Abstract
The ability of environmental factors to promote a phenotype or disease state not only in the individual exposed but also in subsequent progeny for multiple generations is termed transgenerational inheritance. The majority of environmental factors such as nutrition or toxicants such as endocrine disruptors do not promote genetic mutations or alterations in DNA sequence. In contrast, these factors have the capacity to alter the epigenome. Epimutations in the germ line that become permanently programmed can allow transmission of epigenetic transgenerational phenotypes. This review provides an overview of the epigenetics and biology of how environmental factors can promote transgenerational phenotypes and disease.
Keywords: Endocrine Disruptors, Adult Onset Disease, Germ Cells, Epigenetics, Transgenerational
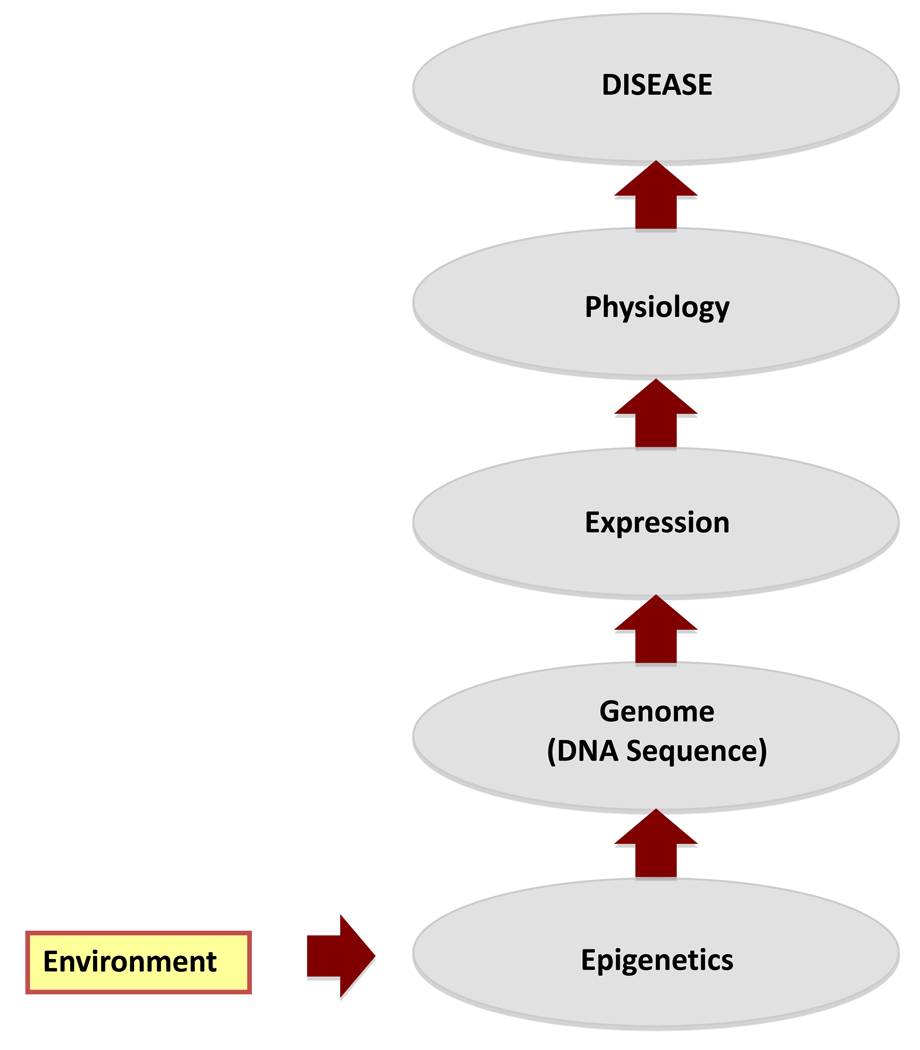
The current paradigm for disease etiology is that the presence of a genetic mutation, polymorphism or chromosomal abnormality promotes disease. Although this is a critical component of disease, the environment is an equally important consideration in disease etiology (Figure 1). Since the genome is evolutionarily and chemically stable, the ability of the environment to influence or promote disease does not generally involve DNA mutations. Therefore, environmental factors must generally regulate genome activity independent of DNA sequence manipulation (e.g. epigenetics). An additional consideration for environmental influences on disease etiology is the developmental stage of exposure. Exposures during a critical time of development can alter genome activity associated with the differentiation program of cells or organ systems. This altered program and gene expression profile can then promote an abnormal physiology and disease at the later adult stage of development.
Figure 1. Proposed etiology of how the environmental impacts on disease.
The cascade of molecular and physiological processes following an environmental exposure to promote disease is shown.
A large number of epidemiology studies suggest the environment is a major factor in disease etiology [1, 2]. Examples include phenomena such as the regional differences in disease frequency, the low frequency of the genetic component of disease, the increase in the majority of specific disease frequencies, the variability in disease frequency between identical twins, and the large number of environmental exposures that promote disease. This review focuses on how environmental factors promote adult onset disease transgenerationally.
Environmental Factors and Disease
Epidemiology research suggests significant environmental impacts on disease. Each geographic region around the world generally has a distinct disease frequency. For example, some regions have high rates of prostate disease and low rates of stomach disease (North America), while others have low rates of prostate disease and high rates of stomach disease (Eastern Asia) [3, 4]. If one is moved early in life from one region to the other, they often develop the new region’s disease frequencies. Interestingly, when identical twins develop in different geographic regions, they also have different disease frequencies [5]. Therefore, although the genetics is nearly identical, disease development is different suggesting an environmental influence [6]. Another example is the dramatic and rapid increase in nearly all disease frequencies over the past several decades that can not be explained through genetics alone. There are also a large number of environmental compounds and toxicants shown to promote disease, but most do not alter the DNA sequence [7]. Therefore, environmental factors are critical in the etiology of disease.
Although numerous environmental factors influence and promote adult onset disease (such as nutrition and stress), the current review will emphasize endocrine disruptors due to this group of environmental compounds being one of the largest we are exposed to daily in society. Endocrine disruptors are environmental chemicals that affect the function of the endocrine system by mimicking or blocking the actions of hormones, altering hormone signaling or disrupting hormone production [8]. Endocrine disruption can have profound consequences due to the crucial role hormones have in development.
A number of disease states are promoted by endocrine disruptors (Table 1). Many endocrine disruptors with reproductive hormone actions (e.g. estrogen or androgen) influence reproduction and fertility including bisphenol-A (BPA), DDT and vinclozolin. Activation of the male and female reproductive systems at an inappropriate time during development by endocrine disruptor chemicals can alter normal physiology [9]. For example, prenatal exposure to diethylstilbestrol (DES) produces several developmental abnormalities in the male mouse reproductive tract and increases tumor incidence [10]. Embryonic exposure to the pesticide methoxychlor during the period of sex determination affects embryonic testis cellular composition and germ cell number and survival [11]. Embryonic testicular cord formation is also affected when embryos are exposed in vitro to vinclozolin. Transient in utero exposure to vinclozolin increases apoptotic germ cell numbers in the testis of pubertal and adult animals which correlates with reduced sperm motility and number in the adult [12]. In utero exposure to the plastic-derived compounds phthalates also disrupts differentiation of androgen-dependent tissues in male rat offspring [13]. A more recent example of an endocrine disruptor is the plastic component BPA that acts as an estrogenic compound causing numerous pathologies including prostate cancer in low doses [14]. Other examples include the plant derived estrogenic compounds (phytoestrogens) such as genistein, which influences a number of reproductive organs [15, 16]; eating aflatoxin-contaminated food has been correlated with the incidence of liver cancer in Asia and Africa [17]; tobacco contains cadmium, which is an estrogenic endocrine disruptor (18); and use of tobacco can cause reproductive problems in addition to carcinogen-induced lung cancer. Heterocyclic amines in well-cooked meat products can result in cancer of colon, breast and stomach in consumers [19]. Abnormalities in mouse testicular Leydig cells are induced by chronic low dose exposure to arsenic [20]. Estrogen receptor-alpha promoter hypomethylation may play a role in induction of hepatocellular carcinoma by arsenic exposure in utero [21]. Therefore, it is apparent that a large number of environmental compounds have endocrine disruptor activity. How an early life exposure to an endocrine disruption can promote an adult onset disease, long after the compound is removed, is presumed to involve in part epigenetic mechanisms that are reviewed below.
Table 1.
Common Endocrine Disruptors and Actions
| Endocrine Disruptor | Effect | Reference |
|---|---|---|
| DDT | Reproductive Failure | [110] |
| Phytoestrogens (e.g. Genistein) |
Impaired Fertility, Reproductive Effects, Breast Cancer Protection |
[15, 16] |
| Diethylstilbestrol (DES) |
Vaginal Cancer in Humans Developmental Toxicity in Hamsters |
[111 – 113] |
| Dicofol | Abnormal Ovarian Follicles, High Plasma Estrogen Levels |
[114] |
| Bisphenol-A (BPA) | Prostate Cancer | [14, 115] |
| Aflatoxin | Liver Cancer | [17] |
| Cadmium | Lung Cancer, Reproductive Problems | [18] |
| Heterocyclic amines | Cancer of Colon, Stomach and Breast | [19] |
| Arsenic | Liver Cancer | [21] |
| Dioxins (TCDD) | Mammary Tumor | [116] |
| Vinclozolin | Impaired Male Fertility | [33] |
| Methoxychlor | Impaired Male Fertility | [117] |
| Phthalates | Impairs Male Reproductive Tract and Testis | [13] |
Epigenetics
Although the history and definition of epigenetics has evolved (Box 1), the majority of the molecular elements of epigenetic regulatory processes have only been recently elucidated [1]. The first epigenetic molecular factor identified was DNA methylation in the 1970’s [22] (Table 2). Significant focus was put on DNA methylation with the analysis of X chromosome inactivation and imprinted genes in the late 1980s and early 1990s [23]. The next epigenetic element identified was histone modifications in the mid 1990s and the appreciation of chromatin structure in the regulation of the genome [24]. This was followed by the identification of non-coding RNA around 2000 and the first whole epigenome analysis in 2005 [25] (Table 2). Epigenetic processes are likely to be expanded in the future. For example, the recent identification of hydroxy-methylcytosine residues in the brain is a new epigenetic mark whose function remains to be elucidated [26]. These epigenetic processes are equally important in regulating genome activity (i.e. gene expression) and DNA sequence (i.e. genetics).
Box 1. Epigenetics
The term epigenetics was coined by Conrad Waddington in the 1940s. Waddington integrated the new knowledge about genes and genetics to embryology. The study of embryological growth and differentiation was commonly known as Epigenesis, a concept that was around since Aristotelian times. The integration of the concepts of Epigenesis and Genetics gave origin to the term Epigenetics [101, 102]. Waddington’s goal with epigenetics was to provide insight into gene-environment interactions that influence development and embryology [101–103]. Pioneering epigenetic experiments from Waddington on Drosophila demonstrated that a temperature shock from hours 17–23 after puparium formation produced cross veinless wings in flies. Flies with this phenotype were culled from the population and only those showing normal wings were used to carry on the line. After an expected initial reduction of the cross wingless phenotype in the population, it surprisingly raised after generation 16 [104]. This phenotype was considered a ‘genetic assimilation’ and dealt with environmental exposures early in development with subsequent consequences on phenotypic inheritance.
The definition of epigenetics has evolved with greater clarity of the molecular mechanisms involved and a better understanding of genetic phenomena. The initial definition of Waddington focused on gene-environment interactions but had no molecular insights to consider [102]. In 1990, Holliday defined epigenetics as “the study of the mechanisms of temporal and spatial control of gene activity during the development of complex organisms”. His definition rescues the original Waddington’s meaning of developmental biology, although does not differentiate between the action of what we currently know as epigenetic mechanisms and the action of genetic regulators of gene expression such as transcription factors [105]. Another early definition, by Riggs and colleagues states that epigenetics is “the study of mitotically and/or meiotically heritable changes in gene function that cannot be explained by changes in DNA sequence” [106]. However, the term heritable is generally in reference to generational inheritance and not associated with growth of cells or tissues. Perhaps a more direct term would be mitotically stable. A more recent definition focuses on molecular elements that influence chromatin, independent of DNA sequence. Bird defines epigenetics as the “structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states” [107]. Since there are a number of epigenetic elements that do not fit into this definition such as non-coding RNA and minor modifications of histones and DNA methylation of promoters, this definition appears not global enough to encompass all of epigenetics. Therefore, we propose a definition that is more global and encompasses all molecular elements and includes the use of the term Epi for around DNA. In this case, epigenetics is defined as “molecular factors and processes around DNA that are mitotically stable and regulate genome activity independent of DNA sequence.”
Table 2.
History of Epigenetics
| 1940s | Conrad Waddington defined epigenetics as environment-gene interactions that induce developmental phenotypes |
| 1975 | Holliday and Pugh identify DNA methylation |
| 1988 | X-chromosome inactivation and DNA methylation |
| 1990s | Imprinted genes, allelic expression and DNA methylation |
| 1995 | Histone modifications and chromatin structure |
| 2000s | Small non-coding RNA’s |
| 2005 | Epigenome mapping |
A special category of genes called imprinted genes are subject to epigenetic programming and can be influenced by environmental exposures. For example, in vitro treatment of preimplantation embryos with the contaminant 2,3,7,8-tetra-chlorodibenzo-p-dioxin alters DNA methylation in the H19 and IGF-2 imprinted genes [27]. From an epigenetic perspective, imprinted genes are a special class of genes because they have relatively unchanged DNA methylation patterns over generations and are not affected by the overall reset in methylation patterns that occur early in development [28]. Imprinted genes carry a molecular memory of their parent of origin allele acquired early in the germ line [29]. This molecular memory is associated with differential methylation patterns between the two alleles, which affect monoallelic gene expression [30]. These allelic differences in methylation are defined in the developing embryo during the establishment of germ line development [28]. Methylation of imprinted genes initiated during germ line development can be completed after fertilization [28, 31]. Some imprinted genes remain imprinted throughout the organism’s life; however, a group of them are imprinted in specific tissues in a temporally specific manner [32]. Interestingly, if external agents altered DNA methylation in these imprinted genes or induced new methylation sites during critical periods of their establishment, such changes could persist transgenerationally [33, 34] (Figure 2). This heritable transmission of environmentally induced phenotypes is referred to as transgenerational inheritance [1, 35].
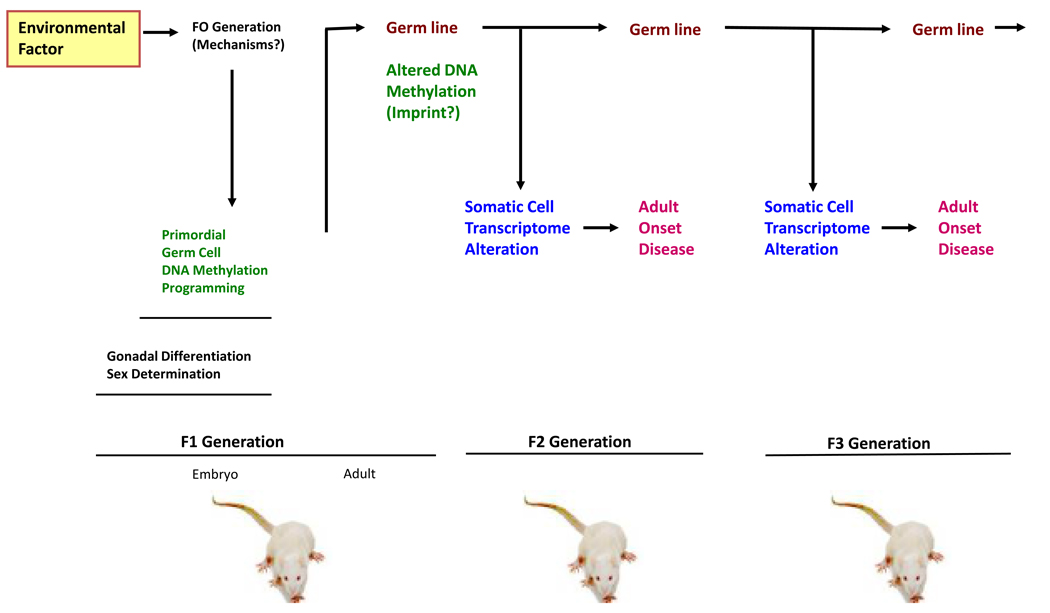
Figure 2. Role of the germ line in epigenetic transgenerational inheritance.
(i) An environmental factor acts on the F0 generation gestating female to influence (ii) the developing F1 generation fetus and alter gonadal development to reprogram the primordial germ cell DNA methylation. (iii) This altered DNA methylation in the germ line becomes permanently programmed similar to an imprinted-like gene and is transferred through the germ line to subsequent generations. The embryo generated from this germ line starts with an altered epigenome that (iv) affects developing somatic cells and tissues to have an altered transcriptome. This altered somatic cell transcriptome can then promote adult onset disease associated with the transgenerational phenotype.
From a human health perspective, a number of disease states exist that have an epigenetic origin. Several diseases and syndromes have abnormal DNA methylation or imprinted gene sites leading to various pathologies [32]. These include Silver-Russell Syndrome [36], Beckwith-Weidemann Syndrome [37], Angelman and Prader-Willi Syndromes [38]. Another epigenetic disease due to abnormal DNA methylation of the X-chromosome is the Fragile X Syndrome [39]. A number of brain disorders such as Autism, Schizophrenia and Rett’s Syndrome also appear to have major epigenetic components [39–41]. Cancer also has an epigenetic component to regulate genome stability and is associated with transformation and disease phenotype [42, 43]. A growing list of diseases with an epigenetic component suggests epigenetics will have a critical role in disease etiology for many disease states (Figure 1).
Epigenetics and Environmental Factors
Initial observations of how the environment can influence epigenetics and phenotype were shown in plants [44]. In animals, many examples associate environmental influences to epigenetic changes. Epigenetic influences have been observed with environmental compounds, nutritional factors [45, 46] such as methyl donors like folate [47, 48], inorganic contaminants such as arsenic [20, 21], airborne polycyclic aromatic hydrocarbons [49], drugs such as cocaine [50], endocrine disruptors such as BPA [14, 51, 52], phytoestrogens [53, 54] and chemicals used as fungicides [33] or pesticides [55] (Table 1). Some studies have also demonstrated behavioral effects on DNA methylation including maternal effects on nursing behavior [56] or depression [57]. Therefore, numerous examples of environmental factors have been shown to alter the epigenome.
Holliday initially proposed a link between hormone action and establishment of DNA methylation in mammalian embryos. He proposed that maternal effects of teratogens might disrupt the normal distribution of DNA methylation in a developing embryo, leading to developmental abnormalities or defects that would appear in the next generations [58]. McLachlan and collaborators [59] proposed that exposure to environmental endocrine disrupting chemicals during early development affects adult stages, potentially involving gene imprinting leading to persistent genetic change at the level of DNA methylation. The first experimental evidence that endocrine disrupting chemicals produce epigenetic changes came from experiments where neonatal exposure to DES produced abnormalities in the demethylation of the lactoferrin promoter [60].
A classic model for studying endocrine and nutritional epigenetic effects is the Agouti mouse, which consists of detecting changes in methylation of the Avy allele. Methylation in this meta-stable allele correlates with changes in coat color, which shifts from yellow-agouti to yellow by decreasing DNA methylation in the intracisternal A particle retrotransposon upstream of the Agouti gene [47]. Maternal methyl donor (i.e. folate) consumption leads to changes in the coat color of offspring correlated with alteration in methylation of the Avy allele [47]. Interestingly, transgenerational exposure of Avy/a mice to an ad libitum diet produces amplification of obesity, an effect that is suppressed when the diet is methyl-supplemented with extra folate [61]. Maternal BPA treatment also decreases the offspring’s CpG DNA methylation in this meta-stable epi-allele, resulting in a coat color change [51]. Dietary supplementation of BPA or genistein treatments with methyl donors inhibits the hypo-methylating effect of BPA or genistein, shifting the coat color of heterozygous yellow-agouti offspring toward pseudo-agouti, which is the same coat color pattern observed in controls [51]. This mouse model has clearly established the ability of environmental factors to influence epigenetics to promote phenotypic changes later in development.
Endocrine disruptors have the ability to alter DNA methylation patterns of key genes that produce related transcriptional changes [1, 7, 62, 63]. Administration of the plant derived endocrine disrupting phytoestrogens, coumestrol and equol, to newborn mice enhances DNA methylation to inactivate the proto-oncogene H-ras [64]. DNA methylation patterns were altered in 8-week-old mice that consumed high doses of the phytoestrogen genistein [65]. Recently, gender-specific changes in Acta1 methylation have been shown as a response to dietary isoflavones in mice [54]. Environmental compounds with endocrine disruptor activity tested for epigenetic effects include the fungicide vinclozolin, the plastic residue BPA and the pharmacological compound DES (Table 1). Exposure to environmentally relevant doses of BPA during the neonatal developmental period in rats produces DNA methylation changes associated with carcinogenic processes [14]. Maternal exposure to BPA has also been shown to alter methylation in the fetal mouse forebrain [52] and to produce changes in behavior responses in the offspring [66]. These findings correlate with other studies showing epigenetic changes due to endocrine disruptor exposure which affect aspects of neuroendocrine systems [67] and behavioral neuroendocrinology [68–70]. Changes in methylation also explain the reappearance of increased susceptibility for tumor formation in F2 generation mice after developmental exposure to DES [71, 72]. Therefore, the actions of a number of endocrine disruptors involve alterations in epigenetic processes.
The implication of these environmentally induced epigenetic effects in evolutionary biology is also a topic of interest. An assumption of new-Darwinian theory is that evolution proceeds based on random DNA sequence mutations and that the environment is not able to alter the occurrence or frequency of these mutations [73]. Epigenetics offers an alternative view regarding the molecular mechanism involved. For example, DNA methylation of CpG sites increases the rate of mutations of methylated cytosines by an order of magnitude [74]. Therefore, in the event DNA methylation patterns are altered due to an environmental stimulus, these CpG sites will be more prone to undergo mutations than sites that are not methylated [34]. If this is transgenerationally maintained in a population, this is an epigenetically controlled mutation frequency. This bias in the mutation rates over generations is environmentally induced. Simulations in the evolution of the BRCA1 gene show that methylation-biased derived mutations are a feasible process [75]. Recent studies highlight the role of environmental compounds on epigenetic mechanisms from an evolutionary and ecological perspective [34, 54, 69, 76].
Epigenetic Transgenerational Phenomena
Since the germ line is required for transmitting genetic information between generations, a permanent epigenetic modification of the germ line can result in transgenerational phenomena (Box 2). Epigenetic programming of the germ line occurs during the migration of the primordial germ cells in the embryo. The migrating primordial germ cells in the genital ridge undergo an erasure of methylation of the DNA during migration and colonize the early bipotential gonad prior to gonadal sex determination [77, 78]. Once gonadal sex determination is initiated, the primordial germ cells develop female or male germ cell lineage and re-methylate the DNA in a male or female specific manner. Therefore, the germ cell epigenetic programming during gonadal sex determination is a sensitive period to environmental factors [77] (Box 3).
Box 2. Transgenerational Phenotype Definition
The majority of the actions of environmental factors or toxicants involve direct exposures of somatic tissues that are important for the exposed individual’s disease, but will not be transmitted to the next generation. In contrast, transgenerational phenotypes and toxicology by definition excludes direct exposure and must be transmitted through multiple generations [1, 108]. For example, exposure of a gestating female provides direct exposure of the F0 generation female, the F1 generation embryo, and the germ-line that will generate the F2 generation [108]. Therefore, a phenotype in the F3 generation is required to have a transgenerational phenomenon or phenotype. The effects observed in the F0 and F1 generations are due to direct exposures, as well as that in the F2 generation germ line [1, 108]. The ability of a direct exposure to influence multiple generations is defined as a multiple generation phenotype and not a transgenerational phenotype. In contrast, a transgenerational phenotype requires the absence of a direct exposure and transmission of the phenotype to minimally the F3 generation [108].
Box 3. Germ Cell Developmental Epigenetics
An important factor to consider with a transgenerational phenotype is the action of environmental factors on the germ line and gonadal development. During embryonic development in mammalian species, the primordial germ cells migrate down the genital ridge towards the developing gonad prior to sex determination [77, 78, 109]. At the time of gonadal sex determination, the germ cell develops into a male or female germ cell lineage at the initial stages of gonadal sex determination. The female germ line then enters meiosis in the developing embryonic ovary, while male germ cells continue to proliferate until immediately prior to birth and then resume proliferation after birth until puberty [77, 78, 109]. In the adult, the female germ line undergoes oogenesis during follicle development to generate oocytes. The male germ line, in turn, develops from spermatogonial stem cells and undergoes spermatogenesis for the production of spermatozoa in the testis. The critical period for epigenetic regulation and modification of the germ line is during the period of primordial germ cell migration and gonadal sex determination. The permanent alteration in the epigenetic programming of the germ line appears to be the mechanism involved in the transgenerational phenotype [1, 33].
Although there are alterations in the male and female germ line epigenomes (i.e. DNA methylation) during gametogenesis in the adult gonads [79], the embryonic period of gonadal sex determination is the most sensitive to environmental insults. During spermatogenesis, the male germ cell replaces the majority of histones with protamines, DNA condensation occurs to eliminate chromatin structure, and the genome is silenced for reduced expression of non-coding RNAs [80]. Although a small percentage of histones are maintained in developmentally important loci [81], the role of histones in sperm remains to be established. Therefore, the primary epigenetic process that is transmitted through the male germ line is DNA methylation.
One of the first studies to demonstrate the ability of an environmental factor to modify the epigenetic programming of the male germ line used the endocrine disruptor vinclozolin. When embryonic rats were exposed through maternal administration to vinclozolin, an anti-androgenic environmental endocrine disruptor, during gonadal sex determination, adult onset disease occurred in the first generation and persisted for four subsequent generations [33] (Figure 2). This phenomenon was found to be due to male germ line changes in DNA methylation, which resulted in heritable changes in transcription in a number of tissues, such as the testis [82], brain [70] and prostate [83]. The pathology of adult onset disease from vinclozolin exposure during embryonic life included testicular, prostate and renal abnormalities and incidence of tumors [33, 84, 85]. A modification of the sperm epigenome appears to have occurred following vinclozolin exposure at the time of gonadal sex determination, and this enabled transgenerational transmission to subsequent generations to promote adult onset disease [1] (Figure 2). This was one of the first reports of an environmental factor promoting epigenetic transgenerational inheritance.
A follow-up study by a company that produces vinclozolin (BASF, Germany) found that oral administration of the same dose used intraperitoneally (IP) [33] did not have transgenerational effects nor major effects in the F1 generation [86]. Previously, we found that a 4-fold decrease in the dose eliminated the vinclozolin effect [84]. For most compounds, oral gavage treatment generally has an order of magnitude lower circulating dose than an IP injection, such that the lack of effect was likely due to insufficient dosing [33]. In regards to toxicology, this study suggests vinclozolin may not be a significant risk factor at the dose used [86]. However, in our studies, we used vinclozolin as a pharmacologic agent to promote the transgenerational phenotype and to study mechanism [33] and did not perform risk assessment or classic toxicology experiments. A second study repeated the vinclozolin experiment [87] using a more inbred CD-Sprague Dawley (Charles River) rat line versus the outbred Harlan Sprague Dawley line [33]. This study did not obtain a dramatic transgenerational phenotype [87]. Previously, we reported that the inbred Fisher rat line did not respond as well as the outbred Harlan Sprague Dawley line [33, 84] and have recently found the CD-Sprague Dawley response is also not as robust. The hypothesis is proposed that the inbred status of the line may be a factor in the efficiency of promoting the phenotype. We recently repeated the original observation [33] with the outbred Harlan Sprague Dawley line [88]. In addition to the outbred status of the line, the exposure timing and duration have been found to be critical. The parameters required to obtain the transgenerational phenotype will likely help reveal aspects of the mechanisms involved. Several other recent studies confirm the ability of environmental agents to promote transgenerational phenotypes [89], and a recent independent study confirmed the epigenetic transgenerational actions of vinclozolin [90].
A number of epigenetic transgenerational phenomena and phenotypes have since been observed in various species and with various environmental factors involved (Table 3). The first non-Mendelian hereditary phenomenon reported in plants was called paramutation [44] and later this transgenerational phenomenon was found to be epigenetic in nature and controlled by DNA methylation [91]. This event was observed in mammals when a similar mode of inheritance was found in mice [92, 93]. Nutrition also promotes a transgenerational adult onset obesity phenotype, as described in the Agouti mouse model [61], and there is also documentation of transgenerational responses to nutrition in humans [94]. A transgenerational mechanism exists which appears to capture an alteration in nutrition in a sensitive period of perinatal development from the previous generation(s). This requires a mechanism for transmitting the change in environmental exposure (epigenetic) that then alters gene expression and phenotype in the next generation (Figure 2). A nutritionally induced transgenerational response has been observed down the male line and implies that the sperm carries the ancestral exposure information. A study by Arai et. al. [95] demonstrates the ability of an animal’s environment to modulate the signaling network that promotes long-term potentiation (LTP) in the hippocampus and to improve contextual fear memory formation across generations. In addition, environment also enhances LTP in their future offspring through adolescence, even if the offspring are not exposed. Stress-induced maternal programming also promotes behavioral changes transgenerationally [96, 97].
Table 3.
Epigenetic Transgenerational Events
| Epigenetic Transgenerational Event, Environmental Factor and Generation |
Reference |
|---|---|
| Paramutation in Maize Modification of plant color (F1–F2) |
[118] |
| Paramutation in Arabidopsis (F1–F4) | [91] |
| Epigenetic (Paramutation) Non-Mendelian change in mouse (F1–F6) | [92, 93] |
| Vinclozolin induced epigenetic transgenerational adult onset disease in rats (F1–F4) |
[33, 86] |
| BPA-induced transgenerational testicular abnormality (F1–F3) | [89] |
| Transgenerational promotion of long term potentiation (F1–F2) by altered environment |
[95] |
| Stress induced behavior alterations (F0–F2) | [96] |
| Nutrition induced transgenerational obesity in mice (F1–F3) | [61] |
| Transgenerational response in longevity to nutrition (F0–F2) | [94] |
| Gender bias in multiple sclerosis following (F1–F2) Epigenetic changes in HLA class III risk haplotypes |
[98] |
| Tumor susceptibility in Drosophila (F1–F3) | [119] |
| Stem cell culture induced adult onset disease (F0–F4) | [99] |
Heritable disease states such as multiple sclerosis (MS) also appear to have an epigenetic origin [98]. Epigenetic modifications differentiate among human leukocyte antigen class II risk haplotypes and are involved in the gender bias in MS [98]. Processes such as embryonic stem cell culture to generate spermatogonial stem cells have been shown to epigenetically alter the germ line and promote abnormalities transgenerationally (F0–F4) in mice [99]. As discussed, environmental toxicants such as vinclozolin [33] and the plastic component BPA promote transgenerational disease. The plasticizer BPA also promotes testicular disease from F1 to F3 generations in rats [89]. Further studies (Box 4) are required to determine the critical time of exposure of environmental toxicants and to identify factors which result in germ line-transmitted adult onset diseases and those that have an epigenetic basis.
Box 4. Future Questions and Considerations
The epigenetic and genetic mechanisms of how the germ line epigenome becomes permanently programmed to transmit a transgenerational phenotype needs to be determined.
A correlation of epigenetic biomarkers with disease needs to be assessed for the potential future development of early stage diagnosis of disease.
A correlation of epigenetic biomarkers with environmental exposures is needed to develop advanced risk and toxicology assessments.
The paradigm that genetics is the primary molecular mechanism involved in biology and medicine needs to be modified to incorporate epigenetics as a critical regulatory factor as well.
Concluding Remarks
Epigenetic transgenerational phenomena generally require the involvement of the germ line to allow the transmission of an epigenetic abnormality between multiple generations. The ability of environmental factors or toxicants to alter the epigenome will be common in somatic tissues, but less common for the germ line due to the limited developmental period the germ line is sensitive to reprogramming. In the event an altered germ line epigenome becomes permanently programmed, an epigenetic transgenerational phenotype would be possible (Figure 2).
The phenomenon of the fetal basis of adult onset disease has been established [1, 100], and epigenetics likely plays a critical role in this process. Transient early life exposures in the exposed individual, or transgenerational exposures if the germ line is involved, are now included as causal factors for adult onset disease. Further investigation into the role of epigenetics in disease etiology is needed to determine how significant early life toxicology is to disease. Elucidating the epigenetic mechanisms involved in transgenerational toxicology will provide insights into the diagnosis of environmental exposures and provide potential therapeutic targets for disease. Although the prevalence of epigenetic transgenerational inheritance needs to be assessed in various disease states, the role of epigenetics will likely be a major factor to consider in toxicology and medicine in the future.
Endocrine disruptors are one of the most prevalent groups of environmental compounds we are exposed to daily. Although these compounds disrupt the endocrine system, it is the long term response of molecular processes like epigenetics that will promote downstream developmental events, adult onset disease (Figure 1). Elucidation of the role of epigenetics in endocrine disruptor actions and in the etiology of disease will undoubtedly provide insights into diagnostics and therapeutics for environmental exposures, risk assessment and adult onset disease (Box 4). In addition to these abnormal endocrine disrupting agents, it is likely that epigenetics will also be critical to consider in normal endocrinology and metabolic events.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jirtle RL, Skinner MK. Environmental epigenomics and disease susceptibility. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szyf M. The dynamic epigenome and its implications in toxicology. Toxicol Sci. 2007;100:7–23. doi: 10.1093/toxsci/kfm177. [DOI] [PubMed] [Google Scholar]
- 3.Haas GP, Sakr WA. Epidemiology of prostate cancer. CA Cancer J Clin. 1997;47:273–287. doi: 10.3322/canjclin.47.5.273. [DOI] [PubMed] [Google Scholar]
- 4.Brenner H, Rothenbacher D, Arndt V. Epidemiology of stomach cancer. Methods Mol Biol. 2009;472:467–477. doi: 10.1007/978-1-60327-492-0_23. [DOI] [PubMed] [Google Scholar]
- 5.Kukreja A, Maclaren NK. NKT cells and type-1 diabetes and the "hygiene hypothesis" to explain the rising incidence rates. Diabetes Technol Ther. 2002;4:323–333. doi: 10.1089/152091502760098465. [DOI] [PubMed] [Google Scholar]
- 6.Williamson DM. Studies of multiple sclerosis in communities concerned about environmental exposures. J Womens Health (Larchmt) 2006;15:810–814. doi: 10.1089/jwh.2006.15.810. [DOI] [PubMed] [Google Scholar]
- 7.Edwards TM, Myers JP. Environmental exposures and gene regulation in disease etiology. Environ Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crisp TM, Clegg ED, Cooper RL, Wood WP, Anderson DG, Baetcke KP, Hoffmann JL, Morrow MS, Rodier DJ, Schaeffer JE, Touart LW, Zeeman MG, Patel YM. Environmental endocrine disruption: an effects assessment and analysis. Environ Health Perspect. 1998;106 Suppl 1:11–56. doi: 10.1289/ehp.98106s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Danzo BJ. The effects of environmental hormones on reproduction. Cell Mol Life Sci. 1998;54:1249–1264. doi: 10.1007/s000180050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bullock BC, Newbold RR, McLachlan JA. Lesions of testis and epididymis associated with prenatal diethylstilbestrol exposure. Environ Health Perspect. 1988;77:29–31. doi: 10.1289/ehp.887729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupp AS, Uzumcu M, Suzuki H, Dirks K, Phillips B, Skinner MK. Effect of transient embryonic in vivo exposure to the endocrine disruptor methoxychlor on embryonic and postnatal testis development. J Androl. 2003;24:736–745. doi: 10.1002/j.1939-4640.2003.tb02736.x. [DOI] [PubMed] [Google Scholar]
- 12.Uzumcu M, Suzuki H, Skinner MK. Effect of the anti-androgenic endocrine disruptor vinclozolin on embryonic testis cord formation and postnatal testis development and function. Reprod Toxicol. 2004;18:765–774. doi: 10.1016/j.reprotox.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Gray LE, Jr, Wolf C, Lambright C, Mann P, Price M, Cooper RL, Ostby J. Administration of potentially antiandrogenic pesticides (procymidone, linuron, iprodione, chlozolinate, p,p'-DDE, and ketoconazole) and toxic substances (dibutyl- and diethylhexyl phthalate, PCB 169, and ethane dimethane sulphonate) during sexual differentiation produces diverse profiles of reproductive malformations in the male rat. Toxicol Ind Health. 1999;15:94–118. doi: 10.1177/074823379901500109. [DOI] [PubMed] [Google Scholar]
- 14.Ho SM, Tang WY, Belmonte de Frausto J, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moutsatsou P. The spectrum of phytoestrogens in nature: our knowledge is expanding. Hormones (Athens) 2007;6:173–193. [PubMed] [Google Scholar]
- 16.Tomar RS, Shiao R. Early life and adult exposure to isoflavones and breast cancer risk. J Environ Sci Health C Environ Carcinog Ecotoxicol Rev. 2008;26:113–173. doi: 10.1080/10590500802074256. [DOI] [PubMed] [Google Scholar]
- 17.IARC; World Health Organization, International Agency for Research on Cancer Multivolume work:V10 60. Present Monographs on the evaluation of the carcinogenic risk of chemicals to man. 1972
- 18.Henson MC, Chedrese PJ. Endocrine disruption by cadmium, a common environmental toxicant with paradoxical effects on reproduction. Exp Biol Med (Maywood) 2004;229:383–392. doi: 10.1177/153537020422900506. [DOI] [PubMed] [Google Scholar]
- 19.Wogan GN, Hecht SS, Felton JS, Conney AH, Loeb LA. Environmental and chemical carcinogenesis. Semin Cancer Biol. 2004;14:473–486. doi: 10.1016/j.semcancer.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 20.Singh KP, DuMond JW., Jr Genetic and epigenetic changes induced by chronic low dose exposure to arsenic of mouse testicular Leydig cells. Int J Oncol. 2007;30:253–260. [PubMed] [Google Scholar]
- 21.Waalkes MP, Liu J, Chen H, Xie Y, Achanzar WE, Zhou YS, Cheng ML, Diwan BA. Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J Natl Cancer Inst. 2004;96:466–474. doi: 10.1093/jnci/djh070. [DOI] [PubMed] [Google Scholar]
- 22.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 23.Chen ZX, Riggs AD. Maintenance and regulation of DNA methylation patterns in mammals. Biochem Cell Biol. 2005;83:438–448. doi: 10.1139/o05-138. [DOI] [PubMed] [Google Scholar]
- 24.Turner BM. Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci. 1998;54:21–31. doi: 10.1007/s000180050122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, Zeitlinger J, Lewitter F, Gifford DK, Young RA. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 26.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu Q, Ohsako S, Ishimura R, Suzuki JS, Tohyama C. Exposure of mouse preimplantation embryos to 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) alters the methylation status of imprinted genes H19 and Igf2. Biol Reprod. 2004;70:1790–1797. doi: 10.1095/biolreprod.103.025387. [DOI] [PubMed] [Google Scholar]
- 28.Constancia M, Pickard B, Kelsey G, Reik W. Imprinting mechanisms. Genome Res. 1998;8:881–900. doi: 10.1101/gr.8.9.881. [DOI] [PubMed] [Google Scholar]
- 29.Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature. 2001;414:122–128. doi: 10.1038/35102186. [DOI] [PubMed] [Google Scholar]
- 30.Costello JF, Plass C. Methylation matters. J Med Genet. 2001;38:285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park CH, Kim HS, Lee SG, Lee CK. Methylation status of differentially methylated regions at Igf2/H19 locus in porcine gametes and preimplantation embryos. Genomics. 2009;93:179–186. doi: 10.1016/j.ygeno.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Ideraabdullah FY, Vigneau S, Bartolomei MS. Genomic imprinting mechanisms in mammals. Mutat Res. 2008;647:77–85. doi: 10.1016/j.mrfmmm.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerrero-Bosagna C, Sabat P, Valladares L. Environmental signaling and evolutionary change: can exposure of pregnant mammals to environmental estrogens lead to epigenetically induced evolutionary changes in embryos? Evol Dev. 2005;7:341–350. doi: 10.1111/j.1525-142X.2005.05033.x. [DOI] [PubMed] [Google Scholar]
- 35.Whitelaw NC, Whitelaw E. Transgenerational epigenetic inheritance in health and disease. Curr Opin Genet Dev. 2008;18:273–279. doi: 10.1016/j.gde.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Yamazawa K, Kagami M, Nagai T, Kondoh T, Onigata K, Maeyama K, Hasegawa T, Hasegawa Y, Yamazaki T, Mizuno S, Miyoshi Y, Miyagawa S, Horikawa R, Matsuoka K, Ogata T. Molecular and clinical findings and their correlations in Silver-Russell syndrome: implications for a positive role of IGF2 in growth determination and differential imprinting regulation of the IGF2-H19 domain in bodies and placentas. J Mol Med. 2008;86:1171–1181. doi: 10.1007/s00109-008-0377-4. [DOI] [PubMed] [Google Scholar]
- 37.Temple IK. Imprinting in human disease with special reference to transient neonatal diabetes and Beckwith-Wiedemann syndrome. Endocr Dev. 2007;12:113–123. doi: 10.1159/000109638. [DOI] [PubMed] [Google Scholar]
- 38.Mann MR, Bartolomei MS. Towards a molecular understanding of Prader-Willi and Angelman syndromes. Hum Mol Genet. 1999;8:1867–1873. doi: 10.1093/hmg/8.10.1867. [DOI] [PubMed] [Google Scholar]
- 39.Walter E, Mazaika PK, Reiss AL. Insights into brain development from neurogenetic syndromes: evidence from fragile X syndrome, Williams syndrome, Turner syndrome and velocardiofacial syndrome. Neuroscience. 2009;164(1):257–271. doi: 10.1016/j.neuroscience.2009.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schanen NC. Epigenetics of autism spectrum disorders. Hum Mol Genet. 2006;15(Spec No 2):R138–R150. doi: 10.1093/hmg/ddl213. [DOI] [PubMed] [Google Scholar]
- 41.Graff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur J Neurosci. 2009;30:1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- 42.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009;8:1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 43.Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9:394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cuzin F, Grandjean V, Rassoulzadegan M. Inherited variation at the epigenetic level: paramutation from the plant to the mouse. Curr Opin Genet Dev. 2008;18:193–196. doi: 10.1016/j.gde.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 45.Bertram C, Khan O, Ohri S, Phillips DI, Matthews SG, Hanson MA. Transgenerational effects of prenatal nutrient restriction on cardiovascular and hypothalamic-pituitary-adrenal function. J Physiol. 2008;586:2217–2229. doi: 10.1113/jphysiol.2007.147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci U S A. 2008;105:17046–17049. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cooney CA, Dave AA, Wolff GL. Maternal methyl supplements in mice affect epigenetic variation and DNA methylation of offspring. J Nutr. 2002;132:2393S–2400S. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 48.Cropley JE, Suter CM, Beckman KB, Martin DI. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proc Natl Acad Sci U S A. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perera F, Tang WY, Herbstman J, Tang D, Levin L, Miller R, Ho SM. Relation of DNA methylation of 5'-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS ONE. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novikova SI, He F, Bai J, Cutrufello NJ, Lidow MS, Undieh AS. Maternal cocaine administration in mice alters DNA methylation and gene expression in hippocampal neurons of neonatal and prepubertal offspring. PLoS ONE. 2008;3:e1919. doi: 10.1371/journal.pone.0001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yaoi T, Itoh K, Nakamura K, Ogi H, Fujiwara Y, Fushiki S. Genome-wide analysis of epigenomic alterations in fetal mouse forebrain after exposure to low doses of bisphenol A. Biochem Biophys Res Commun. 2008;376:563–567. doi: 10.1016/j.bbrc.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol. 2008;8:17. doi: 10.1186/1472-6793-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Andersen HR, Schmidt IM, Grandjean P, Jensen TK, Budtz-Jorgensen E, Kjaerstad MB, Baelum J, Nielsen JB, Skakkebaek NE, Main KM. Impaired reproductive development in sons of women occupationally exposed to pesticides during pregnancy. Environ Health Perspect. 2008;116:566–572. doi: 10.1289/ehp.10790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Champagne FA, Weaver IC, Diorio J, Dymov S, Szyf M, Meaney MJ. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–2915. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- 57.Oberlander TF, Weinberg J, Papsdorf M, Grunau R, Misri S, Devlin AM. Prenatal exposure to maternal depression, neonatal methylation of human glucocorticoid receptor gene (NR3C1) and infant cortisol stress responses. Epigenetics. 2008;3:97–106. doi: 10.4161/epi.3.2.6034. [DOI] [PubMed] [Google Scholar]
- 58.Holliday R. The possibility of epigenetic transmission of defects induced by teratogens. Mutat Res. 1998;422:203–205. doi: 10.1016/s0027-5107(98)00219-x. [DOI] [PubMed] [Google Scholar]
- 59.McLachlan JA. Environmental signaling: what embryos and evolution teach us about endocrine disrupting chemicals. Endocr Rev. 2001;22:319–341. doi: 10.1210/edrv.22.3.0432. [DOI] [PubMed] [Google Scholar]
- 60.Li S, Washburn KA, Moore R, Uno T, Teng C, Newbold RR, McLachlan JA, Negishi M. Developmental exposure to diethylstilbestrol elicits demethylation of estrogen-responsive lactoferrin gene in mouse uterus. Cancer Res. 1997;57:4356–4359. [PubMed] [Google Scholar]
- 61.Waterland RA, Travisano M, Tahiliani KG, Rached MT, Mirza S. Methyl donor supplementation prevents transgenerational amplification of obesity. Int J Obes (Lond) 2008;32:1373–1379. doi: 10.1038/ijo.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guerrero-Bosagna CVL. Endocrine disruptors, epigenetically induced changes, and transgenerational transmission of characters and epigenetic states. In: G A, editor. Endocrine disrupting chemicals: from basic research to clinical practice. Totowa, NJ: Humana Press Inc.; 2007. pp. 175–189. [Google Scholar]
- 63.Li S, Hursting SD, Davis BJ, McLachlan JA, Barrett JC. Environmental exposure, DNA methylation, and gene regulation: lessons from diethylstilbesterol-induced cancers. Ann N Y Acad Sci. 2003;983:161–169. doi: 10.1111/j.1749-6632.2003.tb05971.x. [DOI] [PubMed] [Google Scholar]
- 64.Lyn-Cook BD, Blann E, Payne PW, Bo J, Sheehan D, Medlock K. Methylation profile and amplification of proto-oncogenes in rat pancreas induced with phytoestrogens. Proc Soc Exp Biol Med. 1995;208:116–119. doi: 10.3181/00379727-208-43842. [DOI] [PubMed] [Google Scholar]
- 65.Day JK, Bauer AM, DesBordes C, Zhuang Y, Kim BE, Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, Huang TH, Lubahn DB. Genistein alters methylation patterns in mice. J Nutr. 2002;132:2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 66.Palanza P, Gioiosa L, vom Saal FS, Parmigiani S. Effects of developmental exposure to bisphenol A on brain and behavior in mice. Environ Res. 2008;108:150–157. doi: 10.1016/j.envres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- 67.Gore AC. Developmental programming and endocrine disruptor effects on reproductive neuroendocrine systems. Front Neuroendocrinol. 2008;29:358–374. doi: 10.1016/j.yfrne.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Crews D. Epigenetics and its implications for behavioral neuroendocrinology. Front Neuroendocrinol. 2008;29:344–357. doi: 10.1016/j.yfrne.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crews D, Gore AC, Hsu TS, Dangleben NL, Spinetta M, Schallert T, Anway MD, Skinner MK. Transgenerational epigenetic imprints on mate preference. Proc Natl Acad Sci U S A. 2007;104:5942–5946. doi: 10.1073/pnas.0610410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Skinner MK, Anway MD, Savenkova MI, Gore AC, Crews D. Transgenerational epigenetic programming of the brain transcriptome and anxiety behavior. PLoS ONE. 2008;3:e3745. doi: 10.1371/journal.pone.0003745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. Neonatal diethylstilbestrol exposure induces persistent elevation of c-fos expression and hypomethylation in its exon-4 in mouse uterus. Mol Carcinog. 2003;38:78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- 72.Newbold RR, Hanson RB, Jefferson WN, Bullock BC, Haseman J, McLachlan JA. Proliferative lesions and reproductive tract tumors in male descendants of mice exposed developmentally to diethylstilbestrol. Carcinogenesis. 2000;21:1355–1363. [PubMed] [Google Scholar]
- 73.Lenski RE, Mittler JE. The directed mutation controversy and neo-Darwinism. Science. 1993;259:188–194. doi: 10.1126/science.7678468. [DOI] [PubMed] [Google Scholar]
- 74.Sved J, Bird A. The expected equilibrium of the CpG dinucleotide in vertebrate genomes under a mutation model. Proc Natl Acad Sci U S A. 1990;87:4692–4696. doi: 10.1073/pnas.87.12.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huttley GA. Modeling the impact of DNA methylation on the evolution of BRCA1 in mammals. Mol Biol Evol. 2004;21:1760–1768. doi: 10.1093/molbev/msh187. [DOI] [PubMed] [Google Scholar]
- 76.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- 77.Allegrucci C, Thurston A, Lucas E, Young L. Epigenetics and the germline. Reproduction. 2005;129:137–149. doi: 10.1530/rep.1.00360. [DOI] [PubMed] [Google Scholar]
- 78.Durcova-Hills G, Hajkova P, Sullivan S, Barton S, Surani MA, McLaren A. Influence of sex chromosome constitution on the genomic imprinting of germ cells. Proc Natl Acad Sci U S A. 2006;103:11184–11188. doi: 10.1073/pnas.0602621103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zamudio NM, Chong S, O'Bryan MK. Epigenetic regulation in male germ cells. Reproduction. 2008;136:131–146. doi: 10.1530/REP-07-0576. [DOI] [PubMed] [Google Scholar]
- 80.Godmann M, Lambrot R, Kimmins S. The dynamic epigenetic program in male germ cells: Its role in spermatogenesis, testis cancer, and its response to the environment. Microsc Res Tech. 2009;72:603–619. doi: 10.1002/jemt.20715. [DOI] [PubMed] [Google Scholar]
- 81.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009;460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Anway MD, Rekow SS, Skinner MK. Transgenerational epigenetic programming of the embryonic testis transcriptome. Genomics. 2008;91:30–40. doi: 10.1016/j.ygeno.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Anway MD, Skinner MK. Transgenerational effects of the endocrine disruptor vinclozolin on the prostate transcriptome and adult onset disease. Prostate. 2008;68:517–529. doi: 10.1002/pros.20724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Anway MD, Memon MA, Uzumcu M, Skinner MK. Transgenerational effect of the endocrine disruptor vinclozolin on male spermatogenesis. J Androl. 2006;27:868–879. doi: 10.2164/jandrol.106.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Anway MD, Leathers C, Skinner MK. Endocrine disruptor vinclozolin induced epigenetic transgenerational adult-onset disease. Endocrinology. 2006;147:5515–5523. doi: 10.1210/en.2006-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Schneider S, Kaufmann W, Buesen R, van Ravenzwaay B. Vinclozolin--the lack of a transgenerational effect after oral maternal exposure during organogenesis. Reprod Toxicol. 2008;25:352–360. doi: 10.1016/j.reprotox.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Inawaka K, Kawabe M, Takahashi S, Doi Y, Tomigahara Y, Tarui H, Abe J, Kawamura S, Shirai T. Maternal exposure to anti-androgenic compounds, vinclozolin, flutamide and procymidone, has no effects on spermatogenesis and DNA methylation in male rats of subsequent generations. Toxicol Appl Pharmacol. 2009;237:178–187. doi: 10.1016/j.taap.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 88.Anway MD, Rekow SS, Skinner MK. Comparative anti-androgenic actions of vinclozolin and flutamide on transgenerational adult onset disease and spermatogenesis. Reprod Toxicol. 2008;26:100–106. doi: 10.1016/j.reprotox.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salian S, Doshi T, Vanage G. Impairment in protein expression profile of testicular steroid receptor coregulators in male rat offspring perinatally exposed to Bisphenol A. Life Sci. 2009;85(1–2):11–18. doi: 10.1016/j.lfs.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 90.Stouder C, Paoloni-Giacobino A. Transgenerational effects of the endocrine disruptor vinclozolin on the methylation pattern of imprinted genes in the mouse sperm. Reproduction. 2009 doi: 10.1530/REP-09-0340. In press. [DOI] [PubMed] [Google Scholar]
- 91.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 92.Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 93.Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 94.Kaati G, Bygren LO, Pembrey M, Sjostrom M. Transgenerational response to nutrition, early life circumstances and longevity. Eur J Hum Genet. 2007;15:784–790. doi: 10.1038/sj.ejhg.5201832. [DOI] [PubMed] [Google Scholar]
- 95.Arai JA, Li S, Hartley DM, Feig LA. Transgenerational rescue of a genetic defect in long-term potentiation and memory formation by juvenile enrichment. J Neurosci. 2009;29:1496–1502. doi: 10.1523/JNEUROSCI.5057-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Matthews SG, Phillips DI. Minireview: Transgenerational Inheritance of the Stress Response: A New Frontier in Stress Research. Endocrinology. 2009 doi: 10.1210/en.2009-0916. In press. [DOI] [PubMed] [Google Scholar]
- 97.Roth TL, Lubin FD, Funk AJ, Sweatt JD. Lasting epigenetic influence of early-life adversity on the BDNF gene. Biol Psychiatry. 2009;65:760–769. doi: 10.1016/j.biopsych.2008.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chao MJ, Ramagopalan SV, Herrera BM, Lincoln MR, Dyment DA, Sadovnick AD, Ebers GC. Epigenetics in multiple sclerosis susceptibility: difference in transgenerational risk localizes to the major histocompatibility complex. Hum Mol Genet. 2009;18:261–266. doi: 10.1093/hmg/ddn353. [DOI] [PubMed] [Google Scholar]
- 99.Lee J, Kanatsu-Shinohara M, Ogonuki N, Miki H, Inoue K, Morimoto T, Morimoto H, Ogura A, Shinohara T. Heritable imprinting defect caused by epigenetic abnormalities in mouse spermatogonial stem cells. Biol Reprod. 2009;80:518–527. doi: 10.1095/biolreprod.108.072330. [DOI] [PubMed] [Google Scholar]
- 100.Hanson MA, Gluckman PD. Developmental origins of health and disease: new insights. Basic Clin Pharmacol Toxicol. 2008;102:90–93. doi: 10.1111/j.1742-7843.2007.00186.x. [DOI] [PubMed] [Google Scholar]
- 101.Van Speybroeck L. From epigenesis to epigenetics: the case of C. H. Waddington. Ann N Y Acad Sci. 2002;981:61–81. [PubMed] [Google Scholar]
- 102.Waddington CH. Organisers and Genes. Cambridge: Cambridge Univ. Press; 1940. [Google Scholar]
- 103.Waddington CH. Principles of embryology. London: George Allen & Unwin Ltd; 1956. [Google Scholar]
- 104.Waddington CH. Gene assimilation of an acquired character. Evolution. 1953:118–126. [Google Scholar]
- 105.Holliday R. Mechanisms for the control of gene activity during development. Biol Rev Camb Philos Soc. 1990;65:431–471. doi: 10.1111/j.1469-185x.1990.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 106.Russo VEA, Martienssen RA, Riggs AD. Epigenetic Mechanisms of Gene Regulation. Woodbury: Cold Spring Harbor Laboratory Press; 1996. [Google Scholar]
- 107.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 108.Skinner MK. What is an epigenetic transgenerational phenotype? F3 or F2. Reprod Toxicol. 2008;25:2–6. doi: 10.1016/j.reprotox.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trasler JM. Origin and roles of genomic methylation patterns in male germ cells. Semin Cell Dev Biol. 1998;9:467–474. doi: 10.1006/scdb.1998.0225. [DOI] [PubMed] [Google Scholar]
- 110.Fry DM. Reproductive effects in birds exposed to pesticides and industrial chemicals. Environ Health Perspect. 1995;103 Suppl 7:165–171. doi: 10.1289/ehp.95103s7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Greenwald P, Barlow JJ, Nasca PC, Burnett WS. Vaginal cancer after maternal treatment with synthetic estrogens. N Engl J Med. 1971;285:390–392. doi: 10.1056/NEJM197108122850707. [DOI] [PubMed] [Google Scholar]
- 112.Herbst AL, Ulfelder H, Poskanzer DC. Adenocarcinoma of the vagina. Association of maternal stilbestrol therapy with tumor appearance in young women. N Engl J Med. 1971;284:878–881. doi: 10.1056/NEJM197104222841604. [DOI] [PubMed] [Google Scholar]
- 113.Hendry WJ, 3rd, Sheehan DM, Khan SA, May JV. Developing a laboratory animal model for perinatal endocrine disruption: the hamster chronicles. Exp Biol Med (Maywood) 2002;227:709–723. doi: 10.1177/153537020222700904. [DOI] [PubMed] [Google Scholar]
- 114.Guillette LJ, Jr, Gross TS, Masson GR, Matter JM, Percival HF, Woodward AR. Developmental abnormalities of the gonad and abnormal sex hormone concentrations in juvenile alligators from contaminated and control lakes in Florida. Environ Health Perspect. 1994;102:680–688. doi: 10.1289/ehp.94102680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Prins GS, Tang WY, Belmonte J, Ho SM. Perinatal exposure to oestradiol and bisphenol A alters the prostate epigenome and increases susceptibility to carcinogenesis. Basic Clin Pharmacol Toxicol. 2008;102:134–138. doi: 10.1111/j.1742-7843.2007.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Birnbaum LS, Fenton SE. Cancer and developmental exposure to endocrine disruptors. Environ Health Perspect. 2003;111:389–394. doi: 10.1289/ehp.5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Skinner MK, Anway MD. Seminiferous cord formation and germ-cell programming: epigenetic transgenerational actions of endocrine disruptors. Ann N Y Acad Sci. 2005;1061:18–32. doi: 10.1196/annals.1336.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brink RA. A Genetic Change Associated with the R Locus in Maize Which Is Directed and Potentially Reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xing Y, Shi S, Le L, Lee CA, Silver-Morse L, Li WX. Evidence for transgenerational transmission of epigenetic tumor susceptibility in Drosophila. PLoS Genet. 2007;3:1598–1606. doi: 10.1371/journal.pgen.0030151. [DOI] [PMC free article] [PubMed] [Google Scholar]