Abstract
We have investigated the ability of fetal mesenchymal stem cells (fMSCs) to differentiate into brown and white adipocytes and compared the expression of a number of marker genes and key regulatory factors. We have shown that the expression of key adipocyte regulators and markers during differentiation is similar to that in other human and murine adipocyte models, including induction of PPARγ2 and FABP4. Notably we found that the pre-adipocyte marker Pref-1, is induced early in differentiation and then declines markedly as the process continues, suggesting that fMSCs first acquire pre-adipocyte characteristics as they commit to the adipogenic lineage, prior to their differentiation into mature adipocytes. After adipogenic induction, some stem cell isolates differentiated into cells resembling brown adipocytes and others into white. Detailed investigation of one isolate showed that the novel brown fat determining factor PRDM16 is expressed both before and after differentiation. Importantly these cells exhibited elevated basal UCP-1 expression, which was dependent on the activity of the orphan nuclear receptor ERRα, highlighting a novel role for ERRα in human brown fat. Thus fetal MSCs represent a useful in vitro model for human adipogenesis, and provide opportunities to study the stages prior to commitment to the adipocyte lineage. They also offer invaluable insights into the characteristics of human brown fat.
Keywords: Mesenchymal Stem Cells, Adipogenesis, Adipocytes, Brown, PRDM16, UCP1, ERRalpha
Introduction
The developmental programmes that control the generation of white adipose as a storage depot for triglyceride versus brown fat for adaptive thermogenesis are poorly understood. While they both express common marker genes including Fatty Acid Binding Protein 4 (the human homologue of aP2 in the mouse) brown fat has a higher mitochondrial content and increased expression of many genes involved in adaptive thermogenesis (Cannon and Nedergaard, 2004). The regulation of adipogenesis from pre-adipocytes has been examined extensively in murine models (Farmer, 2006) and led to the identification of PPARγ and C/EBPα as a key transcription factors. Conversely, Pref-1 (Pre-adipocyte factor 1, also identified as Dlk1), a trans-membrane protein related to the EGF (Epidermal Growth Factor) that is highly expressed in pre-adipocytes, blocks adipogenesis, but its levels fall with the onset of differentiation. Exogenous expression of Pref-1 blocks differentiation of 3T3-L1 pre-adipocytes (Smas and Sul, 1993) while overexpression in transgenic mice results in lipodystrophy (Lee et al., 2003); Pref-1 null mice on the other hand have accelerated adiposity (Moon et al., 2002).
It was generally assumed that brown and white fat cells were derived from a common adipogenic progenitor. However, expression profiling indicated that brown fat cells express a large number of muscle related genes (Timmons et al., 2007), and genetic studies suggest that brown adipose tissue may have a distinct origin from brown adipocytes found in white adipose tissue (Timmons et al., 2007). Moreover a population of cells derived from human skeletal muscle can differentiate in vitro into brown adipocytes (Crisan et al., 2008). Recent work indicates that brown adipose tissue and skeletal muscle are derived from common Myf5 expressing progenitor cells and that the transcriptional regulator, PRDM16, controls a brown fat/skeletal muscle switch (Seale et al., 2008). Exogenous PRDM16 expression was able to drive brown adipogenesis, but only if expressed prior to the onset of differentiation (Seale et al., 2007). Nevertheless, brown fat cells that appear in white adipose tissue upon cold exposure do not appear to be derived from these Myf5 expressing progenitor cells (Seale et al., 2008), and so it is conceivable that these brown fat cells are derived from a common adipogenic precursor cell.
Interestingly, manipulation of a number of cofactors for nuclear receptors suggests that it may be possible to switch white fat into brown fat. Thus exogenous expression of PGC1α (Puigserver et al., 1998) in white fat cells or the deletion of the corepressor RIP140 results in the expression of UCP1 (Christian et al., 2005; Leonardsson et al., 2004; Morganstein et al., 2008), a characteristic marker of brown fat cells. The coactivator PGC1β is also important for the expression of many typical brown fat genes (Lelliott et al., 2006).
Mesenchymal stem cells can be expanded in vitro and induced to differentiate into cells of multiple mesenchymal lineages (Pittenger et al., 1999). Human adult mesenchymal stem cells have been shown to differentiate into white adipocytes (Mackay et al., 2006) but their ability to differentiate into brown adipocytes is unclear. One study of human amniotic fluid derived stem cells did address the phenotype of the differentiated cells and found low levels of expression of the brown fat markers UCP-1 and PGC1α. However this expression was lower than that seen in adult sub-cutaneous white adipocytes (Gemmis et al., 2006) which the differentiated stem cells most closely resembled. Mesenchymal stem cells have been isolated from blood, liver and bone marrow of first trimester human fetuses, and these can also differentiate into adipocytes (Campagnoli et al., 2001; Ryden et al., 2003). However, the process of adipogenesis and the functional properties of the derived adipocytes was only minimally characterised. Therefore we have studied first trimester cells to investigate their potential to differentiate into either white or brown adipocytes. We therefore studied the process of adipogenesis in fetal mesenchymal stem cells to determine whether key markers and regulators were expressed in a similar manner to that previously described in other adipocyte models (Farmer, 2006; Morganstein et al., 2008; Nakamura et al., 2003; Tomlinson et al., 2006) and to characterise further the differentiated adipocytes.
Results
Adipogenic Differentiation of fMSCs
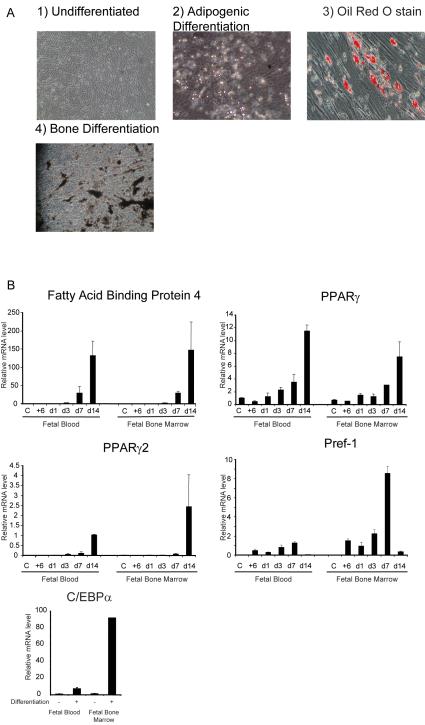
Initially we confirmed previous reports that fetal mesenchymal stem cells were able to differentiate into adipocytes (Campagnoli et al., 2001). After 14 days, droplets were clearly visible in approximately 30% of cells, and staining with Oil Red O confirmed the presence of intracellular lipid (Fig 1 A). To establish the mesenchymal stem cell origin of the isolates, we demonstrated that they were also able to differentiate into bone (Fig 1 A). We next analysed the expression of a number of adipocyte markers and regulatory factors that were expressed during mouse adipogenesis. Expression analysis was performed with two different isolates (one from fetal blood and one from fetal bone marrow from different fetuses) before and after treatment with adipogenic differentiation media. Expression of the adipocyte marker Fatty Acid Binding Protein 4 (the human homologue of the mouse gene aP2) was detected after 3 days and increased progressively with a 150 fold induction after 14 days (Fig 1B), in parallel with lipid accumulation.
Fig 1.
Fetal Mesenchymal stem cells differentiate into adipocytes
A) Fetal Mesenchymal Stem Cells before (1) and after differentiation. into adipocytes and bone as demonstrated by light microscopy (2), Oil Red O staining to demonstrate lipid accumulation (3) or Von Kossa staining to demonstrate mineralisation (4) (400x magnification).
B) Time course of mRNA expression of key markers and regulators of adipocytes by quantitative PCR. C= Confluent cells, +6 = 6 hours post hormonal induction of differentiation, all other time points indicate days post induction. Fetal Blood and Fetal Bone Marrow indicate the source of isolate. Error bars indicate SD. Relative expression in arbitrary units.
The key adipogenic regulator PPARγ was expressed in undifferentiated stem cells but increased throughout differentiation and after 14 days showed an 8-12 fold upregulation compared to untreated cells. PPARγ 2, the adipocyte specific isoform of PPARγ was not expressed in undifferentiated cells but was induced markedly during differentiation (Fig 1 B). Expression of C/EBPα also increased following differentiation (Fig 1 B). Pref-1/Dlk1, which suppresses the differentiation of mouse pre-adipocytes into adipocytes, was not detectable in confluent untreated fMSCs but addition of adipogenic cocktail led to expression within 6 hours, consistent with the appearance of pre-adipocytes (Fig 1 B). Expression levels increased for 7 days but declined markedly by day 14, when a significant proportion of pre-adipocytes had undergone differentiation into mature adipocytes. It seems, therefore, that fMSCs express many markers and regulators of adipocytes at the appropriate times and the pattern of expression of these genes was remarkably similar in the two isolates of fMSCs examined.
Isolates of fMSCs can differentiate into either brown or white adipocytes
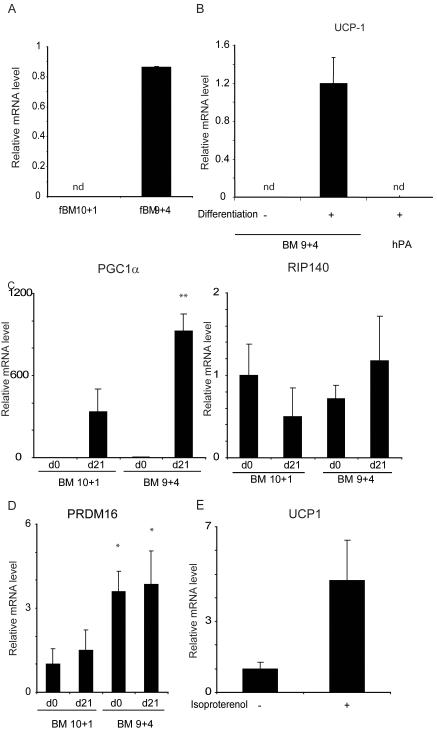
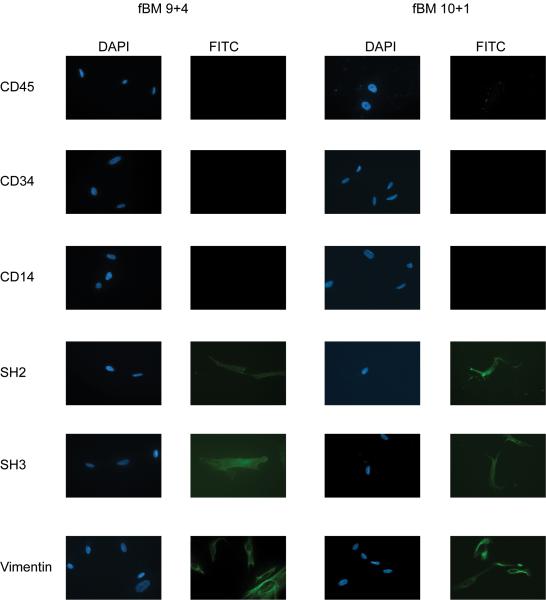
The ability of fMSCs to differentiate into brown fat cells was examined by determining the expression of UCP-1 after adipogenic differentiation using identical differentiation conditions shown to promote adipogenesis in adult MSCs (adapted from (Janderova et al., 2003). Analysis of 5 different isolates indicated a wide range of UCP-1 expression (Supplementary Figure 1) and so we examined two from fetal bone marrow in detail. One obtained at 10 weeks plus 1 day gestation (BM 10+1) gave rise to adipocytes with virtually undetectable expression of UCP1 while the other from a 9 weeks and 4 day gestation fetus (BM 9+4) differentiated into adipocytes that expressed high levels of UCP1 (Fig 2A). The mesenchymal stem cell identity of both isolates was confirmed by characteristic expression of markers: SH2, SH3 and Vimentin positive and CD14, 34, and 45 negative (Fig 3), and by their ability to differentiate into bone (data not shown). Expression of UCP1 was dependent on differentiation, and considerably higher than the levels of UCP1 found in human adult white adipocytes derived from the in vitro differentiation of sub-cutaneous preadipocytes (Fig 2B). Given that UCP1 gene expression is induced during thermogenesis in response to PGC1α expression (Puigserver et al., 1998), we analysed the expression of this co-factor. PGC1α expression was markedly induced in both isolates following differentiation, and so whilst it is expressed at higher levels in the BM 9 week isolate compared to the 10 week cells, it cannot account for the marked difference in UCP1 gene expression. The possibility that differential RIP140 expression might account for the difference in UCP1 expression was also investigated, but its level was similar in both isolates before and after differentiation (Fig 2C). The brown fat regulator PRDM16 was also expressed in both isolates, both before and after differentiation into adipocytes (Fig 2 D), although interestingly it was significantly higher in the 9+4 (brown) fMSCs than in the 10+1 (white) cells. It would therefore appear that certain isolates of fMSCs have a propensity to differentiate into white adipocytes and others into brown adipocytes, despite identical differentiation conditions. It is also noteworthy that basal expression could be further induced by treating with isoproterenol to mimic the effects of β-adrenergic agents (Fig 2 E). Thus the BM 9+4 isolate retained properties expected in brown adipocytes. It should be noted that these two isolates came from fetuses differing by only 4 days in gestational age and examined at similar passage number.
Fig 2.
Two different isolates of fetal mesenchymal stem cells differentiate into adipocytes which resemble white and brown adipocytes respectively.
A) UCP-1 mRNA expression in fMSC 10+1 and 9+4 isolates after 21 days of differentiation.
B) UCP-1 mRNA expression in fMSC 9+4 isolatre before and after 21 days of differentiation, and differentiated human adult sub-cutaneous adipocytes (hPA). nd = not detected.
C and D) mRNA expression of PGC1α and RIP140 (C) and PRDM16 (D) in the two isolates before and after differentiation. ** indicates p<0.001, * p<0.05 for comparison between the two isolates.
E) Expression of UCP-1 mRNA in 9+4 isolate after 21 days differentiation, before and after treatment with 10μM Isoproteranol for 5 hours.
Error bars indicate SD. nd indicates not detected. Relative expression in arbitrary units.
Fig 3.
Immunophenotyping confirms Mesenchymal Stem Cell Charecterisitics
Undifferentiated fMSC 9+4 and 10+1 cells express SH2, SH3 and Vimentin, but not CD14, CD 34 or CD45, confirming there mesenchyme origin.
ERRα is a regulator of UCP1 expression in human fMSC derived brown adipocytes
A number of nuclear receptors have been shown to regulate UCP-1 gene expression (del Mar Gonzalez-Barroso et al., 2000; Puigserver et al., 1998). Therefore differentiated 9+4 cells were treated with agonists or antagonists for relevant nuclear receptors, and the effects on UCP1 mRNA levels determined.
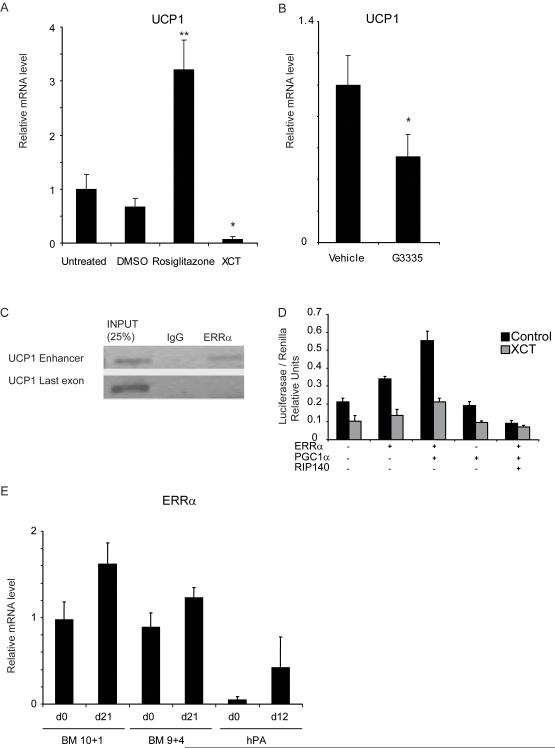
In keeping with the known role of PPARγ in regulating UCP1 expression, its agonist rosiglitazone resulted in a 4 fold induction of UCP1 expression whereas the antagonist G3335 resulted in a 50% inhibition of UCP1 expression (Fig 4 A and B). We postulated that the residual expression might reflect the action of other nuclear receptors. We found that an inverse agonist for ERRα, XCT790 (Busch et al., 2004), resulted in almost complete suppression of UCP1 expression (Fig 4 A). Thus we conclude that while PPARγ activation contributes to the expression of UCP1, ERRα is the predominant nuclear receptor responsible for its expression in the absence of β adrenergic stimulation. Expression of the fatty acid binding protein FABP4 was also examined. This is a well characterised target of PPARγ and accordingly its expression was increased approximately 15 fold following treatment with Rosiglitazone (Supplementary figure 2). However XCT790 did not reduce expression of this gene, and in fact resulted in a small induction. Thus it appears that PPARγ and ERRα control transcription from distinct subsets of genes in these adipocytes.
Fig 4.
UCP-1 expression in fMSC 9+4 adipocytes can be induced by Rosiglitazone treatment, and reduced by the ERRα inverse agonist.
mRNA expression determined by qPCR
A)UCP-1 expression following Rosiglitazone or XCT790 treatment for 48 hours. * p=<0.05, ** = p<0.005.
B)UCP-1 expression after 48 hour treatment with PPARg anatagonist G3335 * = p<0.05.
C) Chromatin Immunoprecipitation showing ERRa bound to the UCP1 enhancer element in differentiated 9+4 fMSCs, but not to a region in the coding sequence of the same gene.
D) Reporter assay using this construct in Cos-1 cells. ERRa activates the enhancer, and its activity is further trans-activated by PGC1a and repressed by RIP140. The ERRa inverse agonist XCT790 prevents this induction.
E) ERRα expression in 10+1 and 9+4 isolates before and after 21 days adipogenic differentiation. mRNA expression by qPCR. Error Bars indicate SD.
Previous analysis of both the mouse and human UCP1 gene has identified an enhancer responsible for mediating the stimulatory effects of a number of nuclear receptors (del Mar Gonzalez-Barroso et al., 2000; Kozak, 1994) and several putative ERRα binding sites were identified (Supplementary Figure 3). We therefore performed Chomatin Immunoprecipitation experiments to investigate the binding of ERRα to the enhancer region of UCP1. We found that ERRα was bound in the vicinity of the UCP1 enhancer element in differentiated adipocytes from the fMSC 9+4 isolate but not the last coding exon, which we tested as a negative control. To confirm that the effects of ERRα were mediated through this site, the 220bp enhancer element of the human UCP1 gene was cloned into the reporter plasmid pGL3-TK, and transiently transfected into Cos-1 cells. We found that the expression of ERRα resulted in a 50% increase in reporter activity and this was further increased by the expression of PGC1α (Fig 4 D). Importantly, the ERRα inverse agonist XCT790, reduced the induction of reporter gene activity by ERRα and PGC1α. PGC1α alone had no effect on activity. Conversely RIP140 expression blocked the ability of ERRα and PGC1α to activate transcription, in keeping with its role as a suppressor of UCP1 transcription in murine adipose tissue.
ERRα mRNA levels were then determined in both the stem cells and adipocytes derived from the two isolates of fMSCs described above. ERRα expression was similar in the two isolates both before and after differentiation, although both lines expressed higher levels than adult sub-cutaneous adipocytes (Fig 4 E). Thus although ERRα is required for the expression of UCP-1 in the 9+4 cells it is not sufficient in the absence of other factors.
Discussion
Our study demonstrates the ability of fMSCs to differentiate into either brown or white adipocytes under identical cell culture conditions. Both regulators such as PPARγ and markers such as FABP4 were upregulated during differentiation in a similar manner to that seen in well described murine and human adipocyte cell models (Farmer, 2006; Hung et al., 2004; Sekiya et al., 2004; Tomlinson et al., 2006). Of note, the adipocyte specific isotype of PPARγ, PPARγ2, was markedly upregulated during adipogenesis in contrast to a previous report in which cells (Gemmis et al., 2006) from second trimester amniotic fluid were used. This difference might reflect the greater proportion of cells undergoing adipogenesis (>30%) in our study, although a fundamental difference between the first trimester MSCs used here and second trimester cells cannot be ruled out. It should be noted that an even higher proportion of cells underwent adipogenic differentiation in previous studies that used single cell clones (Campagnoli et al., 2001). However due to the limited survival of isolated MSCs in culture, we did not feel this would be a viable approach to proceed to fully characterizing the differentiated adipocytes. Of particular interest is the pattern of expression of the pre-adipocyte marker Pref-1, which rises early in the differentiation process, prior to falling in the later stages. The induction of Pref-1 expression suggests that the stem cells first commit to the adipogenic lineage, acquiring the features of pre-adipocytes, prior to undergoing differentiation into mature adipocytes.
The ability of fMSCs to differentiate into brown adipocytes was dependent on the isolate tested and was unaffected by the precise hormonal treatments used to induce differentiation. Interestingly, the BM 9+4 isolate which could undergo differentiation into brown adipocytes was also able to differentiate into other mesenchymal lineages. The discovery of an isolate of fMSCs that differentiated into brown adipocytes suggests that this specialised adipose cell is indeed derived from the mesenchymal lineage, along with white adipocytes. This is in keeping with recent observations that brown adipocytes may share developmental origins with muscle cells, which are also of mesenchymal origin. A comparison of this isolate with BM 10+1 fMSC that differentiates into white adipocytes shows that differences in expression of the nuclear receptor coregulators RIP140 or PGC1 co-activators can not explain the alternative fates of the two isolates, despite the role for these regulators in mature white and brown adipocytes. Interestingly PRDM16 was expressed at a higher level in the 9+4 isolate that was capable of differentiating into brown adipocytes, both before and after differentiation. Nevertheless given that PRDM16 was also expressed in the 10+1 isolate, albeit at lower levels, it seems that PRDM16 is not sufficient to account for the alternative cell fates of these two isolates. It remains unclear if these cells represent an in vivo source of both white and brown adipocytes and what factors may influence stem cell commitment during development.
A number of nuclear receptors have been shown to regulate Ucp1 expression, including PPARγ and the retinoic acid receptor (del Mar Gonzalez-Barroso et al., 2000). In this study analysis of UCP1 expression in the fMSC derived brown adipocytes has shown a novel role for the orphan nuclear receptor ERRα in regulating human UCP1 expression. ERRα is expressed at high levels in brown fat, and is important in the regulation of many genes involved in fatty acid metabolism, such as medium-chain acyl coenzyme A dehydrogenase (Sladek et al., 1997). The role of this receptor in Ucp1 gene regulation in the mouse is unclear. Mice devoid of ERRα express increased amounts of Ucp1 in their white adipose tissue (Luo et al., 2003) suggesting that ERRα or one of its downstream target genes may repress Ucp1 expression in this tissue. However, more recent studies have not shown a role for this receptor in the regulation of Ucp1 in murine brown fat (Villena et al., 2007). Nevertheless, it does regulate many genes important for mitochondrial function and fatty acid oxidation in brown adipocytes so that that ERRα null animals have impaired adaptive thermogenesis. On the other hand ERRα has been shown to be important for the increased expression of Ucp1 found in mouse white adipocytes lacking RIP140 and the elevation of Ucp1 in these cells is not dependent on adrenergic signalling (Debevec et al., 2007). Strikingly brown adipocytes differentiated from fMSCs also express UCP1 the absence of β-adrenergic signalling, and therefore ERRα activation may represent a common pathway for expression of this gene independently of cold stimulation. We have shown that ERRα activity is required for the expression of UCP1 in fMSC derived brown adipocytes, that ERRα can alter the transcriptional activity of the human UCP1 enhancer, and that the receptor binds to the UCP1 enhance in these cells.
Interestingly the one physiological situation where brown fat is present and active prior to cold exposure is in the neonate, particularly the neonates of larger mammals, including humans, who are born with fully functional brown adipose tissue (reviewed in (Cannon and Nedergaard, 2004)). This contrasts with mice, where brown adipose tissue is recruited in response to cold over the first few days of life. Thus it is possible that these fetal mesenchymal stem cells are differentiating into adipocytes resembling the brown fat found in neonates, and may hint at a role for ERRα in this neonatal period. Furthermore the demonstration that ERRα can regulate expression of human UCP1 expression in these cells suggests that increasing ERRα activity may be a viable therapeutic target to induce UCP1, and hence increase uncoupling as a potential therapy for obesity. Thus as well as giving insights into the developmental origin of human brown adipocytes, we have also demonstrated that these cells can be useful in elucidating signaling pathways using classical molecular biological techniques that have been difficult to apply to existing human models.
Conclusions
This study has demonstrated that fetal mesenchymal stem cells are a valid and viable model for studying aspects of adipogenesis that can not be addressed with traditional models. There seems to be heterogeneity among adipocytes derived from different isolates of these cells, and this seems to be due to innate differences in the stem cells. The underlying mechanisms of these differences are unclear and further studies to determine this may be of importance if further understanding determinants of cell fate within the adipocyte lineage. Nevertheless, these studies have given insights not only into human brown adipogenesis, but also revealed novel molecular pathways controlling the expression of key genes within brown adipocytes.
Materials and Methods
Cell Culture
Human fetal mesenchymal stem cells (fMSC) were obtained from normal 1st trimester human fetuses at clinically-indicated termination of pregnancy as described elsewhere (Campagnoli et al., 2001), after written maternal consent and in accordance with institutional ethical approval and national guidelines on the use of fetal tissue in research. Gestational age was determined by crown rump length on intra-operative ultrasound. Briefly, first trimester FB samples (20-800 μl) were obtained by ultrasound-guided cardiac aspiration between 9-11 weeks gestation prior to clinically-indicated surgical termination of pregnancy. Heparinised (by flushing with heparin sodium 1,000 units/ml from CP Pharmaceuticals), disposable siliconised 20G 15-cm needles (COOK (UK) Ltd.) attached to 1-ml syringes were used for cardiocentesis. Fetal gestational age was determined by crown-rump length (CRL) measurement referenced to standard biometry charts (Robinson and Fleming, 1975). First trimester fetal bone marrow tissues were collected after clinically-indicated surgical termination of pregnancy. Fetal BM were flushed from marrows of fetal long bones or spines while FL was diced. Tissues were then washed with phosphate-buffered saline (PBS), and filtered through a 40-μm cell strainer (BD Biosciences) to yield cell suspensions. Harvested cell suspensions are centrifuged at 1,500 rpm (400 ×g) for 5 min and the cell pellet resuspended in high glucose Dulbecco’s modified Eagle’s medium (DMEM; Sigma) supplemented with 10% fetal bovine serum (FBS; BioSera, Batch selected for maximal MSC growth), 2 mM L-glutamine, 50 IU/mL penicillin and 50 μg/mL streptomycin (Invitrogen) (D10 medium). They were then plated in 100mm tissue culture dishes (Corning) dishes at a density of 106 per ml and incubated at 37°C with 5% carbon dioxide (CO2). After 3 days, they were washed in PBS twice to remove non-adherent cells. Established adherent cell colonies were detached with 0.05% Trypsin-EDTA (StemCell Technologies) when they reached 70-80% confluence and replated at 104 / cm2 in 100 mm dishes.
Cells derived from whole isolates of either fetal blood or bone marrow from single fetuses were expanded and passaged in DMEM with 10% FBS. Adipogenic differentiation was induced in early passage cells (n=3-6) by plating the fMSC at 2×104/cm2, allowing the cells to reach confluence and then incubating for a further 48 hours. The media was then changed to DMEM supplemented with 10% FBS as above and the following hormones: Insulin 10μg/ml, Dexamethasone 1 μM, Indomethacin 200μM and Isobutylmethylxanthine (IBMX) 0.5 mM (Campagnoli et al., 2001). Media were changed every 4 days and differentiation continued for 14-21 days.
Bone differentiation was induced as previously described (Campagnoli et al., 2001).
Human adult sub-cutaneous pre-adipocytes were obtained from Cambrex (UK) and differentiated according to the manufacturers instructions.
Oil Red O stain was used to confirm the presence of lipid in cells. Cells were washed with PBS, then fixed in 2% Paraformaldehyde, 0.2% Gluteraldehyde in PBS for 15 minutes, then rinsed with PBS. They were then stained with Oil Red O stain (made up in Isopropanol) for 10 minutes and rinsed in 60% isopropanol then PBS (adapted from (Ramirez-Zacarias et al., 1992)). Lipid droplets stained red were visualised by light microscopy. Von Kossa staining was performed by fixing the cells in 10% Formalin for 1 hour, washing and staining with 2% Silver Nitrate for 10 minutes in the dark, washing and exposing to light.
Ligand experiments were performed by incubating differentiated cells in media supplemented with 10% FBS and Insulin 10μg/alone for 48 hours prior to the addition of the indicated ligand for a further 48 hours. Rosiglitazone was obtained from Alexxis (UK) and XCT709 from Sigma (UK).
Immunocytochemistry
Cells were grown to 60% confluence on glass cover slips, fixed in 4% Paraformaldehyde in 125 mM HEPES (pH 7.6; 10 min, 4°C), re-fixed in 8% Paraformaldehyde in HEPES (50 min, 4°C), and then permeabilised in 0.5% Triton X-100 in PBS with gentle shaking for 30 min. Next, cells were blocked with PBS+ (PBS containing 1% BSA, 0.2% fish gelatin, 0.1% casein at pH 7.6) for 1 hour, incubated overnight (4°C) with the appropriate primary antibody in PBS+, washed for 1.5 hours in PBS+, incubated with secondary antibodies in PBS+ for 1 hour, washed overnight (4°C) in PBS+, washed in PBS and then mounted in 4′,6 diamidino-2-phenylindole (DAPI) Vectashield mounting medium (Vector Laboratories). Slides were viewed under fluorescence microscopy (Zeiss Axioscope I microscope). Images were captured using a photonics digital camera (Hamamatsu) and processed by iPlab (Scanalytics) software. The following primary antibodies were used: rabbit polyclonal IgG laminin (Sigma), mouse monoclonal IgG SH2, SH3 (BD Biosciences), mouse monoclonal IgG CD14, CD34, CD45, and vimentin (Dako). Secondary antibodies for immunofluorescence were donkey anti-mouse IgG conjugated with FITC (Jackson ImmunoResearch Laboratories) and goat anti-rabbit IgG conjugated to Alexafluor 488 (Molecular Probes).
Quantitative RT-PCR
Total RNA from cells was extracted with TRIZOL reagent (Invitrogen) according to manufacturer’s protocols. 1μg of the resulting RNA was treated with DNAse (Sigma) and then reverse transcribed with MMLTV-RT (Sigma) using Oligo-dT primers (Invitrogen). Quantitative Real Time PCR was performed on an Opticon2 cycler (MJ research), using JumpStart Taq ready mix for quantitative PCR (Sigma). Gene expression was normalised to the house keeping gene L19 and fold differences were calculated by the ΔCt method(Bustin et al., 2005). Data are expressed in arbitrary units. Primer sequences are available on request.
Reporter Assays
The 220bp enhancer element of the human UCP-1 gene was amplified by PCR using the following primers: forward caattggtaccgaacttgctgccactcctttg and reverse attggctcgagtctggattccaaggagcagg. The product was ligated into the vector pGL3-TK.
Reporter assays were performed in Cos-1 cells, in 96 well plates in phenol red free DMEM supplemented with 5% Dextrose Charcoal Stripped Serum. 20ng of Reporter construct was transfected along with 5ng of an internal control plasmid expressing Renilla luciferase constitutively, together with the indicated amounts of expression plasmids. Total amount of DNA transfected was kept constant by using varying amounts of an empty pCI vector. Transfections were performed with FuGene6 (Roche) transfection reagent according to manufacturers instructions. Indicated ligands were added after 24 hours. Luciferase activity was assayed 48 hours post transfection with the LucLite reagent according to manufacturers instructions, and the luminescence read on a Victor2 plate reader (Perkin Elmer). Activity was normalised to the Renilla activity.
Chromatin Immunoprecipitation
Cells were incubated in PBS with Dimethyl adipimidate 2.4mg / ml for 30 minutes in the dark at room temperature. They were then fixed in 1% formaldehyde in DMEM for 15min at 37 C. Cross-linked cells were lysed, sonicated, and immunoprecipitated with protein A/G PLUS-agarose (SC-2003; Santa Cruz Biotechnology, Santa Cruz, CA) and antibodies against ERRα (LS-A5402; Lifespan, Seattle, WA) or control normal rabbit IgG (SC-2027;Santa Cruz Biotechnology). DNA fragments were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA) and used as templates in PCRs. The primers used for the UCP1 enhancer were F AACTTGCTGCCACTCCTTTG, R TCTGACAGGCTCTGGGAAGT and the last coding exon as a control F GGGTGAAGCCTCATCTCAAA, R TCGTTTCAGTTGTTCAAAGCA.
Statistics
Data was analysed with GraphPad Prism software (GraphPad software, California, USA). Comparisons were made with the one way ANOVA (Analysis of Variance) with Tukey’s post-hoc analysis.
Supplementary Material
mRNA expression of Ucp-1 in 5 isolates of fetal mesenchymal stem cells after differentiation into adipocytes. fBl = Fetal Blood MSCs, fBM = Fetal Bone Marrow MSC, numbers indicate gestation (weeks + days). Error bars indicate SD.
mRNA expression of FABP4 in differentiated 9+4 fMSCs following treatment with Rosiglitazone, and XCT790 * p =<0.01, ** p=<0.005. Error bars indicate SD.
The human Ucp-1 enhancer element sequence with putative ERRα binding sites marked (boxes). Numbers indicate base pairs from the transcriptional start site.
Acknowledgements
This work was funded by a Wellcome Trust Training Fellowship to DM (074456). PW is supported by a Wellbeing of Women project grant, and NMF acknowledges salary support from the UK National Institutes of Health Research Biomedical Research Centre funding scheme.
Reference List
- 1.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 2.Farmer SR. Transcriptional control of adipocyte formation. Cell Metabolism. 2006;4(4):263–73. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73(4):725–34. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 4.Lee K, Villena JA, Moon YS, et al. Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1) J Clin Invest. 2003;111(4):453–61. doi: 10.1172/JCI15924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon YS, Smas CM, Lee K, et al. Mice Lacking Paternally Expressed Pref-1/Dlk1 Display Growth Retardation and Accelerated Adiposity. Mol Cell Biol. 2002;22(15):5585–92. doi: 10.1128/MCB.22.15.5585-5592.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Timmons JA, Wennmalm K, Larsson O, et al. Myogenic gene expression signature establishes that brown and white adipocytes originate from distinct cell lineages. PNAS. 2007;104(11):4401–6. doi: 10.1073/pnas.0610615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crisan M, Casteilla L, Lehr L, et al. A Reservoir of Brown Adipocyte Progenitors in Human Skeletal Muscle. Stem Cells. 2008;26(9):2425–33. doi: 10.1634/stemcells.2008-0325. [DOI] [PubMed] [Google Scholar]
- 8.Seale P, Bjork B, Yang W, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454(7207):961–7. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seale P, Kajimura S, Yang W, et al. Transcriptional Control of Brown Fat Determination by PRDM16. Cell Metab. 2007;6(1):38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puigserver P, Wu Z, Park CW, et al. A Cold-Inducible Coactivator of Nuclear Receptors Linked to Adaptive Thermogenesis. Cell. 1998;92(6):829–39. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 11.Leonardsson G, Steel JH, Christian M, et al. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc Natl Acad Sci U S A. 2004;101(22):8437–42. doi: 10.1073/pnas.0401013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Christian M, Kiskinis E, Debevec D, et al. RIP140-targeted repression of gene expression in adipocytes. Mol Cell Biol. 2005;25(21):9383–91. doi: 10.1128/MCB.25.21.9383-9391.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morganstein DL, Christian M, Turner JJ, Parker MG, White R. Conditionally immortalized white preadipocytes: a novel adipocyte model. J Lipid Res. 2008;49(3):679–85. doi: 10.1194/jlr.D700029-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Lelliott CJ, Medina-Gomez G, Petrovic N, et al. Ablation of PGC-1β Results in Defective Mitochondrial Activity, Thermogenesis, Hepatic Function, and Cardiac Performance. PLoS Biology. 2006;4(11):e369. doi: 10.1371/journal.pbio.0040369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284(5411):143–7. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 16.Mackay DL, Tesar PJ, Liang LN, Haynesworth SE. Characterizing medullary and human mesenchymal stem cell-derived adipocytes. J Cell Physiol. 2006;207(3):722–8. doi: 10.1002/jcp.20617. [DOI] [PubMed] [Google Scholar]
- 17.Gemmis PD, Lapucci C, Bertelli M, et al. A Real-Time PCR Approach to Evaluate Adipogenic Potential of Amniotic Fluid-Derived Human Mesenchymal Stem Cells. Stem Cells and Development. 2006;15(5):719–28. doi: 10.1089/scd.2006.15.719. [DOI] [PubMed] [Google Scholar]
- 18.Campagnoli C, Roberts IAG, Kumar S, et al. Identification of mesenchymal stem/progenitor cells in human first-trimester fetal blood, liver, and bone marrow. Blood. 2001;98(8):2396–402. doi: 10.1182/blood.v98.8.2396. [DOI] [PubMed] [Google Scholar]
- 19.Ryden M, Dicker A, Gotherstrom C, et al. Functional characterization of human mesenchymal stem cell-derived adipocytes. Biochem Biophys Res Commun. 2003;311(2):391–7. doi: 10.1016/j.bbrc.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 20.Tomlinson JJ, Boudreau A, Wu D, Atlas E, Hache RJG. Modulation of Early Human Preadipocyte Differentiation by Glucocorticoids. Endocrinology. 2006;147(11):5284–93. doi: 10.1210/en.2006-0267. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura T, Shiojima S, Hirai Y, et al. Temporal gene expression changes during adipogenesis in human mesenchymal stem cells. Biochem Biophys Res Commun. 2003;303(1):306–12. doi: 10.1016/s0006-291x(03)00325-5. [DOI] [PubMed] [Google Scholar]
- 22.Janderova L, McNeil M, Murrell AN, Mynatt RL, Smith SR. Human mesenchymal stem cells as an in vitro model for human adipogenesis. Obes Res. 2003;11(1):65–74. doi: 10.1038/oby.2003.11. [DOI] [PubMed] [Google Scholar]
- 23.del Mar Gonzalez-Barroso M, Pecqueur C, Gelly C, et al. Transcriptional Activation of the Human ucp1 Gene in a Rodent Cell Line. SYNERGISM OF RETINOIDS, ISOPROTERENOL, AND THIAZOLIDINEDIONE IS MEDIATED BY A MULTIPARTITE RESPONSE ELEMENT. Journal of Biological Chemistry. 2000;275(41):31722–32. doi: 10.1074/jbc.M001678200. [DOI] [PubMed] [Google Scholar]
- 24.Busch BB, Stevens WC, Martin R, et al. Identification of a Selective Inverse Agonist for the Orphan Nuclear Receptor Estrogen-Related Receptor α. J Med Chem. 2004;47(23):5593–6. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 25.Kozak UC. An upstream enhancer regulating brown-fat-specific expression of the mitochondrial uncoupling protein gene. Mol Cell Biol. 1994;14(1):59. doi: 10.1128/mcb.14.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekiya I, Larson BL, Vuoristo JT, Cui JG, Prockop DJ. Adipogenic differentiation of human adult stem cells from bone marrow stroma (MSCs) J Bone Miner Res. 2004;19(2):256–64. doi: 10.1359/JBMR.0301220. [DOI] [PubMed] [Google Scholar]
- 27.Hung SC, Chang CF, Ma HL, Chen TH, Low-Tone HL. Gene expression profiles of early adipogenesis in human mesenchymal stem cells. Gene. 2004;340(1):141–50. doi: 10.1016/j.gene.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 28.Sladek R, Bader JA, Giguere V. The orphan nuclear receptor estrogen-related receptor alpha is a transcriptional regulator of the human medium-chain acyl coenzyme A dehydrogenase gene. Mol Cell Biol. 1997;17(9):5400–9. doi: 10.1128/mcb.17.9.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo J, Sladek R, Carrier J, et al. Reduced Fat Mass in Mice Lacking Orphan Nuclear Receptor Estrogen-Related Receptor {alpha} Mol Cell Biol. 2003;23(22):7947–56. doi: 10.1128/MCB.23.22.7947-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villena JA, Hock MB, Chang WY, et al. Orphan nuclear receptor estrogen-related receptor {alpha} is essential for adaptive thermogenesis. PNAS. 2007;104(4):1418–23. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Debevec D, Christian M, Morganstein D, et al. Receptor Interacting Protein 140 Regulates Expression of Uncoupling Protein 1 in Adipocytes through Specific Peroxisome Proliferator Activated Receptor Isoforms and Estrogen-Related Receptor {alpha} Mol Endocrinol. 2007;21(7):1581–92. doi: 10.1210/me.2007-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with oil red O. Histochemistry and Cell Biology. 1992;97(6):493–7. doi: 10.1007/BF00316069. [DOI] [PubMed] [Google Scholar]
- 33.Bustin SA, Benes V, Nolan T, Pfaffl MW. Quantitative real-time RT-PCR - a perspective. J Mol Endocrinol. 2005;34(3):597–601. doi: 10.1677/jme.1.01755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA expression of Ucp-1 in 5 isolates of fetal mesenchymal stem cells after differentiation into adipocytes. fBl = Fetal Blood MSCs, fBM = Fetal Bone Marrow MSC, numbers indicate gestation (weeks + days). Error bars indicate SD.
mRNA expression of FABP4 in differentiated 9+4 fMSCs following treatment with Rosiglitazone, and XCT790 * p =<0.01, ** p=<0.005. Error bars indicate SD.
The human Ucp-1 enhancer element sequence with putative ERRα binding sites marked (boxes). Numbers indicate base pairs from the transcriptional start site.