Protein acetylation is a prevalent posttranslational modification (PTM) that modulates diverse biological activities in eukaryotes1a as well as in bacterial pathogenesis1b. In particular, reversible protein acetylation regulated by lysine acetyltransferases (KATs) and lysine deacetylases (KDACs) plays key roles in controlling gene expression and is misregulated in many diseases1a. Identifying the protein substrates of specific KATs is crucial for elucidating the function(s) of acetylation2. Protein acetylation is primarily visualized by employing radiolabeled acetate or acetyl-CoA3, however, autoradiography exhibits low-sensitivity and is hazardous to handle. Alternatively, azide/alkyne-functionalized chemical reporters in conjunction with bioorthogonal ligation methods have afforded new opportunities for imaging or proteomic analysis of PTMs4. Herein, we report alkyne-derivatized chemical reporters that enable rapid detection and identification of acetylated proteins in vitro and in mammalian cells via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC)5 (Figure 1a).
Figure 1.
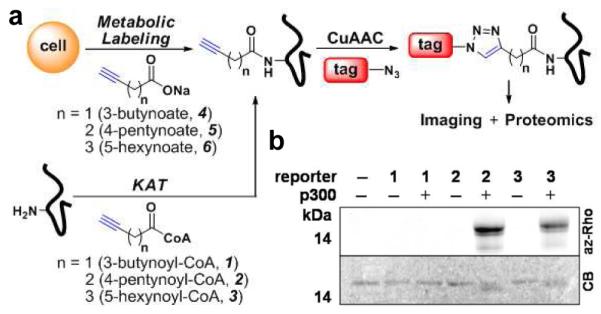
(a) Two-step labeling strategy for detecting protein acetylation in vitro and in cells. (b) In-gel fluorescent detection of p300-catalyzed histone H3 acylation. Coomassie blue (CB) gel shows protein loading control.
We first investigated whether alkynyl-acetyl-CoA could be utilized by KATs in vitro. Three alkynyl-acetyl-CoA analogs, 3-butynoyl-CoA (1), 4-pentynoyl-CoA (2) and 5-hexynoyl-CoA (3), were hence synthesized and evaluated as acyl-CoA donors for full-length p300-KAT6. Mass spectrometry (MS) analysis of p300-acylation reactions with 1-3 using H3 peptide as the acceptor substrate revealed that 2 is readily utilized by p300, while 3 was a less efficient acyl-donor substrate (Figure S1a). Compound 1 did not function as p300 substrate. Examining the stability of these analogs revealed that the half amount of 1 hydrolyzed within 30 minutes, while 2 and 3 remained stable up to 4 hours under p300 reaction conditions (Figure S1b). We hypothesize that the relatively acidic propargylic protons adjacent to the thioester of 1 renders it an unstable acetyl-CoA analog.
To verify that 2 and 3 were transferred onto lysine (Lys) residues, we performed p300-catalyzed acylation with histone H3 protein followed by in-gel trypsin digestion for MS/MS analysis (Figures S1c-d). While alkynyl-acetyl-CoA analogs exhibit low levels of chemical acylation in the absence of KAT, the addition of 4-pentynoyl and 5-hexanoyl groups from 2 and 3, respectively, onto Lys residues significantly increased upon addition of p300 (Table S1). These results suggest that 2 and 3 can be utilized by p300 with similar acceptor specificity in vitro, albeit with reduced efficiency for multiple acylation. These data are consistent with the previous observations of p300 acetyl-donor promiscuity that enables Lys propionylation and butyrylation of histones in vitro and in cells7a,b as well as yeast Gcn5-KAT utilization of chloroacetyl-CoA in vitro7c.
We then evaluated fluorescent detection of KAT activity by CuAAC of the in vitro acylation reactions with azido-rhodamine8 (az-Rho). Both 2 and 3 serve as sensitive reagents for visualizing p300-catalyzed acylation of histone H3 (Figure 1b). In the absence of p300, only minimal fluorescent labeling of histone H3 was observed. Quantifying the observed fluorescent intensities demonstrated that p300-acylation with 2 was time- and enzyme concentration-dependent (Figure S1e-f). Using this fluorescence assay and a standard curve for product formation, we estimated the KM and kcat for 2 is 0.69 ± 0.12 (μM) and 2.10 ± 0.06 (sec−1), respectively (Figure S2). These values are comparable to those determined for acetyl-CoA by radioactivity assay9. These in vitro studies demonstrate that 2 and CuAAC enables rapid fluorescence detection of KAT activity with picomolar sensitivity within minutes compared to days or weeks required for radioactivity.
To investigate whether the alkynyl-acetate analogs can metabolically label proteins in living cells, alkynyl-acetate analogs, 3-butynoate (4), 4-pentynoate (5) and 5-hexynoate (6), were prepared as sodium salts (Figure S3) and examined for incorporation into Jurkat T cells by CuAAC-fluorescent detection (Figure 1a). The results showed that metabolic labeling with 4-6 was both dose- and time-dependent and was optimal at 2.5-10 mM of acetate analogs for 6-8 hours (Figures S4). The selective labeling of core histones and histone H3 derived from metabolically-labeled Jurkat T cells demonstrate that 4-6 can be installed onto known Lys-acetylated proteins (Figure 2a-b). Fluorescent profiling of total cell lysates revealed many proteins labeled by 4-6 (Figure 2c). The majority of selectively labeled proteins were insensitive to cycloheximide treatment and largely distinct from those targeted by longer chain alkynyl-fatty acids analogs8 (Figure S5a-b). Co-incubation with a KDACs inhibitor, suberoylanilide hydroxamic acid (SAHA)10a slightly decreased the addition of 4-6 onto proteins (Figure S5c), which suggests blocking the removal of protein acetylation prevents metabolic incorporation of these acetate analogs. Cells treated with curcumin10b, a p300-inhibitor, reduced the acetate analog labeling of known p300 substrates such as core histones (Figure S5d). Further analysis of total cell lysates showed that not all labeled protein signals were reduce by curcumin (Figure S5d), which suggests these acetate analogs may also be utilized by other KATs, acyltransferases or incorporated by chemical acylation. Nonetheless, these cellular experiments suggest that 4, 5 and 6 can be converted into active alkynyl-acetyl-CoA analogs in cells by promiscuous acetyl-CoA11 or acyl-CoA synthetases and posttranslationally installed onto proteins in living cells.
Figure 2.
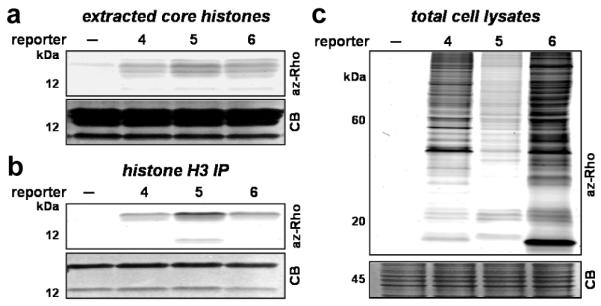
Fluorescent detection of acetate reporter (4, 5 and 6)(labeled (a) core histones, (b) purified histone H3 and (c) total cell lysates from Jurkat T cells.
To identify proteins metabolically labeled by 4, 5 and 6, Jurkat T cell lysates were subjected to CuAAC with the cleavable azido-diazo-biotin tag12 followed by affinity-purification on streptavidin beads (Figure S7). Subsequent treatment of streptavidin beads with sodium dithionite enabled efficient elution of captured proteins for gel-based proteomics using the LTQ-Orbitrap mass spectrometer. A survey of the protein hits from 4, 5 and 6-metabolically labeled cell lysates revealed many reported (86%) as well as new candidate (14%) Lys-acetylated proteins (Table S2). Our in vitro p300-acylation studies, cellular labeling and proteomics data suggest that both 4-pentynoate derivatives (2 and 5) are the optimal chemical reporters for detecting protein acetylation in vitro and in cells. Though 4 and 6 can also metabolically label Lys-acetylated proteins in cells, the CoA derivative of 4, 3-butynoyl-CoA (2) is an unstable substrate for in vitro reactions and 6 may also target long chain fatty-acylated proteins in cells (i.e. transferrin receptor, SNAP-23) as indicated in the MS/MS-identified proteins lists (Table S2). We therefore focused on 5 for additional proteomic studies. From three independent proteomic experiments (Figures S7 and S8), we identified approximately 194 4-pentynoate-labeled proteins from Jurkat T cells (Table S2 and S3), 86% of which were also identified by anti-acetyl-Lys proteomic studies13a,b. We confirmed the enrichment of several MS/MS-identified acetylated proteins, including Ku70, moesin, cofilin, coronin-1A, Hsp90, HMG-1 and adenosine deaminase by Western blotting analysis of the affinity-enriched proteins (Figure S8b). Bioinformatic analysis of our dataset suggest that the majority of acetylated proteins reside in the nucleus and cytoplasm and are associated with diverse cellular functions ranging from metabolism, signal transduction to gene expression (Figure S8c).
To verify that 5 targets Lys residues on proteins in cells, 4-pentynoate-labeled Jurkat T cell lysates were CuAAC-biotinylated, in-solution trypsin digested, purified by streptavidin beads and eluted with Na2S2O4 for MS/MS sequencing. Analysis of recovered peptides demonstrated that 5 is metabolically incorporated onto known sites of Lys acetylation on histone H2B, H3 and H4 (Figure S9). The characteristic marker ion (mass = 259) corresponding to the fragmentation peak of the modified lysine residue (4-pentynoate + CuAAC/Na2S2O4 cleavage adduct) was observed in all MS/MS spectra. These experiments collectively demonstrate the alkynyl-acetate analogs, 2 and 5, function as efficient chemical reporters for protein acetylation in vitro and in cells, respectively. Notably, fluorescent profiling of various mammalian cell lines with 4, 5 and 6 revealed distinct and analog-specific patterns of acetylomes in diverse cell types, which highlights the generality and utility of these bioorthogonal chemical reporters for protein acetylation detection (Figure S6).
Unraveling the functions of protein acetylation remains a challenging task. The bioorthogonal chemical reporters presented here provide readily accessible non-radioactive reagents for fluorescence profiling and large-scale analysis of protein acetylomes. Moreover, alkynyl-acetyl-CoA analogs enable rapid and sensitive detection of KAT activities that should be useful for assigning protein substrates in complex mixtures. This chemical approach provides complementary experimental tools to anti-acetyl-Lys antibodies13a,b, MS/MS13a,b, bioinformatic methods13c as well as affinity-based CoA probes14. The functional analysis of KAT-regulated acetylation will be essential going forward. The incorporation of quantitative proteomic methods13b and bump-hole strategies15 in the future should expand the utility of these chemical tools and facilitate the functional analysis of protein acetylation in physiology and disease.
Supplementary Material
Acknowledgement
We thank members of the Hang laboratory for reagents and advice, Prof. Robert G. Roeder and Dr. Sohail Malik for support and Rockefeller University Proteomics Resource Center for MS analysis. This work was supported by Anderson Cancer Center postdoctoral fellowship at Rockefeller University (Y.-Y.Y.) and NIH/NRSA (1F32DK082140-01A1 - J.M.A.). H.C.H. acknowledges the support from Rockefeller University and NIH/NIDA (1R21DA025751-01).
Footnotes
Supporting Information Available: Experimental procedures and supplemental figures. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1) (a).Yang XJ, Seto E. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Mukherjee S, Hao Y-H, Orth K. Trends Biochem. Sci. 2007;32:210–216. doi: 10.1016/j.tibs.2007.03.007. [DOI] [PubMed] [Google Scholar]
- (2).Lin Y-Y, Lu J-Y, Zhang J, Walter W, Dang W, Wan J, Tao S-C, Qian J, Zhao Y, Boeke JD, Berger SL, Zhu H. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Brownell JE, Allis CD. Proc. Natl. Acad. Sci. U.S.A. 1995;92:6364–6368. doi: 10.1073/pnas.92.14.6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sletten EM, Bertozzi CR. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Meldal M, Tornøe CW. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- (6).An W, Roeder RG. J. Biol. Chem. 2003;278:1504–1510. doi: 10.1074/jbc.M209355200. [DOI] [PubMed] [Google Scholar]
- (7) (a).Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Mol. Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Cheng Z, Tang Y, Chen Y, Kim S, Liu H, Li SSC, Gu W, Zhao Y. Mol. Cell Proteomics. 2009;8:45–52. doi: 10.1074/mcp.M800224-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Yu M, de Carvalho LPS, Sun G, Blanchard JS. J. Am. Chem. Soc. 2006;128:15356–15357. doi: 10.1021/ja066298w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Charron G, Zhang MM, Yount JS, Wilson J, Raghavan AS, Shamir E, Hang HC. J. Am. Chem. Soc. 2009;131:4967–4975. doi: 10.1021/ja810122f. [DOI] [PubMed] [Google Scholar]
- (9).Thompson PR, Kurooka H, nakatani Y, Cole PA. J. Biol. Chem. 2001;276:33721–33729. doi: 10.1074/jbc.M104736200. [DOI] [PubMed] [Google Scholar]
- (10) (a).Marks PA. Oncogene. 2007;26:1351–1356. doi: 10.1038/sj.onc.1210204. [DOI] [PubMed] [Google Scholar]; (b) Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. J. Biol. Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- (11).Ingram-Smith C, Woods BI, Smith KS. Biochemistry. 2006;45:11482–11490. doi: 10.1021/bi061023e. [DOI] [PubMed] [Google Scholar]
- (12).Wilson J, Yang Y-Y, Raghavan A, Charron G, Hang HC. 2009. submitted.
- (13) (a).Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang X-J, Zhao Y. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]; (b) Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]; (c) Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, Allis CD, Hake SB. Proc. Natl. Acad. Sci. U.S.A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hwang Y, Thompson PR, Wang L, Jiang L, Kelleher NL, Cole PA. Angew. Chem. Int. Ed. 2007;46:7621–7624. doi: 10.1002/anie.200702485. [DOI] [PubMed] [Google Scholar]
- (15).Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Proc. Natl. Acad. Sci.U.S.A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


