Abstract
Osteoporosis is prevalent among the elderly and is a major cause of bone fracture in this population. Bone integrity is maintained by the dynamic processes of bone resorption and bone formation (bone remodeling). Osteoporosis results when there is an imbalance of the two counteracting processes. Bone mineral density, measured by dual-energy x-ray absorptiometry has been the primary method to assess fracture risk for decades. Recent studies demonstrated that measurement of bone turnover markers allows for a dynamic assessment of bone remodeling, while imaging techniques, such as dual-energy x-ray absorptiometry, do not. The application of proteomics has permitted discoveries of new, sensitive, bone turnover markers, which provide unique information for clinical diagnosis and treatment of patients with bone diseases. This review summarizes the recent findings of proteomic studies on bone diseases, properties of mesenchymal stem cells with high expansion rates and osteoblast and osteoclast differentiation, with emphasis on the role of quantitative proteomics in the study of signaling dynamics, biomarkers and discovery of therapeutic targets.
Keywords: bone mineral density, bone remodeling, bone turnover marker, osteoporosis, quantitative proteomics
Status of bone disease research
The primary function of the skeleton is to provide structural support for the soft tissues of the body. The strength of bones comprising the skeleton is determined by the individual histological makeup and the mechanical properties determined by the deposition of hydroxylapatite within the collagen matrix. In response to changes in weight-bearing and mechanical stress, there is constant remodeling of bone by the processes of bone resorption and bone formation. The key players in bone remodeling are the osteoclasts, derived from hematopoietic stem cells, and the osteoblasts, derived from bone marrow mesenchymal stem cells (BM-MSCs). Bone resorption is carried out by the osteoclasts, while bone formation is performed by the osteoblasts. Bone structure can be affected by genetic disorders, such as osteogenesis imperfecta, and metabolic diseases, such as vitamin D-resistant rickets. Structural abnormalities in bone result in increased skeletal fragility and risk of fracture. Except for these rare disorders, most bone diseases are the result of an imbalance between bone resorption and formation.
During adult life, resorption and formation are kept in balance and bone mass is maintained at a steady state [1]. From the fifth decade of life, bone resorption begins to exceed bone formation, which leads to bone loss, osteopenia and osteoporosis, and conditions of low bone mass. Three pathogenic reasons for low bone mass are failure to achieve optimal peak bone mass during skeletal growth, which is largely determined by genetic factors, increased bone resorption due to abnormal endocrine regulation, cytokines or other local factors, and inadequate bone formation due to impaired osteoblast function or insufficient stimulus for bone formation [2].
Osteoporosis is the most prevalent bone disease in the USA and other developed countries. It is defined as a metabolic bone disease characterized by low bone mass and deterioration in bone architecture, resulting in enhanced bone fragility and, consequently, increased fracture risk [3]. Based on some surveys, it was estimated that 54% of post-menopausal Caucasian women in the USA have osteopenia and an additional 30% have osteoporosis. As a result, white women alone account for 26 million people who are at risk of fracture. The prevalence of osteopenia and osteoporosis would be comparable to that of hypertension [4] when affected men and nonwhite women are included. The annual cost of osteoporosis-related care has already approached US$17.9 billion annually in the USA and GB£1.7 billion in the UK. This cost for osteoporosis-related care is estimated to double in the first half of this century [5,6]. Osteoporosis is classified as either primary or secondary according to their respective pathogenic mechanisms. Primary osteoporosis frequently occurs in postmenopausal women and older men caused by estrogen deficiency, calcium deficiency and aging. Secondary osteoporosis is associated with diseases, such as conditions of glucocorticoid excess, multiple myelomatosis, hyperparathyroidism and hyperthyroidism [2,7]. Osteoporosis, either primary or secondary, can be the result of either a rise in osteoclast number and/or activity, or a decrease in osteoblast number and/or activity. The differentiation of osteoblasts and osteoclasts from their respective stem cells is affected by circulating cytokines and hormones. When there is an imbalance in osteoblast/osteoclast functions, irreversible bone loss occurs and osteoporosis results.
Despite our increasing knowledge of the individual molecular mechanism of osteoblast/osteoclast activation, how these mechanisms are orchestrated to maintain normal bone structural integrity or to cause osteoporosis is poorly understood [8]. For example, estrogen and parathyroid hormone (PTH) have been shown to influence both osteoclast and osteoblast activities. Estrogen can partly affect the osteoblast by increasing the expression of IGF-1, osteoprotegerin (OPG) and TGF-β, and decreasing the expression of RANKL, and partly affect monocytes by decreasing the expression of IL-1, IL-6 and TNF-α [9–11]. Hyperparathyroidism and aging can significantly increase PTH levels [12,13]. PTH, in addition to acting directly on the osteoblastic lineage, may enhance RANKL expression and, in some cases, inhibit OPG in osteoblastic cells [14,15]. Thus, the role of PTH in stimulating bone formation is more complicated than just increasing osteoblast activity [16,17]. Using the simple model of osteoporosis resulting from an imbalance of osteoblast/osteoclast activities, a number of therapeutic strategies to treat these common conditions are already in use or are under development. For example, estrogen-replacement therapy clearly inhibits bone loss, as well as bone turnover, and increases bone mineral density [18]; bisphosphonates have been shown to be the most effective inhibitors of bone resorption by inactivating osteoclasts and promoting osteoclast apoptosis [19]. Drugs that inhibit the formation or activity of osteoclasts are valuable for treating bone diseases. However, these treatments also have untoward side effects – estrogen is linked to increased risk of breast cancer and bisphosphonates are linked to jaw osteonecrosis [7,20].
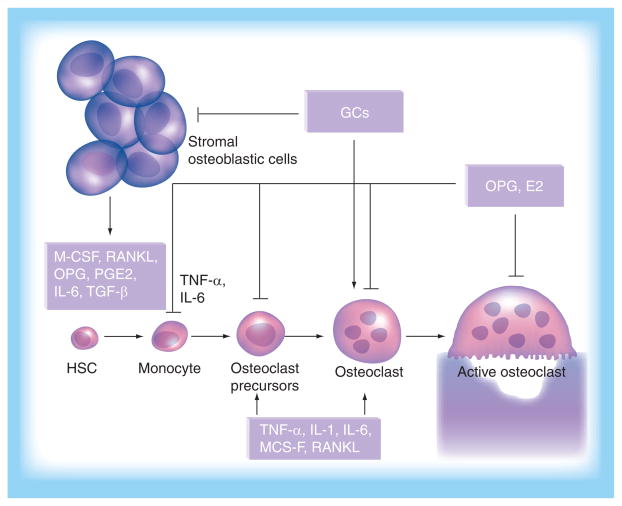
Besides osteoporosis, there are other kinds of bone disease, such as Paget’s disease, bone diseases of cancer and inflammatory bone disease. It is reported that 3% of the population in the UK over the age of 40 years, and a significant number in the Caucasian population of North America, suffer from Paget’s disease [21]. Paget’s disease is caused by increased numbers and activity of osteoblasts, which affects local bone mineral density at multiple sites throughout the whole skeleton. It has been reported that Paget’s disease is caused by viral infection in the nuclei of osteoclasts, and two insertional mutations in exon 1 of the RANK gene have been identified that result in enhanced expression of RANK and increased nuclear factor (NF)-κB signaling and stimulation of osteoclastogenesis [22,23]. Some tumors are known to have significant effects upon the skeleton. Tumor cells need the ability to promote osteoclastogenesis in order to establish growth and metastasis in bone. Either systemic humoral hypercalcemia of malignancy or local bone metastases can cause an increase in osteoclast number and activity [21]. For example, breast cancer cells can increase RANKL formation to promote osteoclastogenesis by producing PTH-related protein (PTHrP), IL-6, IL-11 and COX2. TGF-β, secreted by cancer cells, is also believed to influence the production of bone-resorbing cytokines [24,25]. Inflammatory bone diseases, such as rheumatoid arthritis are characterized by the destruction in articular cartilage for the excessive subchondral osteoclastic bone resorption. The inflammatory factors, including IL-1, IL-6, IL-11, IL-13, IL-17, PTHrP and RANKL, provide the environment of cytokines to stimulate osteoclasto-genesis, which is the primary cause of bone erosion in rheumatoid arthritis [26]. The regulatory process of cytokines/chemokines in osteoclastogenesis is summarized in Figure 1. A major therapy consideration in the treatment of these kinds of bone diseases is to inhibit bone resorption, as with osteoporosis therapy, as well as treatment of the underlying primary cause, such as chemotherapy for cancer and anti-inflammatory therapy for inflammatory bone disease.
Figure 1. Cytokines/chemokines regulate osteoclast development.
Most skeletal osteoclasts originate from circulating monocytes. These cells from osteoporotic patients were reported to have increased levels of bone resorption activity when induced into osteoclasts in vitro. Recruitment of circulating monocytes into bone is mainly conducted by chemokines, such as CCR3 ligand and RANTES. Recruited monocytes in bone are targets of a variety of cytokines, such as RANKL and M-CSF, which also regulates the osteoblast differentiation and function. Therefore, these chemokines and cytokines produced by marrow stromal cells and their derivative osteoblasts play key roles in bone remodeling.
GC: Glucocorticoid; HSC: Hematopoietic stem cell; M-CSF: Macrophage colony-stimulating factor; OPG: Osteoprotegerin.
Current drug therapy for osteoporosis aims to inhibit osteoclastogenesis and osteoclast activity. Such systemic therapy is effective in slowing bone loss and is known to have significant side effects. Some recent discoveries provide new targets for modifying osteoclastic differentiation based on the OPG/RANKL/RANK signal pathways: production of RANKL, interaction of RANKL with RANK and RANK downstream signal activation (Figure 1) [21]. Future advances in the treatment of osteoporosis with drugs specifically targeting these three critical processes of the OPG/RANKL/RANK pathways are greatly anticipated.
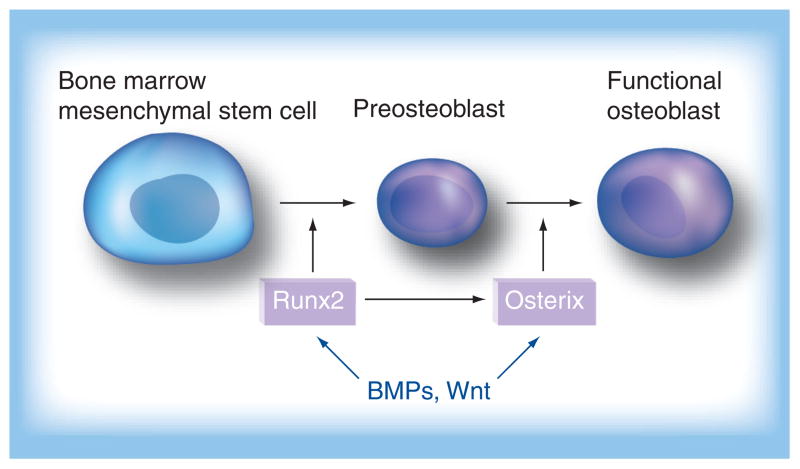
Much work remains to be conducted on the treatment of bone diseases by the stimulation of bone formation. A few pivotal signal pathways involved in osteogenesis in vitro have been found. Osterix, a novel zinc finger-containing transcription factor, plays an essential role in osteoblast differentiation and bone formation [27,28]. Cbfa1/Runx2, as a member of the RUNX family, has been demonstrated to be the key transcriptional factor associated with osteoblast differentiation. Targeted disruption of these two factors results in a complete abortion of bone formation owing to maturational arrest of osteoblast differentiation [28]. Osterix and Runx2 temporally regulate the process of the osteoblast differentiation (Figure 2) [29]. Runx2 plays an important role in the early stage of the differentiation of the BM-MSCs into preosteoblasts (Figure 2). Osterix mainly regulates the process of the preosteoblast differentiation into the functional osteoblast, which leads to overexpression of osteoblast marker genes. Although these two transcriptional factors differentially regulate the process of osteoblast differentiation, Runx2, in general, is considered to be the early regulator and osterix is the late regulator during osteoblast differentiation [28]. On the other hand, these two osteoblastogenic master genes are also regulated by numbers of developmental signal pathways, for example, the canonical Wnt and bone morphogenetic protein (BMP) signaling pathways, which orchestrate the commitment of BM-MSCs to specific cell types (Figure 2). It has been reported that Wnt10b stimulates osteoblastogenesis via activation of the osteogenic transcription factors Runx2 and osterix [30]. It is also reported that BMP-2, BMP-4 and BMP-7 induce osteoblastogenesis via activation of Runx2 and osterix by forming a specific receptor complex, BMPRIA/BMPRIB, and receptor, Smads [28].
Figure 2. BMPs/Wnt signaling-regulated osteoblastogenesis via Runx2 and Osterix.
BMP: Bone morphogenetic protein.
However, there is a big gap between in vitro and in vivo study. Osteogenesis study in vivo is much more complex. Cell–cell and cell–matrix interactions must be considered. Therefore, the whole regulatory network must be carefully studied as a unit and candidate targets should be confirmed in vivo study.
Quantitative proteomics: a powerful tool for bone marker discovery
Proteomics, developed over a decade ago, is now extensively used for directly analyzing protein expression at the post-translational level. The proteome in cells is constantly changing through its biochemical interactions with the genome and the environment. The large increase in protein diversity may be due to alternative splicing [31,32] and post-translational modification [33,34] of proteins. Protein diversity cannot be fully characterized by gene-expression analysis alone, making proteomics a promising tool for characterizing cells and tissues of interest and for biomarker discovery. Proteomic techniques have been developed during the last decade. Owing to the limitation of 2DE for protein profiling, gel-free or liquid chromatography (LC)-based proteomics techniques are now emerging as the choice for quantitatively measuring protein levels with better sensitivity and reproducibility over 2DE-based methods [35,36]. These mass spectrometry (MS)-based proteomics can be generally divided into two approaches: isotope-labeled and label-free MS [37]. An isotope-labeling strategy has been developed that introduces stable isotope tags to proteins via chemical reactions using isotope-coded affinity tags (ICAT) and isobaric tag for relative and absolute quantitation (iTRAQ), enzymatic labeling (e.g., using 18O water for trypsin digestion), or via metabolic labeling (SILAC). With the advances of new instrumentation, computing power and advanced bioinformatics, a series of label-free LC-MS shotgun screening methods, such as multidimensional PIT, have been alternatives for relative and absolute protein quantitation in biological samples. These methods and their limitations have been well discussed previously [37]. Proteomics has been gaining attention in the field of bone disease research for bone biomarker discovery and cell signaling [38]. In the following sections, we will summarize the progress that has been made for the application of proteomics to bone disease research.
Proteomics of bone diseases
Conventional molecular biological approaches examine a limited number of proteins based on signaling or metabolic pathways. Proteomics has emerged as a systematic approach for the qualitative and quantitative mapping of the whole proteome in large-scale studies. In the area of bone diseases, traditional 2DE coupled with MS, a standard method for comparing protein expression profiles between normal and disease states, has been applied to obtain unique protein-expression profiles of cartilage degradation, bone sarcoma, osteoarthritis and femoral head osteonecrosis, and compared with profiles from normal tissues [39–44]. Table 1 summarizes the protein marker candidates discovered in bone diseases using proteomics technology. The major objectives of these works are to discover unique proteins of diseases and provide insights into the mechanisms. However, this analysis yields large amounts of data whose biological relevance is difficult to discern. Proteomic methods introduced into in vitro study are apt to reveal the cellular events and signal transduction of cells in culture [45]. Whether such findings have the same significance in vivo is unknown.
Table 1.
Proteomic studies of bone diseases.
| Protein | Disease | Biological function | Ref. |
|---|---|---|---|
| α crystallin β chain | Osteosarcoma | Antiapoptotic | [39] |
| Ezrin | Osteosarcoma | Cell growth and metastasis | [39] |
| Alcohol dehydrogenase | Osteoarthritis | Glycolysis | [37] |
| Adenylate kinase isoenzyme 1 | Osteoarthritis | Glycolysis | [37] |
| α-enolase | Osteoarthritis | Glycolysis | [37] |
| Pyruvate kinase 3 isoform 2 | Osteoarthritis | Glycolysis | [37] |
| Flavin reductase | Osteoarthritis | Glycolysis | [37] |
| Annexin A1 | Osteoarthritis | Cell proliferation | [37] |
| Phosphatidylethanolamine-binding protein | Osteoarthritis | Signal transduction | [37] |
| Tubby protein homolog | Osteoarthritis | Signal transduction | [37] |
| Peroxiredoxin-3 | Osteoarthritis | Anti-apoptotic, differentiation | [37] |
| Superoxide dismutase | Osteoarthritis | Antioxidant | [37] |
| EGF receptor | Osteosarcoma | Cell growth and differentiation | [40] |
| Ribulose-5-phosphate-epimerase | Osteosarcoma | Catalytic activity | [40] |
| ATP-dependent RNA helicase | Osteosarcoma | Helicase activity | [40] |
| Kelch-like ECH-associated protein 1 | Osteosarcoma | Cell proliferation | [40] |
| Heterogeneous nuclear ribonucleoprotein L | Osteosarcoma | RNA binding | [40] |
| Minichromosome-maintenance protein 7 | Osteosarcoma | Cell proliferation | [40] |
| Erk 2 | Osteosarcoma | Induction of apoptosis | [40] |
| Tissue-type plasminogen activator | Osteosarcoma | Tissue remodeling and degradation, cell migration | [38] |
| Crosslaps | Osteosarcoma | Marker for bone resorption | [38] |
| Anti-p53 antibody | Osteosarcoma | Antiapoptotic | [38] |
| Matrix metalloproteinase 3 | Cartilage degradation | Collagen degradation | [35] |
| Chitinase-3-like protein 1 | Cartilage degradation | Tissue remodeling | [35] |
| Thrombospondin-1 | Cartilage degradation | Cell adhesion | [35] |
| Neutrophil gelatinase-associated lipocalin | Cartilage degradation | Lipophilic substances transport | [35] |
| Gelsolin | Cartilage degradation | Calcium regulation | [35] |
Proteomics of MSCs & osteoblasts
Since bone diseases mainly occur as a result of the disturbance of the bone remodeling rate and the imbalance between the activity of osteoclasts and osteoblasts [1], recent proteomic studies have focused on the differentiation of these cells and their function. Osteoblasts derived from multipotential MSCs can synthesize bone matrix, while osteoclasts derived from monocytes can digest bone [11]. So far, a number of proteomic studies have investigated the self-renewal and differentiation of MSCs and osteoblastogenesis. MSCs, refered to as plastic adherent cells and colony-forming-unit fibroblasts, are pluripotent and have self-renewal capacity [46,47]. Novel molecular and cellular techniques have focused on the quantitation and characterization of STRO-1, CD29, CD44, CD90, CD105, CD166 and MHC-1 as MSC surface markers [48]. However, these biomarkers are not uniquely expressed in stem cells and the molecular mechanism governing MSC self-renewal remains unclear. Proteomic profiles from several clones of BM-MSC at stages of differentiation were obtained using protein separation by 2DE or by 2D LC followed by MALDI MS analysis. These profiling studies indicated that differentiated subcultured MSCs with low-expansion tendency showed differentially expressed proteins in some functional groups: metabolism, signal transduction, cell adhesion and cell growth, cell cytoskeleton, cell–cell interaction, cell cycle, protein degradation and ion transfer (Table 2). In particular, MSCs of high expansion rate (less differentiated) overexpressed calmodulin, T-complex protein 1 α-subunit and tropomyosin compared with those of low expansion rate, while caldesmon and minralocorticoid receptor were downregulated [49–51]. These proteins were reported to be associated with cell cycle and proliferation [50], even contributing to bone turnover [52]. Other studies on osteoblast differentiation from MSCs using 2DE coupled with MALDI TOF MS provide a classic protein profile of differentiating osteoblasts. Some specific differentially expressed proteins, such as chloride intracellular channel 1, have been suggested to play an important role in the process of osteoblast differentiation (Table 3) [53,54].
Table 2.
Proteomic studies of on mesenchymal stem cells.
| Protein | Species | Biological function | Ref. |
|---|---|---|---|
| Vimentin | Mouse | Cell motion | [50] |
| Calreticulin | Mouse | Cell proliferation | [50] |
| Thioredoxin domain containing 7 | Mouse | Electron carrier activity | [50] |
| CD98 | Human | Cell growth | [47] |
| CD147 | Human | Metalloproteinase inducer | [47] |
| CD99 | Human | Cell adhesion | [47] |
| CD47 | Human | Cell adhesion | [47] |
| Calmodulin | Human | Apoptosis | [46] |
| Tropomyosin | Human | Cell division | [46] |
| Corticotropin | Human | Cell proliferation | [46] |
| Caldesmon | Human | Cell division | [46] |
| Annexin-I | Human | Cell proliferation | [46] |
| Annexin-2 | Human | Cell proliferation | [46] |
| Lamin A/C | Human | Embryonic development | [46] |
| Heat-shock protein 27 | Human | Cytoprotection and cell survival | [46] |
| Pyruvate kinase M2 | Human | Cell cycle regulatory | [46] |
| Lecithin cholesterol acyltransferase | Rat | Lipid biosynthesis | [45] |
| Sorbitol dehydrogenase | Rat | Glucose metabolism | [45] |
| Potassium channel 13 | Rat | Ion transport | [45] |
| Mineralocorticoid receptor | Rat | Differentiation | [45] |
| Somatostatin receptor type 5 | Rat | Signal transduction | [45] |
| Adenylate kinase isoenzyme 1 | Rat | Catabolic activity | [45] |
| Cathepsin D | Rat | Proteolysis | [45] |
| T-complex protein 1 α-subunit | Rat | Protein folding | [45] |
| Striatin-3 | Rat | Cell growth | [45] |
Table 3.
Proteomic studies of osteoblast differentiation.
| Protein | Species | Biological function | Ref. |
|---|---|---|---|
| Heat-shock protein 27 | Human | Actin polymerization | [50] |
| Cathepsin D | Human | Proteolysis | [50] |
| Ubiquitin C-terminal hydrolase L1 | Human | Ubiquitin conjugation activity | [50] |
| Phosphoglycerate mutase 1 | Human | Glycolysis | [50] |
| Pyruvate kinase M1 | Human | Cell cycle regulatory | [50] |
| α-enolase | Human | Cell proliferation | [50] |
| ATP synthase | Human | ATP synthesis | [50] |
| Glutathione S-transferase P | Human | Antioxidant | [50] |
| Glutamate dehydrogenase 1 | Human | Antioxidant | [50] |
| Superoxide dismutase | Human | Antioxidant | [50] |
| Chloride intracellular channel 1 | Human | Signal transduction | [49] |
| Acidic ribosomal phosphoprotein P0 | Human | Ribosome biogenesis | [49] |
| Annexin V | Human | Anti-apoptosis | [49] |
| 14-3-3 protein γ | Human | Signal transduction | [49] |
| LIM/homeobox protein | Human | Mesoderm formation | [49] |
| Versican | Human | Cell adhesion | [47] |
| Tenascin | Human | Cell migration | [47] |
| Fatty acid synthase | Human | Lipid biosynthesis | [47] |
Recently, researchers found that some hormones, growth factors and cytokines can regulate the growth, maturation and activity of osteoblasts [21], implying that these circulating factors can affect osteoblastogenesis through specific signal pathways. Extracts from MSCs treated with specific hormones or growth factors were analyzed by 2DE or 2D LC followed by MS analysis in order to find differentially expressed proteins that may play roles in specific signal pathways [55–57]. Of particular applications provided by isotope-labeled quantitative MS, Kratchmarova cultured human MSC with EGF and PDGF in a medium containing distinct forms of arginine either the normal 12C6, 14N4 version or the isotopic variants 13C6, 14N4 (Arg6) or 13C6, 15N4 (Arg10) to metabolically label the entire proteome, making it distinguishable by MS analysis (Table 3). Each treatment matched to one single labeling. Quantitative proteomics can directly compare entire signaling networks in osteoblastogenesis, which are regulated by EGF and PDGF, and discover the critical difference of two factors in regulation of PI3K pathway [58].
Proteomics of osteoclasts
Osteoclasts, members of the monocyte/macrophage family, are bone resorptive cells. Osteoclast proliferation and differentiation is partially driven by osteoblasts, macrophage colony-stimulating factor (M-CSF) and RANKL [11,59]. Researchers have analyzed the whole osteoclast proteome and secreted proteome, as shown in Table 4 [38,60,61]. Czupalla analyzed the protein expression profile of osteoclasts using 2D DIGE coupled with MS, a sensitive and reproducible proteomic technique [61]. By comparing proteins differentially expressed with uniquely expressed gene profile at mRNA levels, two categories of proteins are found. In the first category, proteins differentially expressed were confirmed by the results from mRNA microarray, and in the second category, proteins differentially expressed did not verify their changes in mRNA level. Intriguingly, in the second category, the differentially expressed proteins were detected by using a proteomics approach, but failed to see any change in mRNA level analyzed by gene microarray. The discrepancy in the second category suggests that the proteins encoded by the corresponding genes underwent post-translational modifications, which occurred owing to post-transcriptional interaction and gene–environment interaction. The function of osteoclast and the expression profile of membrane proteins in osteoclasts was studied by Ha et al. using LC-MS/MS [60]. The study focused on the function of differentially expressed Nhedc2 (channel proteins Na+/H+ exchanger domain-containing 2), which was found in osteoclast membrane. Further characterization of Nhedc2 and its family members suggested their key roles in osteoclast fusion during bone resorption. In a recent study from Kubota’s group, they analyzed the secreted proteome of osteoclasts by using both 2DE with MALDI MS/MS analysis and ICAT coupled with quantitative LC MS/MS analysis [38]. Comparison of these two proteomic approaches suggests that these two different methods can produce complementary results, which help elucidate the molecular mechanism of bone resorption and bone formation. Proteins differentially expressed, such as cathepsins, osteopontin, legumain, macrophage and inflammatory protein-1α, were identified, suggesting they are closely related to osteoclast differentiation and bone resorption.
Table 4.
Proteomic studies of osteoclast differentiation.
| Protein | Species | Biological function | Ref. |
|---|---|---|---|
| Vacuolar H-ATPases | Mouse | Proton transport | [57] |
| Cathepsin K | Mouse | Proteolysis | [57] |
| Gelsolin | Mouse | Actin binding | [57] |
| Actin-related protein 2/3 | Mouse | Actin binding | [57] |
| Cofilin | Mouse | Antiapoptosis | [57] |
| Ankylosis | Human | Phosphate transport | [58] |
| Na1/H1 exchanger domain containing 2 | Human | Ion transport | [58] |
| Osteopontin | Mouse | Cell adhesion | [34] |
| Legumain | Mouse | Cysteine-type endopeptidase activity | [34] |
| Macrophage inflammatory protein-1 | Mouse | Immune response | [34] |
| IFNγ-inducible lysosomal thiol reductase | Mouse | Oxidation reduction | [34] |
| Granulin | Mouse | Structural molecule activity | [34] |
To date, (semi)quantitative proteomic approaches have been used successfully in analyzing cellular, membrane and secreted proteins from osteoclasts. The differentially expressed proteins identified by these approaches include known osteoclast markers, such as vacuolar, H-ATPases and cathepsin K, and unknown proteins, such as gelsolin and arp2/3, which play key roles in the maturation of osteoclasts. These proteins respond to the RANKL activation and promote osteoclastogenesis, providing additional information to better understand the molecular mechanism underlying osteoclastogenesis. In addition, the understanding of the OPG/RANKL/RANK signal pathways may provide potential therapeutic targets for the treatment of bone diseases, such as osteoporosis.
Expert commentary
Osteoporosis is prevalent in the elderly population in the USA. It is the result of the imbalance of bone remodeling and the loss of bone mass. Low bone mineral density is one of the strongest risk factors of fracture. Dual-energy x-ray absorptiometry, which measures bone mineral density, is emerging as a better way to monitor the loss of bone mass owing to bone remodeling. However, dual-energy x-ray absorptiometry does not provide information regarding the irreversible process that leads to the loss of bone mass and bone density. Proteomics techniques are useful for dynamic scanning of protein expression in osteoporosis. Bone-turnover markers have proven more powerful than imaging techniques, such as dual-energy x-ray absorptiometry, for predicting the dynamics of bone remodeling [62,63]. Bone turnover markers may provide valuable information of monitoring dynamic bone remodeling. Unfortunately, few bone turnover markers are currently available for early diagnosis, prediction and monitoring of osteoporosis, even though the latest biological study suggests targeting of TGF-β1 signaling could provide an effective therapy for osteoporosis [64]. Recent developments in proteomic technology, particularly quantitative proteomics, provide a great opportunity to discover and validate early bone turnover markers and therapeutic targets.
Five-year view
The use of quantitative proteomics in bone research, namely osteoproteomics, is an emerging field. It is anticipated that such studies will provide insight into the molecular mechanisms of self-renewal and differentiation of MSCs and the transformation of monocytes into osteoclasts. Proteomics, especially quantitative proteomics, hopefully will bring the next generation of diagnostic and therapeutic advances in the management of the very prevalent problem of osteoporosis.
Key issues
Osteoporosis is the result of an imbalance of bone resorption and bone formation.
The key players in bone remodeling are the osteoclasts, derived from hematopoietic stem cells, and the osteoblasts, derived from bone marrow mesenchymal stem cells.
Proteomics has been gaining attention in the field of bone disease research for bone turnover biomarker discovery and cell signaling.
In the area of bone diseases, traditional 2DE coupled with mass spectrometry has been applied to obtain unique protein expression profiles of cartilage degradation, bone sarcoma, osteoarthritis and femoral head osteonecrosis and compared with profiles from normal tissues.
The differentially expressed proteins identified by quantitative proteomic approaches include known osteoclast markers, such as vacuolar, H-ATPases and cathepsin K, and unknown proteins, such as gelsolin and arp2/3, which play key roles in the maturation of osteoclasts.
Mesenchymal stem cells of high expansion rate, overexpress calmodulin, T-complex protein 1 α-subunit and tropomyosin, which are associated with cell cycle and proliferation.
Measurement of protein turnover rates by using quantitative proteomics is a new and powerful approach for discovering of sensitive bone turnover marker in the future.
Footnotes
For reprint orders, please contact reprints@expert-reviews.com
Financial & competing interests disclosure
This work is mainly supported by grants awarded to Gary Guishan Xiao (GGX) from the Bone Biology Program of the Cancer and Smoking Related Disease Research Program and the Nebraska Tobacco Settlement Biomedical Research Program (LB692, LB595, and LB506). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Contributor Information
Hengwei Zhang, Email: hengweizhang@creighton.edu, Genomics & Functional Proteomics Laboratories, Osteoporosis Research Center; Creighton University Medical Center, 601 N 30th Street, Suite 6730, Omaha, NE 68131, USA, Tel.: +1 402 280 4996, Fax: +1 402 280 4284.
Robert Recker, Email: rrecker@creighton.edu, Genomics & Functional Proteomics Laboratories, Osteoporosis Research Center, Creighton University Medical Center, 601 N 30th Street, Suite 6730, Omaha, NE 68131, USA, Tel.: +1 402 280 4471, Fax: +1 402 280 5034.
Wai-Nang Paul Lee, Email: lee@labiomed.org, Metabolomics Core, UCLA Center of Excellence in Pancreatic Diseases, Harbor-UCLA Medical Center, Torrance, CA 90502, USA, Tel.: +1 310 222 6729, Fax: +1 310 222 3887.
Gary Guishan Xiao, Email: gxiao@creighton.edu, Genomics & Functional Proteomics Laboratories, Osteoporosis Research Center, Creighton University Medical Center, 601 N 30th Street, Suite 6730, Omaha, NE 68131, USA, Tel.: +1 402 280 5911, Fax: +1 402 280 4284.
References
Papers of special note have been highlighted as:
• of interest
• • of considerable interest
- 1.Rodan GA. Bone mass homeostasis and bisphosphonate action. Bone. 1997;20:1–4. doi: 10.1016/s8756-3282(96)00318-3. [DOI] [PubMed] [Google Scholar]
- 2.Raisz LG, Rodan GA. Pathogenesis of osteoporosis. Endocrinol Metab Clin North Am. 2003;32:15–24. doi: 10.1016/s0889-8529(02)00055-5. [DOI] [PubMed] [Google Scholar]
- 3.Consensus development conference: diagnosis, prophylaxis, and treatment of osteoporosis. Am J Med. 1993;94:646–650. doi: 10.1016/0002-9343(93)90218-e. [DOI] [PubMed] [Google Scholar]
- 4.Melton LJ., 3rd How many women have osteoporosis now? J Bone Miner Res. 1995;10:175–177. doi: 10.1002/jbmr.5650100202. [DOI] [PubMed] [Google Scholar]
- 5.Advisory Group on Osteoporosis. Report. Department of Health; London, UK: 1994. [Google Scholar]
- 6.Abbott TA, 3rd, Lawrence BJ, Wallach S. Osteoporosis: the need for comprehensive treatment guidelines. Clin Ther. 1996;18:127–149. doi: 10.1016/s0149-2918(96)80186-x. [DOI] [PubMed] [Google Scholar]
- 7.Rodan GA, Martin TJ. Therapeutic approaches to bone diseases. Science. 2000;289:1508–1514. doi: 10.1126/science.289.5484.1508. [DOI] [PubMed] [Google Scholar]
- 8.Riggs BL, Khosla S, Melton LJ., 3rd Sex steroids and the construction and conservation of the adult skeleton. Endocr Rev. 2002;23:279–302. doi: 10.1210/edrv.23.3.0465. [DOI] [PubMed] [Google Scholar]
- 9.Rogers A, Eastell R. Circulating osteoprotegerin and receptor activator for nuclear factor κB ligand: clinical utility in metabolic bone disease assessment. J Clin Endocrinol Metab. 2005;90:6323–6331. doi: 10.1210/jc.2005-0794. [DOI] [PubMed] [Google Scholar]
- 10.Srivastava S, Weitzmann MN, Kimble RB, et al. Estrogen blocks M-CSF gene expression and osteoclast formation by regulating phosphorylation of Egr-1 and its interaction with Sp-1. J Clin Invest. 1998;102:1850–1859. doi: 10.1172/JCI4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teitelbaum SL. Bone resorption by osteoclasts. Science. 2000;289:1504–1508. doi: 10.1126/science.289.5484.1504. [DOI] [PubMed] [Google Scholar]
- 12.Fatayerji D, Mawer EB, Eastell R. The role of insulin-like growth factor I in age-related changes in calcium homeostasis in men. J Clin Endocrinol Metab. 2000;85:4657–4662. doi: 10.1210/jcem.85.12.7031. [DOI] [PubMed] [Google Scholar]
- 13.Seeman E. Invited review: pathogenesis of osteoporosis. J Appl Physiol. 2003;95:2142–2151. doi: 10.1152/japplphysiol.00564.2003. [DOI] [PubMed] [Google Scholar]
- 14.Lee SK, Lorenzo JA. Parathyroid hormone stimulates TRANCE and inhibits osteoprotegerin messenger ribonucleic acid expression in murine bone marrow cultures: correlation with osteoclast-like cell formation. Endocrinology. 1999;140:3552–3561. doi: 10.1210/endo.140.8.6887. [DOI] [PubMed] [Google Scholar]
- 15.Miura M, Tanaka K, Komatsu Y, et al. A novel interaction between thyroid hormones and 1,25(OH)(2)D(3) in osteoclast formation. Biochem Biophys Res Commun. 2002;291:987–994. doi: 10.1006/bbrc.2002.6561. [DOI] [PubMed] [Google Scholar]
- 16.Selye H. On the stimulation of new bone-formation with parathyroid extract and irradiated ergosterol. Endocrinology. 1932;16:547–558. [Google Scholar]
- 17.Lane NE, Sanchez S, Modin GW, Genant HK, Pierini E, Arnaud CD. Parathyroid hormone treatment can reverse corticosteroid-induced osteoporosis. Results of a randomized controlled clinical trial. J Clin Invest. 1998;102:1627–1633. doi: 10.1172/JCI3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hosking D, Chilvers CE, Christiansen C, et al. Prevention of bone loss with alendronate in postmenopausal women under 60 years of age. Early Postmenopausal Intervention Cohort Study Group. N Engl J Med. 1998;338:485–492. doi: 10.1056/NEJM199802193380801. [DOI] [PubMed] [Google Scholar]
- 19.Fisher JE, Rogers MJ, Halasy JM, et al. Alendronate mechanism of action: geranylgeraniol, an intermediate in the mevalonate pathway, prevents inhibition of osteoclast formation, bone resorption, and kinase activation in vitro. Proc Natl Acad Sci USA. 1999;96:133–138. doi: 10.1073/pnas.96.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maerevoet M, Martin C, Duck L. Osteonecrosis of the jaw and bisphosphonates. N Engl J Med. 2005;353:99–102. [PubMed] [Google Scholar]
- 21.Siris ES, Roodman GD. In: Primer on the Metabolic Bone Diseases and Disorders of Mineral Metabolism. 6. Favus MJ, editor. ASBMR; Washington, WA, USA: 2006. pp. 320–330. [Google Scholar]
- 22.Mills BG, Singer FR, Weiner LP, Suffin SC, Stabile E, Holst P. Evidence for both respiratory syncytial virus and measles virus antigens in the osteoclasts of patients with Paget’s disease of bone. Clin Orthop Relat Res. 1984;1(183):303–311. [PubMed] [Google Scholar]
- 23.Hughes AE, Ralston SH, Marken J, et al. Mutations in TNFRSF11A, affecting the signal peptide of RANK, cause familial expansile osteolysis. Nat Genet. 2000;24:45–48. doi: 10.1038/71667. [DOI] [PubMed] [Google Scholar]
- 24.Thomas RJ, Guise TA, Yin JJ, et al. Breast cancer cells interact with osteoblasts to support osteoclast formation. Endocrinology. 1999;140:4451–4458. doi: 10.1210/endo.140.10.7037. [DOI] [PubMed] [Google Scholar]
- 25.Yin JJ, Selander K, Chirgwin JM, et al. TGF-β a signaling blockade inhibits PTHrP secretion by breast cancer cells and bone metastases development. J Clin Invest. 1999;103:197–206. doi: 10.1172/JCI3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romas E, Bakharevski O, Hards DK, et al. Expression of osteoclast differentiation factor at sites of bone erosion in collagen-induced arthritis. Arthritis Rheum. 2000;43:821–826. doi: 10.1002/1529-0131(200004)43:4<821::AID-ANR12>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 27.Komori T, Yagi H, Nomura S, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima K, Zhou X, Kunkel G, et al. The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell. 2002;108:17–29. doi: 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- 29.Galindo M, Pratap J, Young DW, et al. The bone-specific expression of Runx2 oscillates during the cell cycle to support a G1-related antiproliferative function in osteoblasts. J Biol Chem. 2005;280:20274–20285. doi: 10.1074/jbc.M413665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaur T, Lengner CJ, Hovhannisyan H, et al. Canonical WNT signaling promotes osteogenesis by directly stimulating runx2 gene expression. J Biol Chem. 2005;280:33132–33140. doi: 10.1074/jbc.M500608200. [DOI] [PubMed] [Google Scholar]
- 31.Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S. Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics. 2007;6:1000–1006. doi: 10.1074/mcp.M600297-MCP200. [DOI] [PubMed] [Google Scholar]
- 32.Jimenez CR, Spijker S, de Schipper S, et al. Peptidomics of a single identified neuron reveals diversity of multiple neuropeptides with convergent actions on cellular excitability. J Neurosci. 2006;26:518–529. doi: 10.1523/JNEUROSCI.2566-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helmerhorst EJ, Oppenheim FG. Saliva: a dynamic proteome. J Dent Res. 2007;86:680–693. doi: 10.1177/154405910708600802. [DOI] [PubMed] [Google Scholar]
- 34.Kiernan UA. Quantification of target proteins and post-translational modifications in affinity-based proteomic approaches. Expert Rev Proteomics. 2007;4(3):421–428. doi: 10.1586/14789450.4.3.421. [DOI] [PubMed] [Google Scholar]
- 35.Xiao GG, Recker RR, Deng HW. Recent advances in proteomics and cancer biomarker discovery. Clin Med Oncol. 2008;2:1–10. doi: 10.4137/cmo.s539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 37.Zhao Y, Lee WN, Xiao GG. Quantitative proteomics and biomarker discovery in human cancer. Expert Rev Proteomics. 2009;6(2):115–118. doi: 10.1586/epr.09.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota K, Wakabayashi K, Matsuoka T. Proteome analysis of secreted proteins during osteoclast differentiation using two different methods: two-dimensional electrophoresis and isotope-coded affinity tags analysis with two-dimensional chromatography. Proteomics. 2003;3:616–626. doi: 10.1002/pmic.200300410. [DOI] [PubMed] [Google Scholar]
- 39.Wilson R, Belluoccio D, Little CB, Fosang AJ, Bateman JF. Proteomic characterization of mouse cartilage degradation in vitro. Arthritis Rheum. 2008;58:3120–3131. doi: 10.1002/art.23789. [DOI] [PubMed] [Google Scholar]
- 40.Kawai A, Kondo T, Suehara Y, Kikuta K, Hirohashi S. Global protein-expression analysis of bone and soft tissue sarcomas. Clin Orthop Relat Res. 2008;466:2099–2106. doi: 10.1007/s11999-008-0330-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo D, Tan W, Wang F, et al. Proteomic analysis of human articular cartilage: identification of differentially expressed proteins in knee osteoarthritis. Joint Bone Spine. 2008;75:439–444. doi: 10.1016/j.jbspin.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Tan X, Cai D, Wu Y, et al. Comparative analysis of serum proteomes: discovery of proteins associated with osteonecrosis of the femoral head. Transl Res. 2006;148:114–119. doi: 10.1016/j.trsl.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Folio C, Mora MI, Zalacain M, et al. Proteomic analysis of chemonaive pediatric osteosarcomas and corresponding normal bone reveals multiple altered molecular targets. J Proteome Res. 2009;8(8):3882–3888. doi: 10.1021/pr900113w. [DOI] [PubMed] [Google Scholar]
- 44.Kang JH, Park KK, Lee IS, et al. Proteome analysis of responses to ascochlorin in a human osteosarcoma cell line by 2-D gel electrophoresis and MALDI-TOF MS. J Proteome Res. 2006;5:2620–2631. doi: 10.1021/pr060111i. [DOI] [PubMed] [Google Scholar]
- 45.Cubukcuoglu Deniz G, Durdu S, Akar AR, Ozyurda U. Biotechnology and stem cell research: a glance into the future. Anadolu Kardiyol Derg. 2008;8:297–302. [PubMed] [Google Scholar]
- 46.Jiang Y, Jahagirdar BN, Reinhardt RL, et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41–49. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 47.Hassan HT, El-Sheemy M. Adult bone-marrow stem cells and their potential in medicine. J R Soc Med. 2004;97:465–471. doi: 10.1258/jrsm.97.10.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kolf CM, Cho E, Tuan RS. Mesenchymal stromal cells. Biology of adult mesenchymal stem cells: regulation of niche, self-renewal and differentiation. Arthritis Res Ther. 2007;9:204. doi: 10.1186/ar2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49•.Celebi B, Elcin YM. Proteome analysis of rat bone marrow mesenchymal stem cell subcultures. J Proteome Res. 2009;8:2164–2172. doi: 10.1021/pr800590g. Identified differentially regulated proteins during serial subcultures of mesenchymal stem cells. [DOI] [PubMed] [Google Scholar]
- 50.Mareddy S, Broadbent J, Crawford R, Xiao Y. Proteomic profiling of distinct clonal populations of bone marrow mesenchymal stem cells. J Cell Biochem. 2009;106:776–786. doi: 10.1002/jcb.22088. [DOI] [PubMed] [Google Scholar]
- 51.Foster LJ, Zeemann PA, Li C, Mann M, Jensen ON, Kassem M. Differential expression profiling of membrane proteins by quantitative proteomics in a human mesenchymal stem cell line undergoing osteoblast differentiation. Stem Cells. 2005;23:1367–1377. doi: 10.1634/stemcells.2004-0372. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Feng X, McDonald JM. The role of calmodulin in the regulation of osteoclastogenesis. Endocrinology. 2003;144:4536–4543. doi: 10.1210/en.2003-0147. [DOI] [PubMed] [Google Scholar]
- 53.Sun HJ, Bahk YY, Choi YR, et al. A proteomic analysis during serial subculture and osteogenic differentiation of human mesenchymal stem cell. J Orthop Res. 2006;24:2059–2071. doi: 10.1002/jor.20273. [DOI] [PubMed] [Google Scholar]
- 54.Spreafico A, Frediani B, Capperucci C, et al. A proteomic study on human osteoblastic cells proliferation and differentiation. Proteomics. 2006;6:3520–3532. doi: 10.1002/pmic.200500858. [DOI] [PubMed] [Google Scholar]
- 55.Kim SH, Jun S, Jang HS, Lim SK. Identification of parathyroid hormone-regulated proteins in mouse bone marrow cells by proteomics. Biochem Biophys Res Commun. 2005;330:423–429. doi: 10.1016/j.bbrc.2005.02.173. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Khor KA, Sui J, Zhang J, Tan TL, Chen WN. Comparative proteomics profile of osteoblasts cultured on dissimilar hydroxyapatite biomaterials: an iTRAQ-coupled 2-D LC-MS/MS analysis. Proteomics. 2008;8:4249–4258. doi: 10.1002/pmic.200800103. [DOI] [PubMed] [Google Scholar]
- 57.Bennett KP, Bergeron C, Acar E, et al. Proteomics reveals multiple routes to the osteogenic phenotype in mesenchymal stem cells. BMC Genomics. 2007;8:380. doi: 10.1186/1471-2164-8-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58•.Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005;308:1472–1477. doi: 10.1126/science.1107627. Quantitative proteomics compares entire signaling networks and discovers critical differences capable of changing cell fate. [DOI] [PubMed] [Google Scholar]
- 59.Lacey DL, Timms E, Tan HL, et al. Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell. 1998;93:165–176. doi: 10.1016/s0092-8674(00)81569-x. [DOI] [PubMed] [Google Scholar]
- 60.Ha BG, Hong JM, Park JY, et al. Proteomic profile of osteoclast membrane proteins: identification of Na+/H+ exchanger domain containing 2 and its role in osteoclast fusion. Proteomics. 2008;8:2625–2639. doi: 10.1002/pmic.200701192. [DOI] [PubMed] [Google Scholar]
- 61• •.Czupalla C, Mansukoski H, Pursche T, Krause E, Hoflack B. Comparative study of protein and mRNA expression during osteoclastogenesis. Proteomics. 2005;5:3868–3875. doi: 10.1002/pmic.200402059. Analyzed patterns of protein expression during osteoclastogenesis. [DOI] [PubMed] [Google Scholar]
- 62.Kendler DL, Roux C, Benhamou CL, et al. Effects of denosumab on bone mineral density and bone turnover in postmenopausal women transitioning from alendronate therapy. J Bone Miner Res. 2009 doi: 10.1359/jbmr.090716. (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 63.Kostenuik PJ, Nguyen HQ, McCabe J, et al. Denosumab, a fully human monoclonal antibody to RANKL, inhibits bone resorption and increases BMD in knock-in mice that express chimeric (murine/human) RANKL. J Bone Miner Res. 2009;24:182–195. doi: 10.1359/jbmr.081112. [DOI] [PubMed] [Google Scholar]
- 64•.Tang Y, Wu X, Lei W, et al. TGF-β1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med. 2009;15:757–765. doi: 10.1038/nm.1979. Latest issue suggests the new therapy target of TNF-α for osteoporosis. [DOI] [PMC free article] [PubMed] [Google Scholar]