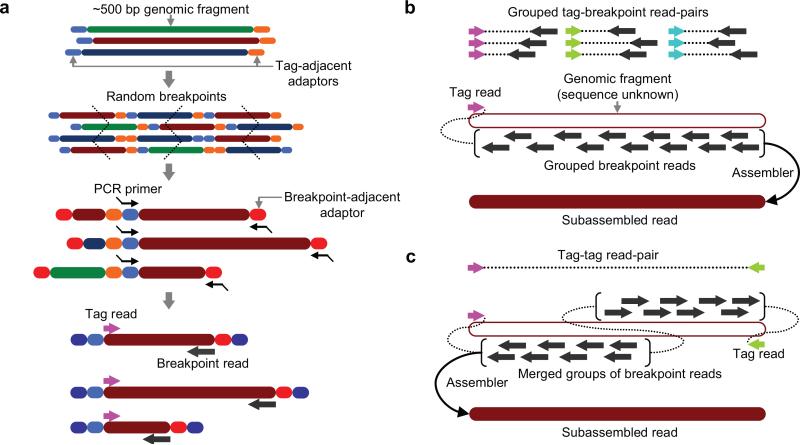
We demonstrate subassembly, an in vitro library construction method that extends the utility of short-read sequencing platforms to applications requiring long, accurate reads. A long DNA fragment library is converted to a population of nested sublibraries, and a tag sequence directs grouping of short reads derived from the same long fragment, enabling localized assembly of long fragment sequences. Subassembly may facilitate accurate de novo genome assembly and metagenome sequencing.
The cost and throughput advantages of massively parallel sequencing are offset by large tradeoffs with respect to read length and accuracy1. Although the availability of reference assemblies renders short reads sufficient for genomic resequencing and digital profiling2,3, other areas such as metagenomics4, de novo assembly of complex genomes5, immunoglobulin diversity profiling6 and molecular haplotyping7 are more challenging. In metagenomics, for example, sequences are derived from a population of related and unrelated genomes with highly varying abundances and a potentially enormous effective complexity. For identifying new open reading frames and for resolving related sequences within such a population, long reads remain indispensable4. Because pyrosequencing produces the longest reads of second-generation platforms8, it largely remains the method of choice for metagenomics4, despite its higher cost and equivalent or higher error rate compared to other second-generation platforms1.
We developed a multiplex, in vitro strategy, termed subassembly, that is conceptually analogous to hierarchical shotgun genome assembly (Fig. 1). In this approach, one of the two reads from a paired-end read serves as a sequence tag that identifies groups of short reads sharing a clonal origin, that is, deriving from the same ~500 base pair (bp) DNA fragment. Each group of short, locally derived reads is then collapsed to a long, subassembled (SA) read. To evaluate performance, we applied this method to two samples: genomic DNA from a (G+C)-rich organism, Pseudomonas aeruginosa strain PAO1, and a previously characterized metagenomic sample from lake sediment9.
Figure 1.
Schematic of subassembly process. (a) Long DNA fragments are ligated to tag-adjacent adaptors, diluted and PCR-amplified. Dilution imposes a complexity bottleneck so that a limited number of long fragments are amplified. Concatemerized PCR products are then sheared by sonication and ligated to a breakpoint-adjacent adaptor. A second PCR amplification prepares amplicons for sequencing; one end of these amplicons corresponds to an end of a long fragment and the other end corresponds to a shearing breakpoint internal to that fragment. (b) Breakpoint reads are grouped in silico based on the sequence of the corresponding tag read. Breakpoint reads within a group, which derive from positions internal to the same parent long fragment, are subjected to local assembly to generate a subassembled read. (c) The metagenomic bottlenecked long-fragment library is subjected directly to paired-end Illumina sequencing to identify pairs of tag reads that were derived from opposite ends of the same original fragment. Two groups of breakpoint reads defined by distinct tag reads are merged and assembled together to generate one or more subassembled reads. In this study, this step was only applied to the metagenomic sample.
For subassembly, we sheared DNA to relatively long lengths (for example, ~500 bp), ligated ‘tag-adjacent’ adaptors to the fragments and then diluted and PCR-amplified these fragments (Fig. 1 and Online Methods). The dilution step before PCR imposed a complexity bottleneck, such that a limited number (~105–107) of long fragments were amplified to high abundance (Supplementary Note 1). The PCR amplicons were concatemerized and then sonicated, and a single ‘breakpoint-adjacent’ adaptor was ligated to the sheared fragments. We performed a second round of PCR in which one primer corresponded to a tag-adjacent adaptor and the other primer corresponded to the breakpoint-adjacent adaptor. The resulting amplicons effectively comprise a population of nested sublibraries derived from the original long-fragment library. The tag-adjacent adaptor provides access to genomic sequence that corresponds to the ends of the long fragments. As this end sequence will be consistent across amplicons derived from the same long fragment, it can serve as a tag to identify molecules that are clonally derived. After paired-end sequencing, the read primed by the tag-adjacent adaptor identifies the original long DNA fragment, and the read primed by the breakpoint-adjacent adaptor represents sequence from a shearing-determined breakpoint in that fragment. As a relatively short read could serve as a unique tag identifier, we obtained paired-end reads of unequal length (20-bp ‘tag read’ and 76-bp ‘breakpoint read’). In the analysis, we used tag reads to group breakpoint reads and separately subjected each tag-defined read group (TDRG) to local assembly with phrap10.
To rigorously assess performance, we applied subassembly to P. aeruginosa strain PAO1. After fragmenting genomic DNA, we size-selected it to ~550 bp (Supplementary Fig. 1) and processed the sample as illustrated in Figure 1. We used Illumina Genome Analyzer II (GA-II) to generate 56.8 million read pairs. We grouped the read pairs into TDRGs by the 20-bp tag (Online Methods) and separately subjected 76-bp breakpoint reads in each TDRG to local assembly with phrap to produce SA reads (Supplementary Table 1). We discarded SA reads not derived from identically oriented breakpoint reads (1.2%) and those failing subassembly entirely (2.7%). For subsequent analyses, we considered only the longest SA read from TDRGs with ≥10 members.
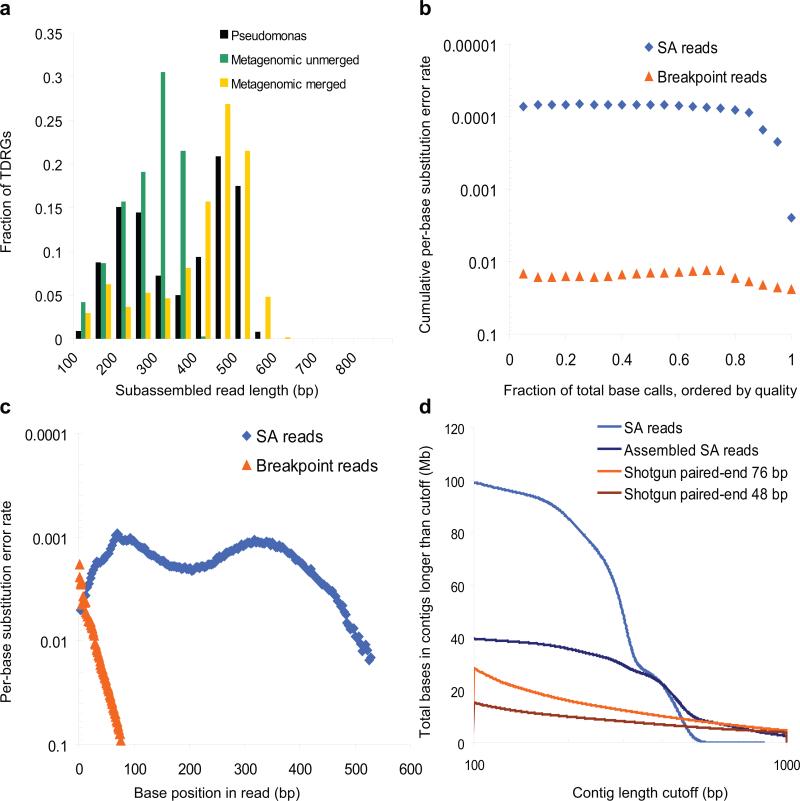
This subset comprised 1.03 million SA reads with a median length of 338 bp (Fig. 2a and Supplementary Table 2). The bimodal distribution may be due to uneven coverage of the original fragment secondary to imperfect size selection (Supplementary Fig. 2). To assess quality, we mapped the SA reads to the P. aeruginosa strain PAO1 reference11 and found that 99.82% had significant (P < 10–6) alignments with basic local alignment search tool (BLAST)12, with 98% of SA reads aligning along ≥95% of their full lengths. Although the contributing Illumina reads had an error rate of 2.4%, the substitution error rate of aligning SA reads was 0.25%. The longest correct SA read was 680 bp, likely an outlier from the gel-based size selection but nonetheless an indicator of the method's potential. We also estimated quality scores for bases in SA reads from the quality scores of contributing breakpoint reads (Online Methods). The 85% of bases in SA reads with the highest estimated quality scores were >99.99% accurate with respect to substitution errors when compared to the P. aeruginosa strain PAO1 reference (Fig. 2b). Finally, we calculated the substitution error rate as a function of position along the SA read. The low overall error rate of one per 400 bp was maintained for hundreds of bases in the SA reads (Fig. 2c).
Figure 2.
Evaluation of subassembly performance. (a) Distribution of subassembled (SA) read length for P. aeruginosa sample and for methylamine metagenomic sample for unmerged and merged pairs of tag-defined read groups. (b) Cumulative per-base substitution error rate of base calls binned as a function of descending base quality in raw and SA reads, or the error rate of the x% of bases with the highest quality scores, after using BLAST to define the corresponding sequence in the reference. (c) Substitution error rate of base calls as a function of base position in raw and SA reads (binned every 3 bases). (d) Total length in sequences longer than a variable cutoff produced from SA reads compared to a standard shotgun library for the 100–1,000 bp range in which metagenomic analyses become possible. SA reads and assembled SA reads were compared to assembly of 48-bp or 76-bp paired-end reads from a standard Illumina shotgun library using Velvet with optimized parameters and an equivalent amount of raw sequence. Assembled SA reads refers to contigs produced by CABOG from SA reads.
Based on alignment with BLAST12, SA reads covered 98.85% of the reference at a mean coverage of 63-fold. We observed bias against regions of extremely high G+C content (>70%) relative to shotgun sequencing (Supplementary Fig. 3), which could be mitigated by optimizing PCR conditions. We also observed slight systematic bias in the distribution of SA read quality scores across the reference that we conclude is unlikely to compromise accuracy at positions with adequate coverage (Supplementary Fig. 3).
To explore the utility of subassembly for de novo genome assembly, we assembled all filtered SA reads using CABOG13, resulting in 708 contigs ≥1 kilobase (kb) with an N50, or the length × such that 50% of the genomic length is in sequences at least × long, of 15 kb (Table 1). The substitution error rate was ~1/14,000, and there was a total of 65 bp of inserted or deleted sequence across 31 contigs. Contigs ≥20 kb, which comprised 2.3 Mb, were more accurate, with a substitution error rate of ~1/250,000 and 20 bp of insertion-deletions across eight contigs. BLAST alignment predicted 11 contigs ranging in size from 1 to 18 kb to contain local misassemblies, but four of these were related to differences between the strain used here and the reference (Supplementary Note 2), leaving only seven true misassemblies. Six of these were very local deletions or expansions of <400 bp (within contigs <20 kb long), and one 1,125 bp contig displayed a more complex BLAST alignment.
Table 1.
De novo assembly of P. aeruginosa genome using subassembled (SA) reads
| Input | Assembly strategy | Number of contigs or scaffolds | Contig or scaffold N50 | Longest contig or scaffold | Total sequence | Physical coverage of reference |
|---|---|---|---|---|---|---|
| SA reads | CABOG | 708 | 15,070 bp | 160,221 bp | 6.07 Mb | 96.2% |
| SA reads plus PE fragment plus jumping mate pair | CABOG and scaffolding | 32 | 444,483 bp | 915,353 bp | 6.11 Mb | 99.3% |
Assembly of SA reads from P. aeruginosa using the CABOG assembler produced long and accurate contigs (≥1 kb) and can be further extended with short (~200 bp) and long (~2.5 kb) mate-pairing data to form scaffolds (≥5 kb).
Shotgun assembly of SA reads therefore resulted in long and highly accurate sequences with contiguity likely limited by sequence content biases. To facilitate scaffolding, we included sequencing data from one lane of a paired-end fragment library (2 × 36 bp; insert size ~200 bp) and one lane of a mate-paired jumping library (2 × 36 bp; insert size ~2.5 kb). Using a custom iterative scaffolding algorithm (Online Methods), we generated 32 scaffolds ≥5 kb, with scaffold N50 of 445 kb, longest scaffold of 915 kb and 99.3% physical coverage of the reference (Table 1). Notably, scaffolding introduced only one misassembly, likely because of the presence of multiple nearly identical phage-like insertions (Supplementary Note 2). Our results, which were generated from a single platform, compare favorably to summary statistics of a published de novo assembly from a related organism that had been generated by combining long-read 454 and short-read Illumina data14 (Supplementary Note 3).
To evaluate subassembly on a complex metagenomic sample, we used total DNA isolated from lake sediment and enriched for methylamine-fixing microbes9. We started with a slightly shorter long-fragment library (~450 bp; Supplementary Figs. 1,4) and imposed a more stringent complexity bottleneck by diluting the long-fragment library to ~105–106 molecules before PCR (Online Methods). We obtained 21.8 million read pairs, which resulted in 262,298 TDRGs, in which the median length of the longest SA read in filtered TDRGs was 256 bp (Supplementary Table 2 and Fig. 2a).
In addition to the nested breakpoint reads that we used to produce SA reads, we also obtained 1.8 million paired-end reads from the original long-fragment library (2 × 20 bp), allowing us to merge TDRGs whose tags were observed as a read pair (Fig. 1). We merged ~68% of the metagenomic TDRGs in this fashion. Subjecting breakpoint reads from merged TDRGs to local assembly yielded SA reads with a median length of 408 bp (Fig. 2a and Supplementary Table 2).
We hypothesized that localized, tag-directed assembly would be particularly useful in the context of metagenomics, for which the highly nonuniform representation of organisms complicates de novo assembly from short reads. To test this, we generated a standard Illumina shotgun paired-end library from the same metagenomic sample and assembled reads from this library with Velvet15 using optimized parameters (Supplementary Table 3 and Supplementary Fig. 5). We evaluated shotgun assemblies from both paired-end 76-bp reads and paired-end 48-bp reads. For both assemblies, we used 2.2 Gb of raw sequence, which was equal to the amount of data used for subassembly.
CABOG assembly of SA reads yielded considerably more total sequence data in longer contigs than direct assembly of shotgun reads, generating greater than twice as much sequence in contigs ≥200 bp (Fig. 2d and Supplementary Table 3). Unassembled SA reads comprised greater than five times as much sequence ≥200 bp. Notably, shotgun assemblies did achieve greater contiguity at the longest lengths (Supplementary Table 3 and Supplementary Fig. 5). These long contigs may be due to deep sampling of the most abundant genomes. However, many are likely to represent misassemblies, as we did not observe long BLAST alignments to the available Sanger sequence data9 or to any sequence in the GenBank nt or env_nt databases.
To conservatively estimate each method's effective coverage, we compared assembled contigs to 37.2 Mb of Sanger sequence data recently reported for the same sample9 (Online Methods and Supplementary Fig. 6). Although the complexity of the metagenomic sample likely remains undersampled, subassembly covered at least 45% more of the Sanger sequence reference when compared to contigs assembled from the paired-end short-read library. In addition, subassembly generated a comparable amount of total sequence as compared to Sanger sequencing data (39.5 Mb versus 37.2 Mb) in somewhat shorter contigs (median of 390 bp versus 835 bp) but with considerably less effort (three Illumina sequencing lanes versus hundreds of Sanger sequencing runs). In summary, subassembly produced substantially more sequence at lengths necessary for accurate phylogenetic classification16 and gene discovery17 than direct assembly from shotgun short reads and did so in better agreement with the available Sanger sequencing data, suggesting that the quality of assembled data may also be higher.
Given that we observed accurate SA reads of nearly 700 bp, optimization of this method in concert with the tag-pairing approach (Fig. 1) could potentially extend the effective length of SA reads to ~1 kb, that is, approaching the maximum length of Sanger sequencing data. One potential concern about the method as described is that tag sequences from different long DNA fragments can occasionally be identical by chance, especially if samples contain repetitive elements at high abundance. A simple modification would be to use a tag-adjacent adaptor containing an embedded degenerate sequence (for example, a randomized 20-bp segment), as this would completely decouple the tag sequence from the sample composition.
Finally, we note that subassembly offers a fundamental advantage in the way that a low error rate is achieved with a second-generation sequencing platform. Accurate assembly of short shotgun reads can be successful, provided that these reads are derived from relatively random sequence and that deep, uniform coverage can be obtained15. Platforms such as Roche 454 offer long reads at a cost that is likely similar to subassembly (Supplementary Note 4) but have error profiles comparable to those of other second-generation sequencing platforms. Therefore, achieving high consensus accuracy also depends on the assumptions of uniform sampling and of a common origin for nearly identical reads. In contrast, because subassembly samples individual long DNA fragments and separately reconstructs a consensus sequence for each one, the production of long, accurate SA reads is insulated from nonuniform representation and sequence relatedness in the sample of interest.
Methods
Data availability
Raw Illumina sequence reads, unfiltered SA reads, P. aeruginosa strain PAO1 contigs from SA reads and P. aeruginosa strain PAO1 scaffolds from SA reads are available at http://krishna.gs.washington.edu/subassembly/.
Subassembly library construction
An overview of library construction steps, including time estimates to aid in planning experiments, is available in Supplementary Protocol 1. Library construction methods are described in detail in Supplementary Protocols 2 and 3. Briefly, library construction proceeded as follows. Source DNA was fragmented by sonication, end-repaired and size-selected to ~550 bp (P. aeruginosa) or ~450 bp (metagenomic sample). Size-selected fragments were A-tailed and ligated to custom adaptors (Supplementary Table 4). Real-time PCR with phosphorylated primers was performed using serial dilutions of adaptor-ligated fragments to impose a complexity bottleneck and generate many copies of a limited number of long fragments. Complexity was estimated from the concentration of input material, the kinetics of PCR amplification and gel electrophoresis of the PCR product. After PCR, the product estimated to have resulted from ~105–107 long fragments was concatemerized to high molecular weight and then fragmented by sonication. Shearing products were end-repaired, A-tailed and ligated to the Illumina Read 2 adaptor. PCR amplification was then performed with one primer corresponding to the Read 2 adaptor and a second primer corresponding to one of the two original adaptors. Finally, the amplification products were size-selected to obtain a uniform distribution of shearing products across the original fragment (Supplementary Fig. 2). For the metagenomic effort, an aliquot of the bottleneck PCR was subjected to an additional round of PCR to prepare the long fragments for paired-end sequencing and subsequently used for tag-pairing and TDRG merging.
Shotgun library construction
P. aeruginosa short insert (~200 bp) and long insert (~2.5 kb), and metagenomic short insert shotgun libraries were constructed according to manufacturer's specifications, except that standard oligonucleotides were obtained from IDT. For the metagenomic library, to conserve source material, size selection to the desired fragment length was performed before A-tailing and adaptor ligation rather than afterward so that the longer size range could be used for subassembly.
Illumina sequencing
For subassembly libraries, an Illumina GA-II instrument was used to collect paired-end reads according to manufacturer's specifications, except that custom sequencing primers (Supplementary Table 4) were used, and asymmetric read lengths were collected (20-bp first read and 76-bp second read). For the tag-pairing metagenomic library, paired-end 36-bp reads were collected according to manufacturer's specifications with custom sequencing primers. For shotgun libraries, paired-end reads were collected according to manufacturer's specifications.
Organizing breakpoint short reads into TDRGs
For all experiments, breakpoint reads paired with identical or nearly identical tag sequences were grouped into TDRGs. As millions of tag reads were involved, an all-against-all comparison to cluster similar tags was not feasible. Instead, a two-step strategy was used to group tag sequences in each experiment. First, perfectly identical tags were collapsed using a simple hash to define a nonredundant set of clusters. From this set, clusters with four or more identical tags were identified as ‘core’ clusters and, in descending order by size, were compared to all other tags. Tags matching a given core cluster with up to one mismatch were grouped with that core cluster (and removed from further consideration if they themselves defined a smaller core cluster). TDRGs with more than 1,000 members were excluded from downstream analysis to limit analysis of adaptors or other low-complexity sequence.
Subassembly of TDRGs
Each TDRG was assembled separately using phrap with the following parameters: “-vector_bound 0 -forcelevel 1 -minscore 12 -minmatch 10 -indexwordsize 8”. Pregrouping reads into TDRGs allowed us to use less stringent parameters than the defaults used in traditional assemblies. Parameters were optimized to balance SA read length and accuracy (Supplementary Table 1). A short-read assembler, Velvet, was also tested but did not produce substantial gains in SA read length relative to phrap (data not shown).
Trimming and filtering of SA reads and assignment of consensus quality scores. SA reads were masked using the cross_match program provided as part of the phrap suite, using the following parameters: “-minmatch 5 -minscore 14 –screen”. Determination of consensus quality scores and further trimming was performed as follows. Because it permits multiple alignments per read, the Bowtie short-read alignment tool18 was used to map contributing 76-bp breakpoint reads to the SA reads to generate consensus quality scores for SA read base calls. Only alignments within TDRGs were allowed (that is, alignments of breakpoint reads to SA reads from another TDRG were ignored). Bowtie was also used to map the 20-bp tag reads back to the SA reads to facilitate end trimming where the SA read had extended into adaptor sequence. Next, SA reads were trimmed using both tag read mapping and adaptor masking information. SA reads were first trimmed from the 3′ end using the mapping location of the tag read; if bases remained that had been masked by cross_match because of the presence of adaptor, the masked bases were removed and the longest remaining continuous sequence was retained. Finally, any sequence containing a base call with quality below 10 within 5% of the 3′ end of the SA read was discarded.
In all subsequent analyses, only SA reads that were at least 77 bp long and were assembled from identically oriented short reads were considered. The read orientation filter was only applicable to SA reads from individual, unmerged TDRGs. In addition, for length and quality analyses, only the longest SA read from each TDRG was analyzed.
Quality assessment
The longest SA read (after trimming as described above) from each TDRG containing at least 10 member reads was aligned to the P. aeruginosa PAO1 reference genome using BLAST with the following parameters: “-p blastn -e 1e-6 -m 8 -F F -a 4”.
Error rate as a function of quality score and position in the SA read was then determined as follows. BLAST alignments containing at least 95% of the length of the SA read query and without any gap openings were used to define the position in the reference of the SA read in question (the BLAST coordinates were extended to encompass the entire length of the SA read). Every base in a SA read whose alignment meets the above criteria was compared to the corresponding reference base. If less than 100% of the SA read aligned, the comparison was forced to extend to the ends of the SA read. From the base-by-base comparison, the error rate as a function of base call quality or position in the SA read was calculated.
We did not perform a base-by-base comparison for cases in which BLAST used a gap opening in making an alignment, which could potentially suppress our error rates if such SA reads were substantially more error-laden. Accuracy of such SA reads within aligned regions was slightly lower (99.56% accurate compared to 99.86% in SA reads without gaps), and such sequences only comprised less than 1% of the sequence being analyzed. We therefore concluded that errors in these sequences that fall outside of aligning regions are unlikely to substantially alter our estimates of error rate as a function of base quality. We performed a similar analysis for SA reads containing larger gaps with respect to the reference (those with a BLAST alignment less than 95% of their length), as we did not perform a base-by-base comparison for such SA reads either. Once again, the accuracy with aligned regions was somewhat lower (99.4% versus 99.86% in those with complete or nearly complete alignments). Such errors probably reflect larger-scale misassemblies owing to repetitive sequence in the true reference sequences. Notably, aggressive trimming substantially reduced the relative abundance of such sequences; only 1.5% of the total number of bases analyzed was contained in such sequences, and only 2.3% of BLAST alignments fell into this category. Once again, forcing the alignment to the very edges of such SA reads was not likely to substantially alter the relationship between error rate and base call quality score.
To analyze quality as a function of raw read base quality, maq was used to align contributing 76-bp breakpoint reads to the reference, Illumina base calls were compared to the reference and, for a randomly chosen subset of 1 million bases, the error rate as a function of Illumina base call quality was determined.
To analyze quality as a function of raw read position, a representative lane of contributing 76-bp breakpoint reads used for the subassembly process was aligned to the reference genome using maq, and the error rate at each position was determined by comparing read base calls to reference bases for each read.
Assembly of SA reads using the Celera assembler (CABOG). For P. aeruginosa and metagenomic samples, all trimmed, orientation- and length-filtered SA reads (not only the longest per TDRG) were subjected to assembly using the Celera assembler. Assembly was guided by consensus quality scores generated as described above. The Celera assembler (CABOG) was run with default parameters and “unitigger=bog”.
Assessment of assembled SA read quality
Contigs produced by the Celera Assembler from SA reads were aligned to the reference using BLAST with the following parameters: “-p blastn -e 1e-6 -m 8 -F F”. Substitution error rate was measured as the number of mismatches within the best BLAST alignment for each contig. To account for a potentially higher error rate in misassembled contigs, if a contig aligned across less than 95% of its length, other BLAST alignments were also considered as long as they comprised at least 10% of the contig length.
Scaffolding of contigs for P. aeruginosa
For de novo assembly of the P. aeruginosa genome, we used independently produced shotgun sequencing libraries to scaffold the contigs produced from SA reads as follows. The resulting contigs were scaffolded using a custom script that used 36-bp shotgun paired-end Illumina reads from one lane each of short-insert (~200 bp) and long-insert (~2.5 kb) libraries. The gap between each pair of adjacent contigs in a scaffold was dynamically estimated based on the distance of the read pairs connecting the two contigs from the ends of the contigs and the expected insert size of the library from which they were derived. Scaffolds were then constructed by separating the contigs by a string of unknown nucleotides (Ns) as long as the estimated gap size. For cases where the expected gap size was close to zero or negative (indicating a possible overlap), the adjacent ends of the two contigs were subjected to a Smith-Waterman alignment and merged accordingly if a match was detected.
TDRG merging algorithm
Paired 36-bp reads were obtained from a sequencing library prepared from bottlenecked, adaptor-ligated metagenomic fragments (Supplementary Protocol 2), then trimmed computationally to 20 bp to correspond to the length of the tag reads that were obtained during sequencing of the subassembly libraries.
To prevent sequencing errors at the ends of the reads from creating spurious tags and tag pairs, we trimmed the reads further to the first 15 bp. If multiple TDRGs (defined by 20-bp tags) could correspond to a single 15-bp tag from a merging read pair, the TDRG with the most members was chosen. In descending order of tag-pair abundance, we defined TDRG pairs, removing tags that had been assigned to TDRG pairs as we proceeded.
Velvet assembly of shotgun metagenomic library. Paired-end shotgun reads constructed according to standard Illumina protocols were assembled using Velvet with the following parameters: “-cov_cutoff 2 -exp_cov [variable] -ins_length 250 -unused_reads yes”.
If exp_cov was set to 1, cov_cutoff was set to 0. As Velvet (along with all other short-read assemblers) is not designed for assembly of metagenomic sequences, considerable effort was made to optimize its performance with respect to length of sequences produced and agreement with the available Sanger sequencing data to make the fairest comparison possible. We found that contig length was sensitive to the exp_cov parameter (Supplementary Fig. 5). However, we observed unpredictable performance with respect to agreement with the Sanger sequencing data when altering this parameter, as agreement improved for the paired-end 76-bp reads but degraded for the paired-end 48-bp reads. We therefore chose an exp_cov value of 100 as the best compromise of sequence length and coverage for the comparator datasets.
Resulting scaffolds were then split into contigs that did not contain Ns, as we reasoned that key goals of metagenomic sequencing such as gene discovery and phylogenetic classification would depend solely on the length of contiguous regions of defined bases.
Comparison to Sanger sequencing data with BLAST. Contigs produced from SA reads with CABOG and contigs produced from shotgun short reads with Velvet were aligned to one another and to the recently collected Sanger sequencing data from the same sample 18. Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Genome Biol. 10, R25 (2009).
(JGI IMG/M Taxon Object ID 2006207002, NCBI accession number ABSR01000000) using BLAST with the following parameters: “-p blastn -e 1e-6 -m 8 -F F”. Two bases were considered to be a shared position between two datasets if they were contained in a BLAST alignment at least 100 bp long and with at least 98% identity. For the Venn diagram (Supplementary Fig. 6), an additional restriction was added so that mappings between the three datasets were not ambiguous: the two bases were required to be in the BLAST alignment with the highest bit score of all the BLAST alignments between the two datasets involving either base.
Supplementary Material
Acknowledgments
We thank L. Chistoserdova and M.G. Kalyuzhnaya (University of Washington) for the gift of the methylamine-enriched metagenomic DNA sample, C. Manoil (University of Washington) for the gift of P. aeruginosa strain PAO1 genomic DNA and P. Green for helpful discussions. J.B.H. is supported by US National Institutes of Health grant T32GM007266 and an Achievement Rewards for College Scientists fellowship.
Footnotes
Accession codes. NCBI Sequence Read Archive: SRA010316.
References
- 1.Shendure J, Ji H. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 2.Hillier LW, et al. Nat. Methods. 2008;5:183–188. doi: 10.1038/nmeth.1179. [DOI] [PubMed] [Google Scholar]
- 3.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 4.Hamady M, Knight R. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Simpson JT, et al. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstein JA, Jiang N, White RA, 3rd, Fisher DS, Quake SR. Science. 2009;324:807–810. doi: 10.1126/science.1170020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentley G, et al. Tissue Antigens. 2009;74:393–403. doi: 10.1111/j.1399-0039.2009.01345.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Margulies M, et al. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalyuzhnaya MG, et al. Nat. Biotechnol. 2008;26:1029–1034. doi: 10.1038/nbt.1488. [DOI] [PubMed] [Google Scholar]
- 10.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 11.Stover CK, et al. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, et al. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myers EW, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 14.Reinhardt JA, et al. Genome Res. 2009;19:294–305. doi: 10.1101/gr.083311.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zerbino DR, Birney E. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brady A, Salzberg SL. Nat. Methods. 2009;6:673–676. doi: 10.1038/nmeth.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw Illumina sequence reads, unfiltered SA reads, P. aeruginosa strain PAO1 contigs from SA reads and P. aeruginosa strain PAO1 scaffolds from SA reads are available at http://krishna.gs.washington.edu/subassembly/.