Abstract
Redox regulation of stress proteins, such as molecular chaperones, guarantees an immediate response to oxidative stress conditions. This review focuses on the two major classes of redox-regulated chaperones, Hsp33 in bacteria and typical 2-Cys peroxiredoxins in eukaryotes. Both proteins employ redox-sensitive cysteines, whose oxidation status directly controls their affinity for unfolding proteins and therefore their chaperone function. We will first discuss Hsp33, whose oxidative stress-induced disulfide bond formation triggers the partial unfolding of the chaperone, which, in turn, leads to the exposure of a high-affinity binding site for unfolded proteins. This rapid mode of activation makes Hsp33 essential for protecting bacteria against severe oxidative stress conditions, such as hypochlorite (i.e., bleach) treatment, which leads to widespread protein unfolding and aggregation. We will compare Hsp33 to the highly abundant eukaryotic typical 2-Cys peroxiredoxin, whose oxidative stress-induced sulfinic acid formation turns the peroxidase into a molecular chaperone in vitro and presumably in vivo. These examples illustrate how proteins use reversible cysteine modifications to rapidly adjust to oxidative stress conditions and demonstrate that redox regulation plays a vital role in protecting organisms against reactive oxygen species-mediated cell death.
Reactive oxygen species (ROS)1 such as superoxide radicals (O2•−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH•−) arise during normal aerobic metabolism. They are constantly generated during electron transfer in the respiratory chain, in peroxisomes, and as products of a growing list of NADPH oxidases (1). Cells cope with ROS by constitutively expressing an arsenal of detoxifying enzymes and proteins involved in maintaining the redox homeostasis of the cell. Superoxide dismutases, peroxiredoxins, and catalases, on the one hand, directly destroy ROS (2). The thioredoxin and glutaredoxin systems, on the other hand, work indirectly by preserving a highly reducing environment in the cytosol using NADPH as the ultimate electron source (3). Cells have thus evolved a fine-tuned system that balances pro-and antioxidants and allows for the presence of low levels of ROS that appear to play important roles in a number of different signaling pathways (3, 4). Once ROS exceed the cell’s antioxidant capacity, however, organisms begin to suffer from a condition that is generally termed oxidative stress.
Oxidative stress can be endogenously caused by defects in energy metabolism or the failure of specific antioxidant systems (5). Alternatively, oxidative stress can be exogenously caused by a variety of sources (i.e., ultraviolet light) or, as illustrated by the innate immune response, when one type of cell seeks to destroy others. Cellular accumulation of ROS leads to oxidative modifications of all macromolecules, including DNA, lipids, and proteins (6). In the case of proteins, the most oxidation-sensitive targets are the sulfur-containing cysteine and methionine residues, which undergo typically reversible modifications in vivo (see below) (7). Most of the irreversible posttranslational protein modifications are additions of reactive carbonyl groups to the side chains of lysine, arginine, and proline residues, a process termed protein carbonylation (8). Oxidative modifications can alter the activity of proteins and can increase their susceptibility to aggregation and degradation (9). This oxidative damage to proteins has often been implicated as one of the leading causes of a variety of pathologies, including neurodegenerative diseases (10).
CYSTEINES AS THE CENTRAL BUILDING BLOCKS OF REDOX SWITCHES IN PROTEINS
To counteract oxidative stress, organisms have developed rapid response systems on both the transcriptional and posttranslational levels; these systems are designed to directly destroy ROS, repair the oxidative damage, and restore the redox homeostasis of the cell. Many of the central players employed in the pro- and eukaryotic oxidative stress defense are proteins with redox-sensitive amino acid side chains (e.g., Cys, Met, and His) or metal centers whose oxidation status is directly tied to the protein’s conformation and ultimately its function (11, 12).
The cysteine residue is by far the most commonly used amino acid in redox-sensing proteins. The reactivity of individual thiol groups is directly determined by the pKa value of their side chain, which is largely controlled by the local amino acid environment within the protein. Typically, the pKa of cysteine thiols is ~8.3, which renders the thiol group protonated (R-SH) and nonreactive at physiological pH (13). In contrast, redox-sensitive thiol groups have significantly lower pKa values (e.g., Gpx3 pKa ~ 5.1) (14) and are therefore present in their deprotonated, thiolate anion (R-S−) state at pH 7.4. Thiolate anions are often stabilized by charge–charge interactions with nearby positively charged amino acids or by aromatic amino acids. The proximity and conservation of these amino acids serve as a good first indication that the thiol group might be redox-sensitive and the protein potentially redox-regulated (3).
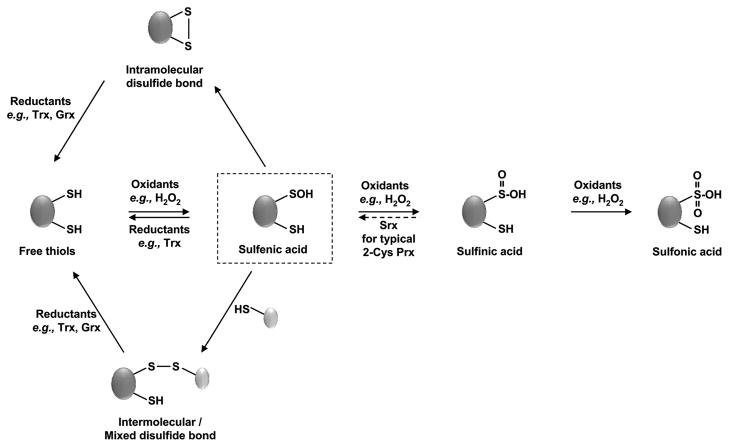
Thiolate anions, which are far more nucleophilic than their protonated counterparts, can undergo a variety of reversible and irreversible oxidation reactions (Figure 1), the nature of which depends largely on the accessibility of the thiol group, the surrounding amino acids, and the type and concentration of oxidant present (15). Reversible thiol oxidation reactions reported to play a role in the redox regulation of proteins include the formation of stable sulfenic acid intermediates (R-SOH) [e.g., GapDH (16)], intra- or intermolecular disulfide bonds (R-S-S-R) [e.g., OxyR, Yap1p, and Hsp33 (17, 18)], mixed disulfides with the small tripeptide glutathione (R-S-S-G) [e.g., α-ketoglutarate dehydrogenase and carbonic anhydrase III (19)], and the overoxidation of cysteine thiols to sulfinic acid (R-SO2H) [e.g., 2-Cys Prx (20)]. Although higher oxidation states, such as sulfonic acid (R-SO3H) or disulfide monoxides (R-S-SO-R), have been reported, these thiol modifications appear to be irreversible in vivo and most likely lead to the degradation of the affected proteins (for review, see ref 21) (Figure 1).
Figure 1.
Oxidative thiol modifications. Exposure of redox-sensitive cysteine residues to oxidants such as H2O2 leads to reversible sulfenic acid formation. Sulfenic acids readily react with nearby thiols of the same protein to form intramolecular disulfide bonds. They can also react with thiols of other proteins or the small tripeptide glutathione to form intermolecular or mixed disulfide bonds. Reduced cysteines can be directly oxidized to disulfide bonds by disulfide exchange reactions with oxidized glutathione (GSSG). These oxidative thiol modifications are reduced by members of the glutaredoxin (Grx) or thioredoxin (Trx) system, which draw their reducing power from cellular NADPH. In the presence of high levels of H2O2, overoxidation to sulfinic acid or sulfonic acid can occur. While sulfiredoxins (Srx) specifically reduce sulfinic acids in 2-Cys peroxiredoxins, no general sulfinic or sulfinic reductases have been identified to date.
Sulfenic Acid Formation (R-SOH)
Reaction of thiolate anions with H2O2, peroxynitrite, or hypochlorite leads to the formation of sulfenic acid, a highly reactive and unstable oxidation product (21). Inmost proteins, cysteine suelfenic acids either rapidly react with other proximal thiol groups to form intramolecular, intermolecular, or mixed disulfide bonds with glutathione or undergo further oxidation to sulfinic or sulfonic acids (Figure 1). So far, few proteins (e.g., NADH peroxidases) have been identified that utilize stabilized sulfenic acids for their catalytic activity (21). Although sulfenic acid formation has been suggested to play a role in the redox regulation of several prokaryotic transcription factors (e.g., OxyR) (22), it remains to be determined whether this product has functional relevance or is simply an oxidation intermediate on the path to disulfide bond formation (23). Progress in answering these questions has recently been facilitated with the development of sulfenic acid-specific detection methods. One example is the dimedone derivative DAz-1, a cell permeable probe that is chemically selective for sulfenic acids. This probe allows one to trap and label sulfenic acids and to identify the sites of oxidative modification (24).
S-Glutathionylation (R-S-S-G)
Several different mechanisms appear to lead to the basal levels of mixed disulfide bonds with glutathione (i.e., S-glutathionylation) that are found under nonstress conditions in vivo (for a review, see ref 25). Because S-glutathionylated proteins accumulate significantly in oxidatively challenged cells, this reversible thiol modification is considered a hallmark of cellular oxidative stress (26). To visualize protein S-glutathionylation in intact cells, a number of different methods have been developed (27). The incorporation of labeled glutathione in pulse–chase experiments with cells challenged with oxidative stress identified several key proteins susceptible to S-glutathionylation (28). Additionally, in situ labeling of mixed disulfides with glutathione has been achieved using a glutaredoxin- 1-dependent trapping technique (29).
Disulfide Bond Formation (R-S-S-R)
Numerous redox-regulated proteins have been shown to use reversible disulfide bond formation as the mechanism of their specific, oxidative stress-mediated activation. Proteins that are activated by disulfide bonds include, among many others, peroxide stress transcription factor OxyR of Escherichia coli, oxidative stress transcription factor Yap1p of Saccharomyces cerevisiae, molecular chaperone Hsp33, and apoptosis factor p66 (Shc) (30–33). Many of these proteins have been shown to play an important role in the oxidative stress defense of the affected organisms. Disulfide-bonded proteins can be detected by “diagonal” two-dimensional gel electrophoresis, in which proteins are first separated under nonreducing conditions followed by separation under reducing conditions (34, 35). While proteins without disulfide bonds will migrate in a diagonal line in the second dimension, proteins that originally contained disulfide bonds will appear as spots above or below this diagonal.
REVERSIBILITY, A CRUCIAL ASPECT OF REDOX REGULATION
A very crucial aspect of all regulatory mechanisms is reversibility. Organisms harbor a number of oxidoreductase systems dedicated to reducing sulfenic acids as well as mixed, intramolecular, and intermolecular disulfide bonds (36). These redox-balancing systems are constitutively expressed to maintain redox homeostasis during nonstress conditions and are an integral part of the oxidative stress response. Oxidative stress-induced over-expression of these oxidoreductases not only helps to quickly restore the cellular redox homeostasis but often functions as a direct feedback mechanism to inactivate redox-regulated stress transcription factors (e.g., OxyR and Yap1p) and shut down the oxidative stress response (18). The two most prominent oxidoreductase systems present in most pro- and eukaryotic species are the glutaredoxin and thioredoxin systems. Both glutaredoxin and thioredoxin make use of their highly reactive Cys-X-Y-Cys motif embedded in a well-conserved thioredoxin fold to undergo direct thiol–disulfide exchange reactions with oxidized protein thiols (36). Regeneration of the small oxidoreductases, which become oxidized in this process, involves use of the small tripeptide glutathione and the NADPH-dependent glutathione reductase (for glutaredoxins), or the direct action of the NADPH-dependent thioredoxin reductase (for thioredoxins).
In addition to being an integral part of regulatory mechanisms, the reversible nature of oxidative thiol modifications also imparts a very important technical advantage. Rapid trapping of reduced cysteines with alkylating agents (e.g., iodoacetamide-IAM, N-ethylmaleimide-NEM, and [12C]ICAT) followed by the reduction of oxidized thiols and their trapping with a modified version of the respective alkylating reagent (e.g., [14C]IAM, [14C]NEM, and [13C]ICAT) provide excellent tools for visualizing oxidative thiol modifications by two-dimensional (2D) gels (35, 37) or for precisely quantifying the extent of oxidative thiol modifications by mass spectrometry (i.e., OxICAT) (15).
Reversible thiol modifications appear to be the mechanism of choice for redox-regulated proteins; for many years, it has also been suggested that these modifications serve an important protective role by limiting the formation of higher, potentially irreversible thiol oxidation states. Although a family of ATP-dependent enzymes (e.g., sulfiredoxin) that are able to reverse higher thiol oxidation states have recently been identified, these proteins appear to be dedicated to resolving sulfinic acid formation in 2-Cys peroxiredoxins only (38). So far, no other enzyme(s) or mechanisms that either generally reverse sulfinic acids or reduce sulfonic acid formation in proteins have been described. However, before we can clearly rule out the possibility that these oxidative modifications are indeed irreversible in vivo, more specific detection methods need to be developed that will allow us to precisely monitor the fate of proteins with overoxidized thiol groups in vivo.
REDOX-REGULATED CHAPERONES
Molecular chaperones constitute a class of highly conserved proteins that assist other polypeptides in acquiring or retaining their functional conformation (for recent reviews, see refs 39–41). The induction of molecular chaperones is an essential part of the universally conserved heat shock response, which allows organisms to survive stress conditions that cause protein unfolding (e.g., elevated temperatures) (42). The peril of accumulating protein folding intermediates lies with their tendency to form large protein aggregates. Most known molecular chaperones use hydrophobic substrate interaction sites to bind and sequester these protein folding intermediates, thereby reducing the propensity of protein aggregation and promoting cell survival (40).
Molecular chaperones are often divided into two main mechanistic groups, chaperone foldases (e.g., Hsp70 and Hsp60) and chaperone holdases (e.g., small heat shock proteins, Hsp33, and HdeA). Chaperone foldases use cycles of ATP binding and hydrolysis to specifically regulate their affinity for unfolding proteins. They support de novo folding of proteins under nonstress conditions, prevent protein aggregation during stress conditions, and promote protein refolding upon recovery from stress (43). In contrast, chaperone holdases are not dependent on ATP. Once activated, chaperone holdases provide high-affinity binding platforms for unfolded proteins and prevent protein aggregation specifically during stress conditions. While the ATP independence of this group of chaperones makes them uniquely suited to prevent protein aggregation in subcellular compartments that lack ATP (e.g., E. coli periplasm) or during stress conditions that decrease cellular ATP levels (e.g., oxidative stress) (44), this mode of action raises several important mechanistic questions. How are substrate binding and release regulated? Moreover, how can the high-affinity binding to unfolding proteins be specifically harnessed during stress conditions and avoided under nonstress conditions where de novo protein folding might be affected? In this review, we focus on two chaperone holdases, prokaryotic Hsp33 and eukaryotic 2-Cys peroxiredoxin, which use reactive oxygen species to activate their chaperone function. Both proteins utilize highly sensitive cysteine residues as regulatory nanoswitches, whose oxidation status directly determines the chaperone’s affinity for unfolded substrate proteins and therefore its ability to act as a first line of defense during oxidative stress conditions that cause protein unfolding.
HSP33, A CHAPERONE SPECIALIZED TO PROTECT AGAINST PROTEIN UNFOLDING OXIDANTS
The redox-regulated chaperone Hsp33 is a well-conserved protein with homologues identified in the vast majority of prokaryotic species as well as in some unicellular eukaryotic parasites (e.g., Trypanosomatidae) (32). Hsp33 functions as a highly specialized chaperone holdase, which protects bacteria against oxidative stress conditions that lead to protein unfolding and aggregation (44, 45). To specifically sense conditions that threaten the organism and respond to them with the activation of its chaperone function, Hsp33 combines a number of regulatory features that make it probably one of the most uniquely regulated chaperones known to date (17).
Hsp33 is tightly regulated on both transcriptional and posttranslational levels. The Hsp33 hslO gene was first identified in E. coli as an inducible heat shock gene under σ32 control (46). It is constitutively expressed under nonstress conditions (steady state concentration of ~1.5 μM), and exposure of bacteria to conditions that trigger the heat shock response (i.e., protein unfolding conditions and solvent stress) leads to massive upregulation of Hsp33’s transcription (46, 47).While this transcriptional regulation controls the cellular amount of Hsp33, posttranslational redox regulation is critical for controlling its activity.
Early on, it became apparent that Hsp33’s in vitro activation as a molecular chaperone requires the simultaneous presence of oxidants, such as H2O2, and elevated temperatures (>40 °C). Neither treatment with peroxide alone nor incubation at heat shock temperatures caused any substantial activation of Hsp33 (32, 48). In vivo studies later confirmed these requirements and provided an attractive physiological explanation of the need for Hsp33’s sophisticated posttranslational regulation (44). Under pure heat shock conditions, bacteria utilize a network of ATP dependent and -independent chaperone machineries (e.g., DnaK and sHsps), which prevent protein aggregation and increase the bacteria’s resistance to stress (44). Pure peroxide stress does not cause widespread protein unfolding or aggregation, making the activation of an effective chaperone holdase like Hsp33 equally unnecessary (17, 44). Peroxide stress does, however, cause a significant drop in intracellular ATP levels in yeast and E. coli, which is largely due to the ROS-mediated inactivation of enzymes central to cellular ATP metabolism (e.g., GapDH) (16, 44). Therefore, when protein unfolding occurs during oxidative stress conditions, cells can no longer depend on ATP-dependent chaperone foldases but require ATP-independent chaperone holdases, such as Hsp33, to conquer the stress (44). While these studies provided an excellent physiological explanation for why Hsp33’s activation is required under these particular stress conditions, they failed to unveil the specific circumstances under which bacteria experience oxidative stress conditions that lead to widespread protein unfolding. The puzzle was solved recently when hypochlorite, the active ingredient of household bleach, was found to function both as an oxidant and as a potent protein unfolding reagent (45). In vivo and in vitro studies demonstrated that hypochlorite causes widespread oxidative protein unfolding and aggregation. Hsp33, which is extremely rapidly activated by very low concentrations of hypochlorite, protects proteins against hypochlorite-induced aggregation and significantly increases the hypochlorite resistance of E. coli and Vibrio cholerae (45). This protective action of Hsp33 in bacteria such as E. coli might become particularly important during bacterial colonization. Evidence suggesting that the hypochlorite-generating enzyme dual oxidase DuOX, which is widely expressed in mucosal barrier epithelia cells, plays an active role in controlling bacterial colonization has recently been presented (49). Within 48 hof knocking down the duox gene in Drosophila melanogaster, the bacterial content in the fly gut increased 300-fold, strongly suggesting that DuOX-mediated hypochlorite production limits the extent of bacterial colonization (49). The protective function of Hsp33 might therefore be directly involved in bacterial colonization.
MECHANISM OF HSP33’S DUAL STRESS SENSING
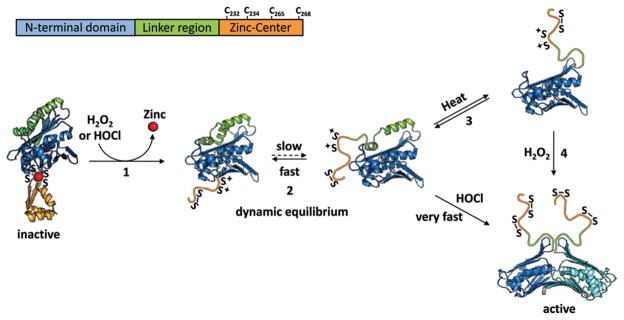
In vivo studies revealed the physiological need for a chaperone like Hsp33. However, the mechanism by which a protein senses and responds to oxidants only under protein unfolding conditions was far from obvious. A combination of structural and functional studies using the slow-acting oxidant hydrogen peroxide at 43 °C for activation has yielded a working model that serves to explain Hsp33’s unique capability to interdependently sense both stress conditions (Figure 2) (17). Hsp33 consists of a compactly folded N-terminal domain (amino acids 1–178) that presumably harbors the substrate binding site, and a C-terminal redox-sensing domain (amino acids 232–294) that accommodates the four absolutely conserved cysteines engaged in the high-affinity binding of one zinc ion (50). The two domains connect via a highly flexible linker region, which has been proposed to serve as the folding sensor in Hsp33, and whose folding status might ultimately control the chaperone function of Hsp33 (Figure 2) (17).
Figure 2.
Model of Hsp33’s activation. Under nonstress conditions, Hsp33 is monomeric and inactive. All four invariant cysteines are reduced and bind one zinc ion (red), forming a compactly folded zinc center (orange). (1) Exposure to oxidants causes the formation of the first disulfide bond connecting Cys265 with Cys268, which triggers zinc release and the unfolding of the zinc binding domain. (2) Unfolding of the zinc binding domain destabilizes the adjacent linker region (green), which is now in a dynamic equilibrium between a folded and partially unfolded conformation. At nonstress temperatures, the equilibrium favors the folded conformation and kinetically slow oxidants, like H2O2, cannot access the cysteines or activate Hsp33. (3) Mild denaturing conditions (e.g., elevated temperatures) shift the equilibrium and allow H2O2 to induce formation of the second disulfide bond between Cys232 and Cys234. (4) This disulfide bond apparently locks the linker region in an unfolded conformation. It causes the exposure of large hydrophobic surfaces on the N-terminal Hsp33 domain (blue), the proposed binding sites for unfolded proteins, and causes the formation of highly chaperone active Hsp33 dimers.
Under nonstress conditions, Hsp33 is present as a reduced monomer with no apparent chaperone activity. The tetrahedral coordination of zinc by the C232XC234X27–32Cys265XXC268 motif (Ka > 1017 M−1 at 25 °C and pH 7.5) in Hsp33’s C-terminus leads to the stabilization of the zinc center as a compact, independent folding unit and apparently protects the four thiolate anions against nonspecific air oxidation (50, 51). The adjacent linker region, which appears to be stably folded in reduced Hsp33, covers ~3800 Å of N-terminal surface area, of which 75% is hydrophobic (52, 53). Peroxide-mediated activation of Hsp33 is initiated by the formation of the distal disulfide bond, connecting Cys265 with Cys268 (Figure 2). Oxidation of two of the four zinc-coordinating cysteines appears to be sufficient to cause zinc release, which, in turn, induces the unfolding of Hsp33’s zinc binding domain, presumably due to its lack of hydrophobic core structure (17, 54). Unfolding of the zinc binding domain destabilizes the upstream linker region and converts the linker into a thermolabile folding sensor. In the absence of additional unfolding conditions, Hsp33 is now present as a chaperone-inactive oxidation intermediate, whose linker region exists in a dynamic equilibrium between a folded and partially unfolded state (Figure 2) (17). At nonstress temperatures, the equilibrium appears to favor the folded state, and kinetically slow oxidants such as hydrogen peroxide or nitric oxide cannot access the cysteines and, thus, are unable to activate Hsp33 (17). Once elevated temperatures or other mild denaturing reagents (e.g., 1M Gdn*HCl) are applied, the equilibrium shifts and slow oxidants are now capable of inducing the formation of the second disulfide bond connecting the two proximal cysteines, Cys232 and Cys234 (15) (Figure 2). This disulfide bond apparently locks the linker region in an unfolded conformation. In vitro studies revealed that linker unfolding precisely parallels the exposure of hydrophobic surfaces in Hsp33, suggesting that the highly hydrophobic surfaces that serve as the interaction site for the linker region under reducing conditions might represent the high-affinity binding site for unfolded proteins under oxidizing conditions (17). This would turn the linker region into an effective redox-and folding-controlled guardian of Hsp33’s substrate-binding site. It remains now to be determined what forces protect these two crucial cysteines against oxidation in the absence of protein unfolding conditions and how these cysteines control the stability of the linker region. The final step in Hsp33’s activation process appears to be the rapid association of two oxidized Hsp33 monomers into kinetically stable homodimers (Kd = 0.6 μM) (48).Dimerization has been demonstrated in a variety of solution studies and was confirmed by the biochemical analysis of Hsp33 mutants with interrupted dimer–dimer interfaces, which were found to be redox-regulated, yet chaperone-inactive in vitro (48).
HYPOCHLORITE, THE PHYSIOLOGICAL ACTIVATOR OF HSP33?
Hypochlorite is an oxidant that was found to combine both oxidizing and protein unfolding effects (45). Incubation ofHsp33 with low concentrations of hypochlorite at nonstress temperatures was found to almost instantaneously activate Hsp33 both in vitro and in vivo. Activation by hypochlorite seems to be based on the extremely fast rate with which chlorine-based bleach interacts with cysteines. With reaction rates that are approximately 7 orders of magnitude higher than the reaction rates of peroxide with free cysteines (6, 55), hypochlorite effectively competes with the refolding of Hsp33’s linker region even at nonstress temperatures, oxidizesHsp33’s proximal cysteines, and converts Hps33 into the activated chaperone (45) (Figure 2). Interestingly, whileHsp33 uses the oxidative unfolding activity of hypochlorite for its specific activation, predominantly thermolabile proteins fall victim to this oxidative insult and aggregate in vivo. Hsp33’s ability to suppress protein aggregation in hypochlorite- treated bacteria appears to be the reason for the increased bleach sensitivity of both E. coli and V. cholerae strains lacking the Hsp33 gene compared to wild-type strains (45).
HSP33, A CENTRAL PLAYER IN A REDOX-REGULATED CHAPERONE NETWORK
The ATP independence ofHsp33’s action in combination with its redox-mediated activation makes Hsp33 uniquely suited to protect proteins against nonspecific aggregation under oxidative stress conditions. These very features, however, pose significant challenges for Hsp33’s substrate proteins once cells return to nonstress conditions. The redox-mediated inactivation of Hsp33 would cause the sudden release of unfolded substrate proteins, while the lack of ATP binding and hydrolysis would prevent Hsp33 from actively participating in the refolding of the released substrate proteins. Aggregation of substrate proteins would be the consequence, the very process that Hsp33 is supposed to prevent. For this reason, it was not surprising to discover that Hsp33’s inactivation is at least as tightly regulated as its activation (56). Analogous to Hsp33 activation, which requires oxidizing and unfolding conditions, inactivation of Hsp33 requires both reducing and refolding conditions. With a return to reducing conditions, Hsp33’s disulfide bonds are rapidly reduced by either the cellular thioredoxin or glutaredoxin system (57). This step is necessary but not sufficient for inactivation. In vitro studies showed that reduction of oxidized Hsp33 dimers generates reduced Hsp33 dimers, which remain in complex with their unfolded substrate proteins. This mechanism avoids the sudden release of substrate proteins upon restoration of reducing conditions. Apparently to connect the substrate release to reducing conditions that are permissive for protein folding, the presence of the functional, ATP-dependent DnaK system is required, which, in turn, supports the refolding of the substrate proteins to their native state (56). The precise mechanism that governs the interaction between substrate-bound Hsp33 and the DnaK system is not fully understood. Analysis of the in vivo substrate binding specificity of Hsp33 confirmed extensive overlap with the substrate binding specificity of the DnaK system, an important prerequisite for the synergistic action of two classes of chaperones (44).
EUKARYOTIC PEROXIREDOXINS HAVE REDOXDEPENDENT DUAL ACTIVITY
Peroxiredoxins form a highly conserved superfamily of ~20–30 kDa antioxidant enzymes. This family, which has apparently evolved from a thioredoxin fold-containing ancestor protein (58), has members in all biological kingdoms. Although most peroxiredoxin homologues are found in the cytosol, isoforms have been identified that are exclusively expressed in mitochondria, chloroplasts, and peroxisomes or are associated with nuclei and membranes (59).
Peroxiredoxins have been shown to detoxify H2O2, peroxynitrite, and various organic hydroperoxides, thereby maintaining the intracellular redox balance and protecting organisms against oxidative stress (60). Moreover, their role in ROS detoxification makes them directly involved in the regulation of peroxide-mediated signaling cascades, NF-κB activity, and apoptosis (61). Their very high cellular abundance appears to compensate for their moderate catalytic efficiency (105 M−1 s−1) compared with those of glutathione peroxidases (108M−1 s−1) and catalases (106M−1 s−1) (59, 62). Peroxiredoxins constitute 0.1–0.8%of all soluble proteins in mammalian cells. They are the third most abundant proteins in erythrocytes and among the ten most highly expressed proteins in E. coli (63, 64). In addition, peroxiredoxins are significantly overexpressed in disease states, such as cancer and neurodegenerative diseases, and appear to play neuroprotective roles in models of Parkinson’s and Alzheimer’s disease (65–67).
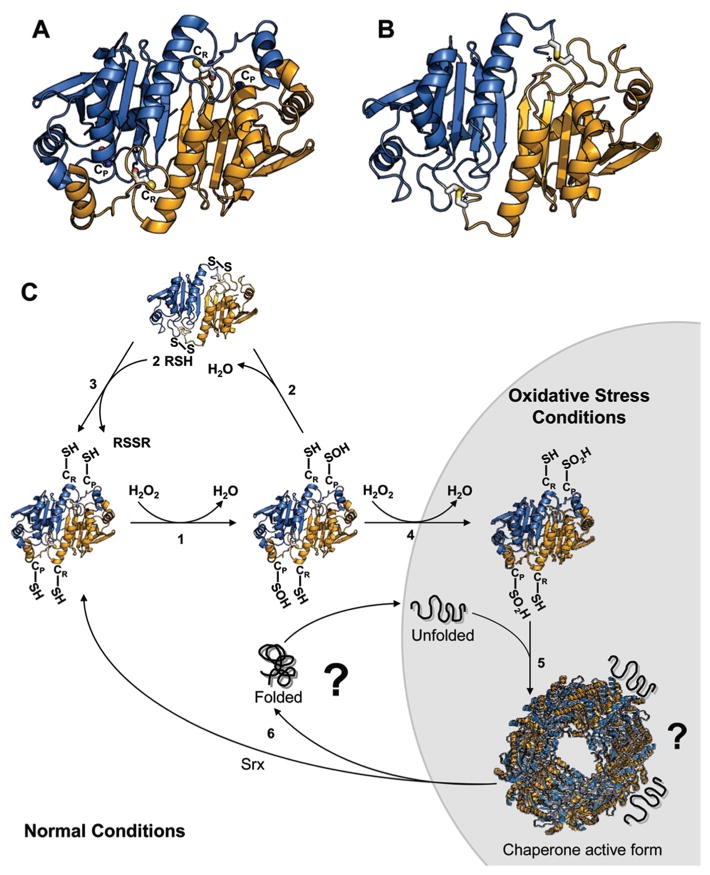
All peroxiredoxins share the same basic catalytic mechanism: a catalytically active cysteine (i.e., peroxidatic cysteine, CP) attacks the O–O bond of the ROOH substrate, thereby forming the ROH product and a sulfenic acid on CP (CP-SOH) (68). The mechanism of CP regeneration, which involves the attack of CP-SOH by a free thiol and the release of water, is different for individual members of the peroxiredoxin family and necessitated classification (expertly reviewed in refs (68–70)). 1-Cys peroxiredoxins (e.g., bacterial BCP, yeast Prx1p, and mammalian PRDX6), which contain only the conserved CP, use a thiol-containing electron donor from other proteins or small molecules for the regeneration of their thiol group (71). In contrast, atypical 2-Cys peroxiredoxins (e.g., bacterial Tpx, yeast Dot5p, and mammalian PRDX5) utilize a highly conserved second cysteine (i.e., resolving cysteine, CR) located at the protein’s C-terminus, which reacts with CP-SOH to form an intramolecular disulfide bond (69). Typical 2-Cys peroxiredoxins, such as bacterial AhpC, yeast Tsa1 (cPrxI) and Tsa2 (cPrxII), Ahp1p, or mammalian PRDX1–4, differ from atypical peroxiredoxins in that they form obligate homodimers in solution. The crystal structure of typical 2-Cys peroxiredoxins reveals that the C-terminus of one subunit reaches across the dimer interface to interact with the other subunit (72). This brings the resolving CR of one subunit into the proximity of CP-SOH of the adjacent subunit and allows the formation of an intermolecular disulfide bond (Figure 3). Reduction of intra- and intermolecular disulfide bonds in atypical and typical peroxiredoxins, respectively, is catalyzed by cell-specific disulfide oxidoreductases, such as thioredoxin or AhpF, ultimately linking peroxiredoxin’s regeneration to the cellular NADPH pool (73).
Figure 3.
Functional switch of 2-Cys peroxiredoxin. Crystal structures of (A) overoxidized, fully folded human PrxII (Protein Data Bank entry 1QMV) and (B) disulfide-bonded, locally unfolded human PrxI (Protein Data Bank entry 1QQS).The two subunits of the homodimer are colored orange and blue, respectively. The peroxidatic (CP) and resolving cysteines (CR) are depicted as balls and sticks. Asterisks mark the ends of the disordered C-termini in panel B. (C)Model of reaction and activation cycle of 2-Cys peroxiredoxins. Under nonstress conditions, (1) Prx catalyzes the reduction of H2O2 to H2O. This leads to sulfenic acid (SOH) formation at the peroxidatic cysteine, CP. (2) The resolving cysteine (CR) of the other subunit attacks CP-SOH, and an intermolecular disulfide bond is formed. (3)The disulfide bond is reduced by the thioredoxin system. Under oxidative stress conditions, (4) CP-SOH reacts with a second H2O2 molecule and a sulfinic acid (CP-SO2H) is formed. Sulfinic acid formation inactivates the peroxidase activity and (5) supports the assembly into chaperone-active high-molecular weight (HMW) complexes, which prevent the aggregation of unfolding proteins in vitro. Neither the precise structure of the HMW complexes nor the binding site for unfolding proteins has been identified. (6) Reduction of overoxidized Prx is catalyzed by sulfiredoxin upon the return to nonstress conditions. The fate of the bound substrate proteins remains to be determined.
During oxidative stress, eukaryotic typical 2-Cys peroxiredoxins have been found to be uniquely susceptible to enzymatic inactivation by the overoxidation of their peroxidatic cysteine to sulfinic acid (74) (Figure 3). This selective inactivation of typical 2-Cys peroxiredoxins has been proposed to serve a regulatory role in eukaryotic peroxide signaling (71). It would allow the enzymes to function as molecular floodgates, which open as the enzymes become inactivated, allowing the intracellular concentration of peroxide to further increase and allowing it to carry out its signaling function. The conclusion that sulfinic acid formation is indeed a specific side reaction and serves an important regulatory role was further strengthened by the discovery of sulfiredoxin, an ATP-dependent enzyme specialized in reducing overoxidized cysteines in 2-Cys peroxiredoxins (38). In vitro overoxidation studies using purified yeast and mammalian 2-Cys peroxiredoxins revealed, moreover, that overoxidation induces the formation of high molecular weight (HMW) oligomers, which function as potent chaperones and prevent protein aggregation (75, 76). These results suggest that typical 2-Cys peroxiredoxins might act as the eukaryotic counterpart of bacterial Hsp33. Instead of using disulfide bond formation as the redox switch, 2-Cys peroxiredoxins apparently use a unique “sulfinic acid switch” to convert from a peroxidase under nonstress conditions to a molecular chaperone under severe oxidative stress conditions (76). Since both susceptibility to overoxidation and ability to function as molecular chaperone have so far been reported for only typical 2-Cys peroxiredoxins, we will for the remainder of this review exclusively focus on this group of eukaryotic peroxiredoxins (75, 76).
SULFINIC ACID FORMATION AS A REDOX SWITCH IN TYPICAL 2-CYS PEROXIREDOXINS
The active site cysteine, CP, of peroxiredoxins is located within the first turn of helix α2 at the bottom of a well-conserved substrate-binding pocket (Figure 3A).Upon reaction with H2O2, the CP-sulfenic acid intermediate is formed. The resolving cysteine, CR, which is located approximately 14 Å from CP, is oriented in the opposite direction and is also partially buried (Figure 3A). Therefore, to effectively undergo disulfide bond formation, significant conformational rearrangements are required, which involve, at a minimum, the local unfolding of the first turn of helix α2. This local unfolding, which now places CP-SOH in a highly exposed loop segment, often termed the CP loop, promotes disulfide bond formation with CR (Figure 3B) (20, 71). The rate at which the local unfolding occurs appears to be determined by the equilibrium constant between the locally unfolded and fully folded conformations and depends on the oxidation state of CP and the type of peroxiredoxin (77). Peroxiredoxins are believed to remain in this locally unfolded conformation until the disulfide bond is reduced, upon which they return to the fully folded conformation. Notably, this proposed series of conformational changes during peroxiredoxin’s catalytic cycle is highly reminiscent of the conformational changes during Hsp33’s activation process (Figure 2). When the active site cysteines are reduced (R-SH), both proteins are proposed to be in a fully folded conformation. Upon initial oxidation of the proteins, an intermediate is formed that exists in a dynamic equilibrium between a folded and locally unfolded conformation. Disulfide bond formation locks both Hsp33 and peroxiredoxin in a locally unfolded conformation that inevitably causes the destabilization of the protein, which directly affects the redox potential of the disulfide bond (78). Increased accessibility combined with an oxidizing redox potential makes the regeneration of both proteins by cellular reductases a kinetically and thermodynamically favored process (45, 71). In contrast to Hsp33’s active site cysteine, however, which, when present in the oxidation intermediate, appears to be resistant to overoxidation, a second and possibly even a third molecule of peroxide can access CP-SOH of peroxiredoxin and cause sulfinic and sulfonic acid formation (20). Although less than 0.1% of peroxiredoxin molecules undergo hyperoxidation to sulfinic acid during the normal peroxide catalysis at low concentrations of peroxide (<1 μM) (79), significant accumulation of overoxidized peroxiredoxin is generated during periods of severe peroxide stress (80).
Disulfide bond formation and overoxidation seem to be competing events in the catalytic cycle of 2-Cys peroxiredoxin. While disulfide bond formation requires the local unfolding of the active site, overoxidation appears to occur in the folded conformation. Therefore, any structural features that stabilize the folded conformation of peroxiredoxin’s active site are predicted to slow disulfide bond formation and increase the rate of overoxidation. Direct structural comparison of overoxidation-sensitive eukaryotic 2-Cys peroxiredoxin with overoxidation-resistant prokaryotic 2-Cys peroxiredoxin in different redox states (i.e., SH, S-S, SOH, SO2H, and SO3H) revealed distinct structural features that likely promote the overoxidation in eukaryotic 2-Cys peroxiredoxin (reviewed in ref 77). The C-terminus of eukaryotic 2-Cys peroxiredoxins contains a “GGLG” loop motif as well as an additional α-helix harboring a conserved “YF” motif. In the reduced state, both the GGLG loop and the extended C-terminus cover the active site helix that contains the catalytic CP. To access CP-SOH for disulfide bond formation now requires at least two local unfolding events: the unfolding of the α-helical loop that contains the active site cysteine, CP, and the unfolding of the extended C-terminus containing the YF motif. Because the additional YF motif is thought to slow the local unfolding event, it increases the kinetic pause between sulfenic acid formation and disulfide bond formation and is likely the ultimate cause for the propensity of eukaryotic 2-Cys peroxiredoxins to become overoxidized (20, 71). In agreement with these structural considerations, mutant studies showed that C-terminally truncated yeast Tsa1 variants are resistant to overoxidation (81).
CLOSING THE OVEROXIDATION CYCLE
Protein sulfinic acids cannot be reduced by the typical cellular oxidoreductases (e.g., thioredoxin) and were long thought to be irreversible oxidative thiol modifications. However, pulse–chase experiments revealed that CP-SO2H generation in peroxiredoxin is fully reversible in the absence of any new protein synthesis (80, 82). Biteau et al. discovered the enzyme responsible for the ATP-dependent reduction of overoxidized yeast Tsa1 and Tsa2 and termed it sulfiredoxin (38). Sulfiredoxins constitute a family of highly conserved proteins that are present only in eukaryotes, which agrees well with the fact that prokaryotic peroxiredoxins are ~100-foldmore resistant to overoxidation (71). Sulfiredoxins are reported to have both phosphotransferase and thioltransferase activity, and their mechanism of action involves an active site cysteine, ATP, Mg2+, and a reducing agent such as glutathione or thioredoxin (83–86). More recently, a second family of potential sulfinic acid reductases termed sestrins was identified, a finding that further underscores the importance of restoring peroxiredoxin’s peroxidase activity in vivo. Sestrins, which are related to the bacterial AhpC reductases (i.e., AhpD), do not exhibit any sequence or structural similarity to sulfiredoxins. Nevertheless, in vitro studies suggested that the sestrin 2 protein Hi95 utilizes a sulfiredoxin-related mechanism for sulfinic acid reduction (87). These conclusions have very recently been challenged by Rhee and co-workers, who were unable to confirm any sulfinic acid reductase activity for recombinant sestrin 2 in vitro or to detect any delay in the recovery of overoxidized Prx1 in embryonic fibroblasts derived from sestrin 2−/− knockout mice (88). It will be interesting to determine the reason(s) behind these different results. In either case, the observation that multicellular organisms like Caenorhabditis elegans, which contain highly overoxidation-prone 2-Cys peroxiredoxins (C. Kumsta and U. Jakob, personal observations), lack any sulfiredoxin homologues suggests the existence of additional enzyme(s) with sulfinic acid activity.
REDOX-MEDIATED CHANGES IN PEROXIREDOXIN’S OLIGOMERIZATION STATE
Typical 2-Cys peroxiredoxins function as head-to-tail arranged homodimers (α2). X-ray crystal structure analysis revealed that at least some typical 2-Cys peroxiredoxins form toroid-shaped decamers, (α2)5 (74, 89). In addition, larger so-called HMW complexes (>600 kDa) have been observed in solution studies (20). Although complex formation is influenced by a variety of solution conditions (e.g., ionic strength, pH, and temperature) and posttranslational modifications (20, 75), the most crucial factor affecting the oligomerization state of peroxiredoxins appears to be the redox state of the active site cysteine, CP. Sedimentation studies revealed that the reduced form of CP stabilizes the decameric forms of peroxiredoxin, whereas the disulfide-bonded form increases the tendency for the decamers to dissociate into dimers (90). Careful comparison of the crystal structures revealed that local unfolding of the CP loop, which occurs concomitant with disulfide bond formation, likely interferes with interactions that stabilize the decamer. This redox-mediated change in the oligomerization state of 2-Cys peroxiredoxins, going from a reduced, fully folded decamer to a disulfide-bonded, partially folded dimer and back, is again highly reminiscent of Hsp33, which cycles between a reduced, fully folded monomer and a disulfide-bonded, partially folded dimer (48, 74).
OVEROXIDATION, OLIGOMER FORMATION, AND CHAPERONE ACTIVITY
The first evidence that the highly abundant 2-Cys peroxiredoxins might serve an additional function came from studies in yeast, which showed that cells deficient in both cytosolic 2-Cys peroxiredoxins, Tsa1 and Tsa2, are highly sensitive to heat shock treatment, a phenotype often associated with organisms that lack crucial chaperones. The temperature-sensitive phenotype of tsa1/2 deletion strains was almost fully rescued by a Tsa1 mutant that missed the resolving cysteine CR and was partially rescued by the expression of a Tsa1 mutant that lacked both active site cysteines (75). This result served as first indication that yeast 2-Cys peroxiredoxins might have an additional cytoprotective function independent of their peroxidase activity. Subsequent in vitro studies confirmed this conclusion by demonstrating that yeast Tsa1 and Tsa2 possess molecular chaperone activity and protect thermally unfolded substrate proteins against nonspecific aggregation (75, 76). Careful functional analysis revealed that the chaperone activity of the peroxiredoxins is linked to their oligomerization state. In addition to the previously mentioned dimers and decamers, which are collectively called the low molecular weight (LMW) species of 2-Cys peroxiredoxins, purified yeast Tsa1 preparations contained a significant proportion of HMW complexes. These HMW complexes were found to be heterogeneous in nature and range from ringlike structures with 5-fold symmetry to spherical 22–28 nm particles (75). In contrast to LMW species of Tsa1, which were shown to predominantly function as peroxidases, HMW complexes lacked the peroxidase activity and exerted high, ATP-independent chaperone holdase activity (75). Analysis of protein aggregates that accumulate in strains lacking tsa1 revealed ribosomal proteins as potential substrate proteins of Tsa1 (91). These results suggested that yeast 2-Cys peroxiredoxins serve dual roles as peroxidases and as molecular chaperones. Subsequent studies in HeLa cells agreed with these initial observations and confirmed that human 2-Cys PrxII also functions as a molecular chaperone when present in HMW complexes (76).
What is the driving force for the formation of these highly chaperone-active HMW complexes? In vivo and in vitro studies revealed that the conversion of peroxidase-active LMW complexes into chaperone-active HMW complexes requires high concentrations of peroxide and the thioredoxin system, conditions known to cause overoxidation of the active site cysteine CP (75). Mutation studies supported this model by demonstrating that assembly into HMW oligomers is dependent on the presence of CP but not of CR. Moreover, C-terminally truncated versions of human PrxII, which are known to be resistant to overoxidation, are unable to form HMW complexes upon treatment with H2O2 (76). Likewise, bacterial 2-Cys peroxiredoxin homologues (e.g., AhpC), which are intrinsically resistant to overoxidation, do not exert detectable chaperone activity. So far, the only exception to this rule appears to be AhpC from Helicobacter pylori, which is more closely related to mammalian peroxiredoxins than to bacterial AhpC. It is prone to overoxidation upon long-term exposure to H2O2 and turns into a molecular chaperone upon oxidative stress-mediated formation of HMW complexes (92). Finally, removal of H2O2 and the presence of Srx1 cause the dissociation of the HMW complexes, restore the peroxidase activity, and lower the chaperone activity to the basal level found under nonstress conditions (75). So far, it is unclear how sulfinic acid formation at the active site cysteine triggers these additional conformational rearrangements that stabilize higher-oligomer formation, cause the exposure of hydrophobic surfaces, and dramatically increase the capacity of peroxiredoxins to recognize and bind unfolding proteins (90). Since the active site of 2-Cys peroxiredoxin appears to be in the fully folded state when CP is overoxidized (71) (Figure 3), it is conceivable that conversion to HMW complexes is simply triggered by the increased level of stabilization of the dimer–dimer interfaces. This would, however, not explain the large additional hydrophobic surface areas that are specifically found in chaperone-active HMW complexes and which are likely the high-affinity binding sites for unfolding proteins (75). It is therefore plausible that additional conformational changes such as domain unfolding occur, which might not be detected in the crystal structures. This is clearly the situation with Hsp33, where many attempts to visualize the structural differences between inactive and active Hsp33 preparations have been made and failed (52, 53). In either case, the results suggest that at low peroxide concentrations, the peroxidatic cysteine CP functions as an active site nucleophile, which attacks and destroys peroxides. In the presence of high peroxide concentrations, however, CP acts as a sulfinic acid switch that confers the conversion of chaperone-inactive LMW complexes into highly chaperone-active HMW complexes.
It is important to note at this point that overoxidation of the active site cysteine is probably not the only mechanism by which chaperone-active HMW complexes can be assembled. Yeast Tsa1 forms chaperone-active HMW complexes in an H2O2- and cysteine-independent manner in response to elevated temperatures, explaining the partial rescue of the heat shock phenotype by Tsa1 mutants lacking both active site cysteines (75). Moreover, site-specific phosphorylation of human PrxI was found to cause the formation of HMW species, which lack peroxidase activity and exhibit significantly higher chaperone activity than the LMW species (93). Finally, irreversible oxidation of the active site cysteine to sulfonic acid has been found to cause formation of HMW complexes with high chaperone activity (94). These results suggest that it is not overoxidation of the active site per se that activates the chaperone function of 2-Cys peroxiredoxins, but the propensity of 2-Cys peroxiredoxins to form stable HMW complexes with high-affinity binding sites for unfolded proteins.
CONCLUSIONS
Redox-mediated activation of ATP-independent chaperone holdases during oxidative stress conditions is an immediate response mechanism aimed to ensure cell survival. The high affinity of chaperone holdases for unfolding proteins is crucial under oxidative stress conditions, when declines in cellular ATP levels incapacitate ATP-dependent chaperone foldases. It likely becomes a hazard, however, under nonstress conditions, when holdases compete with foldases for unfolding proteins and potentially interfere with the regular folding processes in the cell. It is therefore not surprising that activation of these specialized chaperones in a tightly regulated process. In the case of prokaryotic Hsp33, activation involves an intricate dual stress sensing mechanism, which restricts Hsp33’s activation to oxidative stress conditions that lead to protein unfolding. In the case of eukaryotic 2-Cys peroxiredoxins, severe oxidative stress conditions and heat stress conditions convert peroxiredoxin in a molecular chaperone, suggesting that 2-Cys peroxiredoxin might also be capable of simultaneously and potentially interdependently sense both stress conditions. Further support for this hypothesis is given by the astounding similarities in the activation mechanism of these two, completely unrelated proteins. Both proteins sense reactive oxygen species via highly conserved cysteines and undergo oxidative thiol modifications that significantly destabilize structurally important regions of the protein, thereby increasing surface hydrophobicity and affecting the oligomerization status. To improve our understanding of the mechanism of peroxiredoxin activation, it will be important to determine the precise structural changes that lead to the formation of chaperone-active HMW complexes and to identify the binding sites for unfolded proteins. It is obvious that the demand for oxidative stress-specific ATP-independent chaperone holdases like 2-Cys peroxiredoxin is as high in eukaryotes as the demand for Hsp33 in bacteria, yet many questions regarding peroxiredoxin’s cytoprotective chaperone function are still open. What is the precise nature of substrate proteins that bind to 2-Cys peroxiredoxin in vivo? How is substrate release coordinated, and what is the fate of the released substrate proteins? These questions must be answered to unequivocally conclude that 2-Cys peroxiredoxins are indeed, as previously suggested, the eukaryotic counterpart of Hsp33.
Footnotes
Abbreviations: ROS, reactive oxygen species; HMW, high-molecular weight; LMW, low-molecular weight.
References
- 1.Schrader M, Fahimi HD. Peroxisomes and oxidative stress. Biochim Biophys Acta. 2006;1763:1755–1766. doi: 10.1016/j.bbamcr.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Michiels C, Raes M, Toussaint O, Remacle J. Importance of Se-glutathione peroxidase, catalase, and Cu/Zn-SOD for cell survival against oxidative stress. Free Radical Biol Med. 1994;17:235–248. doi: 10.1016/0891-5849(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 3.D’Autreaux B, Toledano MB. ROS as signalling molecules: Mechanisms that generate specificity in ROS homeostasis. Nat Rev Mol Cell Biol. 2007;8:813–824. doi: 10.1038/nrm2256. [DOI] [PubMed] [Google Scholar]
- 4.Rhee SG. Cell signaling. H2O2, a necessary evil for cell signaling. Science. 2006;312:1882–1883. doi: 10.1126/science.1130481. [DOI] [PubMed] [Google Scholar]
- 5.Stadtman ER. Protein oxidation in aging and age-related diseases. Ann NY Acad Sci. 2001;928:22–38. doi: 10.1111/j.1749-6632.2001.tb05632.x. [DOI] [PubMed] [Google Scholar]
- 6.Imlay JA. Pathways of oxidative damage. Annu Rev Microbiol. 2003;57:395–418. doi: 10.1146/annurev.micro.57.030502.090938. [DOI] [PubMed] [Google Scholar]
- 7.Storz G, Imlay JA. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 8.Nystrom T. Role of oxidative carbonylation in protein quality control and senescence. EMBO J. 2005;24:1311–1317. doi: 10.1038/sj.emboj.7600599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan LJ, Sohal RS. Curr Protoc Protein Sci. Unit14. Chapter 14. John Wiley and Sons, Inc; New York: 2001. Analysis of oxidative modification of proteins; p. 14. [DOI] [PubMed] [Google Scholar]
- 10.Aliev G, Smith MA, Seyidov D, Neal ML, Lamb BT, Nunomura A, Gasimov EK, Vinters HV, Perry G, LaManna JC, Friedland RP. The role of oxidative stress in the pathophysiology of cerebrovascular lesions in Alzheimer’s disease. Brain Pathol. 2002;12:21–35. doi: 10.1111/j.1750-3639.2002.tb00419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles NM, Watts AB, Giles GI, Fry FH, Littlechild JA, Jacob C. Metal and redox modulation of cysteine protein function. Chem Biol. 2003;10:677–693. doi: 10.1016/s1074-5521(03)00174-1. [DOI] [PubMed] [Google Scholar]
- 12.Lee JW, Helmann JD. The PerR transcription factor senses H2O2 by metal-catalysed histidine oxidation. Nature. 2006;440:363–367. doi: 10.1038/nature04537. [DOI] [PubMed] [Google Scholar]
- 13.Giles GI, Tasker KM, Jacob C. Hypothesis: The role of reactive sulfur species in oxidative stress. Free Radical Biol Med. 2001;31:1279–1283. doi: 10.1016/s0891-5849(01)00710-9. [DOI] [PubMed] [Google Scholar]
- 14.Ma LH, Takanishi CL, Wood MJ. Molecular mechanism of oxidative stress perception by the Orp1 protein. J Biol Chem. 2007;282:31429–31436. doi: 10.1074/jbc.M705953200. [DOI] [PubMed] [Google Scholar]
- 15.Leichert LI, Gehrke F, Gudiseva HV, Blackwell T, Ilbert M, Walker AK, Strahler JR, Andrews PC, Jakob U. Quantifying changes in the thiol redox proteome upon oxidative stress in vivo. Proc Natl Acad Sci USA. 2008;105:8197–8202. doi: 10.1073/pnas.0707723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenton D, Grant CM. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem J. 2003;374:513–519. doi: 10.1042/BJ20030414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ilbert M, Horst J, Ahrens S, Winter J, Graf PC, Lilie H, Jakob U. The redox-switch domain of Hsp33 functions as dual stress sensor. Nat Struct Mol Biol. 2007;14:556–563. doi: 10.1038/nsmb1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paget MS, Buttner MJ. Thiol-based regulatory switches. Annu Rev Genet. 2003;37:91–121. doi: 10.1146/annurev.genet.37.110801.142538. [DOI] [PubMed] [Google Scholar]
- 19.Cabiscol E, Levine RL. The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc Natl Acad Sci USA. 1996;93:4170–4174. doi: 10.1073/pnas.93.9.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood ZA, Schroder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- 21.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–347. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 22.Kim SO, Merchant K, Nudelman R, Beyer WF, Jr, Keng T, DeAngelo J, Hausladen A, Stamler JS. OxyR: A molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 23.Claiborne A, Miller H, Parsonage D, Ross RP. Protein-sulfenic acid stabilization and function in enzyme catalysis and gene regulation. FASEB J. 1993;7:1483–1490. doi: 10.1096/fasebj.7.15.8262333. [DOI] [PubMed] [Google Scholar]
- 24.Reddie KG, Seo YH, Muse WB, III, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Mol Biosyst. 2008;4:521–531. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dalle-Donne I, Rossi R, Giustarini D, Colombo R, Milzani A. S-Glutathionylation in protein redox regulation. Free Radical Biol Med. 2007;43:883–898. doi: 10.1016/j.freeradbiomed.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Dickinson DA, Forman HJ. Glutathione in defense and signaling: Lessons from a small thiol. Ann NY Acad Sci. 2002;973:488–504. doi: 10.1111/j.1749-6632.2002.tb04690.x. [DOI] [PubMed] [Google Scholar]
- 27.Ghezzi P, Bonetto V. Redox proteomics: Identification of oxidatively modified proteins. Proteomics. 2003;3:1145–1153. doi: 10.1002/pmic.200300435. [DOI] [PubMed] [Google Scholar]
- 28.Casagrande S, Bonetto V, Fratelli M, Gianazza E, Eberini I, Massignan T, Salmona M, Chang G, Holmgren A, Ghezzi P. Glutathionylation of human thioredoxin: A possible crosstalk between the glutathione and thioredoxin systems. Proc Natl Acad Sci USA. 2002;99:9745–9749. doi: 10.1073/pnas.152168599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reynaert NL, Ckless K, Guala AS, Wouters EF, van der Vliet A, Janssen-Heininger YM. In situ detection of S-glutathionylated proteins following glutaredoxin-1 catalyzed cysteine derivatization. Biochim Biophys Acta. 2006;1760:380–387. doi: 10.1016/j.bbagen.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 31.Kuge S, Arita M, Murayama A, Maeta K, Izawa S, Inoue Y, Nomoto A. Regulation of the yeast Yap1p nuclear export signal is mediated by redox signal-induced reversible disulfide bond formation. Mol Cell Biol. 2001;21:6139–6150. doi: 10.1128/MCB.21.18.6139-6150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jakob U, Muse W, Eser M, Bardwell JC. Chaperone activity with a redox switch. Cell. 1999;96:341–352. doi: 10.1016/s0092-8674(00)80547-4. [DOI] [PubMed] [Google Scholar]
- 33.Gertz M, Fischer F, Wolters D, Steegborn C. Activation of the lifespan regulator p66Shc through reversible disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:5705–5709. doi: 10.1073/pnas.0800691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cumming RC, Andon NL, Haynes PA, Park M, Fischer WH, Schubert D. Protein disulfide bond formation in the cytoplasm during oxidative stress. J Biol Chem. 2004;279:21749–21758. doi: 10.1074/jbc.M312267200. [DOI] [PubMed] [Google Scholar]
- 35.Leichert LI, Jakob U. Global methods to monitor the thiol-disulfide state of proteins in vivo. Antioxid Redox Signaling. 2006;8:763–772. doi: 10.1089/ars.2006.8.763. [DOI] [PubMed] [Google Scholar]
- 36.Holmgren A, Johansson C, Berndt C, Lonn ME, Hudemann C, Lillig CH. Thiol redox control via thioredoxin and glutaredoxin systems. Biochem Soc Trans. 2005;33:1375–1377. doi: 10.1042/BST0331375. [DOI] [PubMed] [Google Scholar]
- 37.Le Moan N, Clement G, Le Maout S, Tacnet F, Toledano MB. The Saccharomyces cerevisiae proteome of oxidized protein thiols: Contrasted functions for the thioredoxin and glutathione pathways. J Biol Chem. 2006;281:10420–10430. doi: 10.1074/jbc.M513346200. [DOI] [PubMed] [Google Scholar]
- 38.Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–984. doi: 10.1038/nature02075. [DOI] [PubMed] [Google Scholar]
- 39.Liberek K, Lewandowska A, Zietkiewicz S. Chaperones in control of protein disaggregation. EMBO J. 2008;27:328–335. doi: 10.1038/sj.emboj.7601970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bukau B, Weissman J, Horwich A. Molecular chaperones and protein quality control. Cell. 2006;125:443–451. doi: 10.1016/j.cell.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 41.Hartl FU, Hayer-Hartl M. Molecular chaperones in the cytosol: From nascent chain to folded protein. Science. 2002;295:1852–1858. doi: 10.1126/science.1068408. [DOI] [PubMed] [Google Scholar]
- 42.Straus D, Walter W, Gross CA. DnaK, DnaJ, and GrpE heat shock proteins negatively regulate heat shock gene expression by controlling the synthesis and stability of sigma 32. Genes Dev. 1990;4:2202–2209. doi: 10.1101/gad.4.12a.2202. [DOI] [PubMed] [Google Scholar]
- 43.Deuerling E, Bukau B. Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol. 2004;39:261–277. doi: 10.1080/10409230490892496. [DOI] [PubMed] [Google Scholar]
- 44.Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 45.Winter J, Ilbert M, Graf PC, Ozcelik D, Jakob U. Bleach activates a redox-regulated chaperone by oxidative protein unfolding. Cell. 2008;135:691–701. doi: 10.1016/j.cell.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang SE, Blattner FR. Characterization of twenty-six new heat shock genes of Escherichia coli. J Bacteriol. 1993;175:5242–5252. doi: 10.1128/jb.175.16.5242-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kang HJ, Heo DH, Choi SW, Kim KN, Shim J, Kim CW, Sung HC, Yun CW. Functional characterization of Hsp33 protein from Bacillus psychrosaccharolyticus: Additional function of HSP33 on resistance to solvent stress. Biochem Biophys Res Commun. 2007;358:743–750. doi: 10.1016/j.bbrc.2007.04.184. [DOI] [PubMed] [Google Scholar]
- 48.Graumann J, Lilie H, Tang X, Tucker KA, Hoffmann JH, Vijayalakshmi J, Saper M, Bardwell JC, Jakob U. Activation of the redox-regulated molecular chaperone Hsp33: A two-step mechanism. Structure. 2001;9:377–387. doi: 10.1016/s0969-2126(01)00599-8. [DOI] [PubMed] [Google Scholar]
- 49.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–850. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 50.Jakob U, Eser M, Bardwell JC. Redox switch of hsp33 has a novel zinc-binding motif. J Biol Chem. 2000;275:38302–38310. doi: 10.1074/jbc.M005957200. [DOI] [PubMed] [Google Scholar]
- 51.Graf PC, Martinez-Yamout M, VanHaerents S, Lilie H, Dyson HJ, Jakob U. Activation of the redox-regulated chaperone Hsp33 by domain unfolding. J Biol Chem. 2004;279:20529–20538. doi: 10.1074/jbc.M401764200. [DOI] [PubMed] [Google Scholar]
- 52.Vijayalakshmi J, Mukhergee MK, Graumann J, Jakob U, Saper MA. The 2.2Å crystal structure of Hsp33:Aheat shock protein with redox-regulated chaperone activity. Structure. 2001;9:367–375. doi: 10.1016/s0969-2126(01)00597-4. [DOI] [PubMed] [Google Scholar]
- 53.Janda I, Devedjiev Y, Derewenda U, Dauter Z, Bielnicki J, Cooper DR, Graf PC, Joachimiak A, Jakob U, Derewenda ZS. The crystal structure of the reduced, Zn2+-bound form of the B. subtilis Hsp33 chaperone and its implications for the activation mechanism. Structure. 2004;12:1901–1907. doi: 10.1016/j.str.2004.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Won HS, Low LY, Guzman RD, Martinez-Yamout M, Jakob U, Dyson HJ. The zinc-dependent redox switch domain of the chaperone Hsp33 has a novel fold. J Mol Biol. 2004;341:893–899. doi: 10.1016/j.jmb.2004.06.046. [DOI] [PubMed] [Google Scholar]
- 55.Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radical Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann JH, Linke K, Graf PC, Lilie H, Jakob U. Identification of a redox-regulated chaperone network. EMBO J. 2004;23:160–168. doi: 10.1038/sj.emboj.7600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ilbert M, Graf PC, Jakob U. Zinc center as redox switch: New function for an old motif. Antioxid Redox Signaling. 2006;8:835–846. doi: 10.1089/ars.2006.8.835. [DOI] [PubMed] [Google Scholar]
- 58.Copley SD, Novak WR, Babbitt PC. Divergence of function in the thioredoxin fold suprafamily: Evidence for evolution of peroxiredoxins from a thioredoxin-like ancestor. Biochemistry. 2004;43:13981–13995. doi: 10.1021/bi048947r. [DOI] [PubMed] [Google Scholar]
- 59.Hofmann B, Hecht HJ, Flohe L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- 60.Rhee SG, Chae HZ, Kim K. Peroxiredoxins: A historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radical Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 61.Han YH, Kim HS, Kim JM, Kim SK, Yu DY, Moon EY. Inhibitory role of peroxiredoxin II (Prx II) on cellular senescence. FEBS Lett. 2005;579:4897–4902. doi: 10.1016/j.febslet.2005.07.049. [DOI] [PubMed] [Google Scholar]
- 62.Hillar A, Peters B, Pauls R, Loboda A, Zhang H, Mauk AG, Loewen PC. Modulation of the activities of catalase-peroxidase HPI of Escherichia coli by site-directed mutagenesis. Biochemistry. 2000;39:5868–5875. doi: 10.1021/bi0000059. [DOI] [PubMed] [Google Scholar]
- 63.Link AJ, Robison K, Church GM. Comparing the predicted and observed properties of proteins encoded in the genome of Escherichia coli K-12. Electrophoresis. 1997;18:1259–1313. doi: 10.1002/elps.1150180807. [DOI] [PubMed] [Google Scholar]
- 64.Moore RB, Mankad MV, Shriver SK, Mankad VN, Plishker GA. Reconstitution of Ca2+-dependent K+ transport in erythrocyte membrane vesicles requires a cytoplasmic protein. J Biol Chem. 1991;266:18964–18968. [PubMed] [Google Scholar]
- 65.Kinnula VL, Lehtonen S, Sormunen R, Kaarteenaho-Wiik R, Kang SW, Rhee SG, Soini Y. Overexpression of peroxiredoxins I, II, III, V, and VI in malignant mesothelioma. J Pathol. 2002;196:316–323. doi: 10.1002/path.1042. [DOI] [PubMed] [Google Scholar]
- 66.Krapfenbauer K, Engidawork E, Cairns N, Fountoulakis M, Lubec G. Aberrant expression of peroxiredoxin subtypes in neurodegenerative disorders. Brain Res. 2003;967:152–160. doi: 10.1016/s0006-8993(02)04243-9. [DOI] [PubMed] [Google Scholar]
- 67.Hattori F, Oikawa S. Peroxiredoxins in the central nervous system. Subcell Biochem. 2007;44:357–374. doi: 10.1007/978-1-4020-6051-9_17. [DOI] [PubMed] [Google Scholar]
- 68.Poole LB. The catalytic mechanism of peroxiredoxins. Subcell Biochem. 2007;44:61–81. doi: 10.1007/978-1-4020-6051-9_4. [DOI] [PubMed] [Google Scholar]
- 69.Knoops B, Loumaye E, Van Der Eecken V. Evolution of the peroxiredoxins. Subcell Biochem. 2007;44:27–40. doi: 10.1007/978-1-4020-6051-9_2. [DOI] [PubMed] [Google Scholar]
- 70.Flohe L, Harris JR. Introduction. History of the peroxiredoxins and topical perspectives. Subcell Biochem. 2007;44:1–25. [PubMed] [Google Scholar]
- 71.Wood ZA, Poole LB, Karplus PA. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science. 2003;300:650–653. doi: 10.1126/science.1080405. [DOI] [PubMed] [Google Scholar]
- 72.Hirotsu S, Abe Y, Okada K, Nagahara N, Hori H, Nishino T, Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product. Proc Natl Acad Sci USA. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poole LB, Reynolds CM, Wood ZA, Karplus PA, Ellis HR, Li Calzi M. AhpF and other NADH:peroxiredoxin oxidoreductases, homologues of low Mr thioredoxin reductase. Eur J Biochem. 2000;267:6126–6133. doi: 10.1046/j.1432-1327.2000.01704.x. [DOI] [PubMed] [Google Scholar]
- 74.Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to doughnuts: Redox-sensitive oligomerization of 2-cysteine peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- 75.Jang HH, Lee KO, Chi YH, Jung BG, Park SK, Park JH, Lee JR, Lee SS, Moon JC, Yun JW, Choi YO, Kim WY, Kang JS, Cheong GW, Yun DJ, Rhee SG, Cho MJ, Lee SY. Two enzymes in one: Two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 76.Moon JC, Hah YS, Kim WY, Jung BG, Jang HH, Lee JR, Kim SY, Lee YM, Jeon MG, Kim CW, Cho MJ, Lee SY. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- 77.Karplus PA, Hall A. Structural survey of the peroxiredoxins. Subcell Biochem. 2007;44:41–60. doi: 10.1007/978-1-4020-6051-9_3. [DOI] [PubMed] [Google Scholar]
- 78.Grauschopf U, Winther JR, Korber P, Zander T, Dallinger P, Bardwell JC. Why is DsbA such an oxidizing disulfide catalyst? Cell. 1995;83:947–955. doi: 10.1016/0092-8674(95)90210-4. [DOI] [PubMed] [Google Scholar]
- 79.Yang KS, Kang SW, Woo HA, Hwang SC, Chae HZ, Kim K, Rhee SG. Inactivation of human peroxiredoxin I during catalysis as the result of the oxidation of the catalytic site cysteine to cysteine-sulfinic acid. J Biol Chem. 2002;277:38029–38036. doi: 10.1074/jbc.M206626200. [DOI] [PubMed] [Google Scholar]
- 80.Woo HA, Kang SW, Kim HK, Yang KS, Chae HZ, Rhee SG. Reversible oxidation of the active site cysteine of peroxiredoxins to cysteine sulfinic acid. Immunoblot detection with antibodies specific for the hyperoxidized cysteine-containing sequence. J Biol Chem. 2003;278:47361–47364. doi: 10.1074/jbc.C300428200. [DOI] [PubMed] [Google Scholar]
- 81.Jara M, Vivancos AP, Hidalgo E. C-Terminal truncation of the peroxiredoxin Tpx1 decreases its sensitivity for hydrogen peroxide without compromising its role in signal transduction. Genes Cells. 2008;13:171–179. doi: 10.1111/j.1365-2443.2007.01160.x. [DOI] [PubMed] [Google Scholar]
- 82.Woo HA, Chae HZ, Hwang SC, Yang KS, Kang SW, Kim K, Rhee SG. Reversing the inactivation of peroxiredoxins caused by cysteine sulfinic acid formation. Science. 2003;300:653–656. doi: 10.1126/science.1080273. [DOI] [PubMed] [Google Scholar]
- 83.Jonsson TJ, Tsang AW, Lowther WT, Furdui CM. Identification of intact protein thiosulfinate intermediate in the reduction of cysteine sulfinic acid in peroxiredoxin by human sulfiredoxin. J Biol Chem. 2008;283:22890–22894. doi: 10.1074/jbc.C800124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jonsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrace. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jonsson TJ, Murray MS, Johnson LC, Lowther WT. Reduction of cysteine sulfinic acid in peroxiredoxin by sulfiredoxin proceeds directly through a sulfinic phosphoryl ester intermediate. J Biol Chem. 2008;283:23846–23851. doi: 10.1074/jbc.M803244200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roussel X, Bechade G, Kriznik A, Van Dorsselaer A, Sanglier-Cianferani S, Branlant G, Rahuel-Clermont S. Evidence for the formation of a covalent thiosulfinate intermediate with peroxiredoxin in the catalytic mechanism of sulfiredoxin. J Biol Chem. 2008;283:22371–22382. doi: 10.1074/jbc.M800493200. [DOI] [PubMed] [Google Scholar]
- 87.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 88.Rhee SG, Woo HA, Bae SH, Park S. Sestrin 2 Is Not a Reductase for Cysteine Sulfinic Acid of Peroxiredoxins. Antioxid Redox Signaling. 2008;11(4):739–745. doi: 10.1089/ars.2008.2360. [DOI] [PubMed] [Google Scholar]
- 89.Alphey MS, Bond CS, Tetaud E, Fairlamb AH, Hunter WN. The structure of reduced tryparedoxin peroxidase reveals a decamer and insight into reactivity of 2Cys-peroxiredoxins. J Mol Biol. 2000;300:903–916. doi: 10.1006/jmbi.2000.3881. [DOI] [PubMed] [Google Scholar]
- 90.Schroder E, Littlechild JA, Lebedev AA, Errington N, Vagin AA, Isupov MN. Crystal structure of decameric 2-Cys peroxiredoxin from human erythrocytes at 1.7Å resolution. Structure. 2000;8:605–615. doi: 10.1016/s0969-2126(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 91.Rand JD, Grant CM. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol Biol Cell. 2006;17:387–401. doi: 10.1091/mbc.E05-06-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chuang MH, Wu MS, Lo WL, Lin JT, Wong CH, Chiou SH. The antioxidant protein alkylhydroperoxide reductase of Helicobacter pylori switches from a peroxide reductase to a molecular chaperone function. Proc Natl Acad Sci USA. 2006;103:2552–2557. doi: 10.1073/pnas.0510770103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jang HH, Kim SY, Park SK, Jeon HS, Lee YM, Jung JH, Lee SY, Chae HB, Jung YJ, Lee KO, Lim CO, Chung WS, Bahk JD, Yun DJ, Cho MJ, Lee SY. Phosphorylation and concomitant structural changes in human 2-Cys peroxiredoxin isotype I differentially regulate its peroxidase and molecular chaperone functions. FEBS Lett. 2006;580:351–355. doi: 10.1016/j.febslet.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 94.Lim JC, Choi HI, Park YS, Nam HW, Woo HA, Kwon KS, Kim YS, Rhee SG, Kim K, Chae HZ. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J Biol Chem. 2008;283:28873–28880. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]