Abstract
Rationale
The multi-ligand Receptor for AGE (RAGE) contributes to atherosclerosis in apolipoprotein (ApoE) null mice.
Objective
To delineate the specific mechanisms by which RAGE accelerated atherosclerosis, we performed Affymetrix gene expression arrays on aortas of non-diabetic and diabetic ApoE null mice expressing RAGE or devoid of RAGE at nine weeks of age, as this reflected a time point at which frank atherosclerotic lesions were not yet present, but that we would be able to identify the genes likely involved in diabetes- and RAGE-dependent atherogenesis.
Methods and Results
We report that there is very little overlap of the genes which are differentially expressed both in the onset of diabetes in ApoE null mice, and in the effect of RAGE deletion in diabetic ApoE null mice. Pathway-Express analysis revealed that the Transforming Growth Factor-β pathway (Tgf-β) and focal adhesion pathways might be expected to play a significant role in both the mechanism by which diabetes facilitates the formation of atherosclerotic plaques in ApoE null mice, and the mechanism by which deletion of RAGE ameliorates this effect. Quantitative polymerase chain reaction studies, Western blotting and confocal microscopy in aortic tissue and in primary cultures of murine aortic smooth muscle cells supported these findings.
Conclusions
Taken together, our work suggests that RAGE-dependent acceleration of atherosclerosis in ApoE null mice is dependent, at least in part, on the action of the ROCK1 branch of the Tgf-β pathway.
Keywords: AGE, Atherosclerosis, RAGE
Introduction
The multi-ligand Receptor for AGE (RAGE) contributes to atherosclerosis in ApoE null mice in both the non-diabetic and diabetic states. Previous studies using soluble RAGE or homozygous RAGE null mice showed that blockade or deletion of RAGE resulted in reduction in atherosclerotic lesion area and complexity compared to control animals (1–6). In parallel, significant down-regulation of inflammatory mediators and matrix metalloproteinases was evident in ApoE null mice aortas devoid of RAGE compared to those of RAGE-expressing ApoE null mice.
Although these findings suggested that RAGE modulated inflammatory gene expression in ApoE null mouse aorta, they did not reveal the broader pathways by which RAGE contributed to atherosclerosis. We performed Affymetrix gene expression arrays on aortas of non-diabetic and diabetic ApoE null mice expressing RAGE or devoid of RAGE. Aortas were harvested in the very early stages of atherogenesis, as our goal was not to identify differential genes and products expression in the atherosclerotic lesion, but to identify RAGE-dependent regulatory mechanisms in the aorta prior to the development of frank plaques.
Materials and Methods
(Additional methods are in the supplement to this report)
Animal Studies
Male ApoE null mice in the C57BL/6 background were purchased from Jackson Laboratories (Bar Harbor ME). Homozygous RAGE null mice were backcrossed >12 generations into C57BL/6 prior to crossing with ApoE null mice to generate ApoE null/RAGE null breeding pairs. Mice were maintained on a 12-hour light-dark cycle in a pathogen-free environment with free access to normal rodent chow and water.
Quantification of atherosclerotic lesion area
The frozen sections from aortic roots were fixed in 10% buffered formalin. Six 6-µm sections were collected at 80-µm intervals starting at a 100-µm distance from the appearance of the aortic valves. The sections were stained with Oil Red O and counterstained with hematoxylin. Atherosclerotic lesion areas were quantified using a Zeiss microscope and image analysis system (AxioVision 4.5). Four serial sections each were placed on 6 slides (total 24 sections), and mean lesion areas were calculated by determining the mean lesion area of one section/slide for a total of 6 sections examined. The investigator was blinded to the experimental conditions. The statistical significance of changes in atherosclerotic lesion area between diabetic ApoE null and ApoE /RAGE null mice was determined using the 2 sample t-test.
RNA Isolation and GeneChip Analysis
High-quality RNA samples were extracted from four groups mice at age 9 weeks: diabetic ApoE null (n=4), non-diabetic ApoE null (n=4), diabetic ApoE null/RAGE null (n=4) and non-diabetic ApoE null/RAGE null aortas (n=3). We employed RNA from 3 mice in the last group secondary to failure to generate cRNA from one of the mice. Total aortic RNA was isolated by using TRIzol (Invitrogen, Carlsbad CA) and RNeasy MinElute Cleanup (QIAGEN Inc., Valencia CA) including a DNase step. Total RNA concentration and quality were assessed on a 2100 Bioanalyzer system (Agilent Technologies, Parsippany NJ). All of the samples displayed an RNA integrity score >8, and there was no indication of RNA degradation or contamination with DNA. To prepare for expression analyses, cDNA was in vitro transcribed into biotin-labeled antisense cRNA using an Affymetrix kit according to the standard kit protocol. 1 µg of RNA from each sample was hybridized to Affymetrix Mouse Genome 430 2.0 GeneChips (Gene Expression Omnibus Platform Accession number GPL1261). Arrays were scanned with GeneChip Scanner 3000-7G with GCOS software. Scanning was performed according to the protocol described in the Affymetrix GeneChip® Expression Analysis Technical Manual, November 2004 Edition.
Real-Time RT-PCR validation
The differential expression of especially interesting genes was validated using RT-PCR. Total aortic RNA (0.5 µg) was reverse transcribed with SuperScript II (Invitrogen). After dilution of the cDNA to 50 µl, 1.5 µl of cDNA was amplified by real-time PCR on a sequence-detection system (Prism 7900HTl; ABI, Foster City CA). ABI Assay-on-Demand kits containing primers and probes for mouse transforming growth factor-β2 (mTgfb2) (Mm01321738_m1), mouse thrombospondin-1 (mThbs1) (Mm01335418_m1), and mouse rho-associated protein kinase 1 (mROCK1) (Mm01225244_g1) were used. 18s rRNA was used as an endogenous reference to correct for differences in the amount of RNA.
Western blot analysis
Total lysate from mouse aorta was immunoblotted and probed with antibodies to Thbs1, Tgf-β2 and ROCK1.
Immunohistochemistry
Acetone-fixed cryostat aortic sections were subjected to confocal microscopy for detection and merged images of RAGE, Thbs1, Tgf-β2, and ROCK1 in endothelium and smooth muscle layers using specific antibodies and Bio-Rad Radiance 2000 Confocal System and the Lasersharp 2000 software (Bio-Rad).
ROCK1 activity assays and primary smooth muscle cell studies
Smooth muscle cells were retrieved from the aortas of wild-type and RAGE deficient mice and subjected to ROCK1 activity assays (7–8) and assessment of proliferation and migration as described in Supplementary methods.
Results
We previously established that deletion of RAGE in non-diabetic ApoE null mice reduced atherosclerosis at age 14 weeks (5). To test these concepts in diabetes, we performed studies in RAGE-expressing or ApoE null/RAGE null mice rendered diabetic at age 6 weeks. At 14 weeks of age, mean atherosclerotic lesion area at the aortic root in diabetic ApoE null mice was ≈2.8-fold higher in RAGE-expressing vs. RAGE-deficient ApoE null animals (1.59±0.23 vs. 0.57±0.03 x105 µm2, respectively; p<0.003) (Online Figure IA). Other studies showed that diabetes accelerates atherosclerosis in ApoE null mice after 6, 14 or 20 weeks of hyperglycemia (1, 3, 6).
We examined factors that might account for the beneficial effects of RAGE deletion. Diabetic mice displayed a significantly higher plasma glucose level than non-diabetic mice, and importantly, there was no statistically significant dependence of the glucose concentration of either diabetic or non-diabetic mice on RAGE expression (Online Table I). The plasma cholesterol concentration and body weights of the mice are given in Online Tables II and III and reveal no statistically significant dependence of cholesterol concentration or body weight on either genotype or disease state.
Furthermore, we characterized the cellular lesion content by determining the percent (%) macrophages/lesion area, the % SMC/lesion area, and the % T cells/lesion area. At age 24 weeks (18 weeks of hyperglycemia), an age at which significant lesions formed in RAGE-expressing ApoE null mice (see Online Figure IB), both diabetic and non-diabetic ApoE null/RAGE null mice displayed significantly lower % macrophages/lesion area and % SMCs/lesion area compared to their RAGE-expressing ApoE null cohorts (Online Figure IB). At 24 weeks of age, the % total T cells/lesion area was significantly lower in diabetic ApoE null/RAGE null mice vs. diabetic ApoE null mice (Online Figure IB). Furthermore, non-diabetic ApoE null/RAGE null mice displayed approximately 7% collagen/lesion area, whereas in non-diabetic ApoE null mice lesions, scant collagen was detected (Online Figure IB). In the diabetic state, a nearly two-fold higher % collagen content in ApoE null/RAGE null mice lesions vs. ApoE null mice was observed (Online Figure IB).
Therefore, our data indicate that RAGE contributed importantly to atherosclerosis in ApoE null mice in a manner independent of glucose, cholesterol or body weight. We sought to identify the specific mechanisms by which RAGE contributed to early atherogenesis in ApoE null mice and retrieved entire aortas from non-diabetic and diabetic ApoE null mice at age 9 weeks, a time point at which the mice had not yet developed gross atherosclerotic plaques. Hence, our analyses would not detect genes prevalent in atherosclerotic lesions, but in genes over- or under-represented in early atherogenesis in the aorta as dependent on the state of glycemia and the state of RAGE expression. RNA was prepared from individual aorta samples and subjected to Affymetrix gene arrays. Four comparisons of genome-wide differential expression between conditions were made. Each condition was defined by both its genotype and presence or absence of diabetes. The comparisons were as follows: 1. diabetic ApoE null relative to non-diabetic ApoE null; 2. non-diabetic ApoE null/RAGE null relative to non-diabetic ApoE null; 3. diabetic ApoE null/RAGE null relative to non-diabetic ApoE null/RAGE null; and 4. diabetic ApoE null/RAGE null relative to diabetic ApoE null aorta.
The number of unique genes with the Bayesian log-odds factor B>0 (indicating that the odds of differential expression is greater than 1) are reported. Only genes with Genbank symbols were counted, and genes with more than one probeset were only counted once. Using these parameters, we report that the onset of diabetes affects transcription in ApoE null mice (53 genes, comparison 1) more than in ApoE null/RAGE null mice (3 genes, comparison 3), and that deletion of the RAGE gene in ApoE null mice affects transcription much more if the mice are diabetic (216 genes, comparison 4) than if they are non-diabetic (0 genes, comparison 2). Finally, more genes are affected by deletion of RAGE in diabetic ApoE null mice (216 genes, comparison 4) than by onset of diabetes in ApoE null mice (53 genes, comparison 1). Online Tables IV and V show the log fold changes (log2FC) and B values for all genes with B>0 for comparison 1 and comparison 4 respectively, the two comparisons with a non-negligible number of differentially expressed genes.
We performed a Pathway-Express analysis on the gene lists in Online Tables IV and V to determine the pathways that were most associated with the onset of diabetes in ApoE null mice and the effect of RAGE gene deletion in diabetic ApoE null mice. Statistically significant pathways (with a gamma p-value corrected for false discoveries ) are listed in Online Tables VI and VII. Tgf-β2 and focal adhesion pathways are common to both lists, suggesting that these pathways play a significant role in both the mechanism by which diabetes facilitates the formation of atherosclerotic plaques in ApoE null mice, and the mechanism by which deletion of RAGE ameliorates this effect. Thus, we focused on the Tgf-β pathway, because of the established role for this pathway in atherogenesis (9–15).
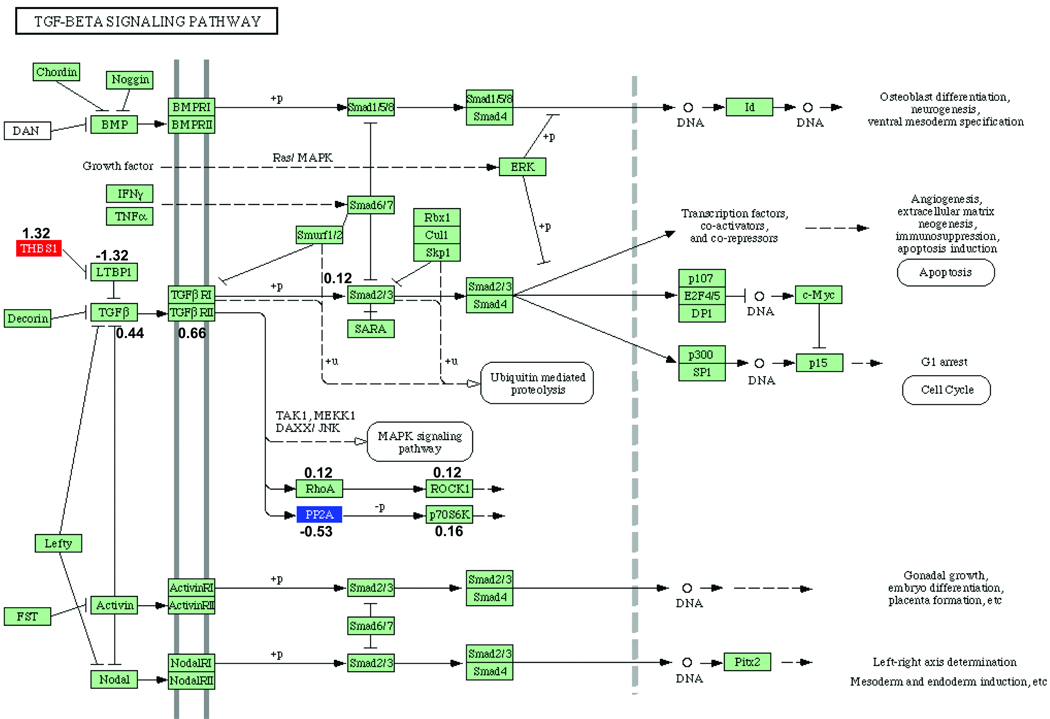
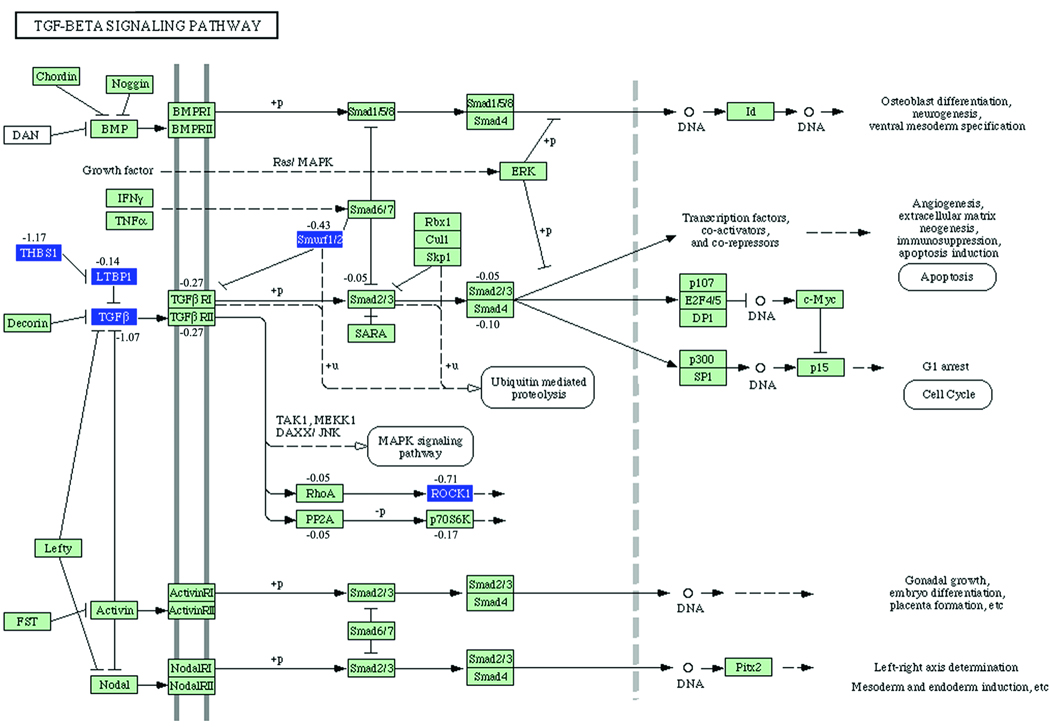
The Tgf- β pathway, with the genes that are differentially expressed indicated for the two comparisons under consideration, are given in Figures 1 and 2. The genes that are differentially expressed in each comparison are given in Online Tables VIII and IX. The genes whose perturbation factors (16) are changed in each comparison are given in Online Tables X and XI. Genes without a statistically significant change may still have non-zero perturbation factor. Perturbation factors are defined briefly in the Supplementary Methods.
Figure 1. Tgf-β KEGG Pathway analysis: effect of diabetes in ApoE null mice.
Gene symbols that are colored reflect genes that are statistically significantly differentially expressed in ApoE null mice with diabetes relative to non-diabetic ApoE null mice. Up-regulated genes are shown in red, and down-regulated genes are shown in blue. Numbers indicate perturbation factors (which may be different in magnitude and even in sign than fold-changes). KEGG Pathways often represent several different and related proteins by a single protein [for example, Tgf-β1, Tgf-β2, and Tgf-β3 are all represented as Tgf-β]). In such a case, the perturbation factor for the product of the gene with non-zero fold change is given.
Figure 2. Tgf-β KEGG Pathway analysis: effect of deletion of RAGE in diabetic ApoE null mice.
Gene symbols that are colored reflect genes that are statistically significantly differentially expressed in diabetic ApoE null/RAGE null mice relative to diabetic ApoE null mice. Up-regulated genes are shown in red, and down-regulated genes are shown in blue. Numbers indicate perturbation factors as in Figure 1.
Online Table VIII shows that expression of Thbs1 mRNA is increased in diabetic ApoE null mice compared to non-diabetic ApoE null mice (comparison 1). Online Table IX shows that expression of Thbs1 mRNA is lower in diabetic ApoE null/RAGE null mice relative to diabetic ApoE null mice (comparison 4). Analysis of Figures 1–2 reveals that Latent transforming growth factor beta binding protein 1 (Ltbp1) is an inhibitor of Tgf-β2 (17–19). Since Thbs1 inhibits the suppressive effect of Ltbp1 on activation of Tgf- β2, our results suggest that in diabetic ApoE null mice, the effect of increased Thbs1 mRNA expression is to activate Tgf-β2 protein. Similarly, Figure 2 suggests that the reduction of Thbs1 expression in diabetic ApoE null/RAGE null mice relative to non-diabetic ApoE null mice deactivates Tgf-β2 protein. Figure 2 and Online Table IX list other genes in the Tgf-β pathway whose expression is reduced in comparison 4. Further, in addition to Thbs1 and Tgf-β2, ROCK1 is also linked to atherogenesis (20–22).
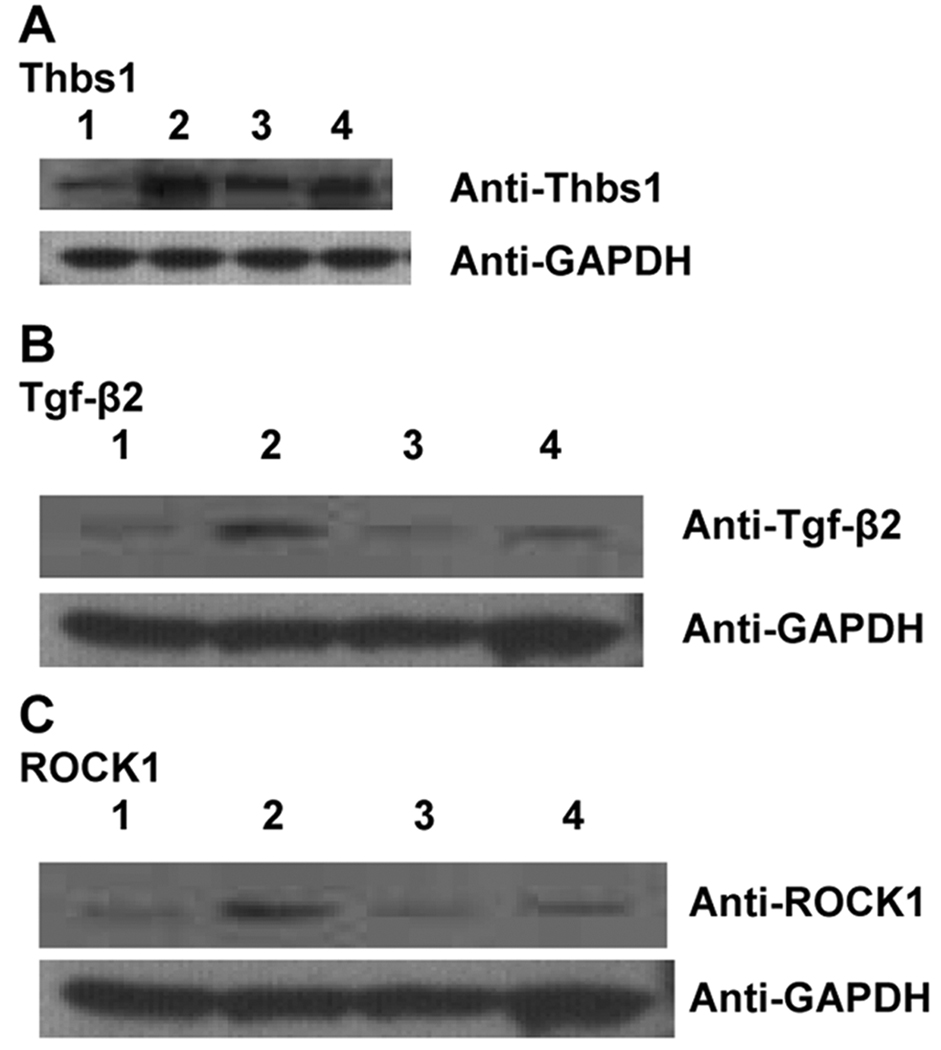
We validated the microarray results for Thbs1, Tgf-β2, and ROCK1 by real-time quantitative PCR followed by Western blotting (Table 1, Figure 3). These data reveal that diabetes increases protein levels of Thbs1, Tgf-β2 and ROCK1 in ApoE null aorta, and that particularly in the diabetic state; deletion of RAGE suppresses diabetes-linked up-regulation of Thbs1, Tgf-β2 and ROCK1 protein in ApoE null aorta.
Table 1.
Change in mRNA Expression by both Microarray and PCR and Protein Expression (Western Blotting) of Key Genes in the ROCK1 Branch of the Tgf-β Pathway
| comparison # | ||||||||||||||
| 1 | Diabetic ApoE null relative to Non-diabetic ApoE null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | 1.32 | 2.50 | 0.22 | 1.50 | Y | 12.34(11.92,12.75) | 11.07(10.89,11.25) | 1.27(0.88,1.66) | 2.41(1.84,3.17) | 8.E-04 | 0.29(0.25,0.33) | 2.0(1.8,2.1) | 7.E-04 | |
| Tgf-Beta2 | 0.45 | 1.37 | 0.40 | −3.19 | N | 11.50(10.18,12.82) | 11.02(9.82,12.22) | 0.48(−0.89,1.86) | 1.39(0.54,3.62) | 0.42 | 0.12(0.03,0.21) | 1.3(1.1,1.6) | 0.03 | |
| ROCK1 | 0.32 | 1.25 | 0.44 | −3.55 | N | 11.52(10.73,12.31) | 11.03(9.96,12.11) | 0.49(−1.02,1.53) | 1.40(0.49,2.90) | 0.29 | 0.42(0.66,1.08) | 2.78(4.52,11.97) | 9.E-04 | |
| 2 | Non-diabetic ApoE null / RAGE null relative to non-diabetic ApoE null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | 0.32 | 1.25 | 1.00 | −4.55 | N | 12.34(11.92,12.75) | 11.69(9.26,14.13) | 0.65(−1.64,2.92) | 1.56(1.19, 7.57) | 0.37 | 0.007(−0.122,0.135) | 1.02(0.76,1.36) | 0.87 | |
| Tgf-Beta2 | −0.25 | 0.84 | 1.00 | −4.50 | N | 11.50(10.18,12.82) | 11.57(10.51,12.62) | −0.06(−1.34,1.20) | 0.95(0.40,2.30) | 0.90 | −0.01(−0.02,0.00) | 0.97(0.95,0.00) | 0.06 | |
| ROCK1 | 0.24 | 1.18 | 1.00 | −4.66 | N | 11.52(10.73,12.31) | 11.27(10.57,11.99) | 0.24(−0.53,1.01) | 1.18(0.69,2.02) | 0.46 | 0.024(0.011,0.038) | 1.06(1.02,1.09) | 0.02 | |
| 3 | Diabetic ApoE null / RAGE null relative to non-diabetic ApoE null / RAGE null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | 0.29 | 1.22 | 0.85 | −4.91 | N | 11.69( 9.26,14.13) | 12.08(11.94,12.22) | −0.38(−2.80,2.02) | 0.77(0.14,4.06) | 0.56 | −0.06(−0.18,0.06) | 0.87(0.66, 1.51) | 0.22 | |
| Tgf-Beta2 | −0.40 | 0.76 | 0.69 | −3.81 | N | 11.57(10.51,12.62) | 12.43(11.83,13.02) | −0.86(−1.71,−0.01) | 0.55(0.31,1.00) | 0.05 | −0.002(0.020,.017) | 1.00(0.95,1.040) | 0.79 | |
| ROCK1 | −1.57 | 0.34 | 0.67 | −2.58 | N | 11.27(10.57,11.99) | 12.54(11.48,13.60) | −1.55(−2.27,−0.25) | 0.34(0.21,0.84) | 2.E-04 | −0.01(−0.02,0.00) | 0.98(0.96,1.01) | 0.11 | |
| 4 | Diabetic ApoE null / RAGE null relative to diabetic ApoE null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | −1.17 | 0.44 | 0.06 | 0.84 | Y | 11.07(10.89,11.25) | 12.08(11.94,12.22) | −1.02(−1.19,−0.84) | 0.49(.56,.81) | 2.E-06 | −0.48(−0.66,−0.29) | 0.33(0.22,0.51) | 6.E-03 | |
| Tgf-Beta2 | −1.09 | 0.47 | 0.07 | 0.36 | Y | 11.02(9.82,12.22) | 12.43(11.83,13.02) | −1.41(−2.54,−0.28) | 0.36(0.17,0.82) | 0.02 | −0.14(−0.23,−0.05) | 0.72(0.59,0.89) | 0.02 | |
| ROCK1 | −0.66 | 0.63 | 0.06 | 0.60 | Y | 11.03(9.96,12.11) | 12.54(11.48,13.60) | −1.50(−2.67, −0.34) | 0.35(0.16,0.79) | 0.02 | −0.27(−0.31,−0.22,) | 0.54(0.49,0.60) | 0.001 | |
| 5 | Diabetic ApoE null / RAGE null relative to non-diabetic Apo E null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | 0.40 | 1.32 | 0.67 | −4.75 | N | 12.34(11.92,12.75) | 12.08(11.94,12.22) | 0.25(−0.14,0.65) | 1.19(0.91,1.57) | 0.14 | −0.05(−0.12,0.01) | 0.88(0.76,1.02) | 0.08 | |
| Tgf-Beta2 | −0.56 | 0.68 | 0.45 | −2.22 | N | 11.50(10.18,12.82) | 12.43(11.83,13.02) | −0.93(−2.17,0.22) | 0.53(0.22,1.25) | 0.11 | −0.01(−0.03, 0.00) | 0.97(0.93,1.01) | 0.11 | |
| ROCK1 | −1.14 | 0.45 | 0.51 | −3.38 | N | 11.52(10.73,12.31) | 12.54(11.48,13.60) | −1.01(−2.06,0.02) | 0.49(0.24,1.02) | 0.05 | 0.017(0.006,0.029) | 1.04(1.01,1.07) | 0.01 | |
| 6 | Non-diabetic Apo E null / RAGE null relative to diabetic ApoE null | |||||||||||||
| Microarray | PCR | Western Blot | ||||||||||||
| ApoE null ND | ApoE null D | ApoE null ND | ApoE null D | |||||||||||
| log2FC | FC | P(BH) | B | sig | ΔCt | ΔCt | log2FC(95%CL) | FC(95%CL) | P | log10FC(95%CL) | FC(95%CL) | P | ||
| Thbs1 | −1.00 | 0.50 | 0.24 | −1.00 | N | 11.07(10.89,11.25) | 11.69(9.26,14.13) | −0.63(−3.02,1.77) | 0.65(0.12,3.47) | 0.38 | −0.42(−0.59,−0.24) | 0.38(0.26,0.57) | 0.003 | |
| Tgf-Beta2 | −0.81 | 0.57 | 0.21 | −0.27 | N | 11.02(9.82,12.22) | 11.57(10.51,12.62) | −0.55(−1.72,0.62) | 0.68(0.30,1.54) | 0.28 | −0.14(−0.23,−0.04) | 0.73(0.58,0.91) | 0.02 | |
| ROCK1 | −0.22 | 0.86 | 0.64 | −4.82 | N | 11.03(9.96,12.11) | 11.27(10.57,11.99) | −0.25(−1.27,0.78) | 0.84(0.42,1.71) | 0.55 | −0.26(−0.31,−0.21) | 0.55(0.49,0.61) | 0.002 | |
Figure 3. Regulation of Thbs1, TGFβ-2 and ROCK1 protein in ApoE null mouse aorta.
Total aorta tissue was lysed and subjected to Western blotting as described above using primary antibodies for detection of Thbs1 (A), Tgf-β2 (B) and ROCK1 (C). After probing with the primary antibody, membranes were stripped and re-probed with antibodies to detect GAPDH. In each case, lane 1 represents non-diabetic ApoE null; lane 2 represents diabetic ApoE null; lane 3 represents non-diabetic ApoE null/RAGE null and lane 4 represents diabetic ApoE null/RAGE null. Statistical analyses are illustrated in Table 1.
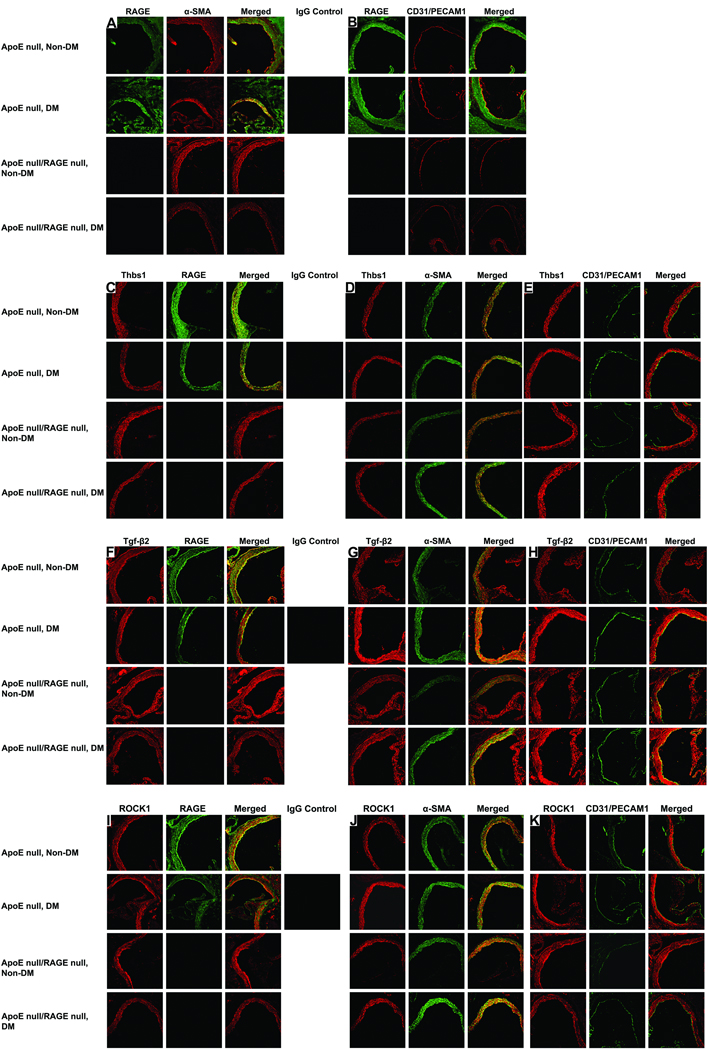
In order to identify the specific histological distribution of the key molecules identified in this model, we immunostained mouse aorta sections from the four groups of mice and subjected these sections to confocal microscopy (Figure 4). First, we examined the expression of RAGE in the aorta of ApoE null mice at age 9 weeks (Figure 4). RAGE is absent in the RAGE null animals, as has been observed previously (5). In both non-diabetic and diabetic ApoE null mice, RAGE (green) is expressed in SMCs (α-smooth muscle actin (red) (Figure 4A), as indicated by the (yellow) merged images in column 3. Furthermore, RAGE (green) and CD31/PECAM1 (red) are co-localized, indicating that RAGE is also expressed in the EC (Figure 4B).
Figure 4. Localization of RAGE, Thbs1, Tgf-β2 and ROCK1 antigens in the aortas of non-diabetic and diabetic ApoE null and ApoE null/RAGE null mice.
Confocal microscopy was performed on aorta tissue and subjected to immunostaining for detection of the indicated antigens. (A). Left column reveals staining with a RAGE specific antibody. Middle column reveals staining with monoclonal mouse smooth muscle actin antibody specific to smooth muscle cell α-actin. Right column reveals the merge of left and right column images. (B). Left column reveals staining with RAGE-specific antibody. Middle column reveals staining with antibody specific for CD31/PECAM1. Right column reveals the merge of left and right column images. Single black square reveals staining with nonimmune IgG control. (C). Left column reveals staining with a Thbs1 specific antibody. Middle column reveals staining with RAGE specific antibody. Right column reveals the merge of left and right column images. (D). Left column reveals staining with Thbs1-specific antibody. Middle column reveals staining with smooth muscle cell specific antibody. Right column reveals the merge of left and right column images. (E). Left column reveals staining with Thbs1 specific antibody. Middle column reveals staining with CD31/PECAM specific antibody. Right column reveals merge of left and right column images. Single black square reveals staining with nonimmune IgG control. (F). Left column reveals staining with a Tgf-β2 specific antibody. Middle column reveals staining with RAGE specific antibody. Right column reveals the merge of left and right column images. (G). Left column reveals staining with Tgf-β2 specific antibody. Middle column reveals staining with smooth muscle cell specific antibody. Right column reveals the merge of left and right column images. (H). Left column reveals staining with Tgf-β2 specific antibody. Middle column reveals staining with CD31/PECAM1 specific antibody. Right column reveals merge of left and right column images. Single black square reveals staining with nonimmune IgG control. (I). Left column reveals staining with a ROCK1 specific antibody. Middle column reveals staining with RAGE specific antibody. Right column reveals the merge of left and right column images. (J). Left column reveals staining with ROCK1 specific antibody. Middle column reveals staining with smooth muscle cell specific antibody. Right column reveals the merge of left and right column images. (K). Left column reveals staining with ROCK1 specific antibody. Middle column reveals staining with CD31/PECAM1 specific antibody. Right column reveals merge of left and right column images. Single black square reveals staining with nonimmune IgG control. Original magnifications: x200.
We next focused on the cellular localization of the three key genes of the Tgf-β pathway identified in these studies. Figure 4C shows co-localization of Thbs1 with RAGE. The Thbs1 and α -SMA images merge under all four conditions, indicating co-localization of the two proteins in the smooth muscle layer (Fig. 4D), consistent with what has been observed previously (23). However, the Thbs1 and CD31/PECAM1 images do not merge under any of the four conditions (Fig. 4E), indicating that Thbs1 is not expressed to appreciable degrees in the endothelial layer of ApoE null mice at age 9 weeks, although Thbs1 expression in ECs has been noted in other settings (24).
Tgf--β2 is co-expressed with RAGE in the aorta (Fig 4F). In all cases, Tgf--β2 merges with RAGE and α-SMA or CD31/PECAM1, with the exception of CD31/PECAM1 in non-diabetic ApoE null mice, indicating that Tgf--β2 is expressed in SMCs in all conditions and in endothelial layers in diabetic but not non-diabetic ApoE null mice aorta (Fig 4G,H). Furthermore, ROCK1 is co-localized with RAGE (Fig 4I). Further, ROCK1 and α-SMA are also co-localized, indicating that ROCK1 is expressed in the smooth muscle layer (Fig. 4J). The images of ROCK1 and CD31/PECAM co-localize only weakly in non-diabetic and diabetic ApoE null mice (Fig. 4K). These findings suggest that ROCK1 is predominantly expressed in the smooth muscle layer in early atherogenesis in ApoE null aorta, but previous studies have noted EC expression as well (25).
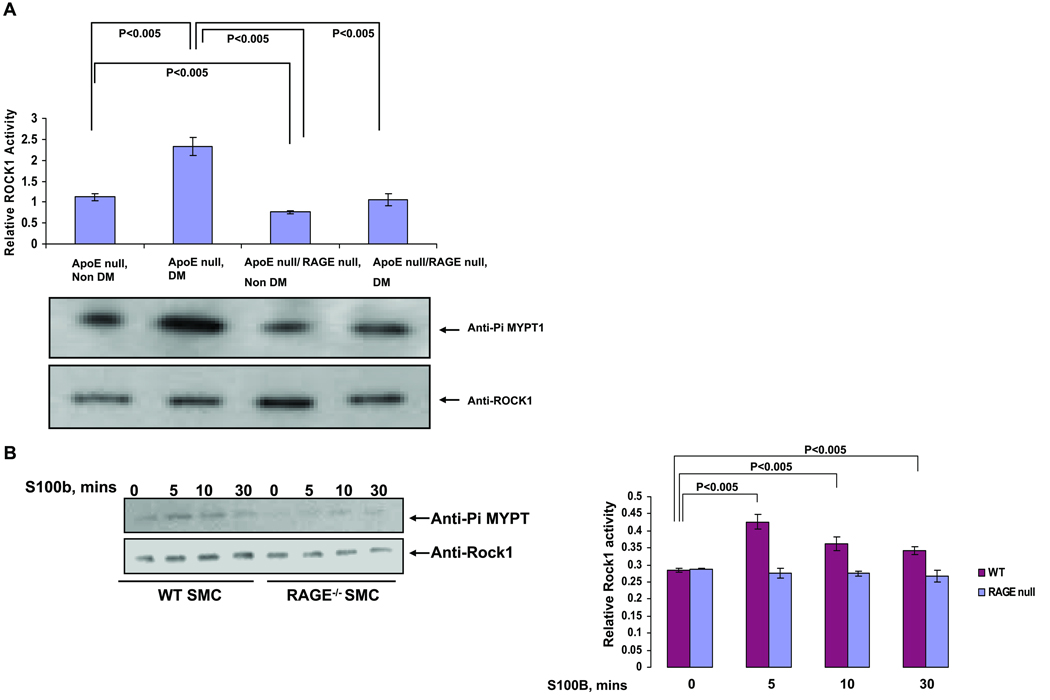
As our analyses suggested that the ROCK1 branch of the Tgf-β pathway is importantly involved in RAGE-dependent atherogenesis, we sought to assess the activation state of ROCK1 in aorta and measured the relative quantity of phosphorylated MYPT1/Ppp1r12a, which is directly phosphorylated by ROCK1. Figure 5A shows the extent of MYPT1/Ppp1r12a phosphorylation increase in diabetic ApoE null mouse aorta relative to non-diabetic ApoE null mice aorta, and diabetic ApoE null/RAGE null mice reveal significantly less MYPT1/Ppp1r12a phosphorylation vs. diabetic ApoE null mice.
Figure 5. ROCK1 activation in ApoE null aorta and primary SMCs: effect of RAGE.
Aortas were retrieved from the indicated mice at age 9 weeks (A) or primary murine aortic SMCs were treated with S100B (10 µg/ml) for the indicated times (B). Lysates were prepared and ROCK1 activity determined. Statistical considerations from at least n=3 distinct experiments are indicated.
Furthermore, as SMCs were the primary cell type expressing ROCK1 in the aorta, we isolated SMCs from the aortas of wild-type and RAGE null mice and treated them with RAGE ligand S100B. Although primary aortic SMCs from wild-type mice displayed increased ROCK1 activity upon incubation with RAGE ligand, S100B, SMCs from RAGE null mice failed to increase ROCK1 activity under these conditions (Figure 5B).
Our data reveal that the observed changes in the Tgf-β pathway are typical of changes in transcription associated with atherogenesis accompanying the onset of diabetes in ApoE null mice, and the effect of RAGE deletion in diabetic ApoE null mice. Online Table XII provides the numbers of differentially expressed unique genes (as distinct from probesets) for each comparison that have Entrez Gene symbols and the numbers with positive and negative log fold changes. In addition, this table gives the numbers of genes resulting from Boolean operations on these genelists. Online Tables XIII-XVII give the lists of genes whose numbers are given in Online Table XII. Online Figure II represents a Venn diagram showing the intersection of comparison 1, diabetic ApoE null relative to non-diabetic ApoE null, with comparison 4, diabetic ApoE null/RAGE null relative to diabetic ApoE null. Although there are 53 genes which are statistically significantly differentially expressed in diabetic ApoE null relative to the non-diabetic ApoE null state, and 216 genes which are statistically significantly differentially expressed in diabetic ApoE null/RAGE null relative to diabetic ApoE null, only 15 of these genes are statistically significantly differentially expressed in both comparisons (Online Tables XII and XIII and Online Figure II). There is very little overlap of the genes which are differentially expressed both in the onset of diabetes in ApoE null mice and in the effect of RAGE deletion in diabetic ApoE null mice.
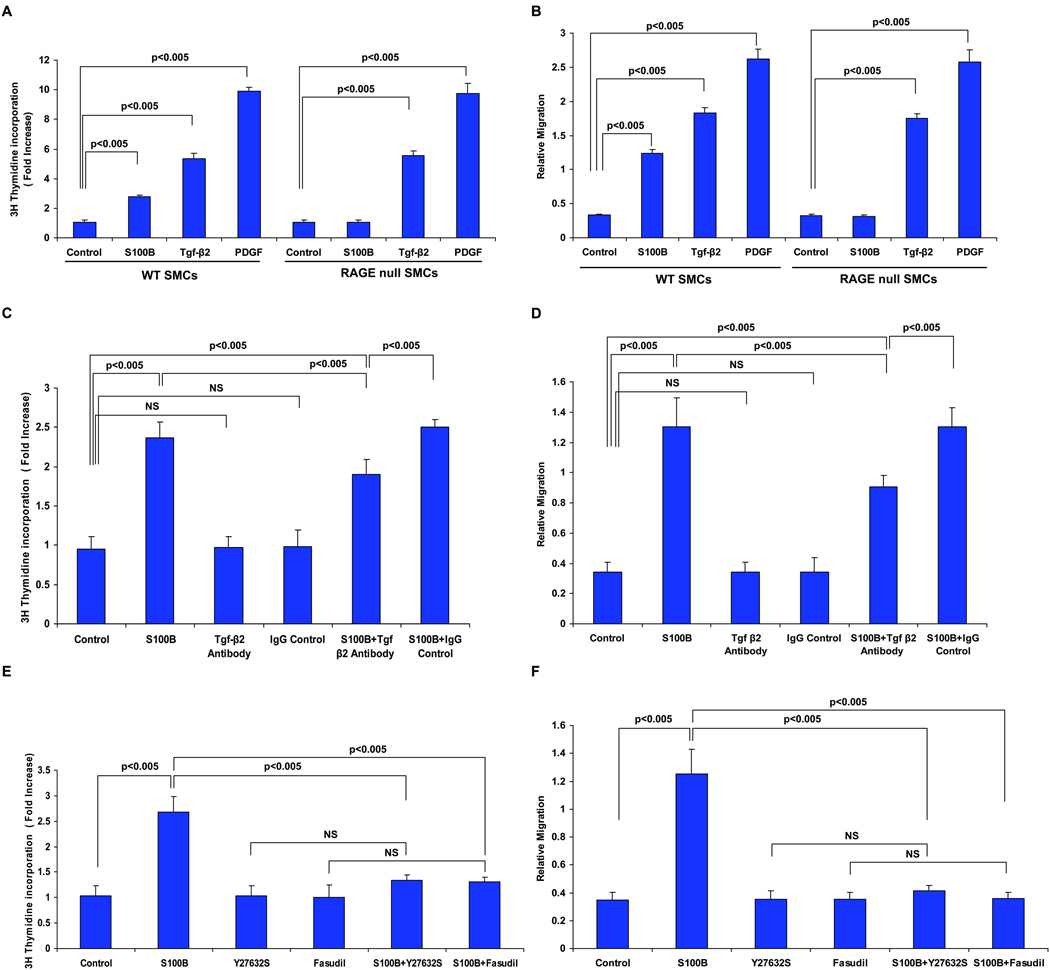
Next, to specifically link RAGE to SMC proliferation and migration, we performed studies in primary SMCs retrieved from RAGE-expressing or RAGE-deficient mouse aortas. As illustrated in figure 6A and 6B, incubation of wild-type SMCs with RAGE ligand S100B resulted in significantly increased proliferation and migration, but S100B failed to stimulate proliferation and migration in RAGE-deficient SMCs. Note that in wild-type and RAGE-deficient SMCs, incubation with Tgf-β2 or a non-RAGE ligand PDGF increased proliferation and migration, suggesting that Tgf-β2 and PDGF are not direct ligands of RAGE, that RAGE deficient SMCs are capable of proliferation and migration, and that exogenous addition of Tgf-β2 to RAGE-deficient cells restores proliferation and migration (Figure 6A and 6B, respectively).
Figure 6. RAGE ligands modulate SMC proliferation and migration: requirement for Tgf-β and ROCK.
A&B. Wild-type and RAGE-deficient SMCs were treated with S100B (10 µg/ml), Tgf-β (10 ng/ml) or PDGF (10 ng/ml) for 5 or 48 hrs and at the end of that time proliferation (A) and migration (B) were assessed. Note that when comparing migration and proliferation responses to S100B between wild-type and RAGE null SMCs; p<0.05. In C&D, wild-type SMCs were pre-treated anti-Tgf-β antibody or irrelevant IgG control antibody (10 µg/ml) followed by assessment of S100B-stimulated proliferation and migration. In E&F, wild-type SMCs were pre-treated with Y27632 (X10 µM) or fasudil (10 µM) followed by S100B (10 µg/ml) and proliferation and migration were monitored. Statistical considerations from at least n=3 distinct assays are shown.
Finally, to establish that RAGE ligand-stimulated SMC proliferation and migration required Tgf-β2 and ROCK1 action, we treated wild-type SMCs with S100B in the presence or absence of Tgf-β2 or ROCK1 inhibitors. Consistent with key roles for Tgf-β2 in S100B-mediated effects on SMCs, pre-treatment of wild-type SMCs with anti-Tgf-β2 antibody resulted in a significant decrease in proliferation and migration compared to treatment with IgG control (Figure 6C&D, respectively). ROCK signaling was implicated in the S100B modulation of SMC properties, as treatment of SMCs with S100B and ROCK inhibitors Y27632 or fasudil significantly reduced S100B-stimulated proliferation and migration (Figure 6E&F, respectively). Note that treatment with anti-Tgf-β2 antibody, Y27632 or fasudil alone had no independent effect on SMC migration or proliferation (Figure 6C, D, E & F).
Discussion
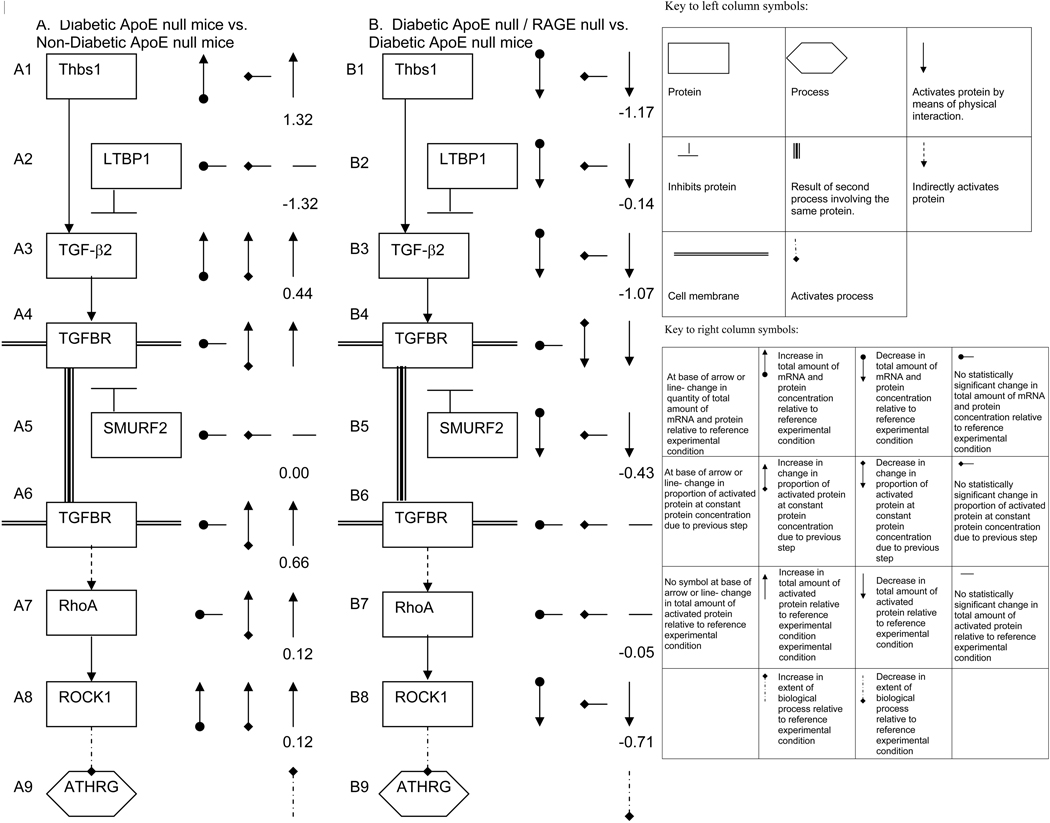
In summary, these findings lead us to speculate on the mechanisms by which diabetes accelerates atherogenesis in ApoE null mice (comparison 1), and by which RAGE deletion slows atherogenesis in diabetic ApoE null mice (comparison 4) as illustrated in Figure 7 (a formal set of rules for the derivation of the model in Figure 7 is given in the Supplementary Methods section). First, we consider the effect of diabetes on ApoE null mice (comparison 1) (Figure 7A). All changes in quantity of mRNA and total and activated protein in this column reflect that in diabetic ApoE null mice vs. non-diabetic ApoE null mice. We infer the mechanism based upon Figs. 1 and 2, Online Tables X and XI as follows: A1: diabetes up-regulates Thbs1; A2: no change in levels of LTBP1 was detected for this comparison; A3: the amount of activated Tgf-β2 may increase because the total amount of Tgf-β2 increases, and because of increased activation due to up-regulation of Thbs1; A4: since Tgf-β2 activates Tgf-βR1&2 complex (TGFBR) and since the amount of activated Tgf-β2 increases, the amount of activated TGFBR increases; A5 and A6: since no change in the amount of SMURF2 mRNA was detected in this comparison, interaction with SMURF2, which targets Tgf-β1 for destruction (26), will not change the amount of total or activated Tgfβ-R; A7: since Tgf-βR complex indirectly activates RhoA (27–28), and since the amount of activated Tgf-βR complex increases, the amount of activated RhoA increases; A8: since RhoA activates ROCK1 (29–30), and since the amount of activated RhoA increases, the amount of activated ROCK1 increases; A9: since ROCK1 accelerates atherogenesis (ATHRG), and since the extent of ROCK1 activation increases, acceleration of atherosclerosis (ATHRG) ensues. It should be noted that although Tgf-β from immune cells has been reported to reduce atherosclerosis, (9–15), Tgf-β action in SMCs has been linked to their proliferation, hypertrophy, migration and production of extracellular matrix (31–33).
Figure 7. Proposed mechanism by which diabetes and RAGE contribute to atherosclerosis in ApoE null mice.
Based on in-depth analysis of microarray findings, we speculate on the mechanisms by which diabetes accelerates atherosclerosis in ApoE null mice (A) and by which RAGE accelerates atherosclerosis in diabetic ApoE null mice (B). In both cases, the left column represents the pathway, and the right column represents the observed change in concentration of mRNA and protein and inferred change in activation of proteins and processes. Numbers accompanying each molecular step are Pathway Express perturbation factors. Note that Tgf-βR appears in two steps for the sake of reliability, but only has one perturbation factor, as any other protein in the pathway.
Next, we address the mechanism by which RAGE deletion delays acceleration of atherosclerosis in diabetic ApoE null mice (Figure 7B). All changes in quantity of mRNA and total and activated protein in this column are for diabetic ApoE null/RAGE null vs. diabetic ApoE null mice (comparison 4) unless otherwise specified. B1: the amount of Thbs1 decreases upon deletion of RAGE; B2: LTBP1 expression decreases; B3: the amount of activated Tgf-β2 decreases as the total amount of Tgf-β2 decreases. (The proportion of activated Tgf-β2 decreases because of the decrease in Thbs1, however, this effect may be cancelled, all or in part, by the decrease in Tgf-β2 deactivation by LTBP1 accompanying the decrease in LTBP1); B4: since Tgf-β2 activates Tgf-βR , and since the amount of activated Tgf-β2 decreases, the amount of activated complex decreases; B5: the amount of SMURF2 decreases; B6: since SMURF2 deactivates Tgf-βR1, and the amount of SMURF2 decreases, the amount of Tgf-βR1 increases, canceling all or part of the effect of decrease in activated Tgf-βR in step B4; B7: since Tgf-βR complex indirectly activates RhoA, and since the amount of activated Tgf-βR is approximately unchanged, the amount of activated ROCK1 is also approximately unchanged; B8: the total amount of ROCK1 decreases (since the amount of activated RhoA remains roughly constant, and the total amount of ROCK1 decreases, the amount of activated ROCK1 decreases); B9: since ROCK1 accelerates atherosclerosis (ATHRG) (18–20), and since the amount of activated ROCK1 decreases, atherosclerosis is reduced.
Studies have implicated the ROCK1 signaling pathway in human vascular cells and atherosclerosis (34–36). Our results in vivo revealed that RAGE-deficient ApoE null mice displayed reduced ROCK1 activity in the aorta and in vitro, stimulation of wild-type SMCs with RAGE ligand S100B mediates proliferation and migration of these cells in a manner suppressed by two distinct inhibitors of ROCK. Of note, a previous study in cultured endothelial cells suggested that antibodies to RAGE blocked AGE-stimulated endothelial hyperpermeability presumably through effects on ROCK and phosphorylation of moesin (37).
It is important to note that roles for RAGE in atherosclerosis have been shown in an additional mouse model, that is, mice deficient in LDL receptor. When these mice were bred into the RAGE null background and fed high-fat diet, reduction of atherosclerosis was observed compared to RAGE-expressing controls (38). The potential effect of RAGE signaling on Tgf-β/ROCK in this model remains to be addressed.
Taken together, these data reveal that suppression of Tgf-β/ROCK1 activity in the atherosclerosis-vulnerable vessel wall, especially in diabetes, but in non-diabetes as well, may underlie the beneficial effects of RAGE antagonism and genetic deletion in ApoE null mice.
Novelty and Significance
“What is known?”
The receptor RAGE (receptor for AGE) contributes to the pathogenesis of non-diabetic and diabetic atherosclerosis in atherosclerosis-vulnerable mouse models.
Previous studies suggested important roles for inflammatory signaling in the role of RAGE in atherosclerosis; however, they did not elucidate the specific signaling pathways by which RAGE contributed to atherosclerosis.
“What new information does this article contribute?”
Remarkably, in vivo and in vitro (the latter in smooth muscle cells), RAGE regulates activity of the ROCK1 branch of the Tgf-β signaling pathway and ROCK inhibitors suppress the effects of RAGE ligands on smooth muscle cell proliferation and migration.
These findings suggest for the first time that RAGE-dependent acceleration of atherosclerosis in ApoE null mice is dependent, at least in part, on the action of the ROCK1 branch of the Tgf-β pathway.
The multi-ligand Receptor for AGE (RAGE) has been shown to contribute to acceleration of atherosclerosis in mouse models, both in the presence and absence of diabetes. However, previous studies did not identify the precise signal transduction pathways by which RAGE contributed to atherosclerosis. We performed unbiased Affymetrix gene expression arrays on aortas of non-diabetic and diabetic ApoE null mice expressing RAGE or devoid of RAGE. Intriguingly, we found very little overlap of the genes which are differentially expressed both in the onset of diabetes in ApoE null mice, and in the effect of RAGE deletion in diabetic ApoE null mice. For the first time we demonstrate by Pathway-Express analysis that the Transforming Growth Factor-β pathway (Tgf-β̣ and particularly its ROCK1 branch are regulated by RAGE, especially in smooth muscle cells both in vivo in aortic tissue, and in vitro in primary smooth muscle cells. Taken together, our work suggests the novel finding that RAGE-dependent acceleration of atherosclerosis in ApoE null mice is dependent, at least in part, on the action of the ROCK1 branch of the Tgf-β pathway. These results may fundamentally impact the selection of complementary therapy strategies for mitigating atherosclerosis, particularly in the diabetic state.
Supplementary Material
Acknowledgements
We thank Dr. Sorin Draghici and his group for making Pathway Express available to us and for assisting us in its use. The authors are grateful to Ms. Latoya Woods for her expertise in preparation of this manuscript.
Sources of Funding
Juvenile Diabetes Research Foundation and the U.S. Public Health Service (HL60901)
Non-Standard Abbreviations and Acronyms
- Apo
apolipoprotein
- ATHRG
atherosclerosis
- EC
endothelial cell
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Ldl
low density lipoprotein
- Ltbp1
Latent transforming growth factor beta binding protein 1
- MYPT1
Myosin phosphatase target subunit 1
- PDGF
platelet derived growth factor
- PECAM
platelet endothelial cell adhesion molecule
- Ppp1r12a
Protein phosphatase 1, regulatory (inhibitor) subunit 12A
- RAGE
receptor for advanced glycation endproducts
- ROCK
rho-associated protein kinase
- SMA
smooth muscle actin
- SMC
smooth muscle cell
- Tgf
transforming growth factor
- TGFBR
transforming growth factor beta receptor
- Thbs
thrombospondin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Subject Codes: [96] Mechanism of Atherosclerosis/Growth Factors, [134] Pathophysiology, [142] Gene Expression, [189] Type 1 Diabetes
Disclosures
None.
Contributor Information
De-xiu Bu, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Vivek Rai, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Xiaoping Shen, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Rosa Rosario, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Yan Lu, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Vivette D’Agati, Department of Pathology, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Shi Fang Yan, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
Richard A. Friedman, Biomedical Informatics Shared Resource, Herbert Irving Comprehensive Cancer Center, Department of Biomedical Informatics, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032
Edem Nuglozeh, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032
Ann Marie Schmidt, Division of Surgical Science, Department of Surgery, College of Physicians & Surgeons, Columbia University, 630 West 168th Street, New York, New York 10032.
References
- 1.Park L, Raman KG, Lee KJ, Lu Y, Ferran LJ, Chow WS, Stern D, Schmidt AM. Suppression of accelerated diabetic atherosclerosis by the soluble receptor for advanced glycation endproducts. Nat Med. 1998;4:1025–1031. doi: 10.1038/2012. [DOI] [PubMed] [Google Scholar]
- 2.Kislinger T, Tanji N, Wendt T, Qu W, Lu Y, Ferran LJ, Jr, Taguchi A, Olson K, Bucciarelli L, Goova M, Hofmann MA, Cataldegirmen G, D'Agati V, Pischetsrieder M, Stern DM, Schmidt AM. Receptor for advanced glycation endproducts mediates inflammation and enhanced expression of tissue factor in vasculature of diabetic apolipoprotein E null mice. Arterioscler Thromb Vasc Biol. 2001;21:905–910. doi: 10.1161/01.atv.21.6.905. [DOI] [PubMed] [Google Scholar]
- 3.Bucciarelli LG, Wendt T, Qu W, Lu Y, Lalla E, Rong LL, Goova MT, Moser B, Kislinger T, Lee DC, Kashypa Y, Stern DM, Schmidt AM. RAGE blockade stabilizes established atherosclerosis in diabetic apolipoprotein E null mice. Circ. 2002;106:2827–2835. doi: 10.1161/01.cir.0000039325.03698.36. [DOI] [PubMed] [Google Scholar]
- 4.Wendt T, Harja E, Bucciarelli L, Qu W, Lu Y, Rong LL, Jenkins DG, Stein G, Schmidt AM, Yan SF. RAGE modulates vascular inflammation and atherosclerosis in a murine model of type 2 diabetes. Atherosclerosis. 2006;185:70–77. doi: 10.1016/j.atherosclerosis.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 5.Harja E, Bu DX, Hudson BI, Chang JS, Shen X, Hallam K, Kalea AZ, Lu Y, Rosario RH, Oruganti S, Nikolla Z, Belov D, Lalla E, Ramasamy R, Yan SF, Schmidt AM. Vascular and inflammatory stresses mediate atherosclerosis via RAGE and its ligands in apoE −/− mice. J Clin Invest. 2008;118:183–194. doi: 10.1172/JCI32703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soro-Paavonen A, Watson AM, Li J, Paavonen K, Koitka A, Calkin AC, Barit D, Coughlan MT, Drew BG, Lancaster GI, Thomas M, Forbes JM, Nawroth PP, Bierhaus A, Cooper ME, Jandeleit-Dahm KA. Receptor for advanced glycation endproducts (RAGE) deficiency attenuates the development of atherosclerosis in diabetes. Diabetes. 2008;57:2461–2469. doi: 10.2337/db07-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura K, Ito M, Amano K, Chihara Y, Fukata M, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 8.Mong PY, Petrulio C, Kaufman HL, Wang Q. Activation of Rho kinase by TNF-alpha is required for JNK activation in human pulmonary microvascular endothelial cells. J Immunol. 2008;180:550–558. doi: 10.4049/jimmunol.180.1.550. [DOI] [PubMed] [Google Scholar]
- 9.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 10.Grainger DJ. Transforming growth factor beta and atherosclerosis: so far, so good for the protective cytokine hypothesis. Arterioscler Thromb Vasc Biol. 2004;24:399–404. doi: 10.1161/01.ATV.0000114567.76772.33. [DOI] [PubMed] [Google Scholar]
- 11.Grainger DJ. TGF-beta and atherosclerosis in man. Cardiovasc Res. 2007;74:213–222. doi: 10.1016/j.cardiores.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 12.Mallat Z, Tedgui A. The role of transforming growth factor beta in atherosclerosis: novel insights and future perspectives. Curr Opin Lipidol. 2002;13:523–529. doi: 10.1097/00041433-200210000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Grainiger DJ, Mosedale DE, Metcalfe JC, Bottinger EP. Dietary fat and reduced levels of TGF beta 1 act synergistically to promote activation of the vascular endothelium and formation of lipid lesions. J Cell Sci. 2000;113:2355–2361. doi: 10.1242/jcs.113.13.2355. [DOI] [PubMed] [Google Scholar]
- 14.Gojova A, Brun V, Esposito B, Cottrez F, Gourdy P, Ardouin P, Tedgui A, Mallat Z, Groux H. Specific abrogation of transforming growth factor-beta signaling in T cells alters atherosclerotic lesion size and composition in mice. Blood. 2003;102:4052–4058. doi: 10.1182/blood-2003-05-1729. [DOI] [PubMed] [Google Scholar]
- 15.Mallat Z, Gojova A, Marchiol-Fournigault C, Esposito B, Kamate C, Merval R, Fradelizi D, Tedgui A. Inhibition of transforming growth factor-beta signaling accelerates atherosclerosis and induces an unstable plaque phenotype in mice. Circ Res. 2001;89:930–934. doi: 10.1161/hh2201.099415. [DOI] [PubMed] [Google Scholar]
- 16.Draghici S, Khatri P, Tarca AL, Amin K, Done A, Voichita C, Georgescu C, Romero RA. systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miyazono K, Hellman U, Hellman C, Wernstedt C, Heldin CH. Latent high molecular weight complex of transforming growth factor beta 1. Purification from human platelets and structural characterization. J Biol Chem. 1998;263:6407–6415. [PubMed] [Google Scholar]
- 18.Saharinen J, Hyytiainen M, Taipale J, Keski-Oja J. Latent transforming growth factor beta binding proteins (LTBPs)-structural extracellular matrix proteins for targeting TGF beta action. Cytokine Growth Factor Rev. 1999;10:99–117. doi: 10.1016/s1359-6101(99)00010-6. [DOI] [PubMed] [Google Scholar]
- 19.Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-beta with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- 20.Mallat Z, Gojova A, Sauzeau V, Brun V, Silvestre JS, Esposito B, Merval R, Groux H, Loirand G, Tedgui A. Rho-associated protein kinase contributes to early atherosclerotic lesion formation in mice. Circ Res. 2003;93:884–888. doi: 10.1161/01.RES.0000099062.55042.9A. [DOI] [PubMed] [Google Scholar]
- 21.Wang HW, Liu PY, Oyama N, Rikitake Y, Kitamoto S, Gitlin J, Liao JK, Boisvert WA. Deficiency of ROCK1 in bone marrow-derived cells protects against atherosclerosis in LDL receptor null mice. FASEB J. 2008;22:3561–3570. doi: 10.1096/fj.08-108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noma K, Rikitake Y, Oyama N, Yan G, Alcaide P, Liu PY, Wang H, Ahl D, Sawada N, Okamoto R, Hiroi Y, Shimizu K, Luscinskas FW, Sun J, Liao JK. ROCK1 mediates leukocyte recruitment and neointima formation following vascular injury. J Clin Invest. 2008;118:1632–1644. doi: 10.1172/JCI29226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riessen R, Kearney M, Lawler J, Isner JM. Immunolocalization of thrombospondin-1 in human atherosclerotic and restenotic arteries. Am Heart J. 1998;135:357–364. doi: 10.1016/s0002-8703(98)70105-x. [DOI] [PubMed] [Google Scholar]
- 24.Lane TF, Iruela-Arispe ML, Sage EH. Regulation of gene expression by SPARC during angiogenesis in vitro. Changes in fibronectin, thrombospondin-1, and plasminogen activator inhibitor-1. J Biol Chem. 1992;267:16736–16745. [PubMed] [Google Scholar]
- 25.Shimokawa H, Rashid M. Development of Rho kinase inhibitors for cardiovascular medicine. Trends Pharmacol Sci. 2007;28:296–302. doi: 10.1016/j.tips.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 26.Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, Wrana JL. Smad7 binds to Smurf2 to form E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–1375. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- 27.Clements Rt, Minnear FL, Singer HA, Keller RS, Vincent PA. Rho A and Rho-kinase dependent and independent signals mediate TGF-beta induced pulmonary endothelial cytoskeletal reorganization and permeability. Am J Physiol Cell Mol Physiol. 2005;288:L294–L306. doi: 10.1152/ajplung.00213.2004. [DOI] [PubMed] [Google Scholar]
- 28.Rosman DS, Phukan S, Huang CC, Pasche B. TGFBR1*6A enhances the migration and invasion of MCF-7 breast cancer cells through RhoA activation. Cancer Res. 2008;68:1319–1328. doi: 10.1158/0008-5472.CAN-07-5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujisawa K, Fujita A, Ishizaki T, Saito Y, Narumiya S. Identification of the Rho-binding domain of p160ROCK, a Rho-associated coiled-coil containing protein kinase. J Biol Chem. 1996;271:23022–23028. doi: 10.1074/jbc.271.38.23022. [DOI] [PubMed] [Google Scholar]
- 30.Ishizaki T, Mackawa M, Fujisawa K, Okawa K, Iwamatsu A, Fujita A, Watanabe N, Saito Y, Kakizuka A, Morii N, Narumiya S. The small GTP-binding protein Rho binds to and activates a 160 kDa Ser/Thr protein kinase homologous to myotonic dystrophy kinase. EMBO J. 1996;15:1885–1893. [PMC free article] [PubMed] [Google Scholar]
- 31.Ruiz-Ortega M, Rodriguez Vita J, Sanchez Lopez E, Carvajal G, Egido J. TGF beta signaling in vascular fibrosis. Cardiovasc Res. 2007;74:196–206. doi: 10.1016/j.cardiores.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 32.Edlin RS, Tsai S, Yamanouchi D, Wang C, Liu B, Kent KC. Characterization of primary and restenotic atherosclerotic plaque from the suprficial femoral artery: potential role of Smad3 in regulation of SMC proliferation. J Vasc Surg. 2009;49:1289–1295. doi: 10.1016/j.jvs.2008.11.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryan ST, Koteliansky VE, Gotwals PJ, Lindner V. Transforming growth factor-beta dependent events in vascular remodeling following arterial injury. J Vasc Res. 2003;40:37–46. doi: 10.1159/000068937. [DOI] [PubMed] [Google Scholar]
- 34.Ming XF, Barandier C, Viswambharan H, Kwak BR, Mach F, Mazzolai L, Hayoz D, Ruffieux J, Rusconi S, Montani JP, Yang Z. Thrombin stimulates human endothelial arginase enzymatic activity via RhoA/ROCK pathway: implications for atherosclerotic endothelial dysfunction. Circ. 2004;110:3708–3714. doi: 10.1161/01.CIR.0000142867.26182.32. [DOI] [PubMed] [Google Scholar]
- 35.Noma K, Goto C, Nishioka K, Jitsuiki D, Umemura T, Jitsuiki D, Nakagawa K, Oshima T, Chayama K, Yoshizumi M, Higashi Y. Roles of rho-associated kinase and oxidative stress in the pathogenesis of aortic stiffness. J Am Coll Cardiol. 2007;49:698–705. doi: 10.1016/j.jacc.2006.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nohria A, Prsic A, Liu PY, Okamoto R, Creager MA, Selwyn A, Liao JK, Ganz P. Statins inhibit rho kinase activity in patients with atherosclerosis. Atherosclerosis. 2009;205:517–521. doi: 10.1016/j.atherosclerosis.2008.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo X, Wang L, Chen B, Li Q, Wang J, Zhao M, Wu W, Zhu P, Huang X, Huang Q. ERM protein moesin is phosphorylated by advanced glycation endproducts and modulates endothelial permeability. Am J Physiol Circ Physiol. 2009;297:H238–H246. doi: 10.1152/ajpheart.00196.2009. [DOI] [PubMed] [Google Scholar]
- 38.Sun L, Ishida T, Yasuda T, Kojima Y, Honjo T, Yamamoto Y, Yamamoto H, Ishibashi S, Hirata K, Hayashi Y. RAGE mediates oxidized LDL-induced pro-inflammatory effects and atherosclerosis in non-diabetic LDL receptor-deficient mice. Cardiovasc Res. 2009;82:371–381. doi: 10.1093/cvr/cvp036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.