Abstract
Background
Carbazole is a recalcitrant compound with a dioxin-like structure and possesses mutagenic and toxic activities. Bacteria respond to a xenobiotic by recruiting exogenous genes to establish a pathway to degrade the xenobiotic, which is necessary for their adaptation and survival. Usually, this process is mediated by mobile genetic elements such as plasmids, transposons, and insertion sequences.
Findings
The genes encoding the enzymes responsible for the degradation of carbazole to catechol via anthranilate were cloned, sequenced, and characterized from a carbazole-degrading Sphingomonas sp. strain XLDN2-5. The car gene cluster (carRAaBaBbCAc) and fdr gene were accompanied on both sides by two copies of IS6100 elements, and organized as IS6100::ISSsp1-ORF1-carRAaBaBbCAc-ORF8-IS6100-fdr-IS6100. Carbazole was converted by carbazole 1,9a-dioxygenase (CARDO, CarAaAcFdr), meta-cleavage enzyme (CarBaBb), and hydrolase (CarC) to anthranilate and 2-hydroxypenta-2,4-dienoate. The fdr gene encoded a novel ferredoxin reductase whose absence resulted in lower transformation activity of carbazole by CarAa and CarAc. The ant gene cluster (antRAcAdAbAa) which was involved in the conversion of anthranilate to catechol was also sandwiched between two IS6100 elements as IS6100-antRAcAdAbAa-IS6100. Anthranilate 1,2-dioxygenase (ANTDO) was composed of a reductase (AntAa), a ferredoxin (AntAb), and a two-subunit terminal oxygenase (AntAcAd). Reverse transcription-PCR results suggested that carAaBaBbCAc gene cluster, fdr, and antRAcAdAbAa gene cluster were induced when strain XLDN2-5 was exposed to carbazole. Expression of both CARDO and ANTDO in Escherichia coli required the presence of the natural reductases for full enzymatic activity.
Conclusions/Significance
We predict that IS6100 might play an important role in the establishment of carbazole-degrading pathway, which endows the host to adapt to novel compounds in the environment. The organization of the car and ant genes in strain XLDN2-5 was unique, which showed strong evolutionary trail of gene recruitment mediated by IS6100 and presented a remarkable example of rearrangements and pathway establishments.
Introduction
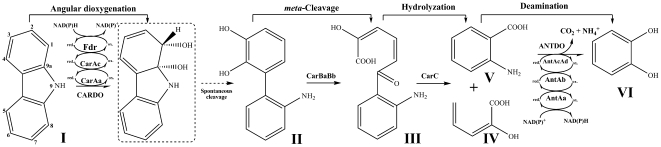
Carbazole is an N-heterocyclic compound that is known to possess mutagenic and toxic activities [1]. To date, a number of bacterial strains capable of degrading carbazole have been isolated and characterized. Phylogenetically, almost all of these strains belong to pseudomonads and sphingomonads. The degradation of carbazole starts with angular dioxygenation in all studied strains so far [2], [3], [4], [5], [6], [7], [8]. In this pathway, carbazole is initially attacked at the 1 and 9a positions by carbazole 1,9a-dioxygenase (CARDO), resulting in the formation of a highly unstable hemiaminal, which gives rise to 2′-aminobiphenyl-2,3-diol after spontaneous cleavage and rearomatization. An extradiol dioxygenase attacks the hydroxylated ring of 2′-aminobiphenyl-2,3-diol at the meta position to generate 2-hydroxy-6-(2′-aminophenyl)-6-oxo-2,4-hexadienoic acid, which is hydrolyzed to produce anthranilic acid and 2-hydroxypenta-2,4-dienoic acid. The resulting metabolite, anthranilate, is converted to catechol in a single step by anthranilate 1,2-dioxygenase (ANTDO) (Figure 1) [9], [10]. In this upper carbazole degradation pathway, CARDO is considered as the key enzyme. In Pseudomonas resinovorans CA10, the most intensively studied pseudomonad, CARDO consists of a terminal oxygenase, a ferredoxin, and a ferredoxin reductase, encoded by carAa, carAc, and carAd, respectively. In pseudomonads, the upper pathway genes, carAaBaBbCAcAd, are in the car cluster, which is transcribed as a single transcriptional unit [6]. Interestingly, unlike the well-organized operons in pseudomonads, the catabolic genes in sphingomonads are often dispersed or not coordinately regulated. For example, plasmid pCAR3 contains multiple gene sets, which are involved in the carbazole degradation pathway in a carbazole-degrader Novosphingobium sp. KA1 (previous Sphingomonas sp. KA1) [10]. These unusual organizations of the degradative genes were often observed in other sphingomonads [11], [12].
Figure 1. Biodegradation of carbazole via the angular pathway by Sphingomonas sp. XLDN2-5 and other carbazole-utilizing bacteria.
The product in dashed square is unstable and has not been detected. Enzyme names: carbazole 1,9a-dioxygenase (carAaAcfdr); meta-cleavage enzyme (carBaBb); hydrolase (carC); anthranilate 1,2-dioxygenase (antAcAdAbAa). Compound I: carbazole; compound II: 2′-aminobiphenyl-2,3-diol; compound III: 2-hydroxy-6-oxo-6-(2′-aminobiphenyl)-hexa-2,4-dienoic acid; compound IV: 2-hydroxypenta-2,4-dienoate; compound V: anthranilic acid; VI: catechol.
Insertion sequences (ISs) are a subject of increasing interests for biodegradation because of the variety of their structures, modes of action, and the biodegradation abilities they confer bacteria [13]. ISs are small and mobile genetic elements that are ubiquitously distributed within bacterial genomes, and play an important role in evolution by facilitating horizontal gene transfers between bacterial populations, which contribute significantly to the diversity of bacteria by enhancing the organisms' adaptive and evolutionary capacities. IS6100 is an important IS that flanks a range of catabolic operons, for example, the operons for metabolism of various aromatic substrates [14], [15]. In this work, we report that two loci coding for the enzymes that convert carbazole to catechol were found to be flanked by IS6100 elements. Evidence was given for the involvement of these genes in the degradation of carbazole in Sphingomonas sp. XLDN2-5.
Results
Screening of the genomic library, DNA sequencing, and genome walking
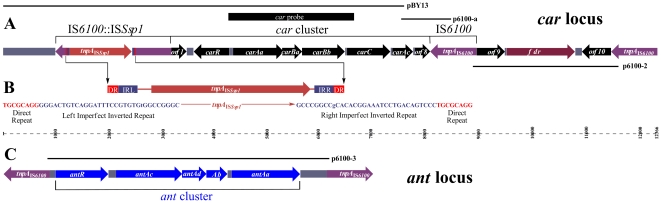
The car probe, labeled with DIG, was used for screening the genomic library, and a positive clone, designated as pBY13 (Figure 2A), was sequenced and analyzed to contain a DNA insert of 6.8 kb. BLAST search results revealed that there were five intact open reading frames (ORFs), carR, carAa, carBa, carBb, and carC, which were found to be 99% identical to the corresponding genes of Novosphingobium sp. strain KA1 [10]. A closer look at the left region suggested that an IS6100, exhibiting 100% identity to that of Mycobacterium fortuitum [16], was interrupted by a novel insertion element ISSsp1. Therefore, the interrupted IS6100 was designated IS6100::ISSsp1. The putative transposase for ISSsp1 was transcribed in the same direction of the car cluster, but in the opposite direction as the interrupted IS6100. ISSsp1 belongs to the IS256 family of prokaryotic ISs, a subgroup of the mutator family of transposases (Pfam00872). ISSsp1 which had imperfect inverted repeats (32-bp in length, one mismatch) was flanked by two copies of 8-bp direct repeat (Figure 2B).
Figure 2. Physical maps of car and ant loci.
(A) Physical map of car locus, which is delimited by IS6100 elements. The upstream IS6100 is interrupted by a novel insert element ISSsp1, and was designated IS6100::ISSsp1. (B) Schematic representation of the main features of the novel ISSsp1 sequence in Sphingomonas sp. XLDN2-5. The orientation of the ISSsp1 is shown by an arrow. The red and blue boxes represent the positions of two direct repeats (DR) and two imperfect, 32 bp, terminal inverted repeats (the left inverted repeat [IRL], and the right inverted repeat [IRR]) with one mismatch, which is indicated by lowercase letters. The nucleotide sequences of DRs and IRs are also given. (C) Physical map of ant cluster which is franked by IS6100 elements along a base pair scale.
At the 3′-end of pBY13, an incomplete ORF (238 bp) showed 100% identity to carAcI of KA1. In order to clone the full gene and its flanking sequences, a 1-kb DNA was amplified by genome walking. After ligation of the fragment to pMD18-T followed by transformation to E. coli DH5α, a positive clone, designated p6100-a, was obtained. DNA sequencing indicated that p6100-a contained the left part of carAc, an ORF8, and interestingly a partial IS6100 sequence. The presence of the partial IS6100 sequence led to the hypothesis that there was a complete IS6100 and might form a transposon, and therefore triggered further study. The presence of an intact IS6100 copy downstream of ORF8 was successfully validated by PCR using carC-sp3 and IS6100-F2 and sequencing. This transposon-like context (IS6100::ISSsp1-ORF1-carRAaBaBbCAc-ORF8-IS6100) was designated TnCar, tentatively.
IS-based PCR
The presence of IS6100 on both sides of the car gene cluster motivated further investigations using IS-based PCR. An IS-based PCR with primers IS6100-F1 and IS6100-R1 targeting the IS6100 element resulted in two distinct PCR products of about 4.3 and 2.8 kb, respectively (Figure S1B, lane 1). The 4.3 and 2.8-kb fragments were ligated to pMD18-T to generate plasmid p6100-1 and p6100-2. DNA sequencing and BLAST search suggested that there were two partial IS6100 at each end as expected in the 4.3 and 2.8-kb fragments. The 4.3-kb fragment contains ORF1carRAaBaBbCAcORF8 from TnCar.
BLAST search suggested that in the 2.8-kb fragment, there were two truncated ORFs and one intact ORF (Fdr) that showed 62% identity to the FdrI of strain KA1 [10]. A sequence comparison showed that the amino acid sequence of Fdr was homologous over its entire length to other members of the FAD-dependent pyridine nucleotide reductase family, containing a flavin binding domain for FAD (consensus sequence TX 6AXGD) and two ADP binding domains (for FAD and NADH, respectively) with the consensus sequence GXGX 2GX 3A [17], [18], [19]. Two truncated ORFs encoded for a putative uncharacterized protein and a transcriptional regulator, were found upstream and downstream of fdr (Table 1). Sequencing of PCR product revealed the existence of entire copies of IS6100 at both ends of the 2.8-kb fragment. These two IS6100 were in the same direction and formed a composite transposon (IS6100-fdr-IS6100) designated TnFdr, tentatively.
Table 1. Coding regions of car locus and ant locus.
| Protein | Position (bp) in sequence (direction) | Length (amino acids) | Putative function | Homologous protein | ||
| % Identity | Protein (accession no.) | Source | ||||
| car locus | ||||||
| TnpAIS6100 | 1116–1263; 2594–3240 (c) | 264 | Transposase of IS6100 | 100 | Tnp (Q79AS6) | Mycobacterium fortuitum |
| TnpAISSsp1 | 1321–2544 (n) | 407 | Transposase, mutator type | 99 | Tnp (A5VHF2) | Sphingomonas wittichii RW1 |
| ORF1 | 3272–3400 (n) | 42 | Integrase family protein (truncated, only C-terminal portion) | 100 | ORF7 (Q84IH1) | Novosphingobium sp. KA1 |
| CarR | 3621–4301 (c) | 226 | Transcriptional regulator of car operon, GntR family | 99 | CarR (Q84IH0) | Novosphingobium sp. KA1 |
| CarAa | 4404–5540 (n) | 378 | Terminal oxygenase component of carbazole 1,9a-dioxygenase | 99 | CarAaI (Q84IG9) | Novosphingobium sp. KA1 |
| 60 | CarAa (Q8G8B6) | Pseudomonas resinovorans CA10 | ||||
| CarBa | 5489–5821 (n) | 110 | small subunit of meta cleavage enzyme | 100 | CarBaI (Q84IG8) | Novosphingobium sp. KA1 |
| CarBb | 5814–6617 (n) | 267 | large subunit of meta cleavage enzyme | 99 | CarBbI (Q84IG7) | Novosphingobium sp. KA1 |
| 42 | CarBb (Q4TTW1) | Pseudomonas sp. XLDN4-9 | ||||
| CarC | 6660–7484 (n) | 274 | Meta cleavage compound hydrolase | 99 | CarCI (Q84IG6) | Novosphingobium sp. KA1 |
| CarAc | 7525–7854 (n) | 109 | Ferredoxin component of carbazole 1,9a-dioxygenase | 100 | CarAcI (Q84IG5) | Novosphingobium sp. KA1 |
| 57 | CarAcII (Q2PFA2) | Novosphingobium sp. KA1 | ||||
| ORF8 | 7896–8117 (n) | 74 | TonB-dependent receptor (truncated, only N-terminal portion) | 100 | ORF35 (Q84IG4) | Novosphingobium sp. KA1 |
| TnpAIS6100 | 8172–8966 (c) | 264 | Transposase of IS6100 | 100 | Tnp (Q79AS6) | Mycobacterium fortuitum |
| ORF9 | 8998–9545 (n) | 182 | Putative uncharacterized protein (truncated, only C-terminal portion) | 34 | Q74F08 | Geobacter sulfurreducens |
| Fdr | 9573–10817 (n) | 407 | Ferredoxin reductase component of carbazole 1,9a-dioxygenase | 62 | FdrI (Q2PF96) | Novosphingobium sp. KA1 |
| 59 | FdrII (Q2PF93) | Novosphingobium sp. KA1 | ||||
| ORF10 | 10936–11486 (c) | 182 | Transcriptional regulator, TetR family (truncated, only C-terminal portion) | 40 | Q1NF20 | Sphingomonas sp. SKA58 |
| TnpAIS6100 | 11541–12335 (c) | 264 | Transposase of IS6100 | 100 | Tnp (Q79AS6) | Mycobacterium fortuitum |
| ant locus | ||||||
| TnpAIS6100 | 55–849 (c) | 264 | Transposase of IS6100 | 100 | Tnp (Q79AS6) | Mycobacterium fortuitum |
| AntR | 1009–1965 (n) | 318 | Transcriptional regulator, AraC family | 100 | AndR (Q0KJU2) | Novosphingobium sp. KA1 |
| 41 | AndR (Q84BZ4) | Burkholderia cepacia DPO1 | ||||
| AntAc | 2163–3446 (n) | 427 | Anthranilate 1,2-dioxygenase large subunit | 100 | AndAc (Q0KJU3) | Novosphingobium sp. KA1 |
| 76 | AndAc (Q84BZ3) | Burkholderia cepacia DPO1 | ||||
| AntAd | 3451–3921 (n) | 156 | Anthranilate 1,2-dioxygenase small subunit | 100 | AndAd (Q0KJU4) | Novosphingobium sp. KA1 |
| 59 | AndAd (Q84BZ2) | Burkholderia cepacia DPO1 | ||||
| AntAb | 3935–4243 (n) | 102 | Ferredoxin component of anthranilate 1,2-dioxygenase | 100 | AndAb (Q0KJU5) | Novosphingobium sp. KA1 |
| 53 | AndAb (Q84BZ1) | Burkholderia cepacia DPO1 | ||||
| AntAa | 4364–5605 (n) | 413 | Ferredoxin reductase component of anthranilate 1,2-dioxygenase | 100 | AndAa (Q0KJU6) | Novosphingobium sp. KA1 |
| 40 | AndAa (Q84BZ0) | Burkholderia cepacia DPO1 | ||||
| TnpAIS6100 | 6183–6977 (n) | 264 | Transposase of IS6100 | 100 | Tnp (Q79AS6) | Mycobacterium fortuitum |
TnCar and TnFdr were each flanked by two copies of IS6100 in tail-to-head configuration. Although tail-to-head was the most abundant case for two copies of IS6100, other configurations were also reported [14]. Early attempts to amplify head-to-head and tail-to-tail configurations proved to be unsuccessful. Considering that the amplification of head-to-head and tail-to-tail configurations may be suppressed by intramolecular hybridization between two IS6100 copies during the primer annealing phase of PCR, TaKaRa LA Taq and GC Buffer I were used in the following experiments. One specific fragment (5.2 kb in size, Figure S1C, lane 3) was amplified using IS6100-R1, while no specific fragment was amplified under the same PCR conditions using IS6100-F1 (Figure S1C, lane 2). The 5.2-kb fragment was gel-purified and cloned to pMD18-T to generate p6100-3, and the nucleotide sequence of the 5.2 kb insert was determined. In the sequenced region, five intact ORFs were found to be almost identical (only one bp mismatch) to andRAcAdAbAa genes of KA1 whose products were expected for catalyzing the conversion of anthranilate to catechol. The five ORFs were designated antR, antAc, antAd, antAb, and antAa (Figure 2C). Strain XLDN2-5 ANTDO was a three-component dioxygenase and composed of a two-subunit oxygenase (antAcAd), a Rieske-type ferredoxin (antAb), and a ferredoxin reductase (antAa). antR, encoding a putative transcriptional activator, was in the same direction of antAcAdAbAa, which was different from that of Burkholderia cepacia DBO1 whose antR is in the opposite direction to its structure genes [20]. In the sequenced region, there were two partial IS6100 at each end as expected. The existence of intact IS6100 elements at both ends was confirmed by PCR and PCR product sequencing. These two IS6100 elements were in the head-to-head configuration and formed a composite transposon designated TnAnt, tentatively.
The positional relation of TnCar, TnFdr and TnAnt
There were two copies of IS6100 elements at both ends of the three transposon-like entities except that the upstream flanking IS6100 on TnCar was disrupted by a novel insertion sequence ISSsp1. It was likely that one IS6100 element was shared by two transposon-like units. In order to analyze the positional relation of TnCar, TnFdr, and TnAnt, five primers were designed outside of the IS6100 elements (Figure S2A). If there was a shared IS6100 element, a 0.9-kb DNA fragment containing IS6100 could be amplified by PCR using different primer pairs. As shown in Figure S2B, the expected 0.9-kb fragment could only be amplified using primers TnCar-F1 and TnFdr-R1. These results suggested that TnFdr shared an IS6100 element with TnCar, but not with TnAnt. The conclusion was confirmed by second-round PCR (Figure S2C and S2D). Thus, we renamed TnCar and TnFdr to the car locus, and TnAnt to the ant locus.
Description of car and ant loci
The structures of car locus and ant locus are depicted in Figure 2 and the ORFs are given in Table 1. Both car cluster and fdr gene were sandwiched between two copies of IS6100. Interestingly, two identical copies of the IS6100 element also flanked the ant gene cluster, possibly making a composite transposon. All five sequenced copies of IS6100 from strain XLDN2-5 were identical over the entire 880 bp. Direct repeats (TGCGCAGG) were found directly upstream and downstream of ISSsp1, whereas no direct repeat was found outside of the IS6100 box. Furthermore, the ISSsp1 consisted of inverted repeats of 32 bp, of which only one base pair was not identical, and a 1224-bp ORF (tnpAISSsp1), encoding a 407 aa putative transposase that showed similarities to transposases of the IS256 family. In order to illustrate that the genes on these two loci were really working in the degradation of carbazole in strain XLDN2-5, transcriptional and functional analyses were performed.
Transcriptional analyses of car and ant genes
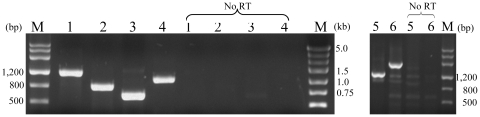
The expression of the genes presenting on the two loci was studied by reverse transcription (RT)-PCR experiments. The primer sets for the carAaBaBbCAc, fdr, and antAcAdAbAa genes could amplify DNA fragments with the expected sizes (Figure 3). No fragments could be amplified using RNA from glucose-grown XLDN2-5 cells as a template (data not shown). These results revealed that carAaBaBbCAc, fdr, and antAcAdAbAa genes were expressed in carbazole grown XLDN2-5 cells, suggesting that the gene products should be involved in the transformation of carbazole to catechol. These results also indicated that carAaBaBbCAc and antAcAdAb gene clusters were operonic. In order to confirm that Fdr was active in the CARDO system, functional analyses were performed.
Figure 3. Electrophoresis results of RT-PCR.
carAaBaBb (lane 1), carBbC (lane 2), carCAc (lane 3), fdr (lane 4), antAa (lane 5) and antAcAdAb (lane 6). Samples containing no reverse transcriptase (No RT) are also shown.
Functional analyses of putative CARDO and ANTDO
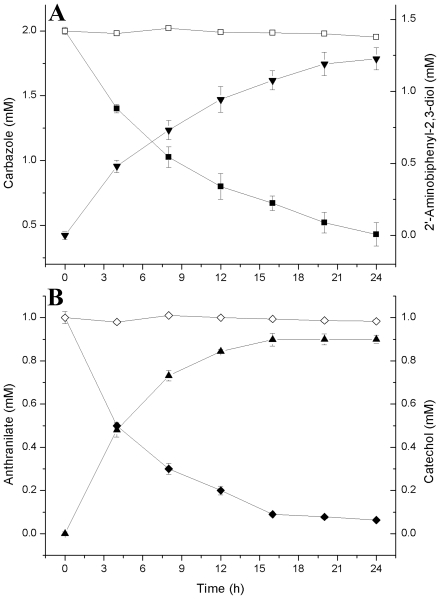
Biotransformation experiments were carried out using E. coli cells expressing putative CARDO components (CarAa, CarAc and Fdr) and ANTDO components (AntAc, AntAd, and AntAaAb) (Figure 4). 2′-Aminobiphenyl-2,3-diol and hydroxycarbazole could be detected for incubations of cell harboring pUcarAaAcfdr (Figure S3). About 80% carbazole was converted to 2′-aminobiphenyl-2,3-diol (Figure 4A). These results clearly indicated that CarAa could catalyze angular dioxygenation and lateral dioxygenation of carbazole using CarAc and Fdr as an electron-transfer system. E. coli DH5α (pUcarAaAc) encoding only the oxygenase and ferredoxin components transformed no more than 0.1% of the carbazole, which suggested that CarAc had the ability to accept electrons from unidentified reductase in E. coli, with a much lower efficiency than the natural reductase. Furthermore, CarAc showed similarity with the cytochrome P 450-type reductase component (RedA2) of dioxin dioxygenase from Sphingomonas wittichii RW1 [17] and putidaredoxin-type ferredoxins (CamA) from Pseudomonas putida [19]. CARDO systems of strain XLDN2-5 can be classified in the class IIA Rieske non-heme iron oxygenase system [10].
Figure 4. Biotransformation of substrates and accumulation of products.
(A) Biotransformation of carbazole (-▪-) and accumulation of 2′-aminobiphenyl-2,3-diol (-▾-) by E. coli DH5α harboring pUcarAaAcfdr. (B) Biotransformation of anthranilate (-♦-) and accumulation of catechol (-▴-) by E. coli DH5α harboring pUantAcdba. E. coli DH5α harboring pUC19 (-□- and -◊-) served as controls. The initial concentrations of carbazole and anthranilate were 2 mM and 1 mM, respectively. Values are means of three replicates ± SD.
The IPTG-induced E. coli cells carrying pUantAcdba or pUantAcdb readily converted anthranilate to catechol. E. coli DH5α (pUAntAcdba) completely transformed 1 mM anthranilate to catechol in 24 h (Figure 4B), while E. coli DH5α (pUAntAcdb) encoding only ferredoxin and oxygenase transformed only 5% of the anthranilate to catechol (data not shown). These results suggested that the expression of CARDO and ANTDO in E. coli requires the presence of the ancillary reductases for full enzymatic activity.
Discussion
Starting from the carAaBaBb fragment isolated from Sphingomonas sp. strain XLDN2-5, a combination of southern blot, genome walking, and IS-based PCR led to the isolation of two loci containing genes involved in the conversion of carbazole from anthranilate to catechol. The DNA fragments obtained are illustrated in Figure 2, and the proposed reaction details are given in Figure 1. All genes were embedded in transposon-like entities, implying the likely involvement of horizontal gene transfer in the evolution of carbazole degradation pathway. Transcriptional and functional analyses suggested that all the genes worked in the degradation of carbazole. The CARDO system catalyzed angular and lateral dioxygenation of carbazole, and ANTDO could transform anthranilate to catechol in a single step. Our research provided useful and important information for the association of IS6100 element with carbazole and anthranilate catabolic genes.
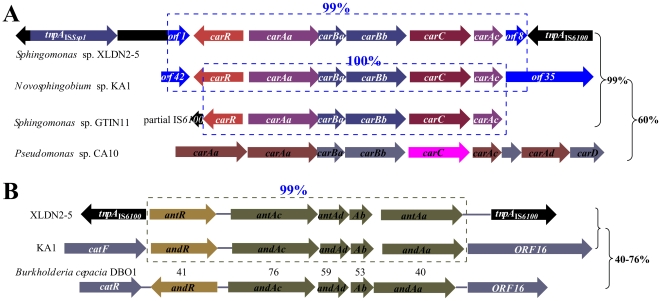
Carbazole-degrading sphingomonads have been isolated in different parts of the world. For example, Sphingomonas. sp. GTIN11 was isolated from USA [3], Novosphingobium sp. KA1 from Japan [10] and strain XLDN2-5 from China [2]. Although nearly identical car and ant genes were found in carbazole-degrading sphingomonad strains isolated from geographically dispersed locations, the organization of these genes in strain XLDN2-5 was unique. Figure 5 shows the organization of the known car genes (Figure 5A) and ant genes (Figure 5B) from different evolutionary origins. There were two truncated ORF1 and ORF8 encoding a putative integrase and a TonB-dependent receptor (Table 1), respectively. ORF1 and ORF8 showed 100% identity to ORF42 and ORF35 in strain KA1 [21], respectively. In strain KA1, ORF42 and ORF35 are intact (Figure 5A), whereas the corresponding ORFs were interrupted by IS6100 in strain XLDN2-5. Furthermore, BLAST search revealed that the terminal 27-bp region of the reported sequence (1–55 bp in AF442494) in strain GTIN11 is identical to the 27-bp left end of IS6100. This suggested that the carR gene in strain GTIN11 might be disrupted by IS6100 element. However, the entire sequence information outside the car genes in strain GTIN11 was not reported.
Figure 5. Comparative analyses of car and ant genes from Sphingomonas sp. XLDN2-5 and other strains.
(A) Comparative analysis of car gene cluster from Sphingomonas sp. XLDN2-5 and related strains. The car genes in the three sphingomonads are more than 99% identity to each other; however, only show 60% identity to that from Pseudomonas sp. CA10. (B) Comparative analysis of ant gene cluster from Sphingomonas sp. XLDN2-5 and related strains. The ant genes in strain XLDN2-5 and KA1 are more than 99% identity to each other; however, only show 40–76% identity to that from Burkholderia cepacia DPO1.
Interestingly, the ant cluster in strain XLDN2-5 was also bordered by copies of IS6100 elements. The organization of ant gene cluster was clearly different from that of KA1, in which catF, the upstream region of and cluster, encodes acetyl-CoA acetyltransferase, while the downstream ORF16 encodes a transposase (Figure 5B). Although there are many ISs belonging to different family, no IS6100 element was found on pCAR3 [21]. The structure and sequence identified in this study suggested that IS6100 element might play an important role in the transfer of carbazole-degrading genes. IS6100 was originally isolated as part of the composite transposon Tn6100 from Mycobacterium fortuitum [16]. Later studies revealed its presence in a wide spectrum of host organisms in distantly related bacterial lineages, e.g. Sphingomonas paucimobilis [14], Arthrobacter sp. [15], Pseudomonas aeruginosa [22], Xanthomonas campestris [23], Salmonella enterica [24], and Corynebacterium glutamicum [25]. The existence of IS6100 in a very broad host range indicates that the IS6100 element plays a vital role in disseminating genes, including catabolic and antibiotic resistance genes, among different bacteria. The sequenced copies of IS6100 from strain XLDN2-5 were 100% identical to those of Mycobacterium fortuitum. This supports the hypothesis that IS6100 elements are distributed widely among microorganisms, whether they are Gram-negative or Gram-positive bacteria [14], [15]. IS6100 belongs to the IS6 family of ISs which exclusively integrate through a cointegrative mechanism [13]. The car and ant genes might become part of strain XLDN2-5 via cointegrative captures. This type of transposition event is consistent with the truncation of the ORF1 and the ORF8 located upstream of carR and carAc, respectively (Figure 2 and Figure 5A). Thus, the genetic structure of the car and ant genes in strain XLDN2-5 has shown strong evolutionary trails of gene recruitment and presents an extraordinary example of rearrangements and pathway evolution.
Although the terminal oxygenase and ferredoxin components of CARDO from XLDN2-5 and KA1 are identical, the reductase components have a much lower level of similarity (62%). In general, the genes encoding the components of the known Rieske oxygenases were found closely together in tightly regulated transcriptional units, as was observed with the car cluster in strain CA10 (Figure 5A) [26]. However, it is becoming increasingly evident that the genes for catabolic pathways in sphingomonads often locate separately from each other. Recently, it is reported that multiple carbazole degradation genes dispersed on four loci (two car loci, one fdxIfdrI locus and one fdrII locus) on pCAR3 in Novosphingbium sp. strain KA1 [10]. The fdxIfdrI locus located 50 and 85 kb downstream of carI and carII gene clusters, while the fdrII gene located about 80 and 115 kb downstream of carI and carII gene clusters. Additional evidence has also been presented for the genes involved in the degradation of PAH by Sphingobium yanoikuyae B1 and Q1, and Novosphingobium aromaticivorans F199 [11], [12], [27], dibenzo-p-dioxin by Sphingomonas wittichii RW1 [28], pentachlorophenol by Sphingmonas chlorophenolica ATCC 39723 [29], and γ-hexachlorocyclohexane by Sphingmonas paucimobilis UT26 [30]. These results suggested that loose association with reductase components might be the characteristic for this type of dioxygenase, which became more independent from the reductase component during evolution.
In the course of cloning car genes, we also discovered a novel insertion sequence ISSsp1 (see results, IS6100::ISSsp1), which insert into the upstream copy of IS6100. ISSsp1 can be classified as the mutator family that consists of transposases from prokayotes and eukaryotes. There was only one example that an IS could insert into another in Comamonas sp. strain JS46 [31]. Thus, it would be interesting to investigate the distribution of ISSsp1 in strain XLDN2-5 and its function in the establishment of the carbazole catabolic gene structure.
Materials and Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are listed in Table 2. Sphingomonas sp. strain XLDN2-5 utilizes carbazole as the sole carbon and nitrogen source [2], [32]. Strain XLDN2-5 was grown in mineral salt medium (MSM) as previously described [33], and carbazole was added as a 200 mM filter-sterilized stock solution in dimethyl sulfoxide (DMSO). Escherichia coli DH5α was used as the recipient strain in all cloning experiments. E. coli strains were grown in Luria-Bertani (LB) broth at 37°C. Ampicillin (Amp), when required, was added to a final concentration of 100 µg ml−1.
Table 2. Bacterial strains and plasmids used.
| Strain or plasmid | Description | source |
| Bacterial strains | ||
| E. coli DH5α | F- endA1 hsdR17 supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF) U169 deoR | TransGen |
| Sphingomonas sp. XLDN2-5 | Aerobic, rod shaped, degrades carbazole | Lab stock |
| Plasmids | ||
| pUC19 | Ampr lacZ, pMB9 replicon, M13IG | TaKaRa |
| pMD18-T | Clone vector | TaKaRa |
| pEASY-Blunt | Clone vector | TransGen |
| pBY13 | Ampr; pUC19 with 7.8-kb fragment that hybridized with car probe | This study |
| p6100-a | Ampr; pMD18-T with 1-kb fragment obtained by genome walking | This study |
| p6100-1 | Ampr; pMD18-T with 4.3-kb fragment obtained by IS-based PCR using pIS6100-F1 and pIS6100-R1 | This study |
| p6100-2 | Ampr; pMD18-T with 2.8-kb fragment obtained by IS-based PCR using pIS6100-F1 and pIS6100-R1 | This study |
| P6100-3 | Ampr; pMD18-T with 5.2-kb fragment obtained by IS-based PCR using pIS6100-R1 | This study |
| pUcarAa | Ampr; pUC19 with 1.2-kb SphI-XbaI fragment containing the carAa gene of strainXLDN2-5 | This study |
| pUcarAc | Ampr; pUC19 with 0.3-kb XbaI-KpnI fragment containing the carAc gene of strain XLDN2-5 | This study |
| pUfdr | Ampr; pUC19 with 1.3-kb KpnI-EcoRI fragment containing the fdr gene of strain XLDN2-5 | This study |
| pEcarAaAc | Ampr; Kar, pEASY-Blunt with 1.5-kb fragment containing the carAa and carAc genes of strain XLDN2-5 | This study |
| pUcarAaAc | Ampr; 1.5-kb SphI-KpnI fragment containing carAaAc from pUcarAaAc cloned into SphI-KpnI site of pUC19 | This study |
| pUcarAaAcfdr | Ampr; 1.5-kb SphI-KpnI fragment containing carAaAc from pUcarAaAc cloned into SphI-KpnI site of pUfdr | This study |
| pUantAcdb | Ampr; pUC19 with 2.1-kb HindIII-EcoRI fragment containing the antAcAdAb genes of strainXLDN2-5 | This study |
| pUantAcdba | Ampr; pUC19 with 3.5-kb HindIII-EcoRI fragment containing the antAcAdAbAa genes of strainXLDN2-5 | This study |
DNA manipulation
Total DNA from pure cultures of Sphingomonas sp. strain XLDN2-5 was extracted using the Wizard® Genomic DNA Purification Kit according to the recommendations of the manufacturer (Promega Corp., Madison, WI). Restriction endonucleases and T4 DNA ligase were used according to the manufacturer's instructions (TaKaRa). Isolations of DNA fragments from agarose gels were accomplished with the Qiaex II Gel Extraction Kit (Qiagen Corp., Germany). Transformations and agarose gel electrophoresis were carried out using standard methods [34].
Construction of genomic library
The genomic DNA of Sphingomonas sp. XLDN2-5 was mechanically sheared. DNA fragments of 6 to 8 kb were gel purified and used for library construction. The fragments were ligated into pUC19 digested with EcoRV, and dephosphorylated with shrimp alkaline phosphatase (Promega). The library was transformed into electrocompetent E. coli DH5α. After incubation at 37°C for 1 h, cells were spread on LB agar plates supplemented with Amp, isopropyl-1-thio-β-D-galactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside. Following overnight incubation at 37°C, plates were scored for white colonies. The white colonies were used for southern blotting using a car probe obtained by PCR.
All PCR amplifications were performed with an Eppendorf Authorized Thermal Cycler (Germany) in 50-µl reaction systems containing 5 µl of 10× buffer, 1.5 mM MgCl2, 200 µM dNTPs, 500 pmol of each primer, 10-100 ng of the template DNA, and 2.5 units of DNA polymerase. TransStart FastPfu polymerase (TransGen Biotech Co. Ltd. China) was used in PCRs whose products were used for the construction of plasmids. TaKaRa LA PCR™ Kit Ver. 2.1 was used for IS-based PCR. Taq polymerase (Generay Biotech Co. Ltd. China) was used in other PCRs. Genome walking was performed using a Genome Walking Kit (TaKaRa) according to the manufacturer's protocol. The amplified products obtained by Taq polymerase were gel purified and ligated into vector pMD18-T, followed transformation to competent E. coli DH5α. All primers used in this study are listed in Table S1.
Southern blot
Primers pcarF and pcarR were designed according to the carRAaBaBb genes of strain KA1 [21]. Products obtained using primers pcarF and pcarR were gel purified and labeled with DIG-11-dUTP, and the DNAs were transferred following the standard protocols [34]. Southern blot was performed using DIG DNA Labeling and Detection Kit (Roche) according to the manufacturer's protocol. Hybridization was performed at 68°C. After hybridization, the membranes were washed twice in a solution containing 2× standard saline citrate (SSC) plus 0.1% sodium dodecyl sulfate (SDS) at 65°C and twice in a solution containing 0.5× SSC plus 0.1% SDS at 68°C.
DNA sequencing and analysis
Sequencing was performed on an ABI sequencer by Shanghai Invitrogen Biotechnology Co., Ltd, China. The sequences were analyzed with Vector NTI DNA analytical software (version 8). Homology searches were performed with the BLAST programs at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/BLAST.html). The deduced amino acid sequences of ORFs were aligned using ClustalW 1.83 [35].
RNA preparation and RT-PCR
After the precultivation of XLDN2-5 in 5 ml of MSM supplemented with 2 mM carbazole at 30°C, cells were harvested by centrifugation at 5,000 g for 5 min and then washed twice using MSM. The washed cells were suspended in 500 ml of MSM. Fifty microliters of the resultant cell suspension was added to 5 ml of MSM supplemented with 5 mM carbazole or with 1 g L−1 glucose and 0.5 g L−1 NH4Cl. After a 2-h incubation with reciprocal shaking (300 strokes/min) at 30°C, the cells were harvested and used for extraction of total RNA using E. Z. N. A™ Total RNA Kit I (Omega Biotech). Finally, the RNA was eluted in 50 µl of RNase-free water. A PrimeScript™ One Step RNA PCR kit (Takara) was used for RT-PCR, in which 100 ng of total RNA was used as a template. Detailed information on the RT-PCR primer sets and the conditions employed for respective gene amplifications are provided in Table S1. Control experiments without the addition of reverse transcriptase were also included.
Construction of plasmids for car and ant genes
The genes, carAa, carAc, and fdr were separately amplified by PCR using the respective primer sets shown in Table S1, which were designed to introduce appropriate restriction sites and the effective ribosome binding sites. In PCR amplification, total DNA of strain XLDN2-5 was used as a template. The amplified products were digested at the introduced restriction sites and ligated into the corresponding sites of pUC19 to produce plasmids (pUcarAa, pUcarAc, and pUfdr) for the expression of single CARDO components. For construction of pEcarAaAc, the carAa gene was amplified by carAa-F1 and carAaAcR, and carAc gene was amplified by carAaAcF and carAc-R1. carAa and carAc were then used as templates, while carAa-F1 and carAc-R1 were added to amplify carAaAc. The fragment obtained was ligated to pEASY-Blunt to generate pEcarAaAc. After their nucleotide sequences were confirmed to be identical to those designed, pEcarAaAc was double-digested using SphI and KpnI and the 1.5-kb fragment containing carAaAc was gel-purified and ligated to the corresponding sites of pUC19 and pUfdr to give pUcarAaAc and pUcarAaAcfdr.
Genes antAcAdAb and antAcAdAbAa were amplified by PCR using the respective primer sets, which were designed to introduce appropriate restriction sites and the effective ribosome binding sites. The amplified fragments were digested with HindIII and EcoRI before being ligated into pUC19 to generate pUAntAcdb and pUAntAcdba.
Biotransformation analysis
The E. coli DH5α strains harboring pUcarAaAcfdr, pUcarAaAc, pUantAcdba or pUantAcdb were cultivated in 5 ml of LB supplemented with Amp at 37°C. Then 100 µl of the culture was transferred to 200 ml of the same medium, and IPTG was added to induce the protein at 37°C for 16 h. Then the cells were harvested by centrifugation (6,000 g, 10 min, 4°C), washed twice with MSM, and resuspended in MSM to an OD600 of 10. Fifty microliters of carbazole (200 mM in DMSO) or anthranilate (200 mM in DMSO) was added to 10 ml of cell suspensions. After incubation on a reciprocal shaker (200 rpm) at 37°C for 20 h, the mixtures were extracted with an equal volume of ethyl acetate. After derivatization with N,O-bis-(trimethylsilyl)trifluoroacetamide (BSTFA) (Sigma) the extracts were analyzed by gas chromatography-mass spectrometry (GC-MS) as described previously [2], [33].
High performance liquid chromatography was carried out to analyze the aqueous samples using an Agilent 1200 series instrument equipped with a variable-wavelength detector and a reversed-phase C18 column (4.6 mm×150 mm, Hewlett-Packard). Residual concentrations of carbazole and anthranilate and the formation of 2′-animobiphenyl-2,3-diol and catechol were determined using a mobile phase of an 80∶20 mixture of methanol and deionized water at a flow rate of 0.5 ml min−1.
Nucleotide sequence accession numbers
The nucleotide sequences of the car locus and ant locus have been deposited in GenBank under accession numbers GU123624 and GU123625, respectively.
Supporting Information
IS6100-based PCR. (A) Schematics of relative configurations of two copies of IS6100. The position of primers IS6100-F1, IS6100-F2, IS6100-R1 and IS6100-R2 are shown by arrows. (B and C) Gel electrophoresis of DNA fragments amplified from XLDN2-5 by IS6100-based PCR using IS6100-F1 and IS6100-R1 (lane 1), single IS6100-F1 (lane 2) and single IS6100-R1 (lane 3). Two fragments (4.3 kb and 2.8 kb in size, Figure S1B, lane 1) were amplified using primers IS6100-F1 and IS6100-R1. One specific fragment (5.2 kb in size, Figure S1C, lane 3) using IS6100-R1 was amplified, while no specific fragment was amplified under the same PCR conditions using IS6100-F1 (lane 2). (D) Second-round PCR (lane 4-6) with primer IS6100-F2 using the first-round PCR product as a template.
(1.12 MB TIF)
Determination of the positional relation of TnCar, TnFdr and TnAnt. (A) The positions of primers TnCar-F1, TnFdr-F1, TnFdr-R1, TnAnt-F1, and TnAnt-R1 with blue arrowheads showing their directions. (B) Agarose gel electrophoresis of DNA fragments amplified from the genomic DNA of XLDN2-5 by PCR using TnCar-F1 and TnFdr-R1 (lane 1), TnCar-F1 and TnAnt-R1 (lane 2), TnCar-F1 and TnAnt-F1 (lane 3), TnFdr-F1 and TnAnt-R1 (lane 4), TnFdr-F1 and TnAnt-F1 (lane 5), TnFdr-R1 and TnAnt-R1 (lane 6), and TnFdr-R1 and TnAnt-F1 (lane 7). (C) PCR results using primers carC-sp3 and fdr-r2 (lane 8-12). (D) PCR results (no specific bands) using primers fdr-f2 and antAc-r2 (lane 13-17), and fdr-f2 and antAa-f2 (lane 18-22).
(1.06 MB TIF)
Mass spectra for the products of carbazole transformed by E. coli DH5α harboring pUcarAaAcfdr. GC-MS analysis was performed after trimethylsilylation with BSTFA. Compound I: 2′-aminobiphenyl-2,3-diol; compound II: hydroxycarbazole.
(0.19 MB TIF)
Oligonucleotides used in this study for the cloning of genes, genome walking, and the construction of plasmids.
(0.05 MB DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was partially supported by the grants from National High Technology Research and Development Program of China (grant number 2007AA10Z401), from National Basic Research Program of China (grant number 2009CB118906), and from National Natural Science Foundation of China (grant numbers 20977061 and 20777051). The authors also acknowledge the financial support from Shanghai Key Basic Research Program of China (grant number 09JC1407700). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jha AM, Bharti MK. Mutagenic profiles of carbazole in the male germ cells of Swiss albino mice. Mutat Res. 2002;500:97–101. doi: 10.1016/s0027-5107(01)00303-7. [DOI] [PubMed] [Google Scholar]
- 2.Gai Z, Yu B, Li L, Wang Y, Ma C, et al. Cometabolic degradation of dibenzofuran and dibenzothiophene by a newly isolated carbazole-degrading Sphingomonas sp. strain. Appl Environ Microbiol. 2007;73:2832–2838. doi: 10.1128/AEM.02704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kilbane JJ, 2nd, Daram A, Abbasian J, Kayser KJ. Isolation and characterization of Sphingomonas sp. GTIN11 capable of carbazole metabolism in petroleum. Biochem Biophys Res Commun. 2002;297:242–248. doi: 10.1016/s0006-291x(02)02183-6. [DOI] [PubMed] [Google Scholar]
- 4.Kirimura K, Nakagawa H, Tsuji K, Matsuda K, Kurane R, et al. Selective and continuous degradation of carbazole contained in petroleum oil by resting cells of Sphingomonas sp. CDH-7. Biosci Biotechnol Biochem. 1999;63:1563–1568. doi: 10.1271/bbb.63.1563. [DOI] [PubMed] [Google Scholar]
- 5.Li L, Li Q, Li F, Shi Q, Yu B, et al. Degradation of carbazole and its derivatives by a Pseudomonas sp. Appl Microbiol Biotechnol. 2006;73:941–948. doi: 10.1007/s00253-006-0530-3. [DOI] [PubMed] [Google Scholar]
- 6.Nojiri H, Sekiguchi H, Maeda K, Urata M, Nakai S, et al. Genetic characterization and evolutionary implications of a car gene cluster in the carbazole degrader Pseudomonas sp. strain CA10. J Bacteriol. 2001;183:3663–3679. doi: 10.1128/JB.183.12.3663-3679.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouchiyama N, Miyachi S, Omori T. Cloning and nucleotide sequence of carbazole catabolic genes from Pseudomonas stutzeri strain OM1, isolated from activated sludge. J Gen Appl Microbiol. 1998;44:57–63. doi: 10.2323/jgam.44.57. [DOI] [PubMed] [Google Scholar]
- 8.Shepherd JM, Lloyd-Jones G. Novel carbazole degradation genes of Sphingomonas CB3: sequence analysis, transcription, and molecular ecology. Biochem Biophys Res Commun. 1998;247:129–135. doi: 10.1006/bbrc.1998.8750. [DOI] [PubMed] [Google Scholar]
- 9.Nojiri H, Nam JW, Kosaka M, Morii KI, Takemura T, et al. Diverse oxygenations catalyzed by carbazole 1,9a-dioxygenase from Pseudomonas sp. Strain CA10. J Bacteriol. 1999;181:3105–3113. doi: 10.1128/jb.181.10.3105-3113.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Urata M, Uchimura H, Noguchi H, Sakaguchi T, Takemura T, et al. Plasmid pCAR3 contains multiple gene sets involved in the conversion of carbazole to anthranilate. Appl Environ Microbiol. 2006;72:3198–3205. doi: 10.1128/AEM.72.5.3198-3205.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinyakong O, Habe H, Omori T. The unique aromatic catabolic genes in sphingomonads degrading polycyclic aromatic hydrocarbons (PAHs). J Gen Appl Microbiol. 2003;49:1–19. doi: 10.2323/jgam.49.1. [DOI] [PubMed] [Google Scholar]
- 12.Romine MF, Stillwell LC, Wong KK, Thurston SJ, Sisk EC, et al. Complete sequence of a 184-kilobase catabolic plasmid from Sphingomonas aromaticivorans F199. J Bacteriol. 1999;181:1585–1602. doi: 10.1128/jb.181.5.1585-1602.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dogra C, Raina V, Pal R, Suar M, Lal S, et al. Organization of lin genes and IS6100 among different strains of hexachlorocyclohexane-degrading Sphingomonas paucimobilis: evidence for horizontal gene transfer. J Bacteriol. 2004;186:2225–2235. doi: 10.1128/JB.186.8.2225-2235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kato K, Ohtsuki K, Mitsuda H, Yomo T, Negoro S, et al. Insertion sequence IS6100 on plasmid pOAD2, which degrades nylon oligomers. J Bacteriol. 1994;176:1197–1200. doi: 10.1128/jb.176.4.1197-1200.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin C, Timm J, Rauzier J, Gomez-Lus R, Davies J, et al. Transposition of an antibiotic resistance element in mycobacteria. Nature. 1990;345:739–743. doi: 10.1038/345739a0. [DOI] [PubMed] [Google Scholar]
- 17.Armengaud J, Timmis KN. The reductase RedA2 of the multi-component dioxin dioxygenase system of Sphingomonas sp. RW1 is related to class-I cytochrome P450-type reductases. Eur J Biochem. 1998;253:437–444. doi: 10.1046/j.1432-1327.1998.2530437.x. [DOI] [PubMed] [Google Scholar]
- 18.Herman PL, Behrens M, Chakraborty S, Chrastil BM, Barycki J, et al. A three-component dicamba O-demethylase from Pseudomonas maltophilia, strain DI-6: gene isolation, characterization, and heterologous expression. J Biol Chem. 2005;280:24759–24767. doi: 10.1074/jbc.M500597200. [DOI] [PubMed] [Google Scholar]
- 19.Peterson JA, Lorence MC, Amarneh B. Putidaredoxin reductase and putidaredoxin. Cloning, sequence determination, and heterologous expression of the proteins. J Biol Chem. 1990;265:6066–6073. [PubMed] [Google Scholar]
- 20.Chang HK, Mohseni P, Zylstra GJ. Characterization and regulation of the genes for a novel anthranilate 1,2-dioxygenase from Burkholderia cepacia DBO1. J Bacteriol. 2003;185:5871–5881. doi: 10.1128/JB.185.19.5871-5881.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shintani M, Urata M, Inoue K, Eto K, Habe H, et al. The Sphingomonas plasmid pCAR3 is involved in complete mineralization of carbazole. J Bacteriol. 2007;189:2007–2020. doi: 10.1128/JB.01486-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall RM, Brown HJ, Brookes DE, Stokes HW. Integrons found in different locations have identical 5′ ends but variable 3′ ends. J Bacteriol. 1994;176:6286–6294. doi: 10.1128/jb.176.20.6286-6294.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sundin GW, Bender CL. Expression of the strA-strB streptomycin resistance genes in Pseudomonas syringae and Xanthomonas campestris and characterization of IS6100 in X. campestris. Appl Environ Microbiol. 1995;61:2891–2897. doi: 10.1128/aem.61.8.2891-2897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boyd DA, Peters GA, Ng L, Mulvey MR. Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium DT104. FEMS Microbiol Lett. 2000;189:285–291. doi: 10.1111/j.1574-6968.2000.tb09245.x. [DOI] [PubMed] [Google Scholar]
- 25.Tauch A, Gotker S, Puhler A, Kalinowski J, Thierbach G. The 27.8-kb R-plasmid pTET3 from Corynebacterium glutamicum encodes the aminoglycoside adenyltransferase gene cassette aadA9 and the regulated tetracycline efflux system Tet 33 flanked by active copies of the widespread insertion sequence IS6100. Plasmid. 2002;48:117–129. doi: 10.1016/s0147-619x(02)00120-8. [DOI] [PubMed] [Google Scholar]
- 26.Sato SI, Nam JW, Kasuga K, Nojiri H, Yamane H, et al. Identification and characterization of genes encoding carbazole 1,9a-dioxygenase in Pseudomonas sp. strain CA10. J Bacteriol. 1997;179:4850–4858. doi: 10.1128/jb.179.15.4850-4858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zylstra GJ, Kim E. Aromatic hydrocarbon degradation by Sphingomonas yanoikuyae B1. J Ind Microbiol Biotechnol. 1997;19:408–414. doi: 10.1038/sj.jim.2900475. [DOI] [PubMed] [Google Scholar]
- 28.Armengaud J, Happe B, Timmis KN. Genetic analysis of dioxin dioxygenase of Sphingomonas sp. strain RW1: catabolic genes dispersed on the genome. J Bacteriol. 1998;180:3954–3966. doi: 10.1128/jb.180.15.3954-3966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cai M, Xun L. Organization and regulation of pentachlorophenol-degrading genes in Sphingobium chlorophenolicum ATCC 39723. J Bacteriol. 2002;184:4672–4680. doi: 10.1128/JB.184.17.4672-4680.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata Y, Miyauchi K, Takagi M. Complete analysis of genes and enzymes for gamma-hexachlorocyclohexane degradation in Sphingomonas paucimobilis UT26. J Ind Microbiol Biotechnol. 1999;23:380–390. doi: 10.1038/sj.jim.2900736. [DOI] [PubMed] [Google Scholar]
- 31.Providenti MA, Shaye RE, Lynes KD, McKenna NT, O'Brien J M, et al. The locus coding for the 3-nitrobenzoate dioxygenase of Comamonas sp. strain JS46 is flanked by IS1071 elements and is subject to deletion and inversion events. Appl Environ Microbiol. 2006;72:2651–2660. doi: 10.1128/AEM.72.4.2651-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X, Gai Z, Yu B, Feng J, Xu C, et al. Degradation of carbazole by microbial cells immobilized in magnetic gellan gum gel beads. Appl Environ Microbiol. 2007;73:6421–6428. doi: 10.1128/AEM.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gai Z, Yu B, Wang X, Deng Z, Xu P. Microbial transformation of benzothiophenes, with carbazole as the auxiliary substrate, by Sphingomonas sp. strain XLDN2-5. Microbiology. 2008;154:3804–3812. doi: 10.1099/mic.0.2008/023176-0. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell D. Cold Spring Harbor, N. Y: Cold Spring Harbor Laboratory Press; 2001. Molecular cloning: A laboratory manual. [Google Scholar]
- 35.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IS6100-based PCR. (A) Schematics of relative configurations of two copies of IS6100. The position of primers IS6100-F1, IS6100-F2, IS6100-R1 and IS6100-R2 are shown by arrows. (B and C) Gel electrophoresis of DNA fragments amplified from XLDN2-5 by IS6100-based PCR using IS6100-F1 and IS6100-R1 (lane 1), single IS6100-F1 (lane 2) and single IS6100-R1 (lane 3). Two fragments (4.3 kb and 2.8 kb in size, Figure S1B, lane 1) were amplified using primers IS6100-F1 and IS6100-R1. One specific fragment (5.2 kb in size, Figure S1C, lane 3) using IS6100-R1 was amplified, while no specific fragment was amplified under the same PCR conditions using IS6100-F1 (lane 2). (D) Second-round PCR (lane 4-6) with primer IS6100-F2 using the first-round PCR product as a template.
(1.12 MB TIF)
Determination of the positional relation of TnCar, TnFdr and TnAnt. (A) The positions of primers TnCar-F1, TnFdr-F1, TnFdr-R1, TnAnt-F1, and TnAnt-R1 with blue arrowheads showing their directions. (B) Agarose gel electrophoresis of DNA fragments amplified from the genomic DNA of XLDN2-5 by PCR using TnCar-F1 and TnFdr-R1 (lane 1), TnCar-F1 and TnAnt-R1 (lane 2), TnCar-F1 and TnAnt-F1 (lane 3), TnFdr-F1 and TnAnt-R1 (lane 4), TnFdr-F1 and TnAnt-F1 (lane 5), TnFdr-R1 and TnAnt-R1 (lane 6), and TnFdr-R1 and TnAnt-F1 (lane 7). (C) PCR results using primers carC-sp3 and fdr-r2 (lane 8-12). (D) PCR results (no specific bands) using primers fdr-f2 and antAc-r2 (lane 13-17), and fdr-f2 and antAa-f2 (lane 18-22).
(1.06 MB TIF)
Mass spectra for the products of carbazole transformed by E. coli DH5α harboring pUcarAaAcfdr. GC-MS analysis was performed after trimethylsilylation with BSTFA. Compound I: 2′-aminobiphenyl-2,3-diol; compound II: hydroxycarbazole.
(0.19 MB TIF)
Oligonucleotides used in this study for the cloning of genes, genome walking, and the construction of plasmids.
(0.05 MB DOC)