product of one-electron oxidation of 8-oxo-7,8-dihydroguanosine. Org. Lett. 2000;2:613–616. doi: 10.1021/ol9913643. [DOI] [PubMed] [Google Scholar]
- (18).Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin is the major product of the 8-oxo-7,8-dihydroguanosine reaction with peroxynitrite in the presence of thiols and guanosine photooxidation by methylene blue. Org. Lett. 2001;3:963–966. [PubMed] [Google Scholar]
- (19).Duarte V, Gasparutto D, Yamaguchi LF, Ravanat JL, Martinez GR, Medeiros MHG, Di Mascio P, Cadet J. Oxaluric acid as the major product of singlet oxygen-mediated oxidation of 8-oxo-7,8-dihydroguanine in DNA. J. Am. Chem. Soc. 2000;122:12622–12628. [Google Scholar]
- (20).Tretyakova NY, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced secondary oxidative lesions at guanine nucleo-bases: Chemical stability and recognition by the Fpg DNA repair enzyme. Chem. Res. Toxicol. 2000;13:658–664. doi: 10.1021/tx000083x. [DOI] [PubMed] [Google Scholar]
- (21).Henderson PT, Neeley WL, Delaney JC, Gu F, Niles JC, Hah SS, Tannenbaum SR, Essigmann JM. Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations. Chem. Res. Toxicol. 2005;18:12–18. doi: 10.1021/tx049757k. [DOI] [PubMed] [Google Scholar]
- (22).Niles JC, Wishnok JS, Tannenbaum SR. A novel nitration product formed during the reaction of peroxynitrite with 2′,3′,5′-tri-O-acetyl-7,8-dihydro-8-oxoguanosine: N-nitro-N′-[1-(2,3,5-tri-O-acetyl-β-D-erythro-pentofuranosyl)-2,4-dioxoimidazolidin-5-ylidene]guanidine. Chem. Res. Toxicol. 2000;13:390–396. doi: 10.1021/tx0000318. [DOI] [PubMed] [Google Scholar]
- (23).Niles JC, Burney S, Singh SP, Wishnok JS, Tannenbaum SR. Peroxynitrite reaction products of 3′,5′-di-O-acetyl-8-oxo-7,8-dihydro-2′-deoxyguanosine. Proc. Natl. Acad. Sci. U.S.A. 1999;96:11729–11734. doi: 10.1073/pnas.96.21.11729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chworos A, Coppel Y, Dubey I, Pratviel G, Meunier B. Guanine oxidation: NMR characterization of a dehydroguanidinohydantoin residue generated by a 2e-oxidation of d(GpT) J. Am. Chem. Soc. 2001;123:5867–5877. doi: 10.1021/ja003945p. [DOI] [PubMed] [Google Scholar]
- (25).Raoul S, Cadet J. Photosensitized reaction of 8-oxo-7,8-dihydro-2′-deoxyguanosine: Identification of 1-(2-deoxy-β-D-erythro-pentofuranosyl)cyanuric acid as the major singlet oxygen oxidation product. J. Am. Chem. Soc. 1996;118:1892–1898. [Google Scholar]
- (26).Niles JC, Wishnok JS, Tannenbaum SR. Spiroiminodihydantoin and guanidinohydantoin are the dominant products of 8-oxoguanosine oxidation at low fluxes of peroxynitrite: Mechanistic studies with 18O. Chem. Res. Toxicol. 2004;17:1510–1519. doi: 10.1021/tx0400048. [DOI] [PubMed] [Google Scholar]
- (27).Niles JC, Wishnok JS, Tannenbaum SR. Mass spectrometric identification of 4-hydroxy-2,5-dioxo-imidazolidine-4-carboxylic acid during oxidation of 8-oxoguanosine by peroxynitrite and KHSO5/CoCl2. Chem. Res. Toxicol. 2004;17:1501–1509. doi: 10.1021/tx040003f. [DOI] [PubMed] [Google Scholar]
- (28).Sheu C, Foote CS. Photosensitized oxygenation of a 7,8-dihydro-8-oxoguanosine derivative: Formation of dioxetane and hydroperoxide intermediates. J. Am. Chem. Soc. 1995;117:4726. [Google Scholar]
- (29).Cadet J, Berger M, Buchko GW, Joshi PC, Raoul S, Ravanat JL. 2,2-Diamino-4-[(3,5-di-O-acetyl-2-deoxy-β-D-erythro-pentofuranosyl)amino]-5-(2H)-oxazolone: A novel and predominant radical oxidation product of 3′,5′-di-O-acetyl-2′-deoxyguanosine. J. Am. Chem. Soc. 1994;116:7403–7404. [Google Scholar]
- (30).Moschel RC, Behrman EJ. Oxidation of nucleic acid bases by potassium peroxodisulfate in alkaline aqueous solution. J. Org. Chem. 1974;39:1983–1989. doi: 10.1021/jo00928a001. [DOI] [PubMed] [Google Scholar]
- (31).Torres MC, Varaprasad CV, Johnson F, Iden CR. Formation of s-triazines during aerial oxidation of 8-oxo-7,8-dihydro-2′-deoxyguanosine in concentrated ammonia. Carcinogenesis. 1999;20:167–172. doi: 10.1093/carcin/20.1.167. [DOI] [PubMed] [Google Scholar]
- (32).Burney S, Niles JC, Dedon PC, Tannenbaum SR. DNA damage in deoxynucleosides and oligonucleotides treated with peroxynitrite. Chem. Res. Toxicol. 1999;12:513–520. doi: 10.1021/tx980254m. [DOI] [PubMed] [Google Scholar]
- (33).Tretyakova NY, Niles JC, Burney S, Wishnok JS, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligonucleotides containing 8-oxoguanine. Chem. Res. Toxicol. 1999;12:459–466. doi: 10.1021/tx980235c. [DOI] [PubMed] [Google Scholar]
- (34).Kornyushyna O, Berges AM, Muller JG, Burrows CJ. In vitro nucleotide misinsertion opposite the oxidized guanosine lesions spiroiminodihydantoin and guanidinohydantoin and DNA synthesis past the lesions using Escherichia coli DNA polymerase I (Klenow fragment) Biochemistry. 2002;41:15304–15314. doi: 10.1021/bi0264925. [DOI] [PubMed] [Google Scholar]
- (35).Delaney S, Neeley WL, Delaney JC, Essigmann JM. The substrate specificity of MutY for hyperoxidized guanine lesions in vivo. Biochemistry. 2007;46:1448–1455. doi: 10.1021/bi061174h. [DOI] [PubMed] [Google Scholar]
- (36).Delaney JC, Smeester L, Wong C, Frick LE, Taghizadeh K, Wishnok JS, Drennan CL, Samson LD, Essigmann JM. AlkB reverses etheno DNA lesions caused by lipid oxidation in vitro and in vivo. Nat. Struct. Mol. Biol. 2005;12:855–860. doi: 10.1038/nsmb996. [DOI] [PubMed] [Google Scholar]
- (37).Delaney JC, Essigmann JM. Assays for determining lesion bypass efficiency and mutagenicity of site-specific DNA lesions in vivo. Methods Enzymol. 2006;408:1–15. doi: 10.1016/S0076-6879(06)08001-3. [DOI] [PubMed] [Google Scholar]
- (38).Chung MH, Kasai H, Jones DS, Inoue H, Ishikawa H, Ohtsuka E, Nishimura S. An endonuclease activity of Escherichia coli that specifically removes 8-hydroxyguanine residues from DNA. Mutat. Res. 1991;254:1–12. doi: 10.1016/0921-8777(91)90035-n. [DOI] [PubMed] [Google Scholar]
- (39).Tchou J, Kasai H, Shibutani S, Chun MH, Laval J, Grollman AP, Nishimura S. 8-Oxoguanine (8-hydroxyguanine) DNA glycosylase and its substrate specificity. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4690–4694. doi: 10.1073/pnas.88.11.4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Cullis PM, Malone ME, Merson-Davies LA. Guanine radical cations are precursors of 7,8-dihydro-8-oxo-2′-deoxyguanosine but are not precursors of immediate strand breaks in DNA. J. Am. Chem. Soc. 1996;118:2775–2781. [Google Scholar]
- (41).Castaing B, Geiger A, Seliger H, Nehls P, Laval J, Zelwer C, Boiteux S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild-type and mutant Fpg proteins. Nucleic Acids Res. 1993;21:2899–2905. doi: 10.1093/nar/21.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Michaels ML, Tchou J, Grollman AP, Miller JH. A repair system for 8-oxo-7,8-dihydrodeoxyguanine. Biochemistry. 1992;31:10964–10968. doi: 10.1021/bi00160a004. [DOI] [PubMed] [Google Scholar]
- (43).Wood ML, Dizdaroglu M, Gajewski E, Essigmann JM. Mechanistic studies of ionizing radiation and oxidative mutagenesis: Genetic effects of a single 8-hydroxyguanine (7-hydro-8-oxoguanine) residue inserted at a unique site in a viral genome. Biochemistry. 1990;29:7024–7032. doi: 10.1021/bi00482a011. [DOI] [PubMed] [Google Scholar]
- (44).Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman AP. Site-specific mutagenesis using a gapped duplex vector: A study of translesion synthesis past 8-oxodeoxyguanosine in Escherichia coli. Mutat. Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- (45).Cheng KC, Cahill DS, Kasai H, Nishimura S, Loeb LA. 8-Hydroxyguanine, an abundant form of oxidative DNA damage, causes G to T and A to C substitutions. J. Biol. Chem. 1992;267:166–172. [PubMed] [Google Scholar]
- (46).Kim MY, Dong M, Dedon PC, Wogan GN. Effects of peroxynitrite dose and dose rate on DNA damage and mutation in the supF shuttle vector. Chem. Res. Toxicol. 2005;18:76–86. doi: 10.1021/tx049777m. [DOI] [PubMed] [Google Scholar]
- (47).Martinez GR, Gasparutto D, Ravanat JL, Cadet J, Medeiros MH, Di MP. Identification of the main oxidation products of 8-methoxy-2′-deoxyguanosine by singlet molecular oxygen. Free Radical Biol. Med. 2005;38:1491–1500. doi: 10.1016/j.freeradbiomed.2005.02.008. [DOI] [PubMed] [Google Scholar]
- (48).Brunelli L, Crow JP, Beckman JS. The comparative toxicity of nitric oxide and peroxynitrite to Escherichia coli. Arch. Biochem. Biophys. 1995;316:327–334. doi: 10.1006/abbi.1995.1044. [DOI] [PubMed] [Google Scholar]
- (49).Motohashi N, Saito Y. Induction of SOS response in Salmonella typhimurium TA4107/pSK1002 by peroxynitrite-generating agent, N-morpholinosydnonimine. Mutat. Res. 2002;502:11–18. doi: 10.1016/s0027-5107(02)00025-8. [DOI] [PubMed] [Google Scholar]
- (50).Lymar SV, Hurst JK. Rapid reaction between peroxonitrite ion and carbon dioxide: Implications for biological activity. J. Am. Chem. Soc. 1995;117:8867–8868. [Google Scholar]
- (51).Denicola A, Freeman BA, Trujillo M, Radi R. Peroxynitrite reaction with carbon dioxide/bicarbonate: Kinetics and influence on peroxynitrite-mediated oxidations. Arch. Biochem. Biophys. 1996;333:49–58. doi: 10.1006/abbi.1996.0363. [DOI] [PubMed] [Google Scholar]
- (52).Leipold MD, Muller JG, Burrows CJ, David SS. Removal of hydantoin products of 8-oxoguanine oxidation by the Escherichia coli DNA repair enzyme, FPG. Biochemistry. 2000;39:14984–14992. doi: 10.1021/bi0017982. [DOI] [PubMed] [Google Scholar]
- (53).Neeley WL, Delaney S, Alekseyev YO, Jarosz DF, Delaney JC, Walker GC, Essigmann JM. DNA polymerase v allows bypass of toxic guanine oxidation products in vivo. J. Biol. Chem. 2007;282:12741–12748. doi: 10.1074/jbc.M700575200. [DOI] [PubMed] [Google Scholar]
- (54).Durandin A, Jia L, Crean C, Kolbanovskiy A, Ding S, Shafirovich V, Broyde S, Geacintov NE. Assignment of absolute configurations of the enantiomeric spiroiminodihydantoin nucleobases by experimental and computational optical rotatory dispersion methods. Chem. Res. Toxicol. 2006;19:908–913. doi: 10.1021/tx060078e. [DOI] [PubMed] [Google Scholar]
- (55).Karwowski B, Dupeyrat F, Bardet M, Ravanat JL, Krajewski P, Cadet J. Nuclear magnetic resonance studies of the 4R and 4S diastereomers of spiroiminodihydantoin 2′-deoxyribonucleosides: Absolute configuration and conformational features. Chem. Res. Toxicol. 2006;19:1357–1365. doi: 10.1021/tx060088f. [DOI] [PubMed] [Google Scholar]
- (56).Hailer MK, Slade PG, Martin BD, Sugden KD. Nei deficient Escherichia coli are sensitive to chromate and accumulate the oxidized guanine lesion spiroiminodihydantoin. Chem. Res. Toxicol. 2005;18:1378–1383. doi: 10.1021/tx0501379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Diaz-Ortiz A, Elguero J, Foces-Foces C, de la Hoz A, Moreno A, Moreno S, Sanchez-Migallon A, Valiente G. Synthesis, structural determination and dynamic behavior of 2-chloro-4,6-bis(pyrazolylamino)-1,3,5-triazines. Org. Biomol. Chem. 2003;1:4451–4457. doi: 10.1039/b310693d. [DOI] [PubMed] [Google Scholar]
- (58).Niles JC, Wishnok JS, Tannenbaum SR. A novel nitroimidazole compound formed during the reaction of peroxynitrite with 2′,3′,5′-tri-O-acetyl-guanosine. J. Am. Chem. Soc. 2001;123:12147–12151. doi: 10.1021/ja004296k. [DOI] [PubMed] [Google Scholar]
- (59).Neeley WL, Henderson PT, Essigmann JM. Efficient synthesis of DNA containing the guanine oxidation-nitration product 5-guanidino-4-nitroimidazole: Generation by a postsynthetic substitution reaction. Org. Lett. 2004;6:245–248. doi: 10.1021/ol036188j. [DOI] [PubMed] [Google Scholar]
- (60).Henderson PT, Delaney JC, Gu F, Tannenbaum SR, Essigmann JM. Oxidation of 7,8-dihydro-8-oxoguanine affords lesions that are potent sources of replication errors in vivo. Biochemistry. 2002;41:914–921. doi: 10.1021/bi0156355. [DOI] [PubMed] [Google Scholar]
- (61).Henderson PT, Delaney JC, Muller JG, Neeley WL, Tannenbaum SR, Burrows CJ, Essigmann JM. The hydantoin lesions formed from oxidation of 7,8-dihydro-8-oxoguanine are potent sources of replication errors in vivo. Biochemistry. 2003;42:9257–9262. doi: 10.1021/bi0347252. [DOI] [PubMed] [Google Scholar]
- (62).Duarte V, Gasparutto D, Jaquinod M, Ravanat J, Cadet J. Repair and mutagenic potential of oxaluric acid, a major product of singlet oxygen-mediated oxidation of 8-oxo-7,8-dihydroguanine. Chem. Res. Toxicol. 2001;14:46–53. doi: 10.1021/tx0001629. [DOI] [PubMed] [Google Scholar]
- (63).Gasparutto D, Da CS, Bourdat AG, Jaquinod M, Cadet J. Synthesis and biochemical properties of cyanuric acid nucleoside-containing DNA oligomers. Chem. Res. Toxicol. 1999;12:630–638. doi: 10.1021/tx980255e. [DOI] [PubMed] [Google Scholar]
- (64).Neeley WL, Delaney JC, Henderson PT, Essigmann JM. In ViVo bypass efficiencies and mutational signatures of the guanine oxidation products 2-aminoimidazolone and 5-guanidino-4-nitroimidazole. J. Biol. Chem. 2004;279:43568–43573. doi: 10.1074/jbc.M407117200. [DOI] [PubMed] [Google Scholar]
- (65).Buchko GW, Cadet J, Morin B, Weinfeld M. Photooxidation of d(TpG) by riboflavin and methylene blue. Isolation and characterization of thymidylyl-(3′,5′)-2-amino-5-[(2-deoxy-β-D-erythro-pentofuranosyl)amino]-4H-imidazol-4-one and its primary decomposition product thymidylyl-(3′,5′)-2,2-diamino-4-[(2-deoxy-β-D-erythro-pentofuranosyl)amino]-5(2H)-oxazolone. Nucleic Acids Res. 1995;23:3954–3961. doi: 10.1093/nar/23.19.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Kino K, Saito I, Sugiyama H. Product analysis of GG-specific photooxidation of DNA via electron transfer: 2-aminoimidazolone as a major guanine oxidation product. J. Am. Chem. Soc. 1998;120:7373–7374. [Google Scholar]
- (67).Kino K, Sugiyama H. Possible cause of G-C to C-G transversion mutation by guanine oxidation product, imidazolone. Chem. Biol. 2001;8:369–378. doi: 10.1016/s1074-5521(01)00019-9. [DOI] [PubMed] [Google Scholar]
- (68).Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Gu F, Stillwell WG, Wishnok JS, Shallop AJ, Jones RA, Tannenbaum SR. Peroxynitrite-induced reactions of synthetic oligo 2′-deoxynucleotides and DNA containing guanine: Formation and stability of a 5-guanidino-4-nitroimidazole lesion. Biochemistry. 2002;41:7508–7518. doi: 10.1021/bi020148q. [DOI] [PubMed] [Google Scholar]
- (70).Kroeger KM, Goodman MF, Greenberg MM. A comprehensive comparison of DNA replication past 2-deoxyribose and its tetrahydrofuran analog in Escherichia coli. Nucleic Acids Res. 2004;32:5480–5485. doi: 10.1093/nar/gkh873. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.