Abstract
The effectiveness of radiotherapy treatment could be significantly improved if tumour cells could be rendered more sensitive to ionizing radiation without altering the sensitivity of normal tissues. However many of the key, therapeutically exploitable mechanisms that determine intrinsic tumour radiosensitivity are largely unknown. We have conducted a siRNA screen of 200 genes involved in DNA damage repair aimed at identifying genes whose knockdown increased tumour radiosensitivity. Parallel siRNA screens were conducted in irradiated and unirradiated tumour cells (SQ20B) and irradiated normal tissue cells (MRC5). Using γH2AX foci at 24 hours after ionising radiation we identified several genes such as BRCA2, Lig IV and XRCC5, whose knockdown is known to cause increased cell radiosensitivity thereby validating the primary screening endpoint. In addition we identified POLQ (DNA polymerase theta) as a potential tumour-specific target. Subsequent investigations demonstrated that POLQ knockdown resulted in radiosensitisation of a panel of tumour cell lines from different primary sites, whilst having little or no effect on normal tissue cell lines. These findings raise the possibility that POLQ inhibition might be used clinically to cause tumour specific radiosensitisation.
Keywords: High-throughput Screen, siRNA, DNA-repair, POLQ, Tumour Radiosensitivity
INTRODUCTION
Radiotherapy is a vital tool in the management of cancer patients. It is often given with curative intent either alone or with chemotherapy in patients with diseases as diverse as head and neck, cervix, bladder, and non-small cell lung cancer. The radiation dose that can safely be delivered to patients is limited by the dose tolerances of surrounding normal tissues(1). It is anticipated that the effectiveness of radiotherapy would be improved if tumour cells could be rendered more sensitive to ionizing radiation (IR) without altering the sensitivity of normal tissues. Such a strategy depends upon exploiting tumour specific molecular targets, many of which remain to be identified. The intrinsic radiosensitivity of tumours differs significantly. Importantly, these variations in radiosensitivity have a large clinical impact as those patients with radioresistant tumours are more likely to develop local recurrences(2-4) and have poorer survival rates(3, 4) than patients with more radiosensitive disease.
A recent trial in patients with locally advanced head and neck cancer compared combination treatment of cetuximab with radiotherapy against radiotherapy alone (5). This trial demonstrated improved locoregional control and overall survival in patients treated with cetuximab and radiotherapy. Importantly the addition of cetuximab was not associated with increased normal tissue toxicity. This important study has demonstrated the potential improvements that can be achieved by specifically rendering tumour cells more sensitive to radiation therapy.
Previous attempts have been made to identify targets involved in radiosensitivity through the screening of small interfering RNA (siRNA) libraries. These studies have assessed radiation sensitivity using short term assays based on cell viability (6, 7). This approach is potentially flawed as it may fail to distinguish between growth inhibition and clonal inactivation. The clonogenic survival assay is the ‘gold standard’ method for assessing intrinsic radiosensitivity in vitro(8). Unfortunately this assay is not suitable for use in large scale siRNA screens due to the highly labour intensive nature of the assay.
The critical role of DNA double-strand breaks (DSB) and chromosome aberrations produced by ionizing radiation in causing cell death has long been recognised (9, 10). DSB formation results in rapid phosphorylation of histone H2AX (γH2AX). Typically, most of these γH2AX foci resolve within a few hours of irradiation. We reasoned that foci persisting at 24 hours may mark sites of delayed repair, and could correspond to the sites likely to lead to chromosome breaks. Previous studies have demonstrated the correlation between intrinsic radiosensitivity and the persistence of γH2AX foci 24 hours after radiation (11, 12).
In the present study we describe a siRNA screen of 200 genes involved in DNA damage repair aimed at identifying genes whose knockdown causes increased tumour radiosensitivity. Using γH2AX foci at 24 hours after IR we identified POLQ (DNA polymerase theta) as a potential tumour-specific target whose knockdown led to tumour cell-specific radiosensitisation.
METHODS
Cell culture
The tumour cells used were the SQ20B (laryngeal), T24 (bladder), PSN1 (pancreas) and HeLa (cervix) lines. The SQ20B cell line was obtained from Dr. Ralph Weichselbaum (University of Chicago, Chicago, IL) and has been described previously(13, 14). The other tumour cell lines were obtained from ATCC. Two types of normal human fibroblast cells (MRC5 and POC) were used at early passage. MRC5 cells were obtained from ATCC. POC cells were established by us and have been described previously (13). Cells were cultured in DMEM containing 4.5g/l glucose (Invitrogen) supplemented with 10% fetal bovine serum. All cultures were maintained at 37°C in water-saturated 5% CO2/95% air. Cells were regularly tested to ensure the absence of Mycoplasma contamination (MycoAlert; Lonza). Cell morphology was regularly checked to ensure the absence of cross contamination of cell lines.
RNAi library and siRNA
A custom designed DNA repair gene library of 200 pools of four siRNA strands (Supplementary Table 1) was used for the screen (siGenome, Dharmacon). In addition to the library wells, each plate contained four replica wells with non-targeting siRNA (NT) and four wells with DNA-PKcs siRNA as negative and positive controls respectively. Individual genes were investigated using both pools and individual siRNAs (ON-TARGET plus, Dharmacon). An initial screen was separately conducted with both tumour cells (SQ20B) and normal tissue cells (MRC5) in order to identify genes whose knockdown caused tumour specific radiosensitisation. SQ20B cells and MRC5 cells were both reverse transfected with siRNA (final concentration 50nM) using Dharmafect 1 as per manufacturer’s instructions in four replica 96 well plates. Forty-eight hours after transfection the medium was replaced with DMEM with 10% fetal bovine serum. For both cell types, two replica plates were treated with 4Gy using an IBL634 caesium irradiator (CIS biointernational) at a dose rate of 0.66 Gy/min and two plates were left unirradiated. Optimisation studies showed that this dose of radiation resulted in sufficiently large differences in γH2AX foci formation between positive and negative controls. The SQ20B cells were irradiated forty-eight hours and the MRC5 cells sixty-six hours after transfection. After irradiation the cells were returned to the incubator for 24 hours. Cells were then fixed using 3% paraformaldehyde diluted in PBS prior to analysis of γH2AX foci.
Analysis of γH2AX foci
The techniques used to quantify γH2AX foci have been described previously (15). Briefly, after fixation the cells were permeabilized and blocked with 0.1% Triton (vol:vol) diluted in PBS containing 1% BSA (Sigma) for 1 hour at room temperature. Cells were incubated with a primary mouse monoclonal antibody to γH2AX (Millipore) 1:1500 overnight at 4°C. Cells were then washed thrice with PBS prior to incubation with Alexafluor 488 conjugate secondary antibody (Invitrogen) 1:1200 for 1 hour at room temperature. Cells were again washed thrice with PBS for 5 min before 4′,6-diamidino-2-phenylindole (DAPI) staining, 0.5 μg/mL diluted with PBS, for 10 min. The DAPI was replaced with PBS before foci were detected using an IN Cell Analyzer 1000 automated epifluorescence microscope (GE Healthcare). Four images were obtained per well. Foci quantitation was accomplished using IN Cell Analyzer Workstation software (v3.5). For unirradiated cells, the read-out was the mean number of γH2AX foci per cell. For irradiated cells, this was the proportion of cells that contained >7 γH2AX foci per cell. This was because optimisation studies showed that analysis based on this technique correlated best with results obtained from clonogenic survival assays.
Clonogenic assay
For the clonogenic assays all cell types were forward transfected in 6 well plates with 50nM siRNA using Dharmafect 1 (Dharmacon). In all clonogenic survival experiments, cells were plated 48 hours after transfection from single-cell suspensions and allowed to adhere to culture dishes before irradiation with an IBL634 caesium irradiator (CIS biointernational) at a dose rate of 0.66 Gy/min. Remaining cells from the transfection were used for qRT-PCR to confirm effective knockdown. Colonies were stained with crystal violet and counted 9 to 16 days after irradiation. Colony counting was primarily accomplished using an Oxford Optronics Colcount. Some primary cells formed diffuse colonies and required manual scoring. Each point on the survival curve represents the mean surviving fraction from four dishes. Clonogenic survival curves are representative of independent replicate experiments.
The surviving fraction was derived using the formula:
Experimental data were fitted with the linear quadratic model (LQ):
where S is the survival probability, D the radiation dose (Gy), α and β are the fit parameters (Gy−1 and Gy−2 respectively).
The sensitisation enhancement ratio (SER) was used to quantify radiosensitisation (the SER10 was deduced from data by using SER10 = Dcontrol/Dtreated, where Dcontrol and Dtreated doses yield 10% survival for controls and treated cells, respectively).
Drug treatment
For clonogenic assays, cells transfected with either POLQ or NT siRNA were allowed to adhere prior to addition of temozolomide (Sigma) at the stated concentrations for 2h. The cells were then washed with PBS prior to the addition of complete medium and incubated for 14 days until colony staining. For γH2AX foci quantification, cells were plated in 96 well plates as described above. In the experiments indicated, forty-eight hours after forward transfection the cells were treated with temozolomide for 2h at which point they were either left unirradiated or treated with 4Gy. One hour after IR the cells were washed thrice with PBS. Complete medium was then added and the cells returned to the incubator until fixation twenty-four hours after IR.
Quantification of gene silencing
RNA was extracted and purified from cells at the times indicated using the RNeasy Mini Kit (Qiagen) as per the manufacturer’s instructions. One step qRT-PCT was performed on 500ng RNA using Superscript III Platinum SYBR Green One-step qRT-PCR kit (Invitrogen). The primers used for each gene are as stated below.
POLQ Forward: TATCTGCTGGAACTTTTGCTGA
POLQ Reverse: CTCACACCATTTCTTTGATGGA
APEX2 Forward: CTGTAAGGACAATGCTACCC
APEX2 Reverse: ACACGTTGATTAGGGTCAAG
GAPDH Forward: CCACCCATGGCAAATTCCATGGCA
GAPDH Reverse: TCTAGACGGCAGGTCAGGTCCACC
qRT-PCR was achieved using a Stratagene Mx3005P system. cDNA synthesis was performed by heating the reagents to 42°C for 30 mins followed by 95°C for 10 minutes. The amplification was performed at the following conditions for all three genes of interest: 95°C for 30 sec, 58°C for 30 sec, 72°C for 60 sec for 40 cycles.
Statistical analysis
Data was analysed using the CellHTS2 package (16) as follows. Sample values from each plate were first normalised using the median of the non-targeting siRNA control wells for each plate. Z-scores for each gene in each replicate were then calculated using the formula: Z-score = Samplenorm – MedianNT) / MADNT, where Samplenorm is the normalised sample value, MedianNT and MADNT are the median and the median absolute deviation (MAD) of all non-targeting control wells across all three library plates, respectively. The final Z-score was then calculated using the mean of the replicate Z-scores for each gene.
For clonogenic assays, unpaired t-tests were conducted at each radiation dose exposure to assess differences in surviving fractions. All tests of significance were two-sided and values of P < 0.05 were considered significant.
RESULTS
A siRNA screen identifies genes potentially involved in tumour cell radiosensitivity
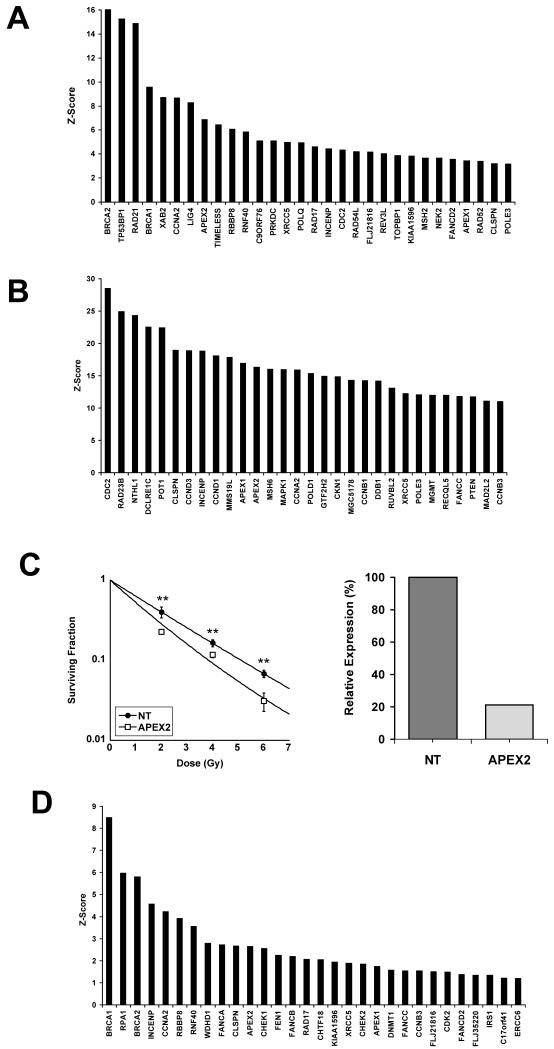
We screened a custom siRNA library of 200 genes involved in DNA repair using the radioresistant SQ20B cell line as well as the MRC5 fibroblast line in order to identify novel tumour-specific radiosensitising targets. The screen of irradiated SQ20B cells was used to compile a list of genes that may be involved in tumour specific radiosensitivity. The magnitude of the Z-scores obtained in each of the screens differed significantly. In view of this we decided the most practical way to define genes of interest was to examine the top 30 genes with the highest Z-scores. The Z-scores of the top 30 genes associated with elevated γH2AX foci in SQ20B cells twenty-four hours after 4Gy radiation are shown in Figure 1A. Several of the genes identified in this screen are already known to increase cell radiosensitivity following knockdown. These included genes involved in homologous recombination such as BRCA1 (17), BRCA2 (18), and RBBP8 (19) as well as genes involved in non-homologous end joining such as Lig IV (20), XRCC5 (Ku80) (21) and PRKDC (DNA-PKcs)(22). Genes involved in DNA damage response such as 53BP1 (23) which has been shown to be involved in cell radiosensitivity were also identified by the screen. Depletion of TIMELESS, which was also associated with a high Z-score, has previously been shown to increase radiosensitivity but through mechanisms that are less clear(24). Of the remaining genes that have not previously been shown to be involved in tumour radiosensitivity, we decided to investigate POLQ, RAD21, APEX2 and XAB2 in more detail as these genes were considered to have clinically exploitable potential if it was shown that depletion of these genes caused tumour specific radiosensitisation.
Figure 1.
Screening of a siRNA library of genes involved in DNA repair in SQ20B and MRC5 cells. A) Irradiated SQ20B cells. Z-scores of the top 30 genes associated with elevated γH2AX foci twenty-four hours after 4Gy radiation. B) Irradiated MRC5 cells. Z-scores of the top 30 genes associated with elevated γH2AX foci twenty-four hours after 4Gy radiation. C) Radiosensitisation of MRC5 cells with APEX2 depletion. Clonogenic assay in MRC5 cells treated with 50nM NT or APEX2 siRNA. **, P < 0.01 unpaired two sided t-test (left). Demonstration of effective knockdown of APEX2 by qRT-PCR. Gene expression normalised to cells treated with NT siRNA (right). D) Unirradiated SQ20B cells. Z-scores of the top 30 genes associated with elevated γH2AX foci in cells transfected with siRNA pools.
The screen of irradiated MRC5 cells was used to filter the list of candidate genes. In order to identify candidate genes whose depletion sensitised tumour cells to radiation without affecting normal tissue radiosensitivity we screened a fibroblast cell line with the same pools of siRNAs that were used to transfect the tumour cells. Figure 1B shows the top 30 genes associated with elevated γH2AX foci in irradiated MRC5 cells. One of the genes identified by this normal tissue screen was APEX2 which also had a high Z-score on the screen of irradiated tumour cells. As APEX2 has not previously been shown to be involved in intrinsic radiosensitivity we performed clonogenic assays with MRC5 cells depleted of APEX2 and found effective knockdown did indeed cause increased cell radiosensitivity (Figure 1C). In view of these findings on a normal tissue line, this gene was not investigated further as a potential tumour specific radiosensitiser. Supplementary Table 2 lists the genes which featured in the top 30 Z-scores of both the irradiated SQ20B and MRC5 cell line screens.
Identification of genes important in tumour cell survival independently of radiation
Of the remaining candidate genes being investigated we tried to use the screen of unirradiated SQ20B cells as a filter to exclude those genes whose knockdown affected cell viability in the absence of exposure to IR. The Z-scores of the top 30 genes associated with elevated γH2AX foci in unirradiated SQ20B cells are shown in figure 1D. The effect of gene knockdown on cell survival has not previously been studied for several of these genes. Colony forming assays performed with unirradiated SQ20B cells transfected with two of the siRNA pools ranked highly in this unirradiated screen (RPA1 and INCENP) siRNA pools resulted in widespread cell death and no colony formation (data not shown). These initial results suggested that the screen could be used as an effective filter for siRNAs which caused cell death in unirradiated conditions. Supplementary Table 3 lists those genes which featured in the top 30 Z-scores of the irradiated SQ20B but not in either the irradiated MRC5 screen or the unirradiated SQ20B screen.
None of the remaining genes being investigated as causing tumour specific radiosensitisation (POLQ, RAD21, and XAB2) were associated with high Z-scores in the screen of unirradiated tumour cells. However colony forming assays performed with a panel of unirradiated cells transfected with both RAD21 and XAB2 siRNAs demonstrated that knockdown of these genes resulted in widespread cell death (Supplementary Table 4). Therefore the absence of elevated γH2AX foci in unirradiated cells transfected with pools of siRNA cannot reliably be used to predict the survival of cells in the absence of IR. However the exclusion of those genes which cause γH2AX foci in the absence of radiation may reduce the number of false positive genes in the screen of irradiated cells. Having excluded three of the four genes initially identified by the screen of irradiated SQ20B cells we investigated the remaining gene, POLQ in more detail.
POLQ knockdown is associated with increased γH2AX foci following IR
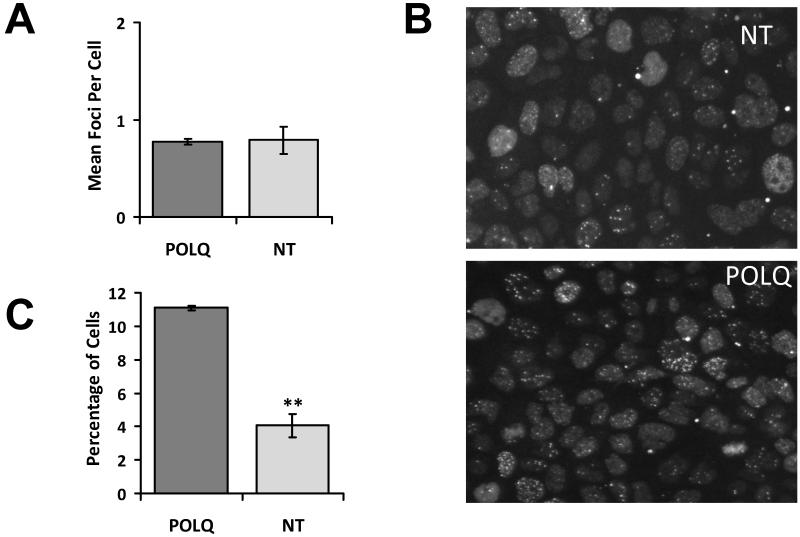
POLQ (DNA Polymerase theta) is a low fidelity DNA Polymerase with limited normal tissue expression whose normal physiological functions are largely unknown. POLQ featured in the top 30 Z-scores of the irradiated SQ20B screen but not in the screens using irradiated MRC5 cells or unirradiated SQ20B cells suggesting it does not affect MRC5 cell survival after irradiation or SQ20B cell viability in the absence of irradiation. As POLQ knockdown has not previously been shown to sensitise human tumour cells to IR this gene was investigated further. First we aimed to replicate the results found in the screen. SQ20B cells were transfected in triplicate wells of two 96 well plates with either NT or POLQ siRNA. One plate was irradiated with 4Gy and the other left unirradiated. Twenty four hours after IR cells were analysed for γH2AX foci. Unirradiated cells with POLQ knockdown did not have increased γH2AX foci compared to negative controls (Figure 2A). However the irradiated cells treated with POLQ siRNA had significantly increased residual γH2AX foci compared to irradiated cells treated with NT siRNA (Figure 2B and C).
Figure 2.
Effects of POLQ knockdown on γH2AX foci. A) In unirradiated SQ20B cells POLQ knockdown has no effect on γH2AX foci formation. B) γH2AX foci in SQ20B cells transfected with either NT or POLQ siRNA fixed 24 hours after receiving 4Gy. C) Increase in proportion of cells with >7 γH2AX foci in irradiated SQ20B cells with POLQ knockdown. **, P < 0.01 unpaired two sided t-test.
POLQ knockdown sensitises several tumour cell lines to IR
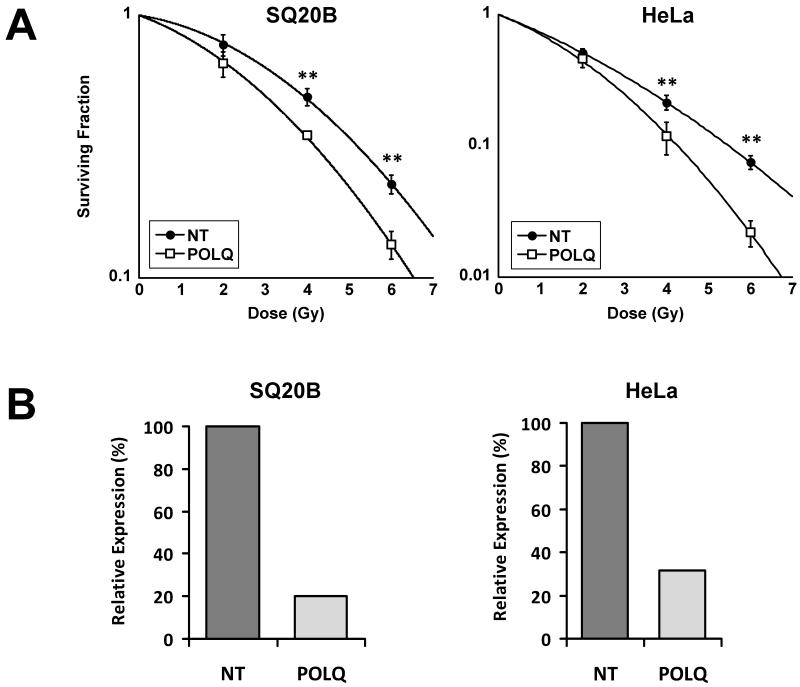
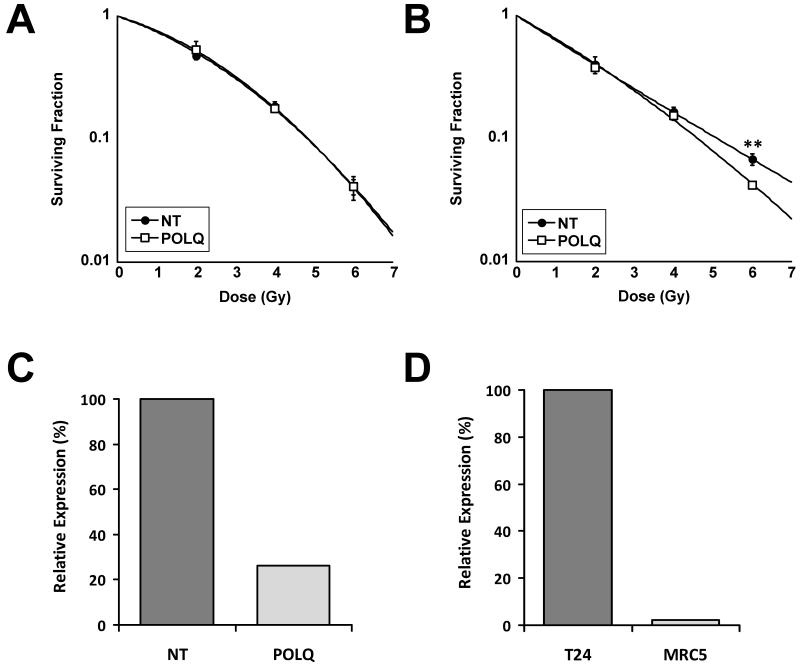
In order to confirm that the observed increase in γH2AX foci associated with irradiated SQ20B cells depleted of POLQ translated to an increase in tumour cell radiosensitivity we performed clonogenic assays with the SQ20B cells used in the primary screen along with a second tumour line (HeLa) following transfection with either NT or POLQ siRNA. Figure 3 confirms that both cell lines were sensitised to IR by POLQ knockdown. Clonogenic assays performed on a third tumour (T24 transitional cell bladder carcinoma) confirmed that the effects of POLQ knockdown were not restricted to squamous cell carcinomas. To confirm that the observed results did not occur as a result of off-target effects, we repeated the experiment with individual siRNAs as well as the siRNA pool (Figure 4A).
Figure 3.
Radiosensitisation of tumour cells following POLQ depletion. A) Radiosensitisation of SQ20B (SER=1.18) and HeLa (SER=1.28) cells following POLQ siRNA transfection. **, P < 0.01 unpaired two sided t-test. B) Effective POLQ knockdown confirmed by qRT-PCR with SQ20B and HeLa cells.
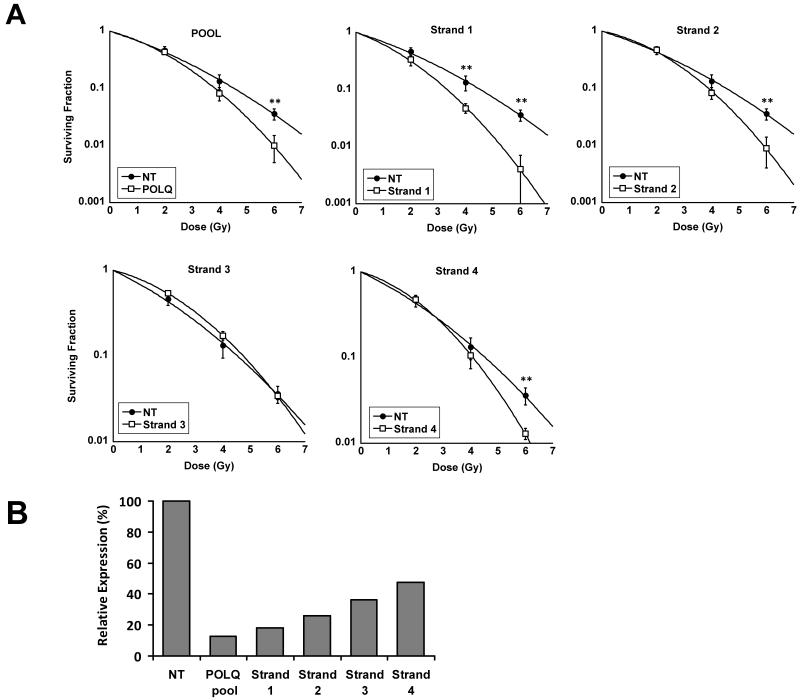
Figure 4.
Demonstration of the absence of off-target effects. A) T24 cells transfected with either 50nM POLQ pool of siRNA (SER=1.18), or 25nM of each individual siRNA strand. **, P < 0.01 unpaired two sided t-test. B) Relative expression of POLQ normalised to cells treated with NT siRNA as determined by qRT-PCR.
The individual siRNA strand transfections were performed with a final concentration of 25nM. The degree of POLQ knockdown was well correlated with the magnitude of associated radiosensitisation strongly suggesting that the observed effects with POLQ siRNA did not occur as a result of off target effects. Of the four individual siRNAs, Strand ‘1’ caused both the most potent silencing and radiosensitisation (Figure 4A and 4B).
POLQ knockdown causes minimal effects on normal tissue radiosensitivity
Previous work has shown that POLQ expression is limited to only a small number of normal tissues (25). In order to confirm that treatment with POLQ siRNA did not significantly alter the radiosensitivity of normal tissue cell lines we performed clonogenic assays on two fibroblast lines, MRC5 and POC cells. The POC cells did not express POLQ (data not shown) and treatment with POLQ siRNA had no radiosensitising effects (Figure 5A). As well as supporting the hypothesis that POLQ knockdown may cause tumour specific radiosensitisation, this also confirms that the observed effects did not occur as a result of off-target effects. In the MRC5 cells, POLQ siRNA treatment caused only a marginal increase in MRC5 radiosensitivity at very high doses of radiation (Figure 5B and C). A comparison between POLQ expression normalised to the expression of a housekeeping gene (GAPDH) in untransfected cells showed that MRC5 cells express POLQ at a level approximately 50 times lower than the T24 tumour cell line (Figure 5D). In unirradiated cells, POLQ knockdown did not consistently reduce colony formation in either the normal tissue or the tumour cell lines (Supplementary Table 5).
Figure 5.
Effect of POLQ siRNA on normal cells. A) POC cells treated with POLQ siRNA were not sensitised to IR. B) MRC5 cells show modest sensitisation to POLQ depletion only at high doses of IR. **, P < 0.01 unpaired two sided t-test. C) Effective knockdown of POLQ in MRC5 cells treated with 50nM POLQ siRNA as determined by qRT-PCR. D) Relative expression of POLQ in untransfected T24 and MRC5 cells as determined by qRT-PCR. The expression of POLQ was expressed as a ratio relative to the presence of GAPDH in each cell line.
POLQ knockdown has no effects on cell response to Temozolomide with or without IR
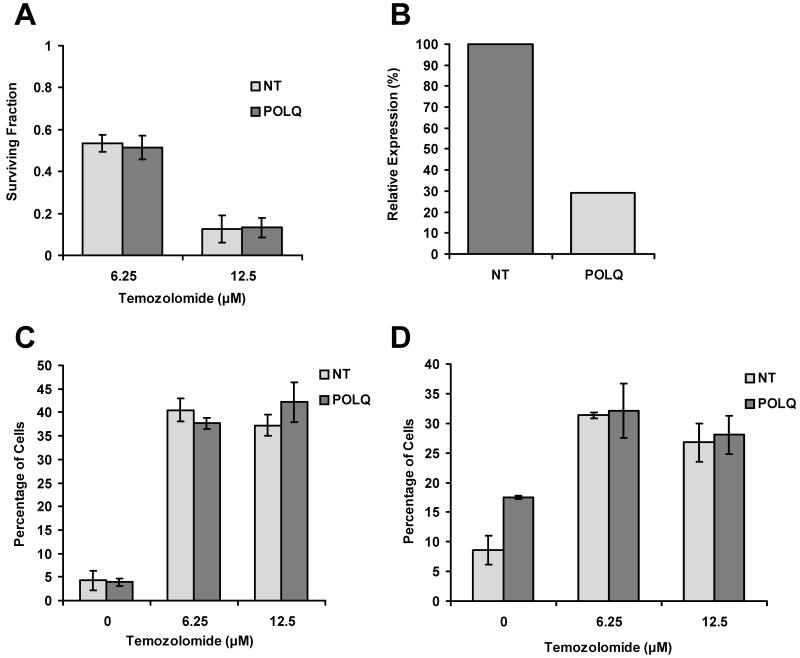
Temozolomide is an orally available alkylating agent that has an established role in the treatment of glioblastomas (26). It has previously been shown that a significant proportion of the DNA damage caused by temozolomide is repaired by the base excision repair (BER) pathway and that cells with deficiencies in the BER pathway have increased sensitivity to temozolomide(27). As it has previously been suggested that POLQ plays a role in BER we examined whether POLQ knockdown rendered cells more sensitive to temozolomide. However, clonogenic assays performed on SQ20B cells treated with either POLQ or NT siRNA showed no difference in sensitivity to drug treatment with temozolomide (Figure 6A and 6B).
Figure 6.
Effect of temozolomide treatment after POLQ knockdown. A) Clonogenic assays performed with SQ20B cells transfected with either NT or POLQ siRNA. Survival following temozolomide treatment expressed as a fraction relative to cells not exposed to temozolomide. B) Confirmation of effective knockdown of POLQ in SQ20B cells as determined by qRT-PCR. C) Effects of temozolomide treatment on the percentage of unirradiated SQ20B cells containing >7 γH2AX foci per cell. D) Percentage of SQ20B cells containing >7 γH2AX foci per cell following 4Gy irradiation.
For both unirradiated (Figure 6C) and irradiated conditions (Figure 6D), SQ20B cells treated with temozolomide were found to have elevated γH2AX foci twenty-four hours after drug exposure compared to cells not exposed to the drug. However, this occurred with the same magnitude regardless of whether cells had been transfected with either NT or POLQ siRNA. These findings would suggest that the mechanism by which POLQ knockdown causes increased radiosensitivity are independent of BER.
DISCUSSION
The known DNA repair defects that produce large differences in radiation sensitivity, are often associated with complex clinical syndromes such as ataxia-telangiectasia, Nijmegan breakage syndrome and Fanconi anaemia(28). The proteins identified by these repair defects do not represent potential therapeutic targets due to lack of tumour specificity. Many of the key, therapeutically exploitable mechanisms that determine intrinsic tumour radiosensitivity are largely unknown. The clinical importance of these mechanisms is shown by the known correlation between increased tumour radioresistance and adverse patient outcomes(2-4).
The EGFR pathway is the most widely studied contributor to tumour cell radioresistance. Recent trials have demonstrated the large benefits that can potentially be derived from biological treatments that selectively render tumour cells more sensitive to radiation by manipulation of this pathway and illustrate the need for greater understanding of the molecular basis for tumour radioresistance(5, 29).
This siRNA screen of genes involved in DNA repair was based on the critical role that unrepaired DSBs play in cell death following IR. Of the genes whose knockdown was associated with increased γH2AX foci in SQ20B cells following IR, several have already been shown to be associated with increased cell radiosensitivity (17-23) thus validating the primary screening endpoint. The experimental design used both irradiated and unirradiated tumour cells as well as parallel siRNA screens in a tumour line and normal tissue line allowing the identification of siRNAs that cause tumour specific radiosensitisation. High throughput screens will inevitably feature both false positive and false negative results. Failure to cause sufficient gene knockdown is one of the most common causes for obtaining false negative results. ATM was one of the genes included in this library which is known to be involved in cell radiosensitivity but which was not in the top 30 Z-scores of the irradiated cell screens. In this case it is probable this occurred because ATM is one of the genes involved in causing H2AX phosphorylation in response to DSB formation(30), and thus is a false negative in this assay.
Of the targets identified by this screen we elected to investigate POLQ further as this gene has not previously been linked to tumour cell radiosensitivity and because a previous study has suggested a potentially exploitable difference in expression between normal tissues and tumour cells (25).
POLQ is a member of the A family of DNA polymerases which, unusually for this class of polymerases, synthesises DNA with very low fidelity(31, 32). The normal physiological functions associated with this protein are currently unclear. It has previously been suggested that POLQ plays a dominant role in the process of somatic hypermutation of immunoglobulin genes. This suggestion arose from the observation that mice deficient in POLQ had a substantially decreased frequency of mutations in immunoglobulin genes(33). A separate group also found a decrease in mutation frequency in POLQ deficient mice but to a lesser degree(34). However a recent study found that mutation types and frequencies were similar in wild type, POLQ−/−, POLH−/−, and POLQ−/− POLH−/− mice(35) Accordingly this group suggested that POLQ does not have a significant role in the hypermutation pathway.
It has been suggested that POLQ has a role in BER but this remains unresolved. Mutation of POLQ in the DT40 chicken B cell lymphocyte line has been shown to increase sensitivity to H2O2. POLQ/POLβ mutants had significantly higher sensitivity to methyl methanesulfonate than either single mutant. Extracts obtained from this cell line were used to show that POLQ mutant cells have markedly reduced single nucleotide BER capacity in vitro and that this reduction was of a similar magnitude to cells deficient in POLβ(36). These findings led to the suggestion that POLQ and POLβ cooperate in BER.
Recent biochemical analysis has looked at the in vitro activity of cloned human POLQ(37). It was shown that full-length POLQ has 5′-deoxyribose phosphate (5′-dRP) lyase activity. A C-terminal fragment of POLQ was shown to carry 5′-dRP lyase activity and this appeared to be independent of polymerase activity. The full-length protein and the C-terminal fragment were shown to have BER activity in vitro. Although these findings have been used to support the argument that POLQ may have a role in BER in vivo it should be noted that the rate of 5′-dRP lyase activity of POLQ is approximately 40 fold slower than that of POLβ. We found that POLQ knockdown did not alter the sensitivity of cells to temozolomide either with or without IR. We interpret this to mean that the mechanism by which POLQ knockdown causes increased sensitivity to IR is independent of base excision repair although it remains possible that POLQ facilitates repair via BER of a lesion that is produced by IR but not by temozolomide.
POLQ expression was previously assessed by RT-PCR in a variety of different normal human tissues(25). Interestingly, expression was primarily limited to lymphoid tissues such as the fetal liver, thymus and bone marrow. Critical normal tissues such as lung, liver, small intestine, kidney, heart, brain and spinal cord that typically limit the radiation dose that can be delivered to patients did not appear to express POLQ. This would imply that inhibition of POLQ would not alter the intrinsic sensitivity of these tissues. Intriguingly this study also found that POLQ was overexpressed in a large proportion of tumours derived from patients with colon, lung and gastric cancer. Given the findings presented here, POLQ inhibition in these tumours would be predicted to reduce their radiation survival.
This difference in expression of POLQ in tumour cells and critical, radiosensitive normal tissues was central to our decision to investigate POLQ further. Although it is possible that depletion of POLQ from the small number of normal tissues that express this protein may render these cells more sensitive to radiation(38), the very restricted normal tissue expression means that POLQ inhibition may improve the therapeutic ratio of radiotherapy.
Our findings confirm that POLQ is overexpressed in tumour cells derived from a variety of primary sites and that POLQ knockdown causes increased intrinsic radiosensitivity. Our results are consistent with the limited normal tissue expression of POLQ, and therefore that depletion of POLQ might cause much less radiosensitisation of normal tissues compared with tumours. These findings raise the possibility that POLQ inhibition could be used clinically to cause tumour specific radiosensitisation. Additionally the technique used in this study successfully identified several other genes already known to play a role in intrinsic radiosensitivity thus validating its use in future screens of larger siRNA libraries.
Supplementary Material
Acknowledgements
This work was supported by grants from Cancer Research UK, the Medical Research Council and the NIHR Biomedical Research Centre, Oxford. GH was supported by a Cancer Research UK/Royal College of Radiologists Clinical Research Fellowship.
Footnotes
Conflict of Interest
The Authors have no conflicts of interest to declare.
References
- 1.Emami B, Lyman J, Brown A, et al. Tolerance of normal tissue to therapeutic irradiation. Int J Radiat Oncol Biol Phys. 1991;21:109–22. doi: 10.1016/0360-3016(91)90171-y. [DOI] [PubMed] [Google Scholar]
- 2.Bjork-Eriksson T, West C, Karlsson E, Mercke C. Tumor radiosensitivity (SF2) is a prognostic factor for local control in head and neck cancers. Int J Radiat Oncol Biol Phys. 2000;46:13–9. doi: 10.1016/s0360-3016(99)00373-9. [DOI] [PubMed] [Google Scholar]
- 3.West CM, Davidson SE, Roberts SA, Hunter RD. The independence of intrinsic radiosensitivity as a prognostic factor for patient response to radiotherapy of carcinoma of the cervix. Br J Cancer. 1997;76:1184–90. doi: 10.1038/bjc.1997.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Girinsky T, Bernheim A, Lubin R, et al. In vitro parameters and treatment outcome in head and neck cancers treated with surgery and/or radiation: cell characterization and correlations with local control and overall survival. Int J Radiat Oncol Biol Phys. 1994;30:789–94. doi: 10.1016/0360-3016(94)90350-6. [DOI] [PubMed] [Google Scholar]
- 5.Bonner JA, Harari PM, Giralt J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–78. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- 6.Zheng M, Morgan-Lappe SE, Yang J, et al. Growth inhibition and radiosensitization of glioblastoma and lung cancer cells by small interfering RNA silencing of tumor necrosis factor receptor-associated factor 2. Cancer Res. 2008;68:7570–8. doi: 10.1158/0008-5472.CAN-08-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sudo H, Tsuji AB, Sugyo A, Imai T, Saga T, Harada YN. A loss of function screen identifies nine new radiation susceptibility genes. Biochem Biophys Res Commun. 2007;364:695–701. doi: 10.1016/j.bbrc.2007.10.074. [DOI] [PubMed] [Google Scholar]
- 8.Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9. doi: 10.1038/nprot.2006.339. [DOI] [PubMed] [Google Scholar]
- 9.Frankenberg-Schwager M, Frankenberg D, Blocher D, Adamczyk C. Effect of dose rate on the induction of DNA double-strand breaks in eucaryotic cells. Radiat Res. 1981;87:710–7. [PubMed] [Google Scholar]
- 10.Bedford JS. Sublethal damage, potentially lethal damage, and chromosomal aberrations in mammalian cells exposed to ionizing radiations. Int J Radiat Oncol Biol Phys. 1991;21:1457–69. doi: 10.1016/0360-3016(91)90320-4. [DOI] [PubMed] [Google Scholar]
- 11.Banath JP, Macphail SH, Olive PL. Radiation sensitivity, H2AX phosphorylation, and kinetics of repair of DNA strand breaks in irradiated cervical cancer cell lines. Cancer Res. 2004;64:7144–9. doi: 10.1158/0008-5472.CAN-04-1433. [DOI] [PubMed] [Google Scholar]
- 12.Taneja N, Davis M, Choy JS, et al. Histone H2AX phosphorylation as a predictor of radiosensitivity and target for radiotherapy. J Biol Chem. 2004;279:2273–80. doi: 10.1074/jbc.M310030200. [DOI] [PubMed] [Google Scholar]
- 13.Kim IA, Bae SS, Fernandes A, et al. Selective inhibition of Ras, phosphoinositide 3 kinase, and Akt isoforms increases the radiosensitivity of human carcinoma cell lines. Cancer Res. 2005;65:7902–10. doi: 10.1158/0008-5472.CAN-05-0513. [DOI] [PubMed] [Google Scholar]
- 14.Kasid U, Pfeifer A, Weichselbaum RR, Dritschilo A, Mark GE. The raf oncogene is associated with a radiation-resistant human laryngeal cancer. Science. 1987;237:1039–41. doi: 10.1126/science.3616625. [DOI] [PubMed] [Google Scholar]
- 15.Prevo R, Deutsch E, Sampson O, et al. Class I PI3 kinase inhibition by the pyridinylfuranopyrimidine inhibitor PI-103 enhances tumor radiosensitivity. Cancer Res. 2008;68:5915–23. doi: 10.1158/0008-5472.CAN-08-0757. [DOI] [PubMed] [Google Scholar]
- 16.Boutros M, Bras LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. doi: 10.1186/gb-2006-7-7-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C, Smith JL, Liu J. Role of BRCA1 in cellular resistance to paclitaxel and ionizing radiation in an ovarian cancer cell line carrying a defective BRCA1. Oncogene. 2003;22:2396–404. doi: 10.1038/sj.onc.1206319. [DOI] [PubMed] [Google Scholar]
- 18.Abbott DW, Freeman ML, Holt JT. Double-strand break repair deficiency and radiation sensitivity in BRCA2 mutant cancer cells. J Natl Cancer Inst. 1998;90:978–85. doi: 10.1093/jnci/90.13.978. [DOI] [PubMed] [Google Scholar]
- 19.Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–3. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adachi N, Ishino T, Ishii Y, Takeda S, Koyama H. DNA ligase IV-deficient cells are more resistant to ionizing radiation in the absence of Ku70: Implications for DNA double-strand break repair. Proc Natl Acad Sci U S A. 2001;98:12109–13. doi: 10.1073/pnas.201271098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnie NJ, Gottlieb TM, Blunt T, Jeggo PA, Jackson SP. DNA-dependent protein kinase activity is absent in xrs-6 cells: implications for site-specific recombination and DNA double-strand break repair. Proc Natl Acad Sci U S A. 1995;92:320–4. doi: 10.1073/pnas.92.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kurimasa A, Ouyang H, Dong LJ, et al. Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc Natl Acad Sci U S A. 1999;96:1403–8. doi: 10.1073/pnas.96.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kao GD, McKenna WG, Guenther MG, Muschel RJ, Lazar MA, Yen TJ. Histone deacetylase 4 interacts with 53BP1 to mediate the DNA damage response. J Cell Biol. 2003;160:1017–27. doi: 10.1083/jcb.200209065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A. 2006;103:18143–7. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawamura K, Bahar R, Seimiya M, et al. DNA polymerase theta is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int J Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 26.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–96. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 27.Trivedi RN, Almeida KH, Fornsaglio JL, Schamus S, Sobol RW. The role of base excision repair in the sensitivity and resistance to temozolomide-mediated cell death. Cancer Res. 2005;65:6394–400. doi: 10.1158/0008-5472.CAN-05-0715. [DOI] [PubMed] [Google Scholar]
- 28.Pollard JM, Gatti RA. Clinical radiation sensitivity with DNA repair disorders: an overview. Int J Radiat Oncol Biol Phys. 2009;74:1323–31. doi: 10.1016/j.ijrobp.2009.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brunner TB, Geiger M, Grabenbauer GG, et al. Phase I trial of the human immunodeficiency virus protease inhibitor nelfinavir and chemoradiation for locally advanced pancreatic cancer. J Clin Oncol. 2008;26:2699–706. doi: 10.1200/JCO.2007.15.2355. [DOI] [PubMed] [Google Scholar]
- 30.McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16:5013–25. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA. Low-fidelity DNA synthesis by human DNA polymerase theta. Nucleic Acids Res. 2008;36:3847–56. doi: 10.1093/nar/gkn310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Seki M, Masutani C, Yang LW, et al. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–94. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zan H, Shima N, Xu Z, et al. The translesion DNA polymerase theta plays a dominant role in immunoglobulin gene somatic hypermutation. EMBO J. 2005;24:3757–69. doi: 10.1038/sj.emboj.7600833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masuda K, Ouchida R, Hikida M, et al. Absence of DNA polymerase theta results in decreased somatic hypermutation frequency and altered mutation patterns in Ig genes. DNA Repair (Amst) 2006;5:1384–91. doi: 10.1016/j.dnarep.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008;7:1603–8. doi: 10.1016/j.dnarep.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoshimura M, Kohzaki M, Nakamura J, et al. Vertebrate POLQ and POLbeta cooperate in base excision repair of oxidative DNA damage. Mol Cell. 2006;24:115–25. doi: 10.1016/j.molcel.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad R, Longley MJ, Sharief FS, Hou EW, Copeland WC, Wilson SH. Human DNA polymerase theta possesses 5′-dRP lyase activity and functions in single-nucleotide base excision repair in vitro. Nucleic Acids Res. 2009;37:1868–77. doi: 10.1093/nar/gkp035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goff JP, Shields DS, Seki M, et al. Lack of DNA Polymerase theta (POLQ) Radiosensitizes Bone Marrow Stromal Cells In Vitro and Increases Reticulocyte Micronuclei after Total-Body Irradiation. Radiat Res. 2009;172:165–74. doi: 10.1667/RR1598.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.