Abstract
Sequence-specific binding by transcription factors (TFs) interprets regulatory information encoded in the genome. Using recently published universal protein binding microarray (PBM) data on the in vitro DNA binding preferences of these proteins for all possible 8-base-pair sequences, we examined the evolutionary conservation and enrichment within putative regulatory regions of the binding sequences of a diverse library of 104 nonredundant mouse TFs spanning 22 different DNA-binding domain structural classes. We found that not only high affinity binding sites, but also numerous moderate and low affinity binding sites, are under negative selection in the mouse genome. These 8-mers occur preferentially in putative regulatory regions of the mouse genome, including CpG islands and non-exonic ultraconserved elements (UCEs). Of TFs whose PBM ‘bound’ 8-mers are enriched within sets of tissue-specific UCEs, many are expressed in the same tissue(s) as the UCE-driven gene expression. Phylogenetically conserved motif occurrences of various TFs were also enriched in the noncoding sequence surrounding numerous gene sets corresponding to Gene Ontology categories and tissue-specific gene expression clusters, suggesting involvement in transcriptional regulation of those genes. Altogether, our results indicate that many of the sequences bound by these proteins in vitro, including lower affinity DNA sequences, are likely to be functionally important in vivo. This study not only provides an initial analysis of the potential regulatory associations of 104 mouse TFs, but also presents an approach for the functional analysis of TFs from any other metazoan genome as their DNA binding preferences are determined by PBMs or other technologies.
Keywords: transcription factors, transcription factor binding sites, protein binding microarrays, conservation, DNA binding site affinities
Introduction
The interactions between transcription factors (TFs) and their DNA binding sites are an integral part of the regulatory networks within cells. Recent computational studies in Saccharomyces cerevisiae [1] and in Drosophila embryonic development have suggested that low affinity TF binding sites are important in gene regulation [2]. However, those studies utilized position weight matrix (PWM) models of TF binding sites, rather than directly measured preferences of TFs for DNA binding sequence ‘words’ (‘k-mers’); PWMs are non-ideal as they typically are based on only a relatively small set of binding site sequences, either inferred or directly measured, and in general do not capture well the full range of binding preferences of TFs [3; 4; 5; 6].
The DNA binding specificities of 104 known and predicted mouse TFs from 22 different DNA binding domain (DBD) structural classes found in metazoan TFs were determined recently [6] using the universal [7] protein binding microarray (PBM) technology [8; 9]. Those PBM data are the first high-resolution binding specificity data for the vast majority of those 104 TFs; universal PBMs provide a complete look-up table of the relative binding preference of a TF for each gapped and ungapped 8-bp sequence variant. For each of the 8-mers in the dataset for each protein, its PBM enrichment score (E-score) [7] was reported [6]. The E-score is related to the area under a receiver operating characteristic curve (AUC) and scales from +0.5 (best) to −0.5 (worst) [7]. Comparisons to Kd data [7; 10] indicated that 8-mers with higher normalized PBM signal intensities, and thus with higher E-scores, are generally bound with higher affinity [6; 7]. From the individual 8-mer data, DNA binding site motifs were derived [6] using the Seed-and-Wobble algorithm [7; 11].
In addition, in that study [6] many TFs were found to bind two distinct sets of high-scoring 8-mers. For each such TF, a secondary motif was identified that captured DNA binding preferences not represented well by the primary motif. A linear regression approach was used to learn weighted combinations of PWMs generated from several different motif finding algorithms; the binding profiles for the vast majority of the proteins were represented best by more than one PWM. PWMs are of practical utility because they are employed by numerous genome scanning tools to identify candidate regulatory elements [12].
While those PBM data provided rich, high-resolution profiles of the in vitro DNA binding specificities of 104 mouse TFs, the in vivo functional relevance of those binding site sequences was unexplored for all but 2 of the TFs [6]. Here, we analyzed those DNA binding specificity data to determine the potential utility of in vitro binding data in computational genome analyses aimed at identifying candidate in vivo regulatory associations [13]. In particular, we examined whether TF-binding 8-mers are evolutionarily conserved and whether they are enriched within regions of the genome that are likely to have cis-regulatory function. We report that:
PBM ‘bound’ 8-mers are enriched within various classes of putative regulatory regions, including CpG islands and non-exonic ultraconserved elements (UCEs) [14].
Non-exonic UCEs exhibit the greatest fold-enrichment of high affinity 8-mers for TFs of the homeodomain and BRIGHT classes. Lower affinity homeodomain and BRIGHT class 8-mers are also enriched within non-exonic UCEs.
Of TFs whose PBM ‘bound’ 8-mers are enriched within sets of tissue-specific UCEs, many are expressed in the same tissue(s) as the UCEs.
Both high and lower affinity 8-mers appear to be under negative selective pressure across mammalian genomes as compared to putatively non-binding 8-mers.
Overall, secondary motif 8-mers are conserved as strongly as primary motif 8-mers in both low and high CpG regions.
Putative target genes of most TFs are enriched for particular Gene Ontology functional categories or for tissue-specific gene expression clusters.
Taken together, our results provide evidence supporting that lower affinity TF binding sites, as determined from PBMs, serve evolutionarily conserved, in vivo regulatory functions.
Results
PBM ‘bound’ 8-mers are enriched within putative regulatory regions, including CpG islands and non-exonic UCEs
We first examined whether 8-mers bound strongly in vitro in PBMs by any of the 104 previously characterized mouse TFs [6] are enriched in putative regulatory regions of the mouse genome, including CpG islands, UTRs, non-exonic UCEs, and intergenic or intronic regions, considering ancestral repeats as a negative control. Considering 8-mers bound (E ≥ 0.40, 0.43, or 0.45) by any of the 104 TFs, the absolute overall enrichment or depletion of TF-binding 8-mer occurrences in these regions was quite minor, with enrichment of these 8-mers in noncoding regulatory regions in general (P < 10−9, Wilcoxon-Mann-Whitney test) and depletion in UTRs (P < 10−9) and ancestral repeats (P < 10−12) (Supplementary Figure S1). We note that these P-values may be inexact because the overlapping 8-mers are not necessarily independent.
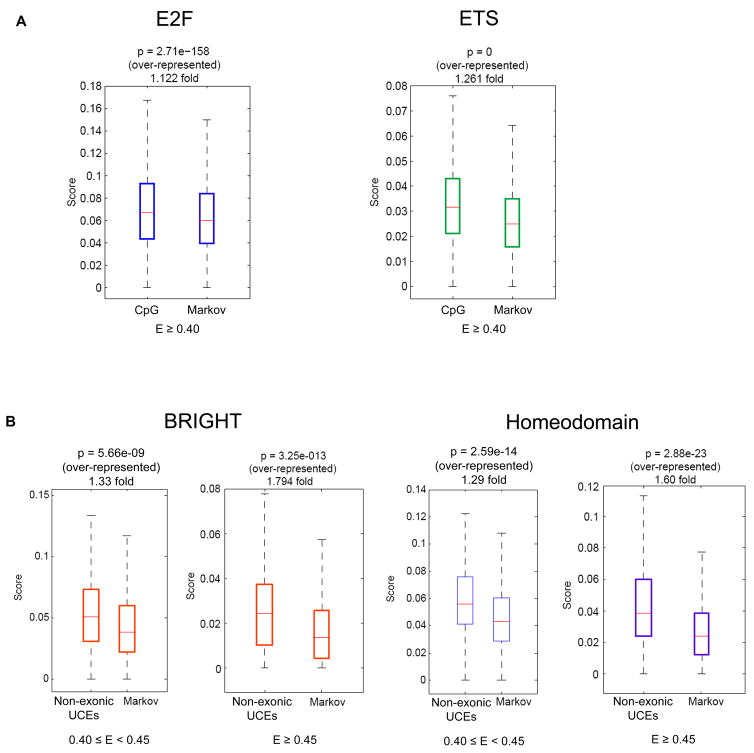
CpG islands were particularly enriched for binding sites for E2F (1.12-fold at E ≥ 0.40) and ETS (1.26-fold at E ≥ 0.40) proteins (Figure 1A, Supplementary Figure S2); see below). Chromatin immunoprecipitation analysis has shown that most in both normal and tumor cell lines E2F1, E2F4, and E2F6 in vivo binding sites are located within 2 kb of a transcription start sites [15], where CpG dinucleotides are enriched [16], and CpG methylation has been shown to differentially regulate the response of distinct E2F elements at various promoters to different E2F family members [17].
Figure 1. Enrichment of particular TFs’ 8-mers within putative regulatory regions.
(A) CpG islands are enriched for PBM ‘bound’ 8-mers for E2F and ETS proteins. Results for 8-mers bound at E ≥ 0.43 or E ≥ 0.45 are shown in Supplementary Figure S2. (B) ‘Moderate’ (0.40 ≤ E < 0.45) and ‘high’ (E ≥ 0.45) affinity 8-mers of TFs in the BRIGHT and homeodomain classes are enriched within non-exonic UCEs as compared to shuffled sequences generated to have the same dinucleotide content. In both panels, “score” and P-values are as described in Supplementary Figure 1.
Within non-exonic UCEs, we found the greatest fold-enrichment of ‘high’ affinity 8-mers (E ≥ 0.45) for TFs of the homeodomain (P < 10−22) and BRIGHT (P < 10−12) classes (Figure 1B); this enrichment is observed even when we considered only the ‘moderate’ affinity 8-mers (0.40 ≤ E < 0.45) (Figure 1B; Supplementary Table S1). Our finding for the homeodomain class supports prior observations of enrichment of matches to a generalized homeodomain motif within UCE-spanning sequences [18; 19].
Previous large-scale testing of these UCEs [20; 21] and human-pufferfish conserved noncoding elements [20] in a transgenic mouse enhancer assay showed that many of them drive tissue-specific gene expression, primarily in the embryonic nervous system [20; 21]. One of these studies also found that noncoding UCEs and extremely constrained human-rodent elements are highly enriched for neighboring genes involved in regulation of transcription, development, and nervous system development [21]. Taken together, these results suggest that UCEs may be fine-tuned to respond to the transcriptional regulatory state of cells, in particular to the levels of homeodomain and BRIGHT class TFs utilized in development and differentiation.
Evidence for functional roles of PBM ‘bound’ 8-mers enriched within highly conserved, putative neuronal regulatory regions
To begin to address the hypothesis that PBM ‘bound’ 8-mers in UCEs have an in vivo regulatory relevance, we examined whether the corresponding TFs are expressed in the same tissue(s) where expression studies of UCE-driven transgenes suggest they may be important. For example, several studies have shown that the Hox genes control motor neuron identity [22]. Consistent with anatomic locations of motor neurons, we observed enrichment (p < 1.2 × 10−5, Fisher’s Exact test) of Hoxa3 PBM ‘bound’ 8-mers (E ≥ 0.45) within UCEs and highly constrained genomic regions [20] driving expression in the cranial nerve, hindbrain, midbrain, heart, limbs, neural tube, trigeminal nerve and dorsal root ganglion, as compared to 10 independent sets of sequences generated by a 1st-order Markov model (i.e., 10 sets of sequences corresponding to the same UCEs shuffled at the dinucleotide level).
To search systematically for correlations between TF and UCE-driven gene expression, for PBM ‘bound’ 8-mers (E ≥ 0.45) enriched within UCEs and highly constrained genomic elements [20], we examined the expression patterns of the 104 TFs, as determined by recent in situ hybridization studies in the mouse embryonic brain [23; 24], within the same tissues as those in which the above constrained genomic elements drove reporter gene expression at E11.5 [20; 21]. We manually annotated the expression of TFs in the developing mouse brain in the Allen Institute’s publicly available developing mouse brain in situ hybridization images [24] and the MGI database (http://www.informatics.jax.org/), where such published annotations were not available. The correlations we found between TF and UCE-driven gene expression, for PBM ‘bound’ 8-mers (E ≥ 0.45) enriched within UCEs and highly constrained genomic elements, are described below. A full listing of PBM 8-mer enrichment results for all 104 TFs and all examined UCE expression patterns is provided in Supplementary Table S2.
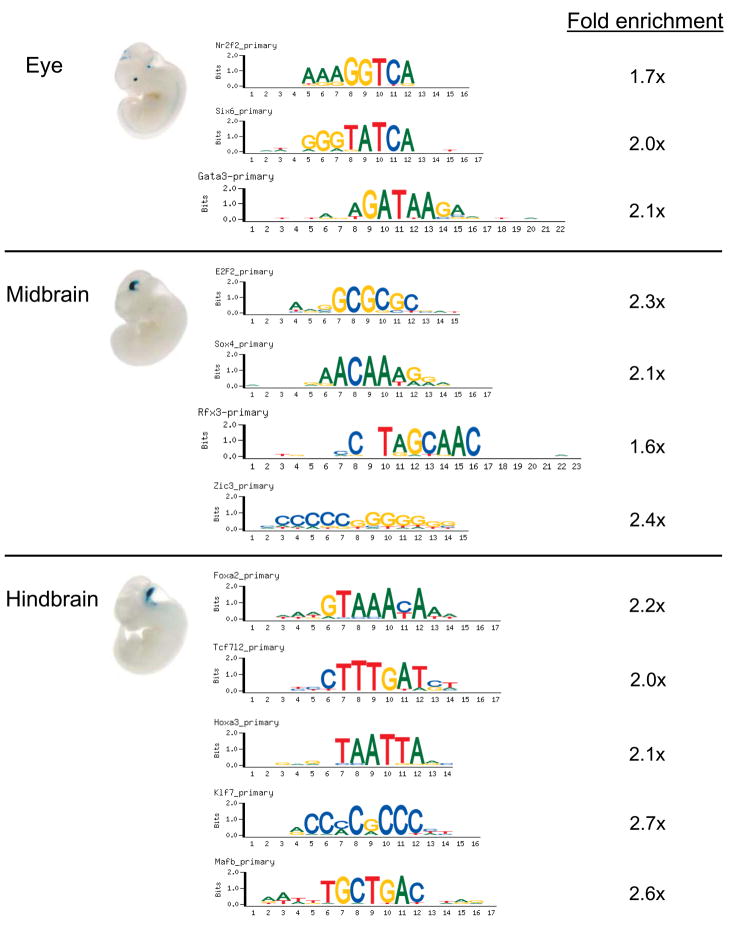
Gata3 is expressed in the lens at E10.5 [23], and its ‘bound’ 8-mers are significantly enriched (p < 0.05, Fisher’s Exact test) in enhancers driving expression in eyes at E11.5 [21] (Figure 2). Nr2f2 and Six6 are expressed in the optic vesicle at E10.5 [23], and their ‘bound’ 8-mers are likewise enriched in enhancers driving expression in eyes at E11.5 [21]. Nr2f2 is also expressed in all neural tissues at E11.5, including the neural tube, dorsal root ganglion, and optic nerve [25], and its ‘bound’ 8-mers are likewise enriched in enhancers driving expression in the neural tube, dorsal root ganglion, forebrain, midbrain, hindbrain, and eye at E11.5. Numerous TFs – E2F2, Foxj1, Foxa2, Foxk1, Gata3, Klf7, Mafb, Mtf1, Nr2f2, Rfx3, Rfx4, Sox4, Sox5, Sox14, Sox21, Tcf3, Tcf7, Tcf7l2, and Zic3 – are expressed in the midbrain at E13.5 [23], and their ‘bound’ 8-mers are enriched in enhancers driving expression in the midbrain at E11.5 (Figure 2). A number of these and other TFs – Foxa2, Foxj1, Gata3, Hoxa3, Klf7, Mafb, Mtf1, Mybl1, Rara, Rfx3, Rfx4, Sox4, Sox5, Sox7, Sox21, Tcf3, Tcf7l2, and Zic3 – are expressed in the hindbrain at E13.5 [23], and their ‘bound’ 8-mers are enriched in enhancers driving expression in the hindbrain at E11.5 [21] (Figure 2). The Allen Institute’s in situ hybridization data support these findings, with expression of Tcf7l2 [26; 27] and Foxa2 in the midbrain at E11.5, and of Foxa2, Hoxa3, and Tcf7l2 in the hindbrain at E11.5 [24]; Allen Institute in situ hybridization images were not available at E11.5 for the other TFs. Prior studies support these correlations for enhancer activity at E11.5 for a number of the other TFs, including Gata3 [28; 29], Sox1 [30], and Zic3 [31] in the midbrain and hindbrain, and for Klf7, which is ubiquitous at E10.5 [32], in the hindbrain.
Figure 2. TFs expressed in same tissues as the UCEs in which their PBM ‘bound’ 8-mers are enriched.
TF binding site sequence logos are presented for graphical convenience; binding sequence enrichment analysis was based on PBM k-mer data. Fold-enrichment of PBM ‘bound’ 8-mers (E ≥ 0.45) within UCEs is calculated as compared to 10 sets of UCE sequences shuffled at the di-nucleotide level. Because of space limitations, only a subset of the expression patterns and TFs are shown. A full listing of PBM 8-mer enrichment results for all TFs and all examined UCE expression patterns is provided in Supplementary Table S2. (UCE reporter assay whole-mount in situ images at mouse E11.5 adapted by permission from Macmillan Publishers Ltd: [Nature] [20], copyright (2006).)
While the enhancer expression data [20; 21] and the TF expression data from Gray et al. [23] were obtained at different embryonic time points (E11.5, and E10.5 or E13.5, respectively), these data suggest that these TFs may regulate gene expression through these UCEs in these embryonic tissues. There is literature evidence supporting at least some of these hypotheses, as follows. Foxa2 homozygous null zebrafish exhibit severe reduction of prospective oligodendrocytes in the midbrain and hindbrain [33], and Foxa2 regulates midbrain dopaminergic neuron development in mouse embryos [34]. Mafb is involved in both segmentation and specification of anteroposterior identity in the hindbrain [35]. AP-2 (Tcfap2a) knockout mice exhibit failure in cranial neural tube closure and defects in cranial ganglia development [36]. Thus, PBM data suggest putative links between transcriptional enhancers and specific TFs, including some with known regulatory functions as described above, that may mediate gene expression by binding within those enhancers. Analysis of over-representation of combinations of TFs’ binding site sequences in the future may suggest combinatorial, co-regulatory TF interactions and associated cis regulatory grammars within UCEs. Further experiments will be required to confirm direct binding of the TFs to their putative target regulatory regions.
Evolutionary conservation properties of PBM ‘bound’ 8-mers
8-mers that score highly in a PBM experiment represent the preferred in vitro DNA binding sites for that TF. We reasoned that if genomic occurrences of those 8-mers are functional in vivo, and if the TFs in our dataset capture a substantial fraction of the diversity in TF binding sites, then selective pressure would result in those 8-mers being evolutionarily conserved as compared to other 8-mers. We therefore examined the conservation properties of 8-mers within the non-protein-coding regions 10 kb upstream to 10 kb downstream of the transcription start sites of RefSeq genes considering sequence alignments of 12 mammalian genomes. We further subdivided the regions into those with high versus low CpG content in their sequence, after inspecting the bimodal distribution generated by the scoring function over all of the RefSeq promoters, as defined by Mikkelsen et al. [37]. We utilized SCONE scores for the substitution rates of individual nucleotides in the genome [38] to calculate substitution rates for all 8-mers within the examined genomic regions separated into low CpG and high CpG regions (Supplementary Figure S3). An 8-mer with a low average substitution rate can be considered to be more conserved within these regions than an 8-mer with a higher average substitution rate.
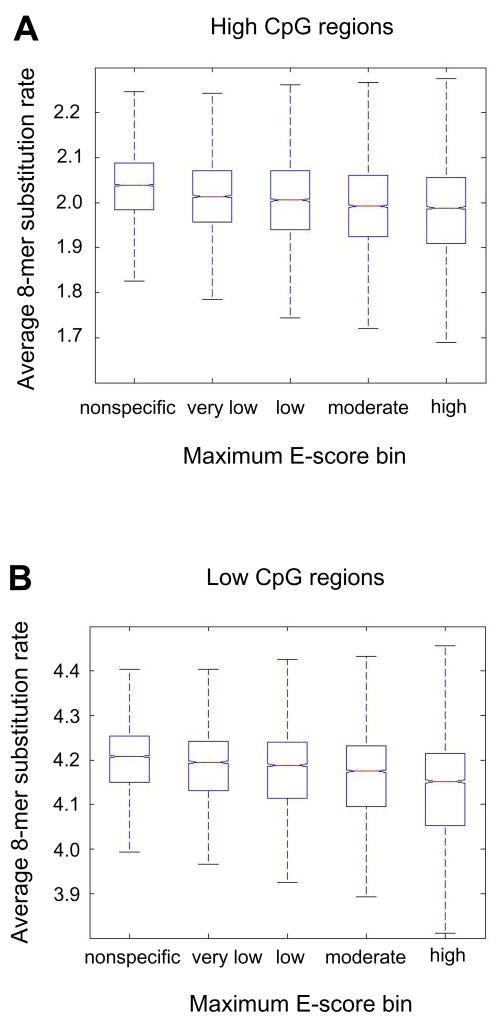
Considering as a whole the set of 8-mers with a maximum E-score above 0.45 for any of the 104 TFs, represented by 111 protein constructs, to be ‘high’ affinity binding sites for at least one TF (i.e., 8-mers that have low E-scores for some TFs but that have E ≥ 0.45 for at least one TF were concerned as ‘high’ affinity binding sites), we found that these 8-mers on average exhibited lower substitution rates than the average of all other 8-mers (P < 10−90 for low CpG regions; P < 10−71 for high CpG regions, two-sample t-test for samples with unequal variances and Satterthwaite’s approximation for the effective degrees of freedom). Moreover, we found that the average substitution rates for 8-mers within the high CpG regions are uniformly lower than the ones within the low CpG regions (Supplementary Figure S3), which suggests that despite the higher mutation rate of CpG dinucleotides [39], on average there has been stronger selective pressure to maintain TF binding sites within high CpG regions. This is consistent with recent observations that CpG islands display high levels of sequence conservation as compared to various other putative regulatory regions [38].
In addition, we observed a trend between 8-mer conservation rates and the relative binding affinities of TFs to those 8-mers. When we binned the 8-mers into ‘high’ (maximum E ≥ 0.45 over all 104 TFs; 4,787 8-mers fell into this bin), ‘moderate’ (0.40 ≤ E < 0.45; 6,678 8-mers fell into this bin), ‘low’ (0.35 ≤ E < 0.40; 6,522 8-mers fell into this bin), ‘very low’ (0.30 ≤ E < 0.35; 5,595 8-mers fell into this bin), and ‘nonspecific’ (−0.50 ≤ E < 0.30; 9,314 8-mers fell into this bin) categories, we found that the within-group mean and median 8-mer substitution rates decrease monotonically with increasing binding site affinity within high CpG regions (Figure 3A) and low CpG regions (Figure 3B). For example, the mean 8-mer substitution rate of the ‘high’ affinity category is significantly lower than that of the ‘moderate’ affinity category (P < 1 × 10−30 for low CpG regions, and P < 1 × 10−8 for high CpG regions; two-sample t-test for samples with unequal variances and Satterthwaite’s approximation for the effective degrees of freedom). Similarly, the ‘very low’ affinity category is significantly different from the ‘nonspecific’ category (P < 1 × 10−15 for low CpG regions, and P < 1 × 10−50 for high CpG regions). These results suggest that not only high affinity binding sites, but also numerous moderate and low affinity binding sites, are under negative selection in the mouse genome. We did not find any mononucleotide composition biases that could explain this observed trend. We did perform this analysis for various genomic sizes around TSS (1 kb upstream, 2 kb upstream, +/− 1 kb, +/− 2 kb) and observed similar trends after likewise subdividing the regions into low and high CpG regions. Alternatively, it is possible that some of the low affinity binding sites putatively under selection for binding by these 104 TFs are actually conserved high affinity binding sites for TFs either not included in, or not highly similar to, those in this set of 104 TFs [6].
Figure 3. The median 8-mer substitution rate decreases monotonically with increasing binding site affinity.
All bins of 8-mers – ‘high’ affinity (0.45 ≤ E ≤ 0.50), ‘moderate’ affinity (0.40 ≤ E < 0.45), ‘low’ affinity (0.35 ≤ E < 0.40), ‘very low’ affinity (0.30 ≤ E < 0.35), and ‘nonspecific’ (E < 0.30) categories – within (A) high CpG regions and (B) low CpG regions exhibited mean substitution rates significantly different from each other (P < 0.05, Tukey’s Honestly Significant Differences test). In each box plot, the central bar indicates the median, the edges of the box indicate the 25th and 75th percentiles, and whiskers extend to the most extreme data points not considered outliers.
A major finding from the recent analysis of the DNA binding preferences of these 104 TFs was the widespread existence of a secondary motif that represented 8-mers bound by the particular TF but not explained by the primary DNA binding site motif. Evidence for the in vivo usage of 8-mers belonging to the primary, and separately the secondary, motifs was provided by their enrichment towards the centers of genomic regions occupied by the corresponding TFs (Hnf4a, Bcl6b) [6]. To investigate the functional importance of secondary motif 8-mers for all 43 proteins which exhibited a significant secondary motif (seed 8-mer had E ≥ 0.45) distinctly different from the primary motif [6] (Supplementary Table S3), here we examined the evolutionary conservation of the top 100 8-mers belonging to the primary versus the secondary Seed-and-Wobble motifs. 8-mers that matched both the primary and the secondary motifs were removed from consideration.
Although the primary motif by definition corresponds to the motif generated from the highest ranking 8-mer in the PBM data, the set of 8-mers represented by the secondary motif can rank quite similarly to those belonging to the primary motif or can be of distinctly lower affinity [6]. To control for potential affinity bias in the set of 8-mers corresponding to the primary versus secondary motifs, we categorized all primary and secondary motif 8-mers into ‘high’, ‘moderate’, ‘low’, and ‘very low’ categories, as described above. Considering 8-mer data for all 43 of these proteins, we found that the secondary motif 8-mers are just as evolutionarily conserved as the primary motif 8-mers in all 4 of these affinity categories (Table 1). Thus, secondary motif 8-mers may be just as biologically important as primary motif 8-mers. In the future, site-directed mutagenesis studies of primary versus secondary motif 8-mers will need to be performed in vivo to dissect the significance and gene regulatory roles of primary versus secondary motif 8-mers.
Table 1. Secondary motif 8-mers are just as evolutionarily conserved as primary motif 8-mers.
The top 100 8-mers corresponding to the primary versus secondary motifs were binned into affinity categories according to E-scores, and assessed separately within low versus high CpG regions for evolutionary conservation. AUC non-parametric rank statistics were calculated to assess whether primary motif 8-mers rank higher than secondary motif 8-mers, with 8-mers ranked according to substitution rates. We found no significant difference in the substitution rates of 8-mers belonging to the primary versus secondary motifs at Bonferroni-corrected P < 0.05; P-values were calculated by the Wilcoxon-Mann-Whitney two-sided rank sum test, with modified Bonferroni correction for multiple hypothesis testing to account for comparisons of primary and secondary motifs across 10 bins.
| Affinity category | Low CpG regions (AUC) | High CpG regions (AUC) |
|---|---|---|
| High (E ≥ 0.45) | 0.512 | 0.511 |
| Moderate (0.40 ≤ E < 0.45) | 0.467 | 0.496 |
| Low (0.35 ≤ E < 0.40) | 0.555 | 0.596 |
| Very low (0.30 ≤ E < 0.35) | 0.537 | 0.536 |
| Nonspecific (E < 0.30) | 0.521 | 0.483 |
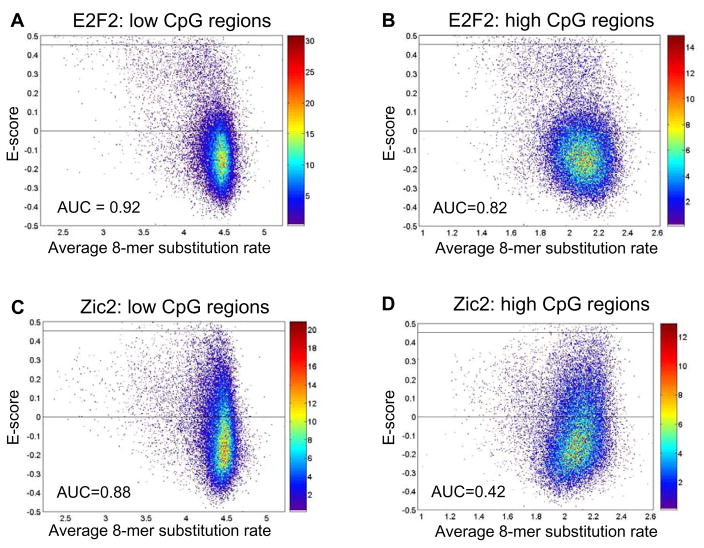
To investigate further the conservation properties of ‘high’ affinity 8-mers, we considered each TF’s ‘high’ affinity 8-mers as compared to those bound most weakly (E ≤ 0) for their conservation within low versus high CpG regions. Although we observed an overall trend that higher affinity 8-mers were more highly conserved within both low and high CpG regions (Figure 3), on an individual TF basis we found that not all TFs’ ‘high’ affinity 8-mers (E ≥ 0.45) were conserved more highly than their most weakly associated 8-mers (E ≤ 0). Within the low CpG regions, for only 51 of the 104 TFs did the ‘high’ affinity 8-mers exhibit greater conservation overall (AUC ≤ 0.5 and P <0.05, Wilcoxon-Mann-Whitney test) than their weakest 8-mers (see E2F2 example in Figure 4A). For 40 of the 104 TFs there was no significant difference between the ‘high’ affinity and the weakest 8-mers, and for the remaining 13 TFs the ‘high’ affinity 8-mers were actually less conserved than their weakest 8-mers (see Zic2 example in Figure 4C). This trend was similar within the high CpG regions (see E2F2 example in Figure 4B, and Zic2 example in Figure 4D). Only 34 TFs, including E2F2, exhibited greater conservation of high affinity 8-mers, as compared to their weakest 8-mers, within both the low and high CpG regions.
Figure 4. Evolutionary conservation properties of 8-mers of different E-scores.
Scatter plot point density is indicated by the color bar in each panel. Horizontal lines at E=0 and E=0.45 are shown in each plot for convenience. For each of 104 TFs examined, the 8-mers belonging to either the ‘high’ affinity category (0.45 ≤ E ≤ 0.50) or those bound most weakly (E < 0) were ranked according to their substitution rates. Significance was assessed by both the area under the receiver operating characteristic curve (AUC > 0.5; shown in each panel) and the Wilcoxon-Mann-Whitney test (P < 0.05); AUC > 0.5 at P < 0.05 indicates that ‘high’ affinity 8-mers are significantly more highly conserved than the most weakly bound 8-mers for a particular TF. For E2F2, the ‘high’ affinity 8-mers exhibited greater conservation than the 8-mers bound most weakly by the TF within both the (A) low and (B) high CpG regions. For Zic2, (C) within the low CpG regions the ‘high’ affinity 8-mers exhibited greater conservation overall (P < 0.05) than the 8-mers bound most weakly, while (D) within high CpG regions the ‘high’ affinity 8-mers were overall less conserved (P < 0.05) than the most weakly bound 8-mers.
Since the presence of 8-mers bound by other TFs among weak 8-mers for the TF in question has the potential to account for a portion of the unexpected high conservation of weak 8-mers, for each TF we next removed from consideration any of its weak 8-mers that had E ≥ 0.45 for any of the other 103 TFs. We then found that within the high CpG regions, for 68 TFs high affinity 8-mers were more conserved than the non-binding 8-mers on average; within the low CpG regions, for 62 TFs high affinity 8-mers were more conserved than the non-binding 8-mers on average. Within the high CpG regions, for 19 TFs the non-binding 8-mers exhibited significantly higher conservation rates on average than the high affinity binding 8-mers; within the low CpG regions, these were 10 TFs. For 25 TFs there was no significant difference (P-value > 0.05) of conservation rates between high affinity and non-binding 8-mers determined within the high CpG regions; there were 40 such TFs within the low CpG regions. 44 TFs exhibited greater conservation of high affinity 8-mers, as compared to their weakest 8-mers, within both the low and high CpG regions. Despite the diversity of TFs present in this collection of 104 TFs, it remains to be seen whether the rest of this signal is due to other, as yet uncharacterized TFs. Thus, analysis of TF binding site conservation as compared to non-binding genomic sequence may underweight the significance of conservation of some TFs’ binding sites.
Association of PBM-derived motifs with specific functional categories of genes
We next sought to further annotate these TFs and to predict their functional categories of target genes, albeit coarsely, by searching for enrichment of phylogenetically conserved motif occurrences in the noncoding sequence surrounding numerous sets of candidate target genes. We used the PhylCRM and Lever algorithms, which predict cis regulatory modules, and infer cis regulatory codes, respectively (i.e., Lever searches for TF binding site motifs enriched within PhylCRM-predicted cis regulatory modules for various input gene sets) [40]. In analysis of the PBM data for the 104 nonredundant TFs we analyzed here, it was found that multiple PWMs frequently capture the binding preferences of a TF better than does a single PWM [6]. Therefore, for Lever analysis is this study, we used the PWMs previously selected as best representing the DNA binding profiles of each of these proteins [6].
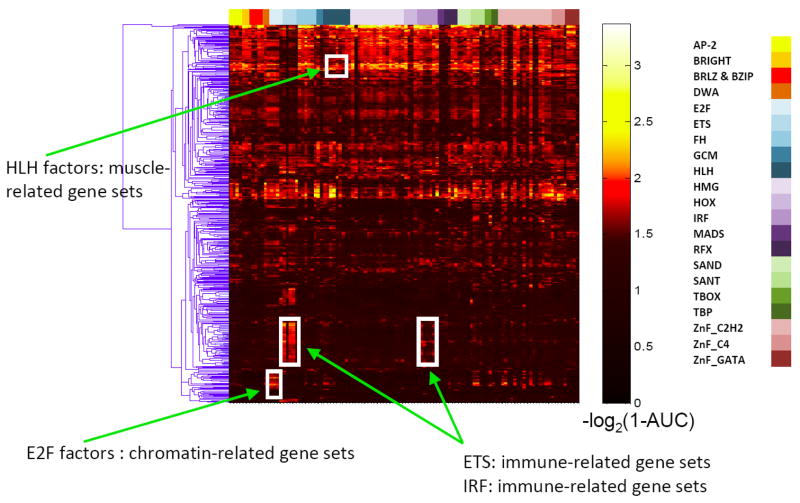
We considered a broad range of mouse tissue-specific gene expression clusters [41] and Gene Ontology (GO) annotation terms [42] in assembling gene sets, which we sorted into low versus high CpG content gene sets, yielding a total of 1,371 gene sets. We considered 20 kb of noncoding sequence surrounding the transcription start site of each gene and 12 mammalian genomes in scoring motif occurrences. Out of the 152,181 pairings of these gene sets with each particular protein, we observed a total of 285 significant pairings, involving a total of 45 gene sets and 88 TFs, at stringent significance thresholds (AUC ≥ 0.8 and Q ≤ 0.01) (Figure 5, Supplementary Table S4). As expected [40], motifs for the myogenic TFs Myf6 and SRF are enriched in various muscle-related GO categories such as ‘sarcomere’ and ‘muscle cell differentiation’ (Figure 5). The Irf proteins are known regulators of interferon response and other aspects of immune response and hematopoiesis [43]; our Lever results revealed Irf3 to be associated with the GO category ‘cytokine biosynthetic process’ (Figure 5), and Irf6 associated with a gene expression cluster appearing in tissues that contain immune cells (e.g. spleen, lymph node, bone marrow, lung, etc.) (Supplementary Figure S4). Other associations represent potentially new findings. For example, the zinc finger protein Sp4 is associated with a gene expression cluster expressed primarily in brain, and reports in the literature [44] indicate that mutants have defects in memory and that the gene itself is expressed primarily in brain. Tcf3, which is best known for its involvement in Wnt signaling [45], is associated with the GO category ‘segmentation’, consistent with a recently described role in restricting induction of the anterior-posterior axis [46]. Zfp161 (ZF5), a repressor of the human fragile X-mental retardation 1 (FMR1) gene [47], is associated with gene expression clusters primarily in the embryo (Supplementary Figure S4).
Figure 5. Lever screen of GO categories.
Heatmap color gradient indicates Lever AUC values. The columns have been restricted such that only those GO categories that exhibit significant enrichment (AUC ≥ 0.8, Q ≤ 0.01) for at least one of the TFs’ binding sites are shown. The rows (TFs) were sorted according to TF structural class, while the columns (GO categories) were clustered hierarchically according to the AUC values calculated by Lever [40].
Other independent lines of evidence also support the biological relevance of our Lever results. First, we observed a tendency for the TF itself to be co-expressed with the gene expression clusters for which its binding sequences are enriched (P < 0.001 at AUC ≥ 0.6, Q ≤ 0.05, for both low and high CpG gene sets; Supplementary Figure S5), likely because transcriptional activation of the TF gene itself is required for transcriptional activation of its targets. There is also a tendency for the GO categories that Lever associates with each TF to be among the categories in which the TF itself is annotated (P < 0.04 and P < 0.005 at AUC ≥ 0.7, Q ≤ 0.05, for low and high CpG gene sets, respectively). Some of the Lever results, however, may correspond to correct associations of motifs to target gene sets, but for related TFs not present in our set of 104 examined mouse TFs but with highly similar DNA binding site motifs; for example, the association of the Myf6 motif with various GO categories pertaining to neuron development may be due to neuronally expressed bHLH factors such as NeuroD, Mash1, Neurog1 or Neurog2 [48].
Discussion
Our analysis of the substitution rates of 8-mers across mammalian genomes suggests that not only high affinity binding sites, but also numerous moderate and low affinity binding sites, are more evolutionarily conserved than nonspecific binding sites. Despite this overall trend, the 8-mers bound most strongly by 27 of the 104 TFs we examined did not exhibit significantly greater conservation in either the low or high CpG regions. Many genomic occurrences of the binding site sequences for such TFs may not be functional, or if functional, may have exhibited significant binding site turnover. Indeed, a recent study of the occupancies of four tissue-specific TFs in human and mouse promoters found that as few as 41% of the binding sites are conserved [49]. Algorithms capable of scoring the conservation of binding site affinity despite sequence differences need to be developed; nucleotide positions with lower information content within the DNA binding site motifs of six out of seven examined S. cerevisiae TFs were found to have a higher rate of substitutions per site comparing four closely related budding yeasts than positions with higher information content [50]. In addition, the sequences not bound by one TF potentially could be genuine binding sites for another TF.
Our analysis of UCEs revealed that groups of UCEs that drive similar gene expression patterns in mouse embryos at E11.5 are enriched for particular TFs’ PBM ‘bound’ 8-mers, and that those TFs are expressed in the same tissue(s) as those in which the UCEs drive gene expression. These results can be utilized as predictions of which TFs may regulate gene expression through particular sites within those UCEs. The enrichment of both high and lower affinity binding sites in UCEs suggests that UCEs may have differential regulatory outputs specific to the nuclear levels of those TFs.
Prior studies typically have focused on high affinity TF binding sites or higher-scoring motif matches. The advent of high-resolution PBM data for these 104 mouse [6] and many other TFs [13; 51; 52] opens a wide avenue for investigation of the regulatory importance of lower affinity sites. In addition, as more TFs’ DNA binding specificities are discovered, much of the genome may appear to be a reservoir of potential regulatory sequences from which binding sites for different TFs may arise or erode.
In our Lever analysis of 1,371 gene sets, we observed a total of 285 significant pairings, involving a total of 45 gene sets and 88 TFs, at stringent significance thresholds (AUC ≥ 0.8 and Q ≤ 0.01). Numerous associations of TFs with GO categories are supported by the literature or by the TF being expressed in the same tissue(s) as the gene set(s) with which its binding sites are enriched, while many others are novel, putative regulatory associations. Evaluation of motif combinations in the future are likely to identify more specific associations of TFs to gene sets [40]. In this analysis we examined 20 kb of noncoding sequence surrounding the transcription start site of each gene for motif occurrences. Our choice of 20 kb was both practical in terms of computational time requirements and also likely somewhat conservative for many TFs. TF binding events in vivo have been observed to be enriched within ~5 kb surrounding transcription start sites, as evidenced by studies employing chromatin immunoprecipitation coupled with microarray readout (ChIP-chip) for a handful of TFs in human cell lines [53; 54]. However, given the limited number of TFs analyzed in those ChIP-chip studies [53; 54] and the abundance of distally located TF binding events in ChIP-chip [53] and numerous validated, distantly located transcriptional enhancers [55], it may be informative for the prediction of cis regulatory modules and codes to expand these Lever analyses in futures studies to consider genomic sequences located further upstream or downstream of the genes [40].
To our knowledge, the Lever analysis we presented here is the first study to employ a multiple motifs model in searching genomic sequence for TF binding site motif matches. This is important, since it was recently shown that multiple motifs often capture the DNA binding preferences of proteins more accurately than does any single motif [6]. Algorithms that can predict cis regulatory modules while scoring the evolutionary conservation of k-mers need to be developed, in particular since TF-specific differences in DNA binding, which may not be captured well by PWMs, are being revealed [51; 52]. Our Lever analysis was limited by potential divergence in TF expression or DNA binding specificity, and potential divergence in binding site composition and locations of orthologous cis regulatory modules [56]. Such k-mer analyses will be important for understanding the ‘grammar’ of how cis regulatory modules in driving particular gene expression patterns, as well as for understanding the evolution of transcriptional regulatory networks. Future studies addressing these issues and considering motif combinations and additional gene expression datasets focused on particular cell types of interest [57], should further specify the context-dependent regulatory roles of these motifs.
Methods
PBM data
We obtained PBM-derived 8-mer data and position weight matrices (PWMs) for 104 nonredundant TFs from a recent study presenting those data [6].
Scanning of genomic regions with 8-mer data
Data collection and generation of control data sets
Human-based ultraconserved conserved sequences [14] and Phastcons syntenic conserved elements [58] were downloaded from the authors’ supplementary websites (http://www.soe.ucsc.edu/%7Ejill/ultra.html and http://compgen.bscb.cornell.edu/~acs/conservation/, respectively). Human CpG island sequences were downloaded from the CpG island track in the hg17 genome build using the UCSC table browser (http://genome.ucsc.edu/cgi-bin/hgTables?command=start) [59]. Control sequences with similar dinucleotide frequencies were generated using 1st order Markov models for every sequence within the data sets, using the program GenRGenS [60]. Two sets of control sequences were generated by: (1) selecting a length-matched set of random sequences across the mm8 build of the mouse genome, and (2) using a 1st and 2nd order Markov model to control for di- and tri-nucleotide frequencies in overlapping 8-mer windows with a step size of 1 bp.
Scanning of sequences genomic regions and promoters
Each sequence in each set of genomic regions and control sequences was scanned in overlapping 8-mer windows with a step size of 1 bp using custom Perl scripts. The fraction of windows with matches to 8-mers with E-scores equal or greater than a threshold value for any TF or any TFs of a particular structural class for each sequence was plotted using MATLAB. Enrichment and statistical significance of scores in real versus control sequence sets were determined using the Wilcoxon-Mann-Whitney test implemented in MATLAB.
Each genomic sequence and the matching control sequences were scanned in overlapping 8-mer windows with a step size of 1 bp for hits against 8-mers with E ≥ 0.40 for each TF using custom Perl scripts. The distribution of the positive 8-mer hits relative to the TSS was plotted as a normalized fraction for each sequence set and each TF using MATLAB.
Evolutionary conservation properties of PBM 8-mer data
We examined the conservation properties of 8-mers of various PBM E-scores by using the SCONE algorithm [38], which calculates a substitution rate for each nucleotide in the whole genome. Using SCONE, the MultiZ 17-way sequence alignments of the vertebrate genomes performed by Genome Bioinformatics Group of UC Santa Cruz and the phylogenetic tree provided by the ENCODE multiple sequence alignment working group [61] that we trimmed down to 12 mammals, we computed the nucleotide substitution rate at every position within the non-protein coding regions from 10 kb upstream to 10 kb downstream of annotated transcription start sites for RefSeq genes with annotated 5′ UTRs. Then, for each sliding window of size 8 nt within the regions for each RefSeq gene, we summed the individual substitution rates for consecutive positions within the 8-nt window. For every possible 8-mer we calculated the average substitution rate over all genomic occurrences within these regions. An 8-mer with a low average substitution rate can be considered to be more conserved genome-wide than an 8-mer with a higher average substitution rate.
Lever analysis
The Lever algorithm was developed previously and is described in a separate paper [40]. The February 2006 mouse (Mus musculus) genome data were obtained from the Build 36 “essentially complete” assembly by NCBI and the Mouse Genome Sequencing Consortium. We incorporated the phylogenetic information from 12 mammals: mouse, rat, human, rabbit, chimp, macaque, cow, dog, armadillo, tenrec, opossum and elephant. We utilized the MultiZ 17-way alignment of: mouse (Feb 2006, mm8); rat (Nov 2004, rn4); rabbit (May 2005, oryCun1); human (Mar. 2006, hg18); chimp (Mar. 2006, panTro2); macaque (Jan 2006, rheMac2); dog (May 2005, canFam2); cow (Mar 2005, bosTau2); armadillo (May 2005, dasNov1); elephant (May 2005, loxAfr1); tenrec (Jul 2005, echTel1); opossum (Jan 2006, monDom4); chicken (Feb 2004, galGal2); frog (Aug 2005, xenTro2); zebrafish (Mar 2006, danRer4); Tetraodon (Feb 2004, tetNig1); Fugu (Aug 2002, fr1). The MultiZ alignment was performed by Genome Bioinformatics Group of UC Santa Cruz. The mouse (mm8) sequence and annotation data were downloaded from the Genome Browser FTP server (ftp://hgdownload.cse.ucsc.edu/goldenPath/mm8/). Repeats from RepeatMasker and Tandem Repeats Finder (with period of 12 or less) were masked out (RepeatMasker January 12 2005 version with RepBase libraries: RepBase Update 9.11, RM database version 20050112). The exon coding regions were also masked out by utilizing the annotation database. For each RefSeq mouse gene and the corresponding 11 alignments, we considered in our Lever analysis the sequence regions from 10 kb upstream through to 10 kb downstream of the annotated transcription start sites for RefSeq genes with annotated 5′ UTRs. Gene sets corresponding to gene expression clusters and GO annotation terms were generated as described below.
Gene expression cluster generation
Probes from Zhang et al. [41] and Su et al. [62] were mapped to MGI gene name identifiers. Expression profiles for probes mapped to the same gene identifier were averaged. Those genes not in the intersection gene set between both studies were excluded from further analysis. The final data contained expression profiles for 12,065 genes. Pair-wise Pearson correlation coefficients between the expression profiles of all genes were obtained for each dataset. Affinity Propagation [63] was used to cluster the data using the sum of the Pearson correlation coefficients between two genes as their similarity metric. This resulted in a total of 392 expression clusters.
GO annotation term gene sets
GO annotations provided by Mouse Genome Informatics (MGI) were downloaded from the Gene Ontology website on Oct. 19, 2007, version 1.686. GO annotations were up-propagated.
We used the batch system provided in MGI for gene naming conversions. Since overlaps in the sequence flanking adjacent genes could create artifactual results, we eliminated any overlaps so that our analysis considers only non-overlapping sequences. We sorted the genes into those with high versus low CpG content in their flanking sequence, after inspecting the bimodal distribution generated by the scoring function over all of the RefSeq promoters, as defined by Mikkelsen et al. [37]. We then eliminated any gene sets that had fewer than 15 or more than 300 genes. Thus, the following gene sets were used in Lever analysis: (a) 25 gene expression clusters filtered to consist solely of low CpG content genes and containing at least 15 and no more than 300 genes; (b) 166 gene expression clusters filtered to consist solely of high CpG content genes and containing at least 15 and no more than 300 genes; (c) 352 Gene Ontology (GO) Biological Process (BP) annotation term gene categories filtered to consist solely of low CpG content genes and containing at least 15 and no more than 300 genes; (d) 628 GO BP annotation term gene categories filtered to consist solely of high CpG content genes and containing at least 15 and no more than 300 genes; (e) 67 GO Cellular Component (CC) annotation term gene categories filtered to consist solely of low CpG content genes and containing at least 15 and no more than 300 genes; (f) 133 GO CC annotation term gene categories filtered to consist solely of high CpG content genes and containing at least 15 and no more than 300 genes.
Lever [40] requires an input set of PWMs to use in the genome sequence searches. The PWMs and corresponding motif match thresholds utilized in Lever analysis are provided in Supplementary Data.
Supplementary Material
Supplementary Table S1: Significant (E ≥ 0.45, ≥ 0.40, ≥ 0.35) PBM 8-mer matches to UCEs.
Supplementary Table S2: Full listing of PBM 8-mer enrichment results for all TFs and all examined UCE expression patterns.
Supplementary Table S3: List of all 43 proteins which exhibited a significant secondary motif (seed 8-mer had E ≥ 0.45) distinctly different from the primary motif.
Supplementary Table S4: Significant (AUC ≥ 0.6 and Q ≤ 0.01) pairings of TF motifs with gene sets from Lever analysis. (a) GO Biological Process high CpG gene sets. (b) GO Biological Process low CpG gene sets. (c) GO Cellular Component high CpG gene sets. (d) GO Cellular Component low CpG gene sets. (e) Tissue-specific gene expression high CpG gene sets. (f) Tissue-specific gene expression low CpG gene sets.
Supplementary Figure S1: Enrichment of TF binding site 8-mers bound at (A) E ≥ 0.40, (B) E ≥ 0.43, or (C) E ≥ 0.45, within various genomic regions.
Supplementary Figure S2: CpG islands are enriched for PBM ‘bound’ 8-mers for E2F and ETS proteins.
Supplementary Figure S3: Distribution of the mean substitution rates of all ungapped 8-mers within (A) low CpG and (B) high CpG regions.
Supplementary Figure S4: Results from Lever screen (AUC ≥ 0.6, Q ≤ 0.05) of tissue gene expression clusters. Gene expression data is from Zhang et al. [41] and Su et al. [62].
Supplementary Figure S5: Relative gene expression levels of TFs and clusters that they are associated with in the results from Lever analysis. Gene expression data is from Zhang et al. [41] and Su et al. [62].
Supplementary Data: PWMs and standard deviation parameter settings used in Lever analysis.
Acknowledgments
This project was supported by funding from the Canadian Institutes of Health Research (MOP-77721), Genome Canada through the Ontario Genomics Institute, the Ontario Research Fund, the Canadian Institute for Advanced Research to T.R.H., and by grant R01 HG003985 from NIH/NHGRI to M.L.B. We thank Saurabh Asthana, Ivan Adzhubey, and Shamil Sunyaev for helpful discussions of k-mer conservation analysis and SCONE scoring, David Beier for helpful discussion about in situ TF expression analysis, Lourdes Pena Castillo for assistance with clustering and GO analysis, and Rachel McCord and Steve Gisselbrecht for critical reading of the manuscript.
List of abbreviations
- AUC
area under receiver operating characteristic curve
- GO
Gene Ontology
- MGI
Mouse Genome Informatics
- PBM
protein binding microarray
- PWM
position weight matrix
- TF
transcription factor
- TSS
transcription start site
- UCE
ultraconserved element
- UTR
untranslated region
Footnotes
Additional Files
Supplementary Figures S1 through S5, Supplementary Tables S1 through S4, and Supplementary Data accompany this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006 doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–40. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 3.Benos PV, Bulyk ML, Stormo GD. Additivity in protein-DNA interactions: how good an approximation is it? Nucleic Acids Res. 2002;30:4442–51. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulyk ML, Johnson PL, Church GM. Nucleotides of transcription factor binding sites exert interdependent effects on the binding affinities of transcription factors. Nucleic Acids Res. 2002;30:1255–61. doi: 10.1093/nar/30.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Man TK, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–8. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, Kuznetsov H, Wang CF, Coburn D, Newburger DE, Morris Q, Hughes TR, Bulyk ML. Diversity and complexity in DNA recognition by transcription factors. Science. 2009 doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulyk ML, Huang X, Choo Y, Church GM. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc Natl Acad Sci USA. 2001;98:7158–63. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–9. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maerkl SJ, Quake SR. A systems approach to measuring the binding energy landscapes of transcription factors. Science. 2007;315:233–7. doi: 10.1126/science.1131007. [DOI] [PubMed] [Google Scholar]
- 11.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulyk ML. Computational prediction of transcription-factor binding site locations. Genome Biol. 2003;5:201. doi: 10.1186/gb-2003-5-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu C, Byers K, McCord R, Shi Z, Berger M, Newburger D, Saulrieta K, Smith Z, Shah M, Radhakrishnan M, Philippakis A, Hu Y, De Masi F, Pacek M, Rolfs A, Murthy T, Labaer J, Bulyk ML. High-resolution DNA binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–66. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–5. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 15.Xu X, Bieda M, Jin VX, Rabinovich A, Oberley MJ, Green R, Farnham PJ. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–61. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–7. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campanero MR, Armstrong MI, Flemington EK. CpG methylation as a mechanism for the regulation of E2F activity. Proc Natl Acad Sci U S A. 2000;97:6481–6. doi: 10.1073/pnas.100340697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailey PJ, Klos JM, Andersson E, Karlen M, Kallstrom M, Ponjavic J, Muhr J, Lenhard B, Sandelin A, Ericson J. A global genomic transcriptional code associated with CNS-expressed genes. Exp Cell Res. 2006;312:3108–19. doi: 10.1016/j.yexcr.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 19.Chiang CW, Derti A, Schwartz D, Chou MF, Hirschhorn JN, Wu CT. Ultraconserved elements: analyses of dosage sensitivity, motifs and boundaries. Genetics. 2008;180:2277–93. doi: 10.1534/genetics.108.096537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, Plajzer-Frick I, Akiyama J, De Val S, Afzal V, Black BL, Couronne O, Eisen MB, Visel A, Rubin EM. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 21.Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–60. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthrie S. Patterning and axon guidance of cranial motor neurons. Nat Rev Neurosci. 2007;8:859–71. doi: 10.1038/nrn2254. [DOI] [PubMed] [Google Scholar]
- 23.Gray PA, Fu H, Luo P, Zhao Q, Yu J, Ferrari A, Tenzen T, Yuk DI, Tsung EF, Cai Z, Alberta JA, Cheng LP, Liu Y, Stenman JM, Valerius MT, Billings N, Kim HA, Greenberg ME, McMahon AP, Rowitch DH, Stiles CD, Ma Q. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–7. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 24.Allen Developing Mouse Brain Atlas. Allen Institute for Brain Science; Seattle, WA: 2009. http://developingmouse.brain-map.org. [Google Scholar]
- 25.Jonk LJ, de Jonge ME, Pals CE, Wissink S, Vervaart JM, Schoorlemmer J, Kruijer W. Cloning and expression during development of three murine members of the COUP family of nuclear orphan receptors. Mech Dev. 1994;47:81–97. doi: 10.1016/0925-4773(94)90098-1. [DOI] [PubMed] [Google Scholar]
- 26.Kurokawa D, Kiyonari H, Nakayama R, Kimura-Yoshida C, Matsuo I, Aizawa S. Regulation of Otx2 expression and its functions in mouse forebrain and midbrain. Development. 2004;131:3319–31. doi: 10.1242/dev.01220. [DOI] [PubMed] [Google Scholar]
- 27.Suda Y, Hossain ZM, Kobayashi C, Hatano O, Yoshida M, Matsuo I, Aizawa S. Emx2 directs the development of diencephalon in cooperation with Otx2. Development. 2001;128:2433–50. doi: 10.1242/dev.128.13.2433. [DOI] [PubMed] [Google Scholar]
- 28.George KM, Leonard MW, Roth ME, Lieuw KH, Kioussis D, Grosveld F, Engel JD. Embryonic expression and cloning of the murine GATA-3 gene. Development. 1994;120:2673–86. doi: 10.1242/dev.120.9.2673. [DOI] [PubMed] [Google Scholar]
- 29.Nardelli J, Thiesson D, Fujiwara Y, Tsai FY, Orkin SH. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev Biol. 1999;210:305–21. doi: 10.1006/dbio.1999.9278. [DOI] [PubMed] [Google Scholar]
- 30.Aubert J, Stavridis MP, Tweedie S, O’Reilly M, Vierlinger K, Li M, Ghazal P, Pratt T, Mason JO, Roy D, Smith A. Screening for mammalian neural genes via fluorescence-activated cell sorter purification of neural precursors from Sox1-gfp knock-in mice. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11836–41. doi: 10.1073/pnas.1734197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purandare SM, Ware SM, Kwan KM, Gebbia M, Bassi MT, Deng JM, Vogel H, Behringer RR, Belmont JW, Casey B. A complex syndrome of left-right axis, central nervous system and axial skeleton defects in Zic3 mutant mice. Development. 2002;129:2293–302. doi: 10.1242/dev.129.9.2293. [DOI] [PubMed] [Google Scholar]
- 32.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–25. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 33.Norton WH, Mangoli M, Lele Z, Pogoda HM, Diamond B, Mercurio S, Russell C, Teraoka H, Stickney HL, Rauch GJ, Heisenberg CP, Houart C, Schilling TF, Frohnhoefer HG, Rastegar S, Neumann CJ, Gardiner RM, Strahle U, Geisler R, Rees M, Talbot WS, Wilson SW. Monorail/Foxa2 regulates floorplate differentiation and specification of oligodendrocytes, serotonergic raphe neurones and cranial motoneurones. Development. 2005;132:645–58. doi: 10.1242/dev.01611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferri AL, Lin W, Mavromatakis YE, Wang JC, Sasaki H, Whitsett JA, Ang SL. Foxa1 and Foxa2 regulate multiple phases of midbrain dopaminergic neuron development in a dosage-dependent manner. Development. 2007;134:2761–9. doi: 10.1242/dev.000141. [DOI] [PubMed] [Google Scholar]
- 35.Giudicelli F, Gilardi-Hebenstreit P, Mechta-Grigoriou F, Poquet C, Charnay P. Novel activities of Mafb underlie its dual role in hindbrain segmentation and regional specification. Dev Biol. 2003;253:150–62. doi: 10.1006/dbio.2002.0864. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Hagopian-Donaldson S, Serbedzija G, Elsemore J, Plehn-Dujowich D, McMahon AP, Flavell RA, Williams T. Neural tube, skeletal and body wall defects in mice lacking transcription factor AP-2. Nature. 1996;381:238–41. doi: 10.1038/381238a0. [DOI] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asthana S, Roytberg M, Stamatoyannopoulos JA, Sunyaev S. Analysis of sequence conservation at nucleotide resolution. PLoS Computational Biology. 2007;3:e254. doi: 10.1371/journal.pcbi.0030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warner J, Philippakis A, Jaeger S, He F, Lin J, Bulyk M. Systematic identification of mammalian regulatory motifs’ target genes and their functions. Nature Methods. 2008;5:347–53. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Morris QD, Chang R, Shai O, Bakowski MA, Mitsakakis N, Mohammad N, Robinson MD, Zirngibl R, Somogyi E, Laurin N, Eftekharpour E, Sat E, Grigull J, Pan Q, Peng WT, Krogan N, Greenblatt J, Fehlings M, van der Kooy D, Aubin J, Bruneau BG, Rossant J, Blencowe BJ, Frey BJ, Hughes TR. The functional landscape of mouse gene expression. J Biol. 2004;3:21. doi: 10.1186/jbiol16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1–14. doi: 10.1016/s0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 44.Zhou X, Long JM, Geyer MA, Masliah E, Kelsoe JR, Wynshaw-Boris A, Chien KR. Reduced expression of the Sp4 gene in mice causes deficits in sensorimotor gating and memory associated with hippocampal vacuolization. Mol Psychiatry. 2005;10:393–406. doi: 10.1038/sj.mp.4001621. [DOI] [PubMed] [Google Scholar]
- 45.Korswagen HC, Clevers HC. Activation and repression of wingless/Wnt target genes by the TCF/LEF-1 family of transcription factors. Cold Spring Harb Symp Quant Biol. 1999;64:141–7. doi: 10.1101/sqb.1999.64.141. [DOI] [PubMed] [Google Scholar]
- 46.Merrill BJ, Pasolli HA, Polak L, Rendl M, Garcia-Garcia MJ, Anderson KV, Fuchs E. Tcf3: a transcriptional regulator of axis induction in the early embryo. Development. 2004;131:263–74. doi: 10.1242/dev.00935. [DOI] [PubMed] [Google Scholar]
- 47.Orlov SV, Kuteykin-Teplyakov KB, Ignatovich IA, Dizhe EB, Mirgorodskaya OA, Grishin AV, Guzhova OB, Prokhortchouk EB, Guliy PV, Perevozchikov AP. Novel repressor of the human FMR1 gene - identification of p56 human (GCC)(n)-binding protein as a Kruppel-like transcription factor ZF5. FEBS J. 2007;274:4848–62. doi: 10.1111/j.1742-4658.2007.06006.x. [DOI] [PubMed] [Google Scholar]
- 48.Ge W, He F, Kim KJ, Blanchi B, Coskun V, Nguyen L, Wu X, Zhao J, Heng JI, Martinowich K, Tao J, Wu H, Castro D, Sobeih MM, Corfas G, Gleeson JG, Greenberg ME, Guillemot F, Sun YE. Coupling of cell migration with neurogenesis by proneural bHLH factors. Proc Natl Acad Sci U S A. 2006;103:1319–24. doi: 10.1073/pnas.0510419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Odom DT, Dowell RD, Jacobsen ES, Gordon W, Danford TW, MacIsaac KD, Rolfe PA, Conboy CM, Gifford DK, Fraenkel E. Tissue-specific transcriptional regulation has diverged significantly between human and mouse. Nat Genet. 2007;39:730–2. doi: 10.1038/ng2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moses AM, Chiang DY, Kellis M, Lander ES, Eisen MB. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol Biol. 2003;3:19. doi: 10.1186/1471-2148-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, Khalid F, Zhang W, Newburger D, Jaeger SA, Morris QD, Bulyk ML, Hughes TR. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–76. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grove CA, De Masi F, Barrasa I, Newburger DE, Alkema MJ, Bulyk ML, Walhout AJM. A multi-parameter network reveals extensive divergence between C. elegans bHLH transcription factors. Cell. doi: 10.1016/j.cell.2009.04.058. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, Kampa D, Piccolboni A, Sementchenko V, Cheng J, Williams AJ, Wheeler R, Wong B, Drenkow J, Yamanaka M, Patel S, Brubaker S, Tammana H, Helt G, Struhl K, Gingeras TR. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- 54.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.West AG, Fraser P. Remote control of gene transcription. Hum Mol Genet. 2005;14(Spec1):R101–11. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 56.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–7. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 57.Philippakis AA, Busser BW, Gisselbrecht SS, He FS, Estrada B, Michelson AM, MLB Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Computational Biology. 2006;2 doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–50. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karolchik E, Baertsch R, Diekhans M, Furey T, Hinrichs A, YTYL, Roskin K, Schwartz M, Sugnet C, Thomas D, Weber R, Haussler D, Kent W, Cruz UoCS. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ponty Y, Termier M, Denise A. GenRGenS: software for generating random genomic sequences and structures. Bioinformatics. 2006;22:1534–5. doi: 10.1093/bioinformatics/btl113. [DOI] [PubMed] [Google Scholar]
- 61.Margulies EH, Cooper GM, Asimenos G, Thomas DJ, Dewey CN, Siepel A, Birney E, Keefe D, Schwartz AS, Hou M, Taylor J, Nikolaev S, Montoya-Burgos JI, Loytynoja A, Whelan S, Pardi F, Massingham T, Brown JB, Bickel P, Holmes I, Mullikin JC, Ureta-Vidal A, Paten B, Stone EA, Rosenbloom KR, Kent WJ, Bouffard GG, Guan X, Hansen NF, Idol JR, Maduro VV, Maskeri B, McDowell JC, Park M, Thomas PJ, Young AC, Blakesley RW, Muzny DM, Sodergren E, Wheeler DA, Worley KC, Jiang H, Weinstock GM, Gibbs RA, Graves T, Fulton R, Mardis ER, Wilson RK, Clamp M, Cuff J, Gnerre S, Jaffe DB, Chang JL, Lindblad-Toh K, Lander ES, Hinrichs A, Trumbower H, Clawson H, Zweig A, Kuhn RM, Barber G, Harte R, Karolchik D, Field MA, Moore RA, Matthewson CA, Schein JE, Marra MA, Antonarakis SE, Batzoglou S, Goldman N, Hardison R, Haussler D, Miller W, Pachter L, Green ED, Sidow A. Analyses of deep mammalian sequence alignments and constraint predictions for 1% of the human genome. Genome Res. 2007;17:760–74. doi: 10.1101/gr.6034307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, Cooke MP, Walker JR, Hogenesch JB. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci USA. 2004;101:6062–7. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Frey BJ, Dueck D. Clustering by passing messages between data points. Science. 2007;315:972–6. doi: 10.1126/science.1136800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Significant (E ≥ 0.45, ≥ 0.40, ≥ 0.35) PBM 8-mer matches to UCEs.
Supplementary Table S2: Full listing of PBM 8-mer enrichment results for all TFs and all examined UCE expression patterns.
Supplementary Table S3: List of all 43 proteins which exhibited a significant secondary motif (seed 8-mer had E ≥ 0.45) distinctly different from the primary motif.
Supplementary Table S4: Significant (AUC ≥ 0.6 and Q ≤ 0.01) pairings of TF motifs with gene sets from Lever analysis. (a) GO Biological Process high CpG gene sets. (b) GO Biological Process low CpG gene sets. (c) GO Cellular Component high CpG gene sets. (d) GO Cellular Component low CpG gene sets. (e) Tissue-specific gene expression high CpG gene sets. (f) Tissue-specific gene expression low CpG gene sets.
Supplementary Figure S1: Enrichment of TF binding site 8-mers bound at (A) E ≥ 0.40, (B) E ≥ 0.43, or (C) E ≥ 0.45, within various genomic regions.
Supplementary Figure S2: CpG islands are enriched for PBM ‘bound’ 8-mers for E2F and ETS proteins.
Supplementary Figure S3: Distribution of the mean substitution rates of all ungapped 8-mers within (A) low CpG and (B) high CpG regions.
Supplementary Figure S4: Results from Lever screen (AUC ≥ 0.6, Q ≤ 0.05) of tissue gene expression clusters. Gene expression data is from Zhang et al. [41] and Su et al. [62].
Supplementary Figure S5: Relative gene expression levels of TFs and clusters that they are associated with in the results from Lever analysis. Gene expression data is from Zhang et al. [41] and Su et al. [62].
Supplementary Data: PWMs and standard deviation parameter settings used in Lever analysis.