Abstract
Cell-based therapy has been widely evaluated in spinal cord injury (SCI) animal models and shown to improve functional recovery. However, host response to cell transplants at gene expression level is rarely discussed. We reported previously that acute transplantation of radial glial cells RG3.6 following SCI promoted early locomotion improvement within 1 week post-injury. To identify rapid molecular changes induced by RG3.6 transplantation in the host tissue, distal spinal cord segments were subjected to microarray analysis. Although RG3.6 transplantation, reduced activity of macrophages as early as 1–2 weeks post-injury, the expression levels of inflammatory genes (e.g., IL-6, MIP-2, MCP-1) were not decreased by RG3.6 treatment as compared to medium or other cell controls at 6–12 h post-injury. However, genes associated with tissue protection (Hsp70 and Hsp32) and neural cell development (Foxg1, Top2a, Sox11, Nkx2.2, Vimentin) were found to be significantly up-regulated by RG3.6 transplants. Foxg1 was the most highly induced gene in the RG3.6-treated spinal cords, and its expression by immunocytochemistry was confirmed in the host tissue. Moreover, RG3.6 treatment boosted the number of Nkx2.2 cells in the spinal cord, and these cells frequently co-expressed NG2, which marks progenitor cells. Taken together, these results demonstrate that radial glial transplants induced rapid and specific gene expression in the injured host tissue, and suggest that these early responses are associated with mechanisms of tissue protection and activation of endogenous neural progenitor cells.
Key words: gene expression, neuroinflammation, radial glia, spinal cord injury
Introduction
Spinal cord injury (SCI) triggers a series of pathophysiological changes that lead to progressive tissue damage called “secondary injury,” which continues for prolonged periods (Beattie et al., 2002; Bramlett et al., 2007). Secondary injury involves immune responses to the primary injury, and may modulate loss of neurons and glia. Molecular analysis revealed distinct patterns of gene expression after SCI in both the injury site and adjacent regions at different times, indicating tissue loss and degenerative events (Carmel et al., 2001; Di Giovanni et al., 2003; Aimone et al., 2004; De Biase et al., 2005). Inflammatory and transcriptional genes were induced within hours after SCI, whereas genes encoding for neuronal structural proteins and ion transport proteins were suppressed (Carmel et al., 2001; Nesic et al., 2002; De Biase et al., 2005). At later times, growth factors, cell proliferation, and angiogenesis-related genes were up-regulated (Bareyre et al., 2003; Velardo et al., 2004), suggesting that tissue repair mechanisms have been initiated.
Gene expression profiles have been widely used to evaluate the efficacy of treatments for SCI, including anti-inflammatory drugs (e.g., Cox2 inhibitor, MP, and MK801) and antibody IN-1 application (Plunkett et al., 2001; Bareyre et al., 2002, 2003; Nesic et al., 2002; Pan et al., 2004). Transplantation of cells acutely following SCI is another approach that may promote recovery; however, little is known about the molecular changes that transplants induce to host tissues during the early phase of SCI. Most cell-based therapies have focused on histological and behavioral improvements that are associated with axonal regeneration and/or remyelination (Enzmann et al., 2006; Oudega, 2007). These processes take place during extended periods after SCI, making it difficult to relate them to underlying mechanisms, particularly at the level of gene expression. The many molecular changes in multiple pathways that have been identified acutely following SCI present a daunting challenge to analyze the effects of potential therapies on secondary injury. The dynamic spread of secondary damage suggests that changes in regions adjacent to the primary injury site may be less complicated to analyze than in the injury site itself (Carmel et al., 2001; De Biase et al., 2005). We found previously that acute transplantation of a radial glial clone RG3.6 cells promoted locomotion improvement during early phases after SCI by comparison to injection of fibroblasts or medium alone (Hasegawa et al., 2005). Functional recovery after RG3.6 treatment was associated with preservation of axons and reduced accumulation of macrophages in and around the injury site after 6 weeks (Hasegawa et al., 2005). The early locomotion improvement, white matter sparing, and suppressed macrophage infiltration suggested that acute RG3.6 treatment may protect tissue by modulating the magnitude of inflammatory signals. In this study, we identified early molecular changes associated with tissue protection by RG3.6 cells within 1 day after transplantation into the injured spinal cord. We did not obtain evidence implicating reduced immune responses at the earliest times studied. Rather, the results suggest that acute radial glia transplantation generates local signals to enhance defense mechanisms and increase numbers of neural precursor cells, which may suppress progression of secondary damage, including the extent of immune cell activation.
Methods
Spinal cord injury and cell transplantation
Fifty-five adult female Sprague-Dawley rats (200–250 g, 77 days old, Taconic, Germantown, NY) were used in this study: laminectomy control (n = 4), laminectomy + RG3.6 transplants (n = 3), SCI + vehicle medium (DMEM+F12; n = 20, 3–4 rats at each time point for histology or RNA preparations), SCI + RG3.6 transplants (n = 22, 3–4 rats at each time point for histology or RNA preparations), SCI + primary neural stem cell (NSC) transplants (n = 3), and SCI + fibroblast transplants (n = 3). SCI procedures were described previously (Hasegawa et al., 2005). Briefly, rats were anesthetized with 40 mg/kg of pentobarbital i.p. (Besse Medical Supply/ASD Specialty Healthcare, Louisville, KY). A 12.5-g-cm MASCIS/NYC contusion injury was induced at spinal segment T10 (Gruner 1992). Within 10 min after injury, a total of 4 μl of RG3.6 cells (200,000 cells/μl in DMEM+F12 medium; 800,000 cells total) were injected to contused spinal cord at three points: 2 μl at the injury epicenter, 1 μl each at 2 mm rostral and caudal to the epicenter. Primary NSC from GFP rat cortices at embryonic 14 day and fibroblasts from GFP neonatal rat skin were used as control cell types for transplantation. For vehicle controls, DMEM+F12 was injected following injury. For non-injury controls, rats were received only T10 laminectomy, or it was followed by RG3.6 transplantation without contusion injury. Each injection was conducted slowly during a period of 10 min (at 1 mm depth) using a sterile glass tip with a diameter of 50 μm. Following injections, muscles and skin were closed separately. Cefazolin (25 mg/kg) and cyclosporine A (10 mg/kg) were administered to all rats to prevent infection and immunorejection. Animal surgery and post-operational care were carefully followed by protocols approved by Rutgers University.
Tissue processing for immunofluorescence
At designated times, animals were anesthetized and perfused intracardially with phosphate-buffered saline (PBS) followed by 4% paraformaldehyde. Spinal cords were removed, post-fixed overnight in the same fixative, and cryoprotected in 25% sucrose for 72 h at 4°C. For coronal sections, three or four 15 mm-spinal cord segments centered at the injury site were aligned and then embedded in OCT compound (Fisher Scientific, Pittsburgh, PA) so that comparable regions could be analyzed on individual slides. Serial 20-μm sections were cut on a cryostat (Hacker), and every 4th section was collected and mounted on Super-frost plus slides (Fisher Scientific, Pittsburgh, PA). A total of 30 slides were collected for each animal. The first 10 slides represented the 5-mm rostral spinal segment to injury site, with each slide containing eight sections at an interval of ∼600 μm. The middle 10 slides represented the injury epicenter segment, and the final 10 slides represented the caudal spinal segment to the injury site. For sagittal sections, 2-cm spinal cord segments centered at the injury site were embedded in OCT compound and serially sectioned at 20 μm on a cryostat. A total of 12 slides were collected for each animal, with each slide containing eight or nine sections with an interval of 240 μm. Quantitative analysis was performed on the three sections surrounding the midline representing ∼500 μm in each of three rats per treatment group.
For immunofluorescence, sections were blocked with 10% normal goat serum/0.3% Triton X-100 in PBS at room temperature for 2 h and incubated at 4°C overnight with primary antibodies as listed in Table 1. Sections then were washed with PBS and incubated with appropriate secondary antibodies at room temperature for 1 h. After washing, sections were counterstained with Hoechst 33342 at 1:2000 and mounted with Gel/Mount. Image analysis was performed using Zeiss 510 confocal laser scanning microscope (LSM).
Table 1.
Antibody List
| Primary antibody | Dilution | Isotype | Source | Secondary antibody | Dilution | Source |
|---|---|---|---|---|---|---|
| Iba1 (ionized calcium-binding adaptor molecule 1) | 1: 1000 | Rabbit Polyclonal IgG | Wako | Alexa568 Goat-anti-rabbit IgG (H + L) | 1: 400 | Molecular Probe |
| ED1 (CD68) | 1: 300 | Mouse Monoclonal IgG | Serotec | Alexa568 Goat-anti-mouse IgG (H + L) | 1: 400 | Molecular Probe |
| Foxg1 | 1: 1000 | Rabbit Polyclonal IgG | Dr. Lorenz Studer | Alexa568 Goat-anti-rabbit IgG (H + L) | 1: 400 | Molecular Probe |
| Nkx2.2 | 1: 20 | Mouse Monoclonal IgG | DSHB | Alexa568 Goat-anti-mouse IgG (H + L) | 1: 400 | Molecular Probe |
| NG2 | 1: 500 | Rabbit Polyclonal IgG | Dr. Joel Levine | Alexa647 (Cy5) Goat-anti-rabbit IgG (H + L) | 1: 400 | Molecular Probe |
| Vimentin | 1: 20 | Mouse Monoclonal IgM | DSHB | Alexa568 Goat-anti-mouse IgM | 1: 400 | Molecular Probe |
Quantitation for immunofluorescence
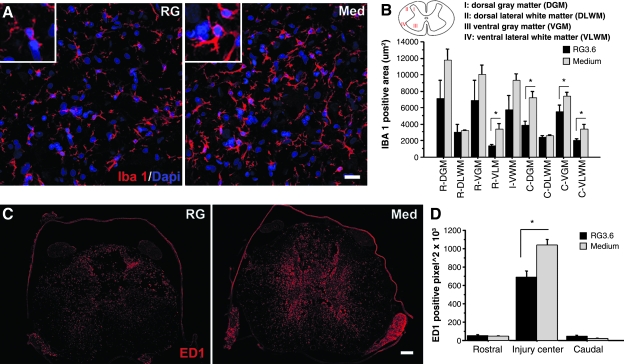
Iba-1 immunostaining was used to examine morphological changes of microglia 7 days after SCI. Higher magnification images were taken through 40 × /1.2 w corr objective lens on Zeiss LSM 510 confocal microscope. Each image consisted of projections of Z stacks of five optical slices of x: 227.8 μm; y: 227.8 μm; z: 4 μm at 1024 × 1024 pixel resolution. Two sections (nos. 3 and 6 of the eight sections on the slide, as described above) from each segment (rostral, injury, and caudal segments) were chosen from each animal for quantitation of comparable regions among different spinal cords. Four regions of the section, which included dorsal gray matter (DGM), dorsal lateral white matter (DLWM), ventral gray matter (VGM), and ventral lateral white matter (VLWM; Fig. 1), were photographed and quantitated. Signal thresholds were determined by mean value of all selected images. Areas displaying expression levels higher than threshold were recorded.
FIG. 1.
Radial glia transplantation reduces neuroinflammation. Microglia were immunolabeled and quantitated by Iba-1 antibody at 7 days post-SCI (A,B). Similar numbers of Iba-1-positive cells were observed in RG3.6 cells (RG) and medium (Med)–treated spinal cords. Microglia in RG3.6-treated spinal cord displayed ramified morphology with thin processes and small cell body (left inset in A). In vehicle medium controls, microglia displayed reactive morphology with thickened processes and hypertrophied cell body (right inset in A). Iba-1 expression was quantitated in injured spinal cords that contained injury epicenter and its adjacent rostral and caudal segments. Four regions were quantitated in each cross section (illustrated in B). RG3.6-treated spinal cords showed significantly lower Iba-1 expression than medium controls (B). Macrophages and activated microglia were immunolabeled and quantitated by ED1 antibody at 14 days post-SCI (C,D). ED1 expression was quantitated in serial sections of injury epicenter (5 mm) and adjacent segments (rostral and caudal 5 mm). RG3.6 transplants significantly reduced ED1 expression at the injury center (D). Scale bar = 20 μm (A), 200 μm (C). Data represent mean ± SEM. *p < 0.05 (B), *p < 0.01 (D), t-test.
ED1 (CD68) immunostaining was used to examine expression of activated microglia and infiltrating macrophages 14 days after SCI. ED1-positive cells exhibit phagocytic morphology and were quantitated using tiled images covering the entire cross-section of the spinal cord taken using a 10 ×objective lens on Zeiss LSM510 confocal microscope. In view of extensive distribution and infiltration of macrophages at this time post-SCI, a wide region was chosen for quantitation for a better understanding on transplants' effect on neuroinflammation; each cross-sectional image was a montage of 16 individual pictures at 512 × 512 pixel resolution. Six sections (nos. 2–7 of eight sections on the slide, as described above) from each segment (rostral, injury, and caudal segments) were chosen from each animal for quantitation. Images were inverted into black (ED1 positive) and white images in NIH Image J program for measuring positive pixels. Regions of meninges and roots were excluded for quantitation. Positive pixels in sections within the same segment (e.g., rostral segment, injury segment, or caudal segment) were summed to represent ED1 expression.
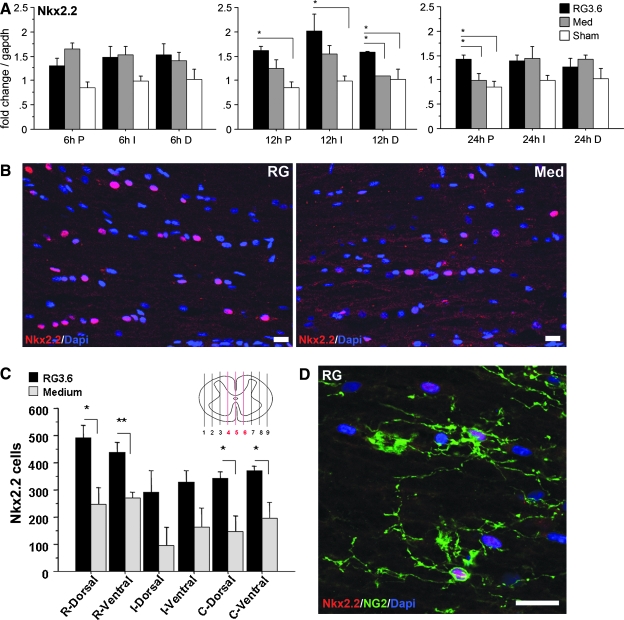
Quantitation of Nkx2.2 immunostaining was conducted on sagittal section at 1 day after SCI. Three para-sagittal sections close to the mid-line were used for quantitation (Fig. 7C) in each of three rats per treatment group. Each para-sagittal section was divided into a 5-mm injury (I) epicenter flanked by rostral (R) and caudal (C) 5-mm segments. Because most of the Nkx2.2-positive cells were found in the white matter (WM), each of the spinal segments were further divided into dorsal and ventral WM regions. Six images (x: 342.8 μm, y: 342.8 μm at 512 × 512 pixel resolution taken by 25 × 0.8 imm corr DIC objective on Zeiss LSM 510 confocal microscope) were chosen consecutively from each defined region on each section for Nkx2.2 counting. Antibodies are listed in Table 1. Differences between RG3.6- and medium-treated spinal cords were analyzed by Student's t-test.
FIG. 7.
Spatial and temporal expression of Nkx2.2 following SCI. Nkx2.2 was upregulated after SCI; the level was higher in RG3.6 than in control treated spinal cord at 12 h post-injury (A). Immunostaining of Nkx2.2 antibody at 24 h post-SCI showed significantly higher numbers of Nkx2.2-positive cells in RG3.6-treated spinal cords than with medium treatment (B). Nkx2.2-positive cells were quantitated in dorsal and ventral white matter in para-sagittal sections close to the midline (red lines 4, 5, 6 in the schematic spinal cord; C). Spinal cords with RG3.6 transplants showed significantly higher numbers of Nkx2.2 cells in rostral and caudal white matter than with medium treatment (C). In RG3.6-treated spinal cords, 54% of Nkx2.2 cells were NG2 positive (Cy5 shown in green) (D). Expression levels in A represent fold change after normalizing to GAPDH. Values are means ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ANOVA. Values in C are means ± SEM, *p < 0.05, **p < 0.01, t-test (C). Scale bar = 20 μm (B,D).
Total cellular RNA preparation and quantitative reverse transcription polymerase chain reaction
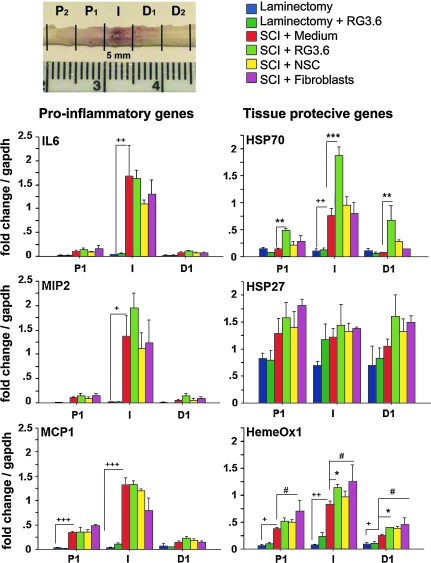
To isolate RNAs, animals were euthanized with pentobarbital at designated times (6 h, 12 h, and 24 h post-SCI). Spinal columns were quickly removed and frozen on dry ice powder. A 5-mm spinal cord segment centered at injury epicenter was collected and labeled “I” for injury site. The adjacent 5-mm segments were collected and labeled “P1” for proximal site (rostral to epicenter) and “D1” for distal site (caudal to epicenter; Fig. 2). Spinal segments were homogenized on ice-cold Trizol, chloroform was added, and the aqueous phase was collected after centrifugation and used to prepare RNA following the Qiagen RNeasy Mini protocol (Qiagen, Valencia, CA). RNA quality was confirmed using Agilent Bioanalyzer (Agilent, Palo Alto, CA), and all samples demonstrated sharp ribosomal RNA bands. 1 μg of total RNA was used for first-strand cDNA with SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) primed by Oligo dT. The polymerase chain reaction (PCR) was performed on 10 ng of cDNA using 50 nM of primers and SYBR Green master mix (Applied Biosystems, Foster City, CA) in 10-μl reactions using Applied Biosystems 7900HT machine. The PCR products were quantified based on standard curves that were developed from pooled cDNA samples (the primers used for each molecule are listed in Table 2). The expression value of each gene was normalized to the amount of GAPDH cDNA to calculate a relative amount of RNA present in each sample. One-way analysis of variance (ANOVA) followed by Fisher's protected least significant difference (PLSD) was used to determine statistical differences.
FIG. 2.
Analysis of gene expression changes using Q-RT-PCR at 12 h post SCI. Spinal cord segments, including 5-mm injury epicenter (I) and its adjacent (P1 and D1) 5-mm segments were subjected to Q-RT-PCR analysis. Pro-inflammatory mediators IL-6 and MIP-2 were significantly increased in the I segments, while MCP-1 was increased in both P1 and I segments by comparison to non-injured controls. RG3.6 transplants, like other treatment groups here, did not reduce the level of these pro-inflammatory genes. Tissue protective genes Hsp70, Hsp27, and Hsp32 (HemeOx1) were also significantly increased in the spinal cord after injury by comparison to non-injured controls. In RG3.6 treated group, the increase of Hsp70 was further enhanced in I and in P1 and D1 as compared to other treatments after SCI. Note that in uninjured spinal cord, RG3.6 did not increase Hsp70. Fibroblasts and RG3.6-treated spinal cords showed small but statistically increased up-regulation of HemeOx1 in comparison with medium treated controls. The increased level of Hsp27 after injury is similar in all treatments tested here. Scale represents fold change after normalizing expression to GAPDH. Values are means ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, +p < 0.05, ++p < 0.01, +++p < 0.001, #p < 0.05. *RG3.6 transplant versus medium; +, medium versus sham; #, fibroblast transplants versus medium, ANOVA.
Table 2.
Primers for Q-RT-PCR
| Gene ID | Forward | Reverse | |
|---|---|---|---|
| NM_017008 | GAPDH | 5′AAATGATACCCCACCGTGTGA3′ | 5′GCTGGCACTGCACAAGAAGAT3′ |
| U55762 | EGFP | 5′CTGAGCAAAGACCCCAACGA3′ | 5′GAACTCCAGGACCATGTG3′ |
| NM_012589 | IL-6 | 5′ATTCTGTCTCGAGCCCACCA3′ | 5′CTGAAGGGCAGATGGAGTTGA3′ |
| U45965 | MIP-2 | 5′ACCTCAACGGGCAGAATCAA3′ | 5′GCTTCCTGGGTGCAGTTTGT3′ |
| M57441 | MCP-1 | 5′CTTCCTCCACCACTATGCAGG3′ | 5′TGAACAACAGGCCCAGAAGC3′ |
| NM_031971 | Hsp70 | 5′TTCAATATGAAGAGCGCCGTG3′ | 5′GCTGATCTTGCCCTTGAGACC3′ |
| NM_031970 | Hsp27 | 5′TGCCCAAAGCAGTCACACAA3′ | 5′CGAAAGTGACCGGAATGGTG3′ |
| NM_012580 | HemeOx1 | 5′CCATCCCTTACACACCAGCC3′ | 5′CCTCGTGGAGACGCTTTACG3′ |
| BM_385445 | Top2a | 5′GGGAGGCAGGACTGTTGACA3′ | 5′ACGGATCACACTTCCTGGCT3′ |
| NM_012560 | Foxg1 | 5′CGTTCAGCTACAACGCGCT3′ | 5′TCTCGGGACTCTGCCTGATG3′ |
| BE_115061 | Lef1 | 5′GACGGATTGCCAAACGTGAC3′ | 5′AAACAGGTAGCCAAGCCCACT3′ |
| NM_031140 | Vimentin | 5′GAGCACCCTGCAGTCATTCA3′ | 5′CGTGCCAGAGAAGCATTGTC3′ |
| AA_925143 | Nkx2.2 | 5′GGCGACACAGGCCCATC3′ | 5′TCGCTAGTGATCATCGTTGCC3′ |
| BE_107070 | Sox11 | 5′GCTGATGTCTTCTATGCATCCG3′ | 5′TTTTTCAAGCTCCCTGCAGTTTA3′ |
| NM_019139 | GDNF | 5′GGTCACCAGATAAACAAGCGG3′ | 5′GCCGGTTCCTCTCTCTTCG3′ |
| NM_012513 | BDNF | 5′AGGCACTGGAACTCGCAATG3′ | 5′AAGGGCCCGAACATACGATT3′ |
| NM_031073 | NT3 | 5′GATATTTTGGCCGGAGGGAA3′ | 5′CCTCAAAAGGGCTGGGTTCT3′ |
| NM_031523 | rNGF | 5′TGCTCCTGCATGCCTGTTAC3′ | 5′CAGGGCGAGGAACAGGATC3′ |
Microarray processing and data analysis
100 ng of total RNA from each sample was used as input material for the 3′-initiated Ovation RNA amplification System V2 (NuGEN, San Carlos, CA). Briefly, first and second strand cDNA synthesis were performed according to the recommended protocol. Amplified cDNA was produced during the Ribo-SPIA amplification and purified. 3.75 μg of amplified cDNA was used as input for the FL-Ovation cDNA Biotinylation kit (NuGEN). cDNAs were chemically and enzymatically fragmented to 50–100-bp fragments. Fragments were subsequently 3′-labeled via addition of a biotinylated nucleotide to the free hydroxyl group generated during fragmentation. Labeled cDNAs were applied to Affymetrix Rat 230.2 microarrays and hybridized according to manufacturer's recommendations.
Data from 6 Affymetrix Rat 230.2 chips (3 for RG, 3 for Med) were imported into R (www.r-project.org) and preprocessed using several packages from Bioconductor (www.bioconductor.org), an open-source bioinformatics package. Briefly, data were background corrected and normalized using the Robust Multi-Array Average (RMA) technique. A Student's t-test was conducted, assuming equal variances, to select differentially expressed genes between the two groups. The p-values were adjusted for multiple comparisons using the Benjamin-Hochberg method. The resulting list contained 845 genes at a p-value false discovery rate (FDR) cutoff of ≤ 5%. Among the 845 genes, 60 genes showed fold changes more than 1.5-fold and were exported to Ingenuity Pathway Analysis (www.ingenuity.com) for classification/functional and pathway analysis.
Results
Transplantation of RG3.6 cells inhibited accumulation of immune cells in SCI
We reported previously that acute transplantation of RG3.6 cells reduced infiltration of macrophages at 6 weeks following SCI (Hasegawa et al., 2005). In the present study, we found that, at 1 week after SCI, activation of local microglia immunolabeled by Iba-1 antibody was decreased by acute RG3.6 transplantation (Fig. 1A,B). There did not appear to be a difference in the number of Iba-1- positive cells, but microglial cells exhibited hypertrophied morphology and stronger Iba-1 staining in medium controls than with RG3.6 treatment. Quantitation of super-threshold regions of staining in the injury as well as in rostral and caudal regions suggested that RG3.6 treatment suppresses activation of microglia significantly at least in the ventrolateral WM region (Fig. 1B). By 2 weeks after spinal cord contusion when macrophage infiltration is extensive (Popovich et al., 1997), ED1-positive macrophages were abundant in the injury site (I) of medium controls, while animals that received RG3.6 transplants exhibited significantly fewer ED1 cells in comparable regions of injured spinal cords (Fig. 1C). The ED1 staining corresponded mostly to macrophages, which appear as phagocytic round-shaped cells. Interestingly, there was abundant ED1 staining in the ventral roots, which are known routes for macrophage infiltration into the central nervous system (CNS) (Popovich et al., 1997), and this was less prominent in the RG3.6 transplanted spinal cords (Fig. 1C). Quantitation of superthreshold regions of ED1 staining indicated that averages were significantly lower in the injured spinal cords with RG3.6 transplants (Fig. 1D). The combined results suggest that RG3.6 treatment inhibits activation of microglia and infiltration of macrophages in the first 2 weeks following SCI.
RG3.6 cells enhanced expression of genes associated with tissue protection but not pro-inflammatory genes
SCI-induced gene expression changes revealed that pro-inflammatory mediators are up-regulated maximally at 12–24 h post-injury before they gradually return to the baseline levels found in sham animals (Carmel et al., 2001; Bareyre et al., 2003; Aimone et al., 2004). Given that we found reduced activation of microglia and macrophages by RG3.6 treatment (Fig. 1), we performed quantitative reverse transcription polymerase chain reaction (Q-RT-PCR) analysis to examine whether the levels of pro-inflammatory genes were modulated by RG3.6 transplantation in SCI. Pro-inflammatory molecules interleukin 6 (IL-6), macrophages inflammatory protein 2 (MIP-2), and monocyte chemoattractant protein 1 (MCP-1) are important for microglial proliferation and activation (Lacroix et al., 2002; Babcock et al., 2003; Krady et al., 2008) and are highly up-regulated acutely following SCI (Carmel et al., 2001; Jones et al., 2005). Our data confirmed significant activation of these pro-inflammatory molecules in all treatment groups following SCI by comparison to the non-injured controls (Carmel et al., 2001; Aimone et al., 2004) (Fig. 2). However, neither RG3.6 nor other cell transplants tested lowered the induction of IL-6, MIP-2, and MCP-1 mRNAs by SCI (Fig. 2). The data suggest that these molecules may not be involved in the early response to RG3.6 treatment.
Several other genes that are associated with tissue protection, including heat shock proteins (Hsp), are also up-regulated following SCI (Carmel et al., 2001; Bareyre et al., 2003) and were examined by Q-RT-PCR at 12 h post-injury (Fig. 2). The induction of Hsp70 was significantly enhanced in the injury center (I) as well as in adjacent spinal segments (P1 and D1) of animals that received RG3.6 transplants as compared to other treatment controls. The up-regulation of Hsp70, however, was not found in non-injured spinal cords that received RG3.6 cells, suggesting this response is injury-dependent (Fig. 2C). Although primary NSC share many similar properties with radial glial cells, they did not further increase Hsp70 in the injured spinal cords by comparison with medium controls (Fig. 2). This suggests that radial glial clone RG3.6 may have unique properties in modulating the injury environment. We also found increased levels of Hsp32 (also known as heme oxygenase 1: HemeOx1) in both fibroblast and RG3.6-treated spinal cords (Fig. 2C). However, the increase was not as prominent as Hsp70 in RG3.6-treated injured spinal cords. There was no significant change in Hsp27 in all treatments for injured spinal cords in comparison to medium controls at 12 h post-injury.
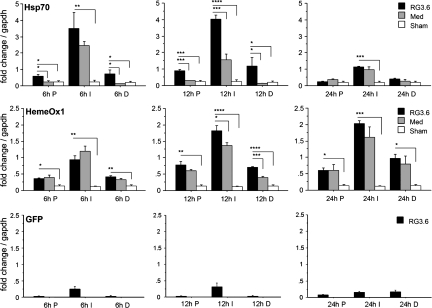
To analyze the timing of these responses by RG3.6 transplants after injury, Hsp70 and HemeOx1 expression was measured at 6, 12, and 24 h post-injury using Q-RT-PCR. Hsp70 peaked at 12 h and declined at 24 h post-injury (Fig. 3). RG3.6 treatment induced statistically higher levels of Hsp70 in the injury center and adjacent segments at 6 and 12 h than in medium controls. HemeOx1 expression appeared to plateau by 24 h post-injury and only showed a significant increase in RG3.6 over medium-treated spinal cords at 12 h (Fig. 3).
FIG. 3.
Spatial and temporal expression of Hsp70, HemeOx1, and GFP following SCI using Q-RT-PCR. Hsp70 expression was measured in the P1, I, and D1 segments at 6, 12, and 24 h post-injury. RG3.6-treated spinal cord showed significant up-regulation of Hsp70 as early as 6 h following SCI and the expression declined at 24 h after injury. HemeOx1 was increased in I and D1 segments of RG3.6-treated spinal cord compared to medium controls at 12 h. The distribution of RG3.6 cells was measured by GFP expression. GFP was detected in I segments, while very limited signal was found in P1 or D1 segments at 6–12 h following SCI. By 24 h post-injury, GFP signals were distributed to P1 and D1 segments. Scale represents fold change after normalizing expression to GAPDH. Values are means ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ANOVA.
Interpretation of the responses in different tissue regions may be complicated by direct contributions of the RG3.6 cells that were transplanted into the injury site immediately following contusion. We therefore evaluated spatial and temporal distributions of the RG3.6 cells since they express GFP (Hasegawa et al., 2005). The results indicate that, at 6–12 h, GFP signals are almost exclusively restricted to the injury site where the cells were injected, but GFP signals were only found at significant levels outside the injury site at 24 h (Fig. 3), suggesting that RG3.6 cells remained in the injury center and did not migrate to P1 or D1 regions until ∼24 h post-injury. Therefore, mRNA measures in the injury center (I) represent changes contributed from both host and implanted RG3.6 cells at 6–12 h post-injury, whereas measures in adjacent (P1 and D1) regions are likely to represent only contributions of the host tissue in response to injury, which may be modulated by signals that the RG3.6 transplants generated.
RG3.6 cells enhanced expression of developmentally regulated genes
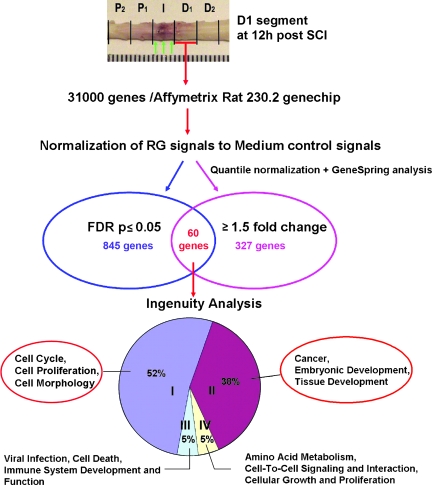
To identify more gene expression changes that are associated with host response to RG3.6 transplants, we subjected D1 segments at 12 h post-SCI to microarray analysis (Fig. 4). This region and time were chosen to limit direct contributions by the RG3.6 cells and because this segment exhibited the highest fold increase for Hsp70 (Fig. 2C). Host response to RG3.6 was obtained by normalizing expression of RG3.6-treated samples over that of medium controls. We did not use other cells as “controls” since none of the candidates are ideal insofar as each may modulate changes in expression that may obscure detecting changes related to RG3.6 cells. A total of 845 genes were obtained after t-test with multiple corrections (FDR p-cut-off of ≤ 0.05), of which 60 genes showed more than 1.5-fold changes in RG3.6-treated samples over medium controls (Fig. 4, Table 3). Functions and classifications of these 60 genes were analyzed using Ingenuity Pathway Analysis (www.ingenuity.com) to search for functionally related genes that may reveal which cellular processes are most responsive to RG3.6 transplants in SCI.
FIG. 4.
RNAs from distal spinal cord segments at 12 h post-injury were subjected to Affymetrix Rat 230.2 genechips. After data normalization, signals from RG3.6-treated samples were compared to medium controls for differential expression. A total of 845 genes reached statistical significance after unpaired t-test with multiple corrections and 60 of them showed ≥ 1.5-fold changes (listed in Table 3). After Ingenuity pathway analysis, four biological networks (I, II, III, IV) were identified with two highlighted, i.e., genes involved in cell proliferation (I) and tissue development (II).
Table 3.
Gene List after FDR p-cut-off ≤ 0.05 and Differential Expression ≥ 1.5 Fold Change
| Genbank ID | RG/Med ratio | Common | Description | *Network |
|---|---|---|---|---|
| NM_012560 | 15.50 | BF1A; Foxo1; | forkhead box G1(Foxg1) | I |
| BM385445 | 3.43 | Top2a | Topoisomerase (DNA) 2 alpha | II |
| BF284168 | 3.09 | Transcribed locus | ||
| BE107070 | 3.03 | Sox11 | SRY-box containing gene 11 | II |
| AA944326 | 2.88 | EST199825 Normalized rat embryo, Bento Soares Rattus sp. cDNA clone REMAF43 3' end, mRNA sequence. | ||
| AI012949 | 2.84 | Transcribed locus, moderately similar to XP_489350.1 RIKEN cDNA 2700063P19 [Mus musculus] | ||
| AI172110 | 2.82 | EST218105 Normalized rat muscle, Bento Soares Rattus sp. cDNA clone RMUBV06 3' end, mRNA sequence. | ||
| AW524041 | 2.33 | Zbtb9 | Zinc finger and BTB domain containing 9 | |
| BF554576 | 2.27 | Sox11 | SRY-box containing gene 11 | II |
| BF408872 | 2.25 | Imp1 | Insulin-like growth factor 2, binding protein 1 | II |
| BE107098 | 2.19 | Transcribed locus | ||
| BE113173 | 2.16 | Tubb2b | microtubule-based movement protein polymerization Tubulin, beta 2b | II |
| BM385870 | 2.13 | Zbtb 10 | Zinc finger and BTB domain containing 10 | III |
| AW144239 | 2.09 | Fstl3 | Follistatin-like 3 | I |
| BI285065 | 2.07 | Tgfb1i4 | Transforming growth factor beta 1 induced transcript 4 | II |
| AA956727 | 1.99 | Splicing factor, arginine/serine-rich 3 (SRp20) (predicted) | I | |
| AI180454 | 1.94 | Similar to IGF-II mRNA-binding protein 2 (predicted) | II | |
| M24024 | 1.91 | RT1Aw2 | RT1 class Ib, locus Aw2 (RT1-Aw2) | I |
| BE113443 | 1.89 | Kinesin family member 23 (predicted) | II | |
| AF030088 | 1.85 | Vesl-1; HOMER1F | homer homolog 1 (Drosophila) (Homer1) | I |
| BF398677 | 1.84 | Arhgef12 | Rho guanine nucleotide exchange factor (GEF) 12 | I |
| BF409715 | 1.81 | Similar to 106 kDa O-GlcNAc transferase-interacting protein (predicted) | ||
| AA800639 | 1.80 | Transcribed locus | ||
| AI058451 | 1.80 | Similar to IGF-II mRNA-binding protein 2 (predicted) | II | |
| BI289386 | 1.78 | Similar to BCoR protein (BCL-6 corepressor) | ||
| AI598485 | 1.78 | Transcribed locus | ||
| AI712582 | 1.77 | Glutamine and serine rich 1 (predicted) | ||
| BE119432 | 1.77 | Transcribed locus | ||
| AA956336 | 1.73 | Anaphase promoting complex subunit 10 (predicted) | ||
| AI072161 | 1.73 | UI-R-C2-mz-f-11-0-UI.s1 UI-R-C2 Rattus norvegicus cDNA clone UI-R-C2-mz-f-11-0-UI 3', mRNA sequence. | ||
| U06434 | 1.71 | Scya4; Mip1-b | chemokine (C-C motif ) ligand 4 (Ccl4) | I |
| BE114260 | 1.70 | Transcribed locus | ||
| BF408816 | 1.68 | UI-R-BT1-bne-g-03-0-UI.s1 UI-R-BT1 Rattus norvegicus cDNA clone UI-R-BT1-bne-g-03-0-UI 3', mRNA sequence. | ||
| BI276223 | 1.64 | Similar to mKIAA0215 protein | ||
| BG374178 | 1.64 | Kitl | Kit ligand | I |
| AI639117 | 1.64 | cBf | complement factor B | II |
| BF419397 | 1.63 | Transcribed locus | ||
| BI282932 | 1.62 | C1qr1 | Complement component 1, q subcomponent, receptor 1; Lymphocyte antigen 68 | IV |
| BI275740 | 1.62 | Similar to triggering receptor expressed on myeloid cells-like 1 | ||
| AI044343 | 1.62 | Similar to cofactor required for Sp1 transcriptional activation Subunit 2, 150kDa(predicted) | ||
| AA963477 | 1.60 | TSP-2 | Thrombospondin-2 | I |
| BE107208 | 1.60 | UI-R-BS1-ayr-b-04-0-UI.s1 UI-R-BS1 Rattus norvegicus cDNA clone UI-R-BS1-ayr-b-04-0-UI 3', mRNA sequence. | ||
| BF404982 | 1.59 | Pnpt1 | Polyribonucleotide nucleotidyltransferase 1 | |
| AI172172 | 1.59 | Aprin | Androgen-induced proliferation inhibitor (predicted) | |
| BF416058 | 1.58 | UI-R-CA0-bkh-f-11-0-UI.s1 UI-R-CA0 Rattus norvegicus cDNA clone UI-R-CA0-bkh-f-11-0-UI 3', mRNA sequence. | ||
| AA892240 | 1.58 | Hypothetical LOC361786 | ||
| AI169080 | 1.57 | Transcribed locus | ||
| AA858605 | 1.56 | Similar to Wolf-Hirschhorn syndrome candidate 1 protein isoform 1 | ||
| AW434912 | 1.54 | Transcribed locus | ||
| AA893192 | 1.54 | EST196995 Normalized rat kidney, Bento Soares Rattus sp. cDNA clone RKIBD36 3' end, mRNA sequence. | ||
| BI274401 | 1.54 | P4ha1 | Procollagen-proline, 2-oxoglutarate 4-dioxygenase (proline 4-hydroxylase), alpha 1 polypeptide | I |
| BF564309 | 1.53 | UI-R-C4-aln-a-11-0-UI.r1 UI-R-C4 Rattus norvegicus cDNA clone UI-R-C4-aln-a-11-0-UI 5', mRNA sequence. | ||
| AI175791 | 1.52 | Transcribed locus | ||
| BE117291 | 1.52 | Similar to chromosome X open reading frame 23 | ||
| AI072424 | 1.51 | Synaptotagmin binding, cytoplasmic RNA interacting protein | ||
| AI010254 | 1.51 | Cachd_1 | Cache domain containing 1 (predicted) | |
| BF546659 | 1.50 | Syncrip | Synaptotagmin binding, cytoplasmic RNA interacting protein | |
| BE115061 | 1.50 | Lef1 | Lymphoid enhancer binding factor 1 | II |
| AI111816 | 0.66 | Transcribed locus | ||
| BF556812 | 0.31 | Transcribed locus |
Network I: Cell cycle, Cell proliferation, morphology. II: Cancer, Embryo and Tissue development. III: Cell death, Immune response. IV: Cell-cell signal, Cell growth and Proliferation.
Among these 60 genes, 40% (23 genes) were classified into functional networks (I, II, III, and IV in Fig. 4), 40% (25 genes) did not fall into any network, and 20% (12 genes) were identified as a transcribed locus (Table 3). Of the genes that clustered into networks, 90% clustered into two groups: network I (52%, 12 genes) defined as cell cycle, proliferation, and morphology; and network II (38%, 9 genes) defined as tissue development. These results suggest that a discrete set of developmentally regulated genes was selectively enhanced in the spinal cord when RG3.6 cells were transplanted in the injury epicenter. To validate these observations, several genes of interest were selected and confirmed by Q-RT-PCR.
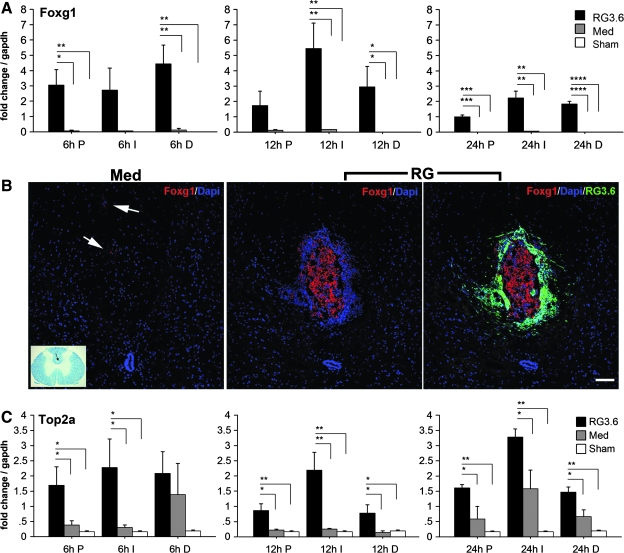
Foxg1, a forkhead-box transcription factor essential for neural progenitor proliferation and differentiation during development (Xuan et al., 1995; Hanashima et al., 2002; Regad et al., 2007), was the candidate most up-regulated in the microarray data (Tables 3 and 4). We then examined spatial and temporal expression pattern of Foxg1 by Q-RT-PCR in P1, I, and D1 spinal cord segments at 6, 12, and 24 h post-injury (Fig. 5A). As early as 6 h post-injury, Foxg1 was specifically increased in RG3.6-treated spinal cord and there were no significant signals for Foxg1 in the control spinal cords even after injury, suggesting that it is not expressed in the adult spinal cord and not induced in response to contusion injury. Foxg1 displayed intracellular expression by immunofluorescence in the dorsal column white matter in RG3.6-treated spinal cords, while very little or no expression was found in medium controls (Fig. 5B). Interestingly, the expression of Foxg1 was not localized in RG3.6 cells; rather it was restricted to a subset of the host spinal cord cells adjacent to the transplants (Fig. 5B), suggesting that injured cells in the spinal cord respond to the neighboring RG3.6 transplants.
Table 4.
Genes of Interest and their Expression at D1 Segment at 12 h after SCI: Analysis by Q-RT-PCR and Genechip
| GenBank ID | Common name | Q-PCR RG/Med ratio | Q-PCR, ANOVA p<0.05 | Genechip RG/Med ratio | Genechip t-test FDR5% |
|---|---|---|---|---|---|
| NM_012560 | foxg1 | 146 | + | 15.5 | + |
| BM385445 | top2a | 5.4 | + | 4.6 | + |
| BE107070 | sox11 | 5.2 | + | 3 | + |
| NM_031971 | hspa1a (Hsp70) | 2.8 | + | 3.5 | − |
| NM_012580 | hmox1 | 1.8 | + | 1.4 | − |
| NM_031140 | vim | 1.7 | + | 1.3 | − |
| BE115061 | lef1 | 1.54 | + | 1.5 | + |
| AA925143 | nkx2.2 | 1.45 | + | 1.3 | − |
+: p ≤ 0.05, −: p > 0.05.
FIG. 5.
Spatial and temporal expression of Foxg1 and Top2a following SCI by Q-RTPCR. Transcription factor Foxg1 showed specific and significant up-regulation in RG3.6-treated spinal cord; the expression lasted up to 24 h post-injury (A). Foxg1 immunostaining at 24 h SCI showed strong expression at the dorsal white matter (∼2.5 mm distal to injury epicenter) and did not overlap with RG3.6 cells (B) shown in green. Medium-treated spinal cords showed very weak Foxg1 expression (arrows in B). DNA replication enzyme Top2a was also up-regulated significantly in RG3.6-treated spinal cords by 6 h post-injury by comparison to medium controls in which the expression was not significantly induced until 24 h post-injury (C). Expression levels represent fold change after normalizing to GAPDH. Values are means ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ANOVA. Scale bar = 100 μm (B); Luxol fast blue stained cross-section illustrates Foxg1 staining region.
Topoisomerase 2a (Top2a), an essential enzyme for DNA replication during cell proliferation (Wang 1996), was another notably increased gene in the injured spinal cord after RG3.6 transplantation in the array result (Tables 3 and 4). Q-RT-PCR confirmed that Top2a, like Foxg1, was quickly and highly induced in RG3.6-treated spinal cord in the time frame we studied (6–24 h post-injury; Fig. 5C). Levels of Top2a in the medium controls, however, did not increase significantly over non-injured controls until 24 h after injury (Fig. 5C).
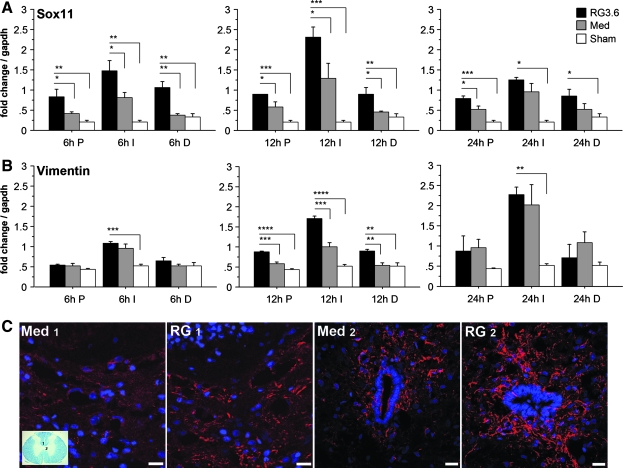
SRY-box containing gene 11 (Sox11), a HMG-box containing transcription factor expressed during CNS development and involved in cell differentiation (Uwanogho et al., 1995; Kuhlbrodt et al., 1998), was significantly increased in RG3.6-treated spinal cord following injury in our microarray analysis (Tables 3 and 4). Q-RT-PCR measurements of Sox11 also indicated rapid increases in response to SCI, and larger increases were found when RG3.6 cells were transplanted (Fig. 6A).
FIG. 6.
Spatial and temporal expression of Sox11 and Vimentin following SCI by QRT-PCR. Sox11 was significantly up-regulated in most regions (P, I, and D) in RG3.6-treated spinal cords by comparison to medium controls at 6, 12, and 24 h post-injury (A). Vimentin expression also showed significant increases in spinal cords with RG3.6 transplants at 12 h post-injury (B). Immunostaining for vimentin at 24 h post-injury showed stronger expression in RG3.6- treated spinal cords than in medium controls (C). Images presented here are from regions below dorsal column (region 1 illustrated in luxol fast blue stained section) and around central canal (region 2 illustrated in luxol fast blue stained section) from spinal cord segment 2.5 mm caudal to the injury epicenter. Scale bar = 20 μm. Expression levels in A and B represent fold change after normalizing to GAPDH. Values are means ± SEM, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ANOVA.
Confirmation by Q-RT-PCR of the three highly induced genes (Foxg1, Top2a, Sox11) by RG3.6 transplants from the networks provided support for the accuracy of the microarray analyses. It is interesting that the selectively induced genes in these two networks (networks I and II; Fig. 4) can broadly be defined as developmentally regulated genes that have been re-induced in the adult CNS. Several lines of evidence indicate that injury activates endogenous NSC in the adult spinal cord (Zai et al., 2005; Horky et al., 2006). To test the emerging hypothesis that RG3.6 transplants activate NSC in the injured spinal cord, we examined other genes that are known to be expressed in NSC and progenitor cells, which did not survive the selection process (Fig. 4) that yielded the genes listed in Table 3. For example, vimentin has been widely regarded as a developmentally regulated marker for NSC and radial glia (Alvarez-Buylla et al., 1987; Noctor et al., 2002), and its expression was weakly up-regulated on the microarray (Table 4). Q-RT-PCR of vimentin showed a statistically significant but moderate increase in RG3.6 over medium-treated spinal cords at 12 h post-injury (Fig. 6B). This transient induction with the RG3.6 transplants was not apparent at 24 h when vimentin signals were higher in the injury site with or without RG3.6 cells by comparison to uninjured controls. There was more vimentin immunoactivity in the ventromedial white matter and gray matter surrounding the central canal (i.e., ependymal region) and in dorsal columns of RG3.6-treated spinal cord than in comparable regions of medium controls at 24 h post-injury (Fig. 6C).
Another developmentally regulated gene identified on the microarrays is the homeodomain transcription factor Nkx2.2, which has multiple biological roles in CNS development and injury (Talbott et al., 2005; Ohori et al., 2006). Q-RT-PCR analysis indicated up-regulation of Nkx2.2 following SCI in medium controls in several regions tested, and at 12 h, the signals in RG3.6-treated spinal cord were at significantly higher levels than medium controls (Fig. 7). Consistent with other work (Talbott et al., 2005), Nkx2.2- positive cells were found primarily in WM, and we confirmed up-regulation of Nkx2.2 following SCI. In several regions of the spinal cord, the number of Nkx2.2-positive cells was higher in RG3.6 transplanted spinal cords than in medium controls at 24 h post-injury (Fig. 7C). The nuclear localization of Nkx2.2 facilitated quantitation of the density of Nkx2.2 + nuclei, which was significantly higher in rostral and caudal regions in RG3.6 transplanted spinal cords by comparison to medium controls (Fig. 7B,C). However, the injury site had fewer cells in general, and the differences measured were not significant. Interestingly, we found that 54% (n = 204) of the Nkx2.2-positive cells were also positive for NG2 (Fig. 7D). NG2 chondroitin sulfate proteoglycan has been recognized as a marker for progenitors in CNS, and NG2-positive cells were reported to proliferate actively after SCI (McTigue et al., 2001; Zai et al., 2005; Horky et al., 2006). Our results suggest that implantation of RG3.6 cells may stimulate neural precursor proliferation and/or differentiation as early as 24 h following SCI. Moreover, the significant stimulation in the number of Nkx2.2 + cells in SCI by RG3.6 cells provides support for the idea that the transplants can rapidly induce signaling to NSC in the host spinal cord.
Discussion
Previously, we reported that acute transplantation of radial glia RG3.6 cells promoted functional improvement possibly through early tissue protection (Hasegawa et al., 2005). The current work provides evidence that transplantation of RG3.6 cells changes gene expression profiles in host tissue early after SCI, which might be one of the underlying mechanisms for functional recovery. The response of the injured spinal cord to RG3.6 transplants was analyzed using microarrays and was characterized by up-regulation of two groups of genes involved in (1) cell cycle, proliferation, and morphology, and (2) cancer and development. Although reduced microglia and macrophages were observed as early as 1 week after RG3.6 transplantation in SCI, we did not find that this treatment suppressed expression of pro-inflammatory genes during the early time window (6–24 h after SCI). It remains controversial whether pro-inflammatory mediators are beneficial or detrimental to the tissue during the acute phase of SCI since pro-inflammatory cytokines/chemokines are essential for recruiting immune cells to clear up debris of dead cells (Schwartz, 2003). The lack of acute suppression of inflammatory mediators at the level of gene expression suggests that RG3.6 transplants may have an indirect role in regulating inflammatory pathways that lead to changes in microglia/macrophages responses 1–2 weeks post-SCI. Rather, the evidence suggests that genes involved in tissue protection and promotion of proliferation and/or differentiation of neural progenitor cells in the host tissue are modulated acutely in RG3.6 treated SCI.
Heat shock proteins are a family of conserved proteins induced in response to stress (e.g., heat, toxins, ischemia, and inflammation) that protect injured cells by refolding damaged proteins and preventing apoptosis. In ischemia/reperfusion models, brief induction of HSPs significantly protects cells from subsequent insults and increases the tolerance to stress (Yenari, 2002). Several HSPs are up-regulated after SCI, including Hsp70, Hsp27, and Hsp32 (HemeOx1) (Sharp et al., 1999; Mautes et al., 2000; Carmel et al., 2001). Here we found that acute transplantation of RG3.6 cells significantly increased the level of Hsp70 not only in the injury site where the cells were injected but also in adjacent spinal cord segments. Given that the RG3.6 cells are restricted to the injury center segment within 6–12 h post-injury, the Hsp70 up-regulation in this region may be a combination of host tissue and RG3.6 transplant responses. However, signals in the P1 and D1 segments should only represent responses by host tissue itself. Like sham animals, normal spinal cords that received RG3.6 transplants did not show induction of Hsp70, suggesting that RG3.6 cells induce certain signals that modulate gene expression in host tissue only after injury. In preliminary experiments, Hsp70 protein expression was examined using immunohistochemistry and immunoblotting; however, we did not detect significant changes between RG3.6 and medium-treated spinal cords (data not shown). Further investigation is needed to determine whether Hsp70 mRNA was translated into functional protein and whether the insignificance of Hsp70 expression at protein level was due to low protein concentration or fast degradation in the injured spinal cord.
To understand host response to injury and RG3.6 transplants, we studied gene expression at D1 segments using microarrays at 12 h post-injury. This region was selected in part because it avoids the injury site where the changes are complicated by the injury and the transplanted RG3.6 cells. Thus, the D1 segment, which contained no direct contribution from RG3.6 cells at 12 h post-injury, should only reflect responses associated with secondary injury that may be altered indirectly by the transplants that were restricted to the injury center. After statistical and functional analysis, developmentally associated genes were found to be specifically up-regulated by RG3.6 transplantation after SCI. Foxg1 was induced most dramatically by RG3.6 transplants. This is particularly interesting since Foxg1 is not normally expressed in the spinal cord. Foxg1 (also known as brain factor-1 [BF1]) is expressed in neuroepithelial cells and is important in neurogenesis in the developing telencephalon (Tao et al., 1992; Xuan et al., 1995). Nuclear Foxg1 acts as a transcriptional repressor to prevent progenitor cells from differentiating and it maintains them in a proliferating state (Dou et al., 1999; Hanashima et al., 2002). Cytoplasmic and axonal Foxg1 has been identified recently in differentiating neurons in which FGF-2 was involved in translocating Foxg1 from nucleus to cytoplasm (Regad et al., 2007). Here we found specific induction of Foxg1 in the injured spinal cord that received RG3.6 transplants, and Foxg1 appeared to be located intracellularly in the descending axons of the dorsal column. Since Foxg1 was not co-localized with RG3.6 cells, we postulate that a developmental program was reactivated perhaps in axons descending from the brain by brain-derived RG3.6 transplants in the host injured spinal cord. Alternatively, these Foxg1 positive cells may represent endogenous progenitor cells responding to a combination of injury and RG3.6 transplants. It is of great interest to investigate what factors/signals from RG3.6 cells are required to activate development machinery in the host tissue after SCI.
RG3.6 transplants induced several other developmentally regulated genes in the host tissue, including Top2a, Sox11, and Nkx2.2 (Tables 3 and 4). The increase of Top2a suggests that RG3.6 treatment rapidly activated DNA synthesis and/or cellular division in the injured spinal cord (Kuan et al., 2004). Transcription factors Sox11 and Nkx2.2 are both important for glial-restricted precursors and/or oligodendrocytes differentiation (Kuhlbrodt et al., 1998; Cheung et al., 2000; Marquardt et al., 2001; Han et al., 2004; Talbott et al., 2005). Moreover, Sox11 has been classified as one of the regeneration-associated genes (Tanabe et al., 2003) and is crucial for neuronal survival and neurite growth (Jankowski et al., 2006). Nkx2.2 promotes proliferation of glial-restricted precursors (Han et al., 2004; Talbott et al., 2005) and differentiation of oligodendrocyte progenitors in the injured spinal cord (Ohori et al., 2006). Thus, the up-regulation of these genes suggests that transplantation of RG3.6 promotes survival, proliferation, and differentiation of neural progenitors in the injured spinal cord.
It has been proposed that stem cells may have multiple applications in neurodegenerative disease and CNS trauma besides replacement of specific cell types (Goldman, 2005). It is believed that stem cells may help host tissue by modulating its immune response and secreting a wide range of trophic factors (Pluchino et al., 2005; Martino et al., 2006). Our previous study showed that acute transplantation of radial glia RG3.6 has a tissue-protective function and decreased macrophage accumulation at 6 weeks after SCI (Hasegawa et al., 2005). Although in the present work we did not find very early suppression of pro-inflammatory genes, evidence for reduced neuroinflammation was found at 1–2 weeks post-SCI. It is possible that RG3.6 cells promoted functional recovery by rapid activation of factors (e.g., Foxg1, Top2a, Sox11, Nkx2.2) in host cells and tissue protective mechanisms (e.g., Hsp70, HemeOx1). The reduced neuroinflammation during sub-acute phase of SCI therefore might be the result of indirect effects of RG3.6 transplants mediated through signaling to host neural progenitor cells to down-regulate the immune response.
Acknowledgments
We thank Dr. Bor Tom Ng, Hock Ng, Sean O'Leary, and Pui Tom for technical support, and Dr. Joel Levine and Dr. Lorenz Studer for antibodies. This work was supported by grants from NIH, the New Jersey Commission on Spinal Cord Research, and the New York State CORE grant C-019772. Y.W.C. was a graduate fellow of the New Jersey Commission on Spinal Cord Research and a Fred Ferarri scholar.
Author Disclosure Statement
No competing financial interests exist.
References
- Aimone J.B. Leasure J.L. Perreau V.M. Thallmair M. Spatial and temporal gene expression profiling of the contused rat spinal cord. Exp. Neurol. 2004;189:204–221. doi: 10.1016/j.expneurol.2004.05.042. [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla A. Buskirk D.R. Nottebohm F. Monoclonal antibody reveals radial glia in adult avian brain. J. Comp. Neurol. 1987;264:159–170. doi: 10.1002/cne.902640203. [DOI] [PubMed] [Google Scholar]
- Babcock A.A. Kuziel W.A. Rivest S. Owens T. Chemokine expression by glial cells directs leukocytes to sites of axonal injury in the CNS. J. Neurosci. 2003;23:7922–7930. doi: 10.1523/JNEUROSCI.23-21-07922.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre F.M. Haudenschild B. Schwab M.E. Long-lasting sprouting and gene expression changes induced by the monoclonal antibody IN-1 in the adult spinal cord. J. Neurosci. 2002;22:7097–7110. doi: 10.1523/JNEUROSCI.22-16-07097.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareyre F.M. Schwab M.E. Inflammation, degeneration and regeneration in the injured spinal cord: insights from DNA microarrays. Trends Neurosci. 2003;26:555–563. doi: 10.1016/j.tins.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Beattie M.S. Hermann G.E. Rogers R.C. Bresnahan J.C. Cell death in models of spinal cord injury. Prog. Brain Res. 2002;137:37–47. doi: 10.1016/s0079-6123(02)37006-7. [DOI] [PubMed] [Google Scholar]
- Bramlett H.M. Dietrich W.D. Progressive damage after brain and spinal cord injury: pathomechanisms and treatment strategies. Prog. Brain Res. 2007;161:125–141. doi: 10.1016/S0079-6123(06)61009-1. [DOI] [PubMed] [Google Scholar]
- Carmel J.B. Galante A. Soteropoulos P. Tolias P. Recce M. Young W. Hart R.P. Gene expression profiling of acute spinal cord injury reveals spreading inflammatory signals and neuron loss. Physiol. Genomics. 2001;7:201–213. doi: 10.1152/physiolgenomics.00074.2001. [DOI] [PubMed] [Google Scholar]
- Cheung M. Abu-Elmagd M. Clevers H. Scotting P.J. Roles of Sox4 in central nervous system development. Brain Res. Mol. Brain Res. 2000;79:180–191. doi: 10.1016/s0169-328x(00)00109-1. [DOI] [PubMed] [Google Scholar]
- De Biase A. Knoblach S.M. Di Giovanni S. Fan C. Molon A. Hoffman E.P. Faden A.I. Gene expression profiling of experimental traumatic spinal cord injury as a function of distance from impact site and injury severity. Physiol. Genomics. 2005;22:368–381. doi: 10.1152/physiolgenomics.00081.2005. [DOI] [PubMed] [Google Scholar]
- Di Giovanni S. Knoblach S.M. Brandoli C. Aden S.A. Hoffman E.P. Faden A.I. Gene profiling in spinal cord injury shows role of cell cycle in neuronal death. Ann. Neurol. 2003;53:454–468. doi: 10.1002/ana.10472. [DOI] [PubMed] [Google Scholar]
- Dou C.L. Li S. Lai E. Dual role of brain factor-1 in regulating growth and patterning of the cerebral hemispheres. Cereb. Cortex. 1999;9:543–550. doi: 10.1093/cercor/9.6.543. [DOI] [PubMed] [Google Scholar]
- Enzmann G.U. Benton R.L. Talbott J.F. Cao Q. Whittemore S.R. Functional considerations of stem cell transplantation therapy for spinal cord repair. J. Neurotrauma. 2006;23:479–495. doi: 10.1089/neu.2006.23.479. [DOI] [PubMed] [Google Scholar]
- Goldman S. Stem and progenitor cell-based therapy of the human central nervous system. Nat. Biotechnol. 2005;23:862–871. doi: 10.1038/nbt1119. [DOI] [PubMed] [Google Scholar]
- Gruner J.A. A monitored contusion model of spinal cord injury in the rat. J. Neurotrauma. 1992;9:123–128. doi: 10.1089/neu.1992.9.123. [DOI] [PubMed] [Google Scholar]
- Han S.S. Liu Y. Tyler-Polsz C. Rao M.S. Fischer I. Transplantation of glial-restricted precursor cells into the adult spinal cord: survival, glial-specific differentiation, and preferential migration in white matter. Glia. 2004;45:1–16. doi: 10.1002/glia.10282. [DOI] [PubMed] [Google Scholar]
- Hanashima C. Shen L. Li S.C. Lai E. Brain factor-1 controls the proliferation and differentiation of neocortical progenitor cells through independent mechanisms. J. Neurosci. 2002;22:6526–6536. doi: 10.1523/JNEUROSCI.22-15-06526.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K. Chang Y.W. Li H. Berlin Y. Ikeda O. Kane-Goldsmith N. Grumet M. Embryonic radial glia bridge spinal cord lesions and promote functional recovery following spinal cord injury. Exp. Neurol. 2005;193:394–410. doi: 10.1016/j.expneurol.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Horky L.L. Galimi F. Gage F.H. Horner P.J. Fate of endogenous stem/progenitor cells following spinal cord injury. J. Comp. Neurol. 2006;498:525–538. doi: 10.1002/cne.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowski M.P. Cornuet P.K. McIlwrath S. Koerber H.R. Albers K.M. SRY-box containing gene 11 (Sox11) transcription factor is required for neuron survival and neurite growth. Neuroscience. 2006;143:501–514. doi: 10.1016/j.neuroscience.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.B. McDaniel E.E. Popovich P.G. Inflammatory-mediated injury and repair in the traumatically injured spinal cord. Curr. Pharm. Des. 2005;11:1223–1236. doi: 10.2174/1381612053507468. [DOI] [PubMed] [Google Scholar]
- Krady J.K. Lin H.W. Liberto C.M. Basu A. Kremlev S.G. Levison S.W. Ciliary neurotrophic factor and interleukin-6 differentially activate microglia. J. Neurosci. Res. 2008;86:1538–1547. doi: 10.1002/jnr.21620. [DOI] [PubMed] [Google Scholar]
- Kuan C.Y. Schloemer A.J. Lu A. Burns K.A. Weng W.L. Williams M.T. Strauss K.I. Vorhees C.V. Flavell R.A. Davis R.J. Sharp F.R. Rakic P. Hypoxia-ischemia induces DNA synthesis without cell proliferation in dying neurons in adult rodent brain. J. Neurosci. 2004;24:10763–10772. doi: 10.1523/JNEUROSCI.3883-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K. Herbarth B. Sock E. Enderich J. Hermans-Borgmeyer I. Wegner M. Cooperative function of POU proteins and SOX proteins in glial cells. J. Biol. Chem. 1998;273:16050–16057. doi: 10.1074/jbc.273.26.16050. [DOI] [PubMed] [Google Scholar]
- Lacroix S. Chang L. Rose-John S. Tuszynski M.H. Delivery of hyper-interleukin-6 to the injured spinal cord increases neutrophil and macrophage infiltration and inhibits axonal growth. J. Comp. Neurol. 2002;454:213–228. doi: 10.1002/cne.10407. [DOI] [PubMed] [Google Scholar]
- Marquardt T. Pfaff S.L. Cracking the transcriptional code for cell specification in the neural tube. Cell. 2001;106:651–654. doi: 10.1016/s0092-8674(01)00499-8. [DOI] [PubMed] [Google Scholar]
- Martino G. Pluchino S. The therapeutic potential of neural stem cells. Nat. Rev. Neurosci. 2006;7:395–406. doi: 10.1038/nrn1908. [DOI] [PubMed] [Google Scholar]
- Mautes A.E. Noble L.J. Co-induction of HSP70 and heme oxygenase-1 in macrophages and glia after spinal cord contusion in the rat. Brain Res. 2000;883:233–237. doi: 10.1016/s0006-8993(00)02846-8. [DOI] [PubMed] [Google Scholar]
- McTigue D.M. Wei P. Stokes B.T. Proliferation of NG2-positive cells and altered oligodendrocyte numbers in the contused rat spinal cord. J. Neurosci. 2001;21:3392–3400. doi: 10.1523/JNEUROSCI.21-10-03392.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesic O. Svrakic N.M. Xu G.Y. McAdoo D. Westlund K.N. Hulsebosch C.E. Ye Z. Galante A. Soteropoulos P. Tolias P. Young W. Hart R.P. Perez-Polo J.R. DNA microarray analysis of the contused spinal cord: effect of NMDA receptor inhibition. J. Neurosci. Res. 2002;68:406–423. doi: 10.1002/jnr.10171. [DOI] [PubMed] [Google Scholar]
- Noctor S.C. Flint A.C. Weissman T.A. Wong W.S. Clinton B.K. Kriegstein A.R. Dividing precursor cells of the embryonic cortical ventricular zone have morphological and molecular characteristics of radial glia. J. Neurosci. 2002;22:3161–3173. doi: 10.1523/JNEUROSCI.22-08-03161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohori Y. Yamamoto S. Nagao M. Sugimori M. Yamamoto N. Nakamura K. Nakafuku M. Growth factor treatment and genetic manipulation stimulate neurogenesis and oligodendrogenesis by endogenous neural progenitors in the injured adult spinal cord. J. Neurosci. 2006;26:11948–11960. doi: 10.1523/JNEUROSCI.3127-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudega M. Schwann cell and olfactory ensheathing cell implantation for repair of the contused spinal cord. Acta Physiol. (Oxf.) 2007;189:181–189. doi: 10.1111/j.1748-1716.2006.01658.x. [DOI] [PubMed] [Google Scholar]
- Pan J.Z. Jornsten R. Hart R.P. Screening anti-inflammatory compounds in injured spinal cord with microarrays: a comparison of bioinformatics analysis approaches. Physiol. Genomics. 2004;17:201–214. doi: 10.1152/physiolgenomics.00177.2003. [DOI] [PubMed] [Google Scholar]
- Pluchino S. Zanotti L. Rossi B. Brambilla E. Ottoboni L. Salani G. Martinello M. Cattalini A. Bergami A. Furlan R. Comi G. Constantin G. Martino G. Neurosphere-derived multipotent precursors promote neuroprotection by an immunomodulatory mechanism. Nature. 2005;436:266–271. doi: 10.1038/nature03889. [DOI] [PubMed] [Google Scholar]
- Plunkett J.A. Yu C.G. Easton J.M. Bethea J.R. Yezierski R.P. Effects of interleukin-10 (IL-10) on pain behavior and gene expression following excitotoxic spinal cord injury in the rat. Exp. Neurol. 2001;168:144–154. doi: 10.1006/exnr.2000.7604. [DOI] [PubMed] [Google Scholar]
- Popovich P.G. Wei P. Stokes B.T. Cellular inflammatory response after spinal cord injury in Sprague-Dawley and Lewis rats. J. Comp. Neurol. 1997;377:443–464. doi: 10.1002/(sici)1096-9861(19970120)377:3<443::aid-cne10>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Regad T. Roth M. Bredenkamp N. Illing N. Papalopulu N. The neural progenitor-specifying activity of FoxG1 is antagonistically regulated by CKI and FGF. Nat. Cell. Biol. 2007;9:531–540. doi: 10.1038/ncb1573. [DOI] [PubMed] [Google Scholar]
- Schwartz M. Macrophages and microglia in central nervous system injury: are they helpful or harmful? J. Cereb. Blood Flow Metab. 2003;23:385–394. doi: 10.1097/01.WCB.0000061881.75234.5E. [DOI] [PubMed] [Google Scholar]
- Sharp F.R. Massa S.M. Swanson R.A. Heat-shock protein protection. Trends Neurosci. 1999;22:97–99. doi: 10.1016/s0166-2236(98)01392-7. [DOI] [PubMed] [Google Scholar]
- Talbott J.F. Loy D.N. Liu Y. Qiu M.S. Bunge M.B. Rao M.S. Whittemore S.R. Endogenous Nkx2.2 + /Olig2 + oligodendrocyte precursor cells fail to remyelinate the demyelinated adult rat spinal cord in the absence of astrocytes. Exp. Neurol. 2005;192:11–24. doi: 10.1016/j.expneurol.2004.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe K. Bonilla I. Winkles J.A. Strittmatter S.M. Fibroblast growth factor-inducible-14 is induced in axotomized neurons and promotes neurite outgrowth. J. Neurosci. 2003;23:9675–9686. doi: 10.1523/JNEUROSCI.23-29-09675.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao W. Lai E. Telencephalon-restricted expression of BF-1, a new member of the HNF-3/fork head gene family, in the developing rat brain. Neuron. 1992;8:957–966. doi: 10.1016/0896-6273(92)90210-5. [DOI] [PubMed] [Google Scholar]
- Uwanogho D. Rex M. Cartwright E.J. Pearl G. Healy C. Scotting P.J. Sharpe P.T. Embryonic expression of the chicken Sox2, Sox3 and Sox11 genes suggests an interactive role in neuronal development. Mech. Dev. 1995;49:23–36. doi: 10.1016/0925-4773(94)00299-3. [DOI] [PubMed] [Google Scholar]
- Velardo M.J. Burger C. Williams P.R. Baker H.V. Lopez M.C. Mareci T.H. White T.E. Muzyczka N. Reier P.J. Patterns of gene expression reveal a temporally orchestrated wound healing response in the injured spinal cord. J. Neurosci. 2004;24:8562–8576. doi: 10.1523/JNEUROSCI.3316-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- Xuan S. Baptista C.A. Balas G. Tao W. Soares V.C. Lai E. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141–1152. doi: 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- Yenari M.A. Heat shock proteins and neuroprotection. Adv. Exp. Med. Biol. 2002;513:281–299. doi: 10.1007/978-1-4615-0123-7_10. [DOI] [PubMed] [Google Scholar]
- Zai L.J. Wrathall J.R. Cell proliferation and replacement following contusive spinal cord injury. Glia. 2005;50:247–257. doi: 10.1002/glia.20176. [DOI] [PubMed] [Google Scholar]