Abstract
Cancer genomic studies that rely on analysis of biopsies from primary tumors may not fully identify the molecular events associated with tumor progression. We hypothesized that characterizing the transcriptome during tumor progression in the TH-MYCN transgenic model would identify oncogenic drivers that would be targetable therapeutically. We quantified expression of 32,381 murine genes in 9 hyperplastic ganglia harvested at 3 time points, and 4 tumor cohorts of progressively larger size in mice homozygous for the TH-MYCN transgene. We found 93 genes that showed a linearly increasing or decreasing pattern of expression from the preneoplastic ganglia to end stage tumors. Cross-species integration identified 24 genes that were highly expressed in human MYCN amplified neuroblastomas. The genes prioritized were not exclusively driven by increasing Myc transactivation or proliferative rate. We prioritized 3 targets (Cenpe, Gpr49, Impdh2) with previously determined roles in cancer. Using siRNA knockdown in human neuroblastoma cell lines, we further prioritized CENPE due to inhibition of cellular proliferation. Targeting CENPE with the small molecular inhibitor GSK923295 showed inhibition of in vitro proliferation of 19 neuroblastoma cell lines (median IC50=41 nM; range 27–266 nM), and delayed tumor growth in 3 xenograft models (p-values ranged from p<0.0001 to p=0.018). We provide preclinical validation that serial transcriptome analysis of a transgenic mouse model followed by cross-species integration is a useful method to identify therapeutic targets, and identify CENPE as a novel therapeutic candidate in neuroblastoma.
Introduction
Neuroblastoma is a pediatric cancer that arises from the developing sympathetic nervous system. 40% of cases present with a “high-risk” phenotype characterized by wide dissemination at diagnosis, unfavorable biology, and a high risk of relapse and treatment failure (1). Despite intensification of chemoradiotherapy, high-risk tumors historically have overall survival under 40%. (2, 3), Further intensification risks adding negligible benefit at a significant cost (4, 5). A phase III trial targeting the ganglioside GD2 glycolipid, with chimeric monoclonal antibody (ch14.18) and cytokines, recently reported the first substantive improvement in survival by reducing relapse by 20% in high-risk disease (6). This suggests that incorporation of targeted therapeutics may improve treatment outcomes for children with high-risk neuroblastoma.
High-risk neuroblastomas acquire copy-number aberrations that are distinct from lowand intermediate-risk cases, suggesting regions of recurrent somatic alterations harbor candidate genes (7–10). The seminal example of this is MYCN oncogene amplification, which is highly predictive for adverse outcome (11, 12), but MYCN is not yet pharmacologically tractable. An ongoing genome-wide association study has identified multiple single nucleotide polymorphisms (13, 14)} and copy number variations (15) associated with sporadic neuroblastoma. Total attributable risk from these loci remains modest. Genome-wide linkage analysis of familial neuroblastoma pedigrees identified germline mutations in anaplastic lymphoma kinase (ALK) as the major familial neuroblastoma predisposition gene (16). While inhibition ALK is a highly attractive therapeutic target for this subset of patients, tractable molecular targets have not been identified for the majority of high-risk patients.
Here, we utilize a transgenic model of high-risk neuroblastoma to discover somatic transcriptional alterations in murine tumors that appear to be critical for progression. By filtering this list with both human neuroblastoma transcriptional data and matching the resultant gene list with anti-cancer drugs currently in development, we show the utility of this strategy in the prioritization of early phase clinical trial planning.
Materials and Methods
Homozygous TH-MYCN +/+ mice
TH-MYCN mice bred to homozygosity (TH-MYCN+/+) develop tumors with near complete penetrance(17–19). TH-MYCN+/+ mice were sacrificed at birth, day 7, or day 14 to harvest superior cervical and celiac sympathetic ganglia. Ultrasonography (Vevo770 Visual Sonics) was performed three times per week. Mice were sacrificed when tumors were within one of four predetermined size ranges (6 mice per cohort). Specimens were cryopreserved for nucleic acid extraction, or preserved in paraffin and OCT for histological evaluation. Mice were maintained under the protocols and conditions approved by the Institutional Animal Care and Use Committee.
Immunohistochemistry
We stained with endothelial cell marker CD34 (Abcam, ab8158, Cambridge, MA) at 1:50 dilution to quantify vessels. Sections were incubated with a biotinylated goat anti-mouse secondary antibody, and tertiary staining was with horseradish peroxidase-conjugated streptavidin. The immune complex was visualized by using liquid 3,3-diaaminobenzidine (DAB) as a chromagen (DAKO, LSAB 2 System, K0673, North American, Inc, Carpinteria, CA).
Microarray
RNA was extracted from 10μm frozen sections using Qiagen RNeasy Micro Kit (Qiagen, Valencia, CA). Tumor total RNA (76ng-1.6μg) was used to generate cDNA targets that were hybridized to the Applied Biosystems Mouse Genome Survey Microarray version 1.0, and ganglia samples (which became available later in the experimental plan) were hybridized to version 2.0 of the array. Two tumors were hybridized to both microarray versions for quality control. All microarray data are available at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17740.
Differential expression in ganglia and tumor progression
We analyzed tumor and ganglia datasets separately as they were analyzed on different microarray versions, and ganglia specimens required additional genome amplification prior to hybridization. Transcripts were excluded if they were 1) not present on both microarray versions, 2) had no human orthologs, or 3) if transcripts were not expressed in at least three tumor specimens within any cohort. To identify genes with serially increasing or decreasing expression across the ganglia and tumor samples, we calculated non-parametric Spearman's correlation coefficients for each transcript. We used a permutation test to evaluate the false discovery rate of identified genes.
Cross-species integration
We carried out cross-species integration to identify transcripts that were overexpressed with murine neuroblastoma progression and human high-risk MYCN amplified neuroblastomas. We tested human gene expression by comparing 28 low-risk and 20 MYCN-amplified high-risk neuroblastoma transcriptomes, previously assayed using the Human Genome U95v2 expression microarray platform (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE3960) (20). The murine genes were mapped to Affymetrix U95 probe sets using the NCBI Homologene database. Genes with concordant overexpression in murine and human neuroblastomas, proceeded to manual curation prioritizing druggable targets based on literature review. PANTHER classification system was used to classify biological function of genes with concordant expression (21).
Quantitative RT-PCR
Real-time quantitative RT-PCR was performed on TH-MYCN +/+ tumors (Applied Biosystems, Foster, CA). Relative expression of target genes was determined by normalization to Beta-2-microglobulin (B2M), TATA box binding protein (Tbp) and Hypoxanthine phosphoribosyl-transferase (Hprt) using a standard curve. All experiments were performed in triplicate.
MYC transcriptional activity during tumor progression
We evaluated expression of the murine Mycn and Mycc, and human MYCN during tumor progression using quantitative RTPCR. We quantified Myc transcriptional activity using a clinically validated a priori defined gene expression signature (22) based on the Myc target gene database (23) (http://www.myccancergene.org/index.asp). High Myc transcriptional activity was indicated by upregulation or downregulation of Mycc, Mycn and Mycl target genes.
siRNA validation
Functional validation of manually curated genes in a human neuroblastoma derived cell line (NB1643) was performed using transient transfection of siRNA against TACSTD1, IMPDH2, GPR49 and CENPE (Dharmacon, Thermo Scientific catalogue numbers: L-004568-01, L-004330-00, L-004552-00 and L-003252-00). CENPE siRNA knockdown was also performed in NB-EBc1, Kelly, SKNAS and NGP. Cell proliferation was monitored using the RT-CES (ACEA Biosciences, San Diego) that measures substrate adherent growth in real time, as described previously (24). Each experiment was performed in triplicate.
Cell cycle analyses
Cells were seeded at 50% confluence and cultured overnight to allow cell attachment. Cells were treated with inhibitor 24 hours after seeding. At each treatment timepoint media was removed and cells were harvested using 0.05% Trypsin-EDTA. Media and trypsin fractions were then combined and spun. Cells were fixed with ice-cold ethanol overnight. Membranes were permeablized by incubating in phosphate-citric acid buffer for 5 minutes. Cells were spun and incubated with propidium iodide (50 μg/mL) and RNase A (250 μg/mL) for 30 minutes and analyzed by flow cytometry using a FACSCalibur (BD Bioscience, Franklin Lakes NJ).
Pharmacologic CENPE inhibition
In vitro activity of GSK923295 dissolved in dimethyl sulfoxide (DMSO) was evaluated in 19 neuroblastoma cell lines using the RT-CES system. IC50 was calculated from the area under the curve (AUC) across a 4 log dose range (1–10,000 nM).
Xenograft studies
CB17 scid mice (Taconic Farms, NY) were used to propagate subcutaneously implanted neuroblastoma tumors. Tumor diameters were measured using calipers. Tumor volumes were calculated using the formula, (π/6)*diameter3. Once tumor volume exceeded 200 mm3, mice were randomized (n=10 per arm) to receive either GSK923295 125 mg/kg IP or vehicle (96% acidified water, 2% DMAC, 2% CREM) for a total of 6 doses using a 3 days on, 4 days off, 3 days on regimen.
Proliferative signature during tumor progression
We utilized an a priori-defined “proliferative signature” that identified genes that are upregulated in rapidly proliferating cells, both malignant and non-malignant (25). We compared the distribution of non-parametric Spearman's correlation coefficients of the “proliferative signature” transcripts to murine transcripts with human orthologs that were not contained within the proliferative signature.
Results
Modeling tumor progression
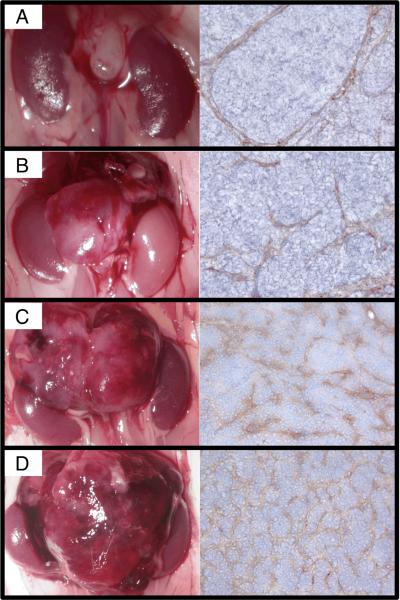
We sacrificed TH-MYCN+/+ mice at day 0 (n=5), day 7 (n=2) and day 14 (N=2) of life to harvest sympathetic ganglia containing foci of neuroblast hyperplasia. To model progression of macroscopic tumors we monitored tumor growth in TH-MYCN+/+ mice using ultrasound. Mice were sacrificed when tumor dimensions were within predetermined ranges (Figure 1). Increasing tumor volume on ultrasound corresponded to increasing tumor weight: Group A, mean weight 0.08±0.02 grams; Group B, 0.35±0.09 grams; Group C, 0.94±0.22 grams; and Group D, 2.01±0.59 grams. We confirmed that harvesting tumors at different sizes provides a model of neuroblastoma progression by comparing cohorts for macroscopic and histological vascularity, locoregional invasion, infiltration of intratumoral vasculature, and metastases. The smallest tumors were asymptomatic, avascular, and had no evidence of metastasis. With increasing tumor size there was increasing displacement and invasion of local structures and increasing vascularity (Figure 1A–D). The two larger tumor sizes (group C and D) invaded tumor vasculature, and in the largest tumors (group D) distant metastases were detected (Supplementary Table 1).
Figure 1. Harvested murine neuroblastomas represented clinically relevant stages of disease.
Ultrasonography was used to monitor tumor growth. Tumors were harvested within four predetermined cohorts (n=6/cohort). Group A tumors were harvested once detected on ultrasound (median 0.08 ± 0.02 grams) and were macroscopically avascular with scant vascularity on CD34 staining (A). Group B tumors (0.35 ± 0.09 grams) displaced local structures, showed increased vascularity (B), but neither Group A or B tumors showed invasion of vasculature or metastases. Group C tumors (0.94 ± 0.22 grams) showed local invasion (C) but no metastasis, although 5/6 tumors had intratumoral invasion of vasculature. Group D (2.01g ± 0.59 grams) showed extensive local invasion (D) and intratumoral invasion of vasculature, with distant metastases in 3/6 animals. Group C and D tumors showed increasing vascularity that was grossly disorganized in the Group D tumors. CD34 immunohistochemistry images shown at 200×.
Identification of differentially expressed genes associated with neuroblastoma progression
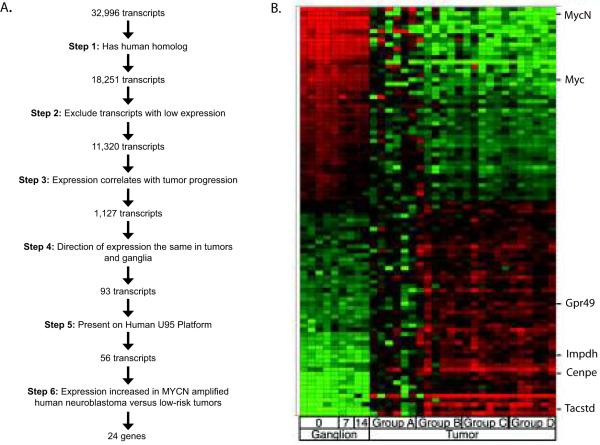
We assayed each harvested sample with a murine-specific microarray. Our algorithmic approach is shown in Figure 2A. The murine specific microarray interrogated 32,381 genes, of which 18,251 genes had human orthologs. The exclusion of transcripts with low expression in any tumor cohort yielded 11,320 transcripts for further consideration. We next identified transcripts showing the strongest correlation of mRNA quantity to tumor progression using Spearman's method (Supplementary Table 2). Utilizing a conservative false discovery rate of 0.05 we obtained 1127 transcripts. We filtered the 1127 candidates by determining which transcripts showed the same direction of expression in murine sympathetic ganglia across day 0, day 7 and day 14 of life. Of 1127 candidates, 93 transcripts showed a consistent alteration in mRNA quantity (one-tailed p value ≤0.01) with progression from hyperplastic ganglia to advanced neuroblastomas (Figure 2B; Supplementary Table 2).
Figure 2.
(A) Algorithm for identifying 24 candidates from ~32,000 transcripts. Transcripts showing a continuous increase or decrease in expression at clinically relevant stages of tumor progression, and in hyperplastic ganglia at day 0, 7 and 14 were identified using Spearman's method. Microscopic ganglia required an additional round of amplification so were analyzed separately. Of the 93 transcripts, cross-species integration was used to prioritize 24 biologically important genes that were overexpressed in human high-risk MYCN amplified neuroblastomas. (B) Heat map representation of 93 differentially expressed murine genes. Samples are grouped into ganglion samples (day 0, day 7, and day 14) and tumor samples (Groups A–D) along the X-axis. The 93 murine transcripts showing the most significant differential expression are shown along the Y-axis with red indicating greater expression, and green less expression. Four genes selected for functional evaluation are indicated. Note that murine Mycc and Mycn showed decreased expression with tumor progression. The full 93 gene set is described in Supplementary Table 2.
Cross species integration of gene expression
Of the 93 murine genes with differential expression, 56 had homologs present on the human microarray platform and 32 of these had increasing expression with tumor progression. We focused on genes with increasing expression, as potential oncogenes provide a more tractable therapeutic target than potential tumor suppressors. Of the 32 murine transcripts, we found 24 unique genes that were overexpressed in human high-risk MYCN amplified neuroblastoma compared to low-risk, non-amplified neuroblastomas (Supplementary Table 2). The 24 genes with concordant overexpression in murine and human MYCN amplified neuroblastoma were classified using PANTHER biological function (21).
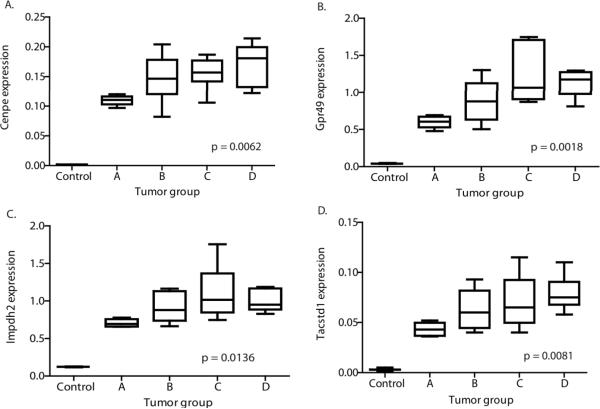
Manual curation of the 24 differentially expressed transcripts prioritized 3 genes with previously described roles in cancer and potential for therapeutic inhibition (Gpr49, Impdh2, and Cenpe). To act as a control for cross species integration, we identified the transcript with the largest increase in expression across murine tumor progression that was not overexpressed in human MYCN amplified neuroblastomas. This transcript was tumor-associated calcium signal transducer-1 gene (Tacstd1). Quantitative PCR was used to confirm that the three candidate transcripts and the control transcript had increased expression with tumor progression (Figure 3A–D).
Figure 3. Prioritized transcripts showed increased expression with tumor progression but Myc transcriptional activity did not increase.
Real-time PCR confirmed that three genes prioritized using a literature search (A) Cenpe, (B) Gpr49, (C) Impdh2 and a control gene not overexpressed in human MYCN amplified neuroblastoma (D) Tacstd1, showed a significant linear increase in expression with tumor size.
MYCN transactivation did not increase with tumor progression
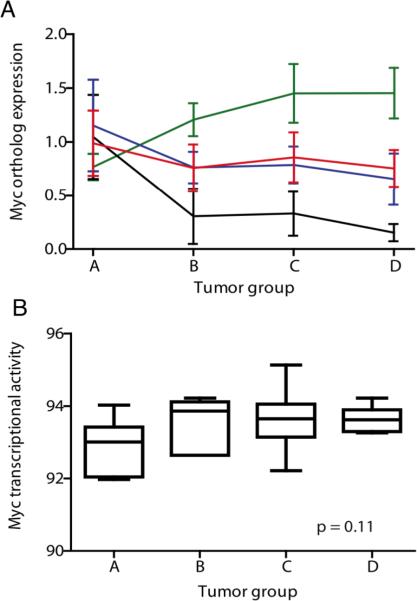
To ensure that our candidate genes were not simply the result of the basal myc activity, we investigated whether the human MYCN transgene was driving alterations in gene expression with tumor progression. Human MYCN transgene expression increased with tumor progression, but murine Mycn and Myc showed a decrease in expression. Overall, the average expression of MYCN, Mycc, and Mycn remained unchanged from small avascular tumors to large metastasizing tumors (Figure 4A). Myc transcriptional activity as indicated by upregulation or downregulation of Myc, Mycn and Myc target genes did not show a significant increase with tumor progression (Figure 4B). Cenpe, Gpr49, and Impdh2 showed a significant linear increase in expression with tumor progression (Figure 3A–D), but Myc transcriptional activity did not (Figure 4B).
Figure 4. Gene expression signatures not driven by basal myc expression.
(A) Murine neuroblastoma expression of human MYCN transgene mRNA (green) increased with tumor progression while murine Mycn (black) and Mycc (blue) expression decreased. Overall, the average MYCN, Mycn and Mycc expression did not increase with tumor progression (red). (B) Myc transcriptional activity was quantified using a clinically validated a priori defined gene expression signature (27) with a higher score indicating greater upregulation or downregulation of Myc targets. Myc transcriptional activity did not show a significant linear increase with tumor expression suggesting that alterations in MYCN transactivation did not exclusively drive differential gene expression.
Transcripts with increased expression were not exclusively composed of genes involved in proliferation
Following prioritization of the mitotic kinesin Cenpe, we sought to characterize if there was an excessive prevalence of other genes with cell cycle-related functions that were overexpressed in the larger murine tumors. Using an a priori defined gene expression signature we established that genes identified as being upregulated in rapidly dividing cells (30) were more likely to show upregulation in larger tumors compared to ~18,000 other murine transcripts (Supplementary Figure 1). Amongst the 93 murine transcripts showing the greatest differential expression with tumor progression, there were 49 where expression increased with tumor progression. Of the 49 murine transcripts showing the greatest increase in expression with tumor progression, only 4 were directly involved with DNA replication, 2 were in the kinetochore, and 2 were involved with purine synthesis.
Transcription of MYC target genes did not increase with tumor progression
To ensure that our candidate genes were not simply the result of the basal myc activity, we investigated whether the human MYCN transgene was driving alterations in gene expression with tumor progression. Human MYCN transgene expression increased with tumor progression, but murine Mycn and Myc showed a parallel decrease in expression. Overall, the average expression of MYCN, Mycc, and Mycn remained unchanged from small avascular tumors to large metastasizing tumors (Figure 4A).
To evaluate expression of Myc target genes in murine tumors, we used a pre-defined gene expression signature of Myc transcriptional targets shown to predict relapse and death from low, intermediate and high-risk human neuroblastoma. This signature quantifies the overall pattern of transcription of Myc targets rather than limiting the evaluation to single transcripts. Overall transcription of the Myc target gene signature did not show a significant increase with tumor progression (Figure 4B).
Functional validation prioritizes CENPE for further investigation
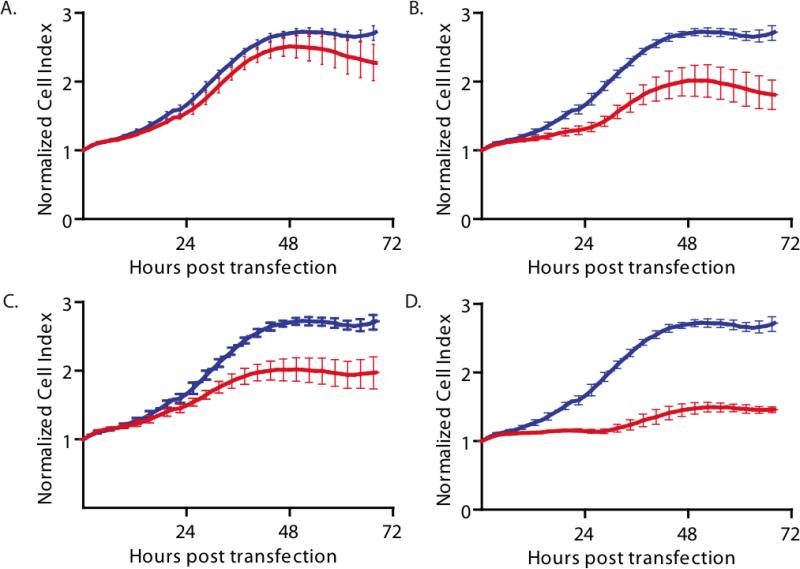
To functionally validate and prioritize the three candidate neuroblastoma targets (CENPE, GPR49, IMPDH2) we performed siRNA knockdown in a MYCN amplified human neuroblastoma derived cell line (NB-1643). We also knocked down the control gene (TACSTD1) which was overexpressed with tumor progression, but not overexpressed by human high-risk MYCN amplified neuroblastomas. siRNA knockdown resulted in variable effects on cellular proliferation. Transient knockdown of TACSTD1 resulted in no significant decrease in growth velocity when compared to a targeted control, GAPDH (Figure 5A). Knockdown of GRP49 (Figure 5B) and IMPDH2 (Figure 5C) showed modest inhibition of proliferation, but knockdown of CENPE resulted in the greatest inhibition prioritizing CENPE for further evaluation (Figure 5D). Finally, to assure that the CENPE siRNA inhibition was not specific to the human neuroblastoma cell line NB1643, we performed additional transient siRNA knockdown experiments in three additional MCYN amplified human neuroblastoma-derived cell lines (Kelly NGP, and NBEbC1) as well as one non-amplified line (SKNAS). There was significant inhibition of cell growth in all three amplified lines, while the non-amplified line showed little effect. (Supplemental Figure 2C)
Figure 5. siRNA knockdown prioritizes CENPE for further functional evaluation.
We monitored proliferation of NB-1643, a MYCN-amplified human neuroblastoma cell line, using the RT-CES microelectronic cell sensor system. (A) The control gene, TACSTD1, which was not overexpressed in human MYCN amplified neuroblastomas showed minimal inhibition of proliferation with knockdown. Inhibition was modest for (B) GPR49 and (C) IMPDH2. (D) CENPE siRNA caused the greatest inhibition. The gene of interest is red and GAPDH (negative control) is blue. All experiments performed in triplicate, shown as mean ± SEM. PLK (positive control) showed 100% cell kill at 72 hours post transfection in all cases.
CENPE knockdown-induced suppression of neuroblastoma cell proliferation results from mitotic arrest
Following confirmation of knockdown by RT-PCR (Supplemental Figure 2B), we examined cell cycle inhibition in two human neuroblastoma cell lines (SKNAS and NGP) using flow cytometry. At twenty-four hours, there was an accumulating cell population in the G2/M phase of the cell cycle. Notably, there is no sub-2N population to suggest apoptotic cell death. (Supplemental Figure 2A).
Pharmacologic inhibition of Cenpe results in anti-tumor activity in vitro and in vivo
We sought validation that pharmacological CENPE inhibition was efficacious in preclinical models. A potent CENPE Inhibitor, GSK923295, demonstrated broad efficacy against a panel of 19 human neuroblastoma derived cell lines with an average growth IC50 of 41 nM (range 27–266 nM; Supplementary Table 3). Growth IC50 did not correlate to cell line MYCN amplification status (p = 0.51), CENPE copy number gain (p = 0.29), in vitro cell line doubling time (r = 0.41, p = 0.0805), or CENPE mRNA expression as previously assayed by the Illumina Human-6 v2 expression BeadChip (r = − 0.12, p = 0.62).
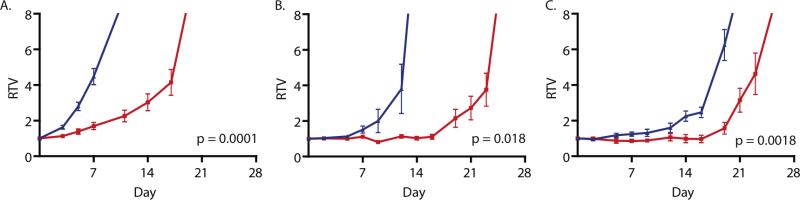
We selected three cells with a spectrum of in vitro growth IC50 (NB-EBc1: 34 nM; NB1643: 57 nM; NB1691: 103 nM) to assess in vivo GSK293295 activity. Xenografts of mice treated with GSK293295A showed significant tumor growth delay compared to the control arm (NB-EBc1 p<0.0001; NB-1643 p = 0.018; NB-1691 p = 0.0018; Figure 6). GSK293295A does not target murine Cenpe so no toxicity was seen, and a therapeutic index could not be estimated.
Figure 6. Pharmacological CENPE inhibition caused tumor growth delay in three human neuroblastoma xenografts.
Mice were randomized to receive either GSK923295 125 mg/kg IP (red) or vehicle (blue: 96% acidified water, 2% DMAC, 2% CREM). Relative tumor volume (RTV), is the measured tumor volume divided by the enrolment tumor volume displayed as mean ± SEM. Linear mixed-effects analysis demonstrated smaller relative tumor volume in the treatment arm in all three xenografts. (A) NB-EBc1,p = 0.0001; (B) NB-1643, p = 0.018; and (C) NB-1691, p = 0.0018.
Discussion
Emerging evidence suggests that neuroblastoma initiation and progression occur due to a combination of inherited variants and mutations (13–16), and stepwise somatic accumulation of drivers (10, 26). Diagnostic specimens provide a snapshot at one time point and the molecular events associated with tumor evolution may not be fully identified. We hypothesized that characterizing the evolution of the neuroblastoma transcriptome at multiple time points would augment existing cancer gene discovery. We serially sacrificed TH-MYCN mice to obtain specimens to provide a model of human neuroblastoma progression. Utilizing a transgenic system meant we could use cross-species integration to reduce the number of candidate genes prior to manual curation. Mouse tumors with chromosomal instability acquire genomic aberrations that are orthologous with their respective human cancers suggesting that selection pressures are tissue specific (27). Cross species integration has been utilized with the transcriptome as expression signatures of oncogenic KRAS2 mutations were identified using cross species integration transgenic and human cancers (28). We predicted that biologically relevant alterations in gene expression would also be conserved. By identifying commonalities in gene expression we posited we would be more likely to identify critical driver genes that could be inhibited therapeutically.
MYCN overexpression is responsible for tumor initiation in the TH-MYCN model, but it was unknown whether increasing MYCN transcriptional activity accounted for differential gene expression with tumor progression. Overall expression of Myc homologs and Myc transcriptional activity remained constant with tumor progression, suggesting that increasing MYCN transactivation was not exclusively driving alterations in gene expression. These findings are consistent with the hypothesis that the latent period prior to murine neuroblastoma initiation is due to the time needed to acquire cooperating mutations (17) which may contribute to differential gene expression. Following prioritization of the mitotic protein, CENPE, we queried how many other genes identified had cell cycle related functions. We established that genes upregulated in rapidly proliferating cells (25) showed greater upregulation with tumor progression than ~18,000 other murine transcripts. However, of the 93 murine genes with the greatest differential expression, 49 of those had increasing expression, and of those only 4 were directly involved with DNA replication, 2 were in the kinetochore, and 2 were involved with purine synthesis. Therefore, upregulation of the remaining transcripts was a not direct result of increased proliferative rate. These validations increase our confidence that we have not purely identified candidate genes that are artifacts of the model system, but additional important functional mediators of tumorigenesis.
We identified 93 genes whose expression continuously increased or decreased across ganglia and tumor progression (Figure 1; Supplementary table 2). We utilized cross-species integration to reduce the number of transcript candidates by selecting those genes whose expression was greater in high-risk MYCN amplified neuroblastomas relative to low-risk tumors without amplification of MYCN (Supplementary Table 2). The resultant 24 genes were prioritized based on whether we could inhibit them pharmacologically, and identified three target genes. The G-protein-coupled-receptor (GPR49/LGR5) is overexpressed in carcinomas (36–38). Inosine monophosphate dehydrogenase type II (IMPDH2) is an enzyme involved in purine synthesis. Functional validation using siRNA-mediated knockdown against two target candidates, IMPDH2 and GPR49, resulted in moderate inhibition. siRNA-mediated knockdown caused low or no inhibition in the control gene, TACSTD1, that was not overexpressed in human high-risk disease. It is possible that further analysis of the candidate transcripts in cell lines with differing genomic profiles or different model systems will establish a greater therapeutic benefit.
Knockdown of centromere-associated protein E (CENPE) demonstrated the greatest growth inhibition in vitro. CENPE is a kinesin motor protein, included in the kinetochore protein complex, whose motor activity is required for correct chromosomal alignment during metaphase. In order to satisfy the spindle assembly checkpoint prior to anaphase (33, 34), CENPE must be simultaneously bound to both the kinetochore and spindle microtubule to prevent asymmetric separation of sister chromatids (34). We focused our initial studies on CENPE as knockdown demonstrated the greatest inhibition, mitotic kinesins are druggable targets and inhibition of mitosis is a proven cancer paradigm. The traditional microtubule binding drugs, vinca alkaloids and taxanes, have documented anti-tumor activity against a broad spectrum of malignancies including neuroblastoma. Microtubule disruption during mitosis inhibits the mitotic spindle resulting in mitotic arrest in tumor cells, followed in some cases by cell death. CENPE's known functions are restricted to cell division suggesting that on-target toxicity will be limited to dividing cells, theoretically providing an advantageous therapeutic index over microtubule binding agents.
GSK923295 is a potent and selective small molecule inhibitor of human CENPE with a Ki of 3.2 nM. It directly inhibits microtubule stimulated ATPase activity of the human CENPE motor domain, resulting in irreversible binding of CENPE to microtubules and mitotic arrest. GSK923295 has demonstrated in vitro and in vivo activity against a broad spectrum of malignancies, A phase I study of GSK923295 is currently nearing completion in adults with refractory solid cancers, and early results have been promising in terms of safety (35) The identification of CENPE in our cross-species integrative genomics study of transcriptome evolution suggests it is also a rational target for neuroblastoma. We show that GSK923295 has significant activity against a panel of human neuroblastoma-derived cell lines in vitro and in vivo. Our findings suggest that a transgenic model recapitulating the MYCN amplified subset of neuroblastoma can discover downstream oncogenic mediators common to tumors without MYCN amplification. Importantly, this inhibitor is relatively inactive against murine Cenpe meaning we were unable to test the inhibitors against TH-MYCN neuroblastomas used for target discovery, and nor could we estimate a therapeutic index in xenograft or transgenic models. Interestingly, although the identification of CENPE included differential expression between MYCN amplified and non-amplified human tumors, GSK023295 activity did not correlate with MYC status or CENPE expression. This suggests the presence of as yet unidentified variables important in predicting CENPE activity.
The mechanism for increased CENPE expression in neuroblastoma remains unclear. CENPE is not a known Myc target gene (http://www.myccancergene.org/index.asp). We analyzed CENPE copy number on 599 primary neuroblastoma tumors previously assayed using the Illumina HumanHap550 SNP microarray. Relative copy number loss occurred in 66/599 (11%), and gain in 48/599 (8%) of tumors. Copy-number gains were mostly whole chromosome gains and were not significantly associated with CENPE overexpression (p = 0.89). This indicates that CENPE would be unlikely to have been prioritized as target utilizing genomic data obtained at diagnosis, but it was identified through serial transcriptomic analysis of neuroblastoma progression. This suggests serial transcriptome analysis can augment existing integrative genomic strategies.
In summary, we utilized serial transcriptome analysis and cross species integration to identify molecular targets associated with tumor progression. Our gene set includes many targets with no previously established role in tumor progression and numerous candidates suitable for pathway analysis. Future follow-up studies with these genes may uncover novel biological roles in neuroblastoma oncogenesis. The focus of this study was to identify molecular targets from this gene set that have the potential to be quickly translated into therapies for high-risk neuroblastoma. This led to the preclinical evaluation of a CENPE inhibitor that is currently in an adult phase 1 trial. Other genes identified in this study may prove to be equally strong candidates for therapeutic targeting. Likewise, analysis of transcripts with decreasing expression may aid identification of bona fide tumor suppressor genes, which are yet to be identified in sporadic neuroblastoma. Future studies will focus on the transcriptional and post-transcriptional regulation of CENPE expression in neuroblastoma cells. In conclusion, we have provided strong preclinical rationale that serial transcriptome profiling of a spontaneous tumor model with cross-species integration can identify and prioritize novel therapeutic targets in neuroblastoma. This strategy may have utility in other tumor types and further utility in neuroblastoma for target discovery.
Supplementary Material
Acknowledgments
This work was supported in part by NIH Grants R01-CA78545 (JMM), R01-CA87847 (JMM), P01 CA97323 (JMM), NH&MRC Australia (GMM), Cancer Institute New South Wales (GMM), Cancer Council New South Wales (GMM), the Alex's Lemonade Stand Foundation (JMM), a research collaborative agreement with GlaxoSmithKline, and the Abramson Family Cancer Research Institute (JMM).
Footnotes
None of the authors had a financial other conflict of interest.
References
- 1.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 2.Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children's Cancer Group. New England Journal of Medicine. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- 3.Berthold F, Boos J, Burdach S, et al. Myeloablative megatherapy with autologous stem-cell rescue versus oral maintenance chemotherapy as consolidation treatment in patients with high-risk neuroblastoma: a randomised controlled trial. Lancet Oncol. 2005;6:649–58. doi: 10.1016/S1470-2045(05)70291-6. [DOI] [PubMed] [Google Scholar]
- 4.Oeffinger KC, Mertens AC, Sklar CA, et al. Chronic health conditions in adult survivors of childhood cancer. N Engl J Med. 2006;355:1572–82. doi: 10.1056/NEJMsa060185. [DOI] [PubMed] [Google Scholar]
- 5.Hobbie WL, Moshang T, Carlson CA, et al. Late effects in survivors of tandem peripheral blood stem cell transplant for high-risk neuroblastoma. Pediatr Blood Cancer. 2008;51:679–83. doi: 10.1002/pbc.21683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu AL, Gilman AL, Ozkaynak MF, et al. A phase III randomized trial of the chimeric anti-GD2 antibody ch14.18 with GM-CSF and IL2 as immunotherapy following dose intensive chemotherapy for high-risk neuroblastoma: Children's Oncology Group (COG) study ANBL0032. Journal of Clinical Oncology. 2009;27 Abstr 10067z. [Google Scholar]
- 7.Mosse YP, Diskin SJ, Wasserman N, et al. Neuroblastomas have distinct genomic DNA profiles that predict clinical phenotype and regional gene expression. Genes Chromosomes Cancer. 2007;46:936–49. doi: 10.1002/gcc.20477. [DOI] [PubMed] [Google Scholar]
- 8.Spitz R, Oberthuer A, Zapatka M, et al. Oligonucleotide array-based comparative genomic hybridization (aCGH) of 90 neuroblastomas reveals aberration patterns closely associated with relapse pattern and outcome. Genes Chromosomes Cancer. 2006;45:1130–42. doi: 10.1002/gcc.20376. [DOI] [PubMed] [Google Scholar]
- 9.Vandesompele J, Baudis M, De Preter K, et al. Unequivocal delineation of clinicogenetic subgroups and development of a new model for improved outcome prediction in neuroblastoma. J Clin Oncol. 2005;23:2280–99. doi: 10.1200/JCO.2005.06.104. [DOI] [PubMed] [Google Scholar]
- 10.Janoueix-Lerosey I, Schleiermacher G, Michels E, et al. Overall genomic pattern is a predictor of outcome in neuroblastoma. J Clin Oncol. 2009;27:1026–33. doi: 10.1200/JCO.2008.16.0630. [DOI] [PubMed] [Google Scholar]
- 11.Brodeur G, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–4. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 12.Seeger RC, Brodeur GM, Sather H, et al. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. New England Journal of Medicine. 1985;313:1111–6. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- 13.Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;41:718–23. doi: 10.1038/ng.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;358:2585–93. doi: 10.1056/NEJMoa0708698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;459:987–91. doi: 10.1038/nature08035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosse YP, Laudenslager M, Longo L, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–5. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO Journal. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansford LM, Thomas WD, Keating JM, et al. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci U S A. 2004;101:12664–9. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chesler L, Goldenberg DD, Seales IT, et al. Malignant progression and blockade of angiogenesis in a murine transgenic model of neuroblastoma. Cancer Res. 2007;67:9435–42. doi: 10.1158/0008-5472.CAN-07-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Diskin S, Rappaport E, et al. Integrative genomics identifies distinct molecular classes of neuroblastoma and shows that multiple genes are targeted by regional alterations in DNA copy number. Cancer Res. 2006;66:6050–62. doi: 10.1158/0008-5472.CAN-05-4618. [DOI] [PubMed] [Google Scholar]
- 21.Thomas PD, Campbell MJ, Kejariwal A, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–41. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fredlund E, Ringner M, Maris JM, Pahlman S. High Myc pathway activity and low stage of neuronal differentiation associate with poor outcome in neuroblastoma. Proc Natl Acad Sci U S A. 2008 doi: 10.1073/pnas.0804455105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol. 2003;4:R69. doi: 10.1186/gb-2003-4-10-r69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cole KA, Attiyeh EF, Mosse YP, et al. A Functional Screen Identifies miR-34a as a Candidate Neuroblastoma Tumor Suppressor Gene. Mol Cancer Res. 2008;6:735–42. doi: 10.1158/1541-7786.MCR-07-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 26.Van Roy N, De Preter K, Hoebeeck J, et al. The emerging molecular pathogenesis of neuroblastoma: implications for improved risk assessment and targeted therapy. Genome Med. 2009;1:74. doi: 10.1186/gm74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett CS, Hodgson JG, Law ME, et al. Genome-wide array CGH analysis of murine neuroblastoma reveals distinct genomic aberrations which parallel those in human tumors. Cancer Res. 2003;63:5266–73. [PubMed] [Google Scholar]
- 28.Sweet-Cordero A, Mukherjee S, Subramanian A, et al. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- 29.Louis SF, Vermolen BJ, Garini Y, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci U S A. 2005;102:9613–8. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto Y, Sakamoto M, Fujii G, et al. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528–33. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- 31.Tanese K, Fukuma M, Yamada T, et al. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol. 2008;173:835–43. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McClanahan T, Koseoglu S, Smith K, et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–26. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 33.Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–93. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- 34.Wood KW, Chua P, Sutton D, Jackson JR. Centromere-associated protein E: a motor that puts the brakes on the mitotic checkpoint. Clin Cancer Res. 2008;14:7588–92. doi: 10.1158/1078-0432.CCR-07-4443. [DOI] [PubMed] [Google Scholar]
- 35.Hu ZKW, Das D, Ziyad S, Gu S, Bhattacharya S, Wyrobek A, Wang NJ, Felier HS, Wooster R, Weber B, Gray JW. Small Molecular Inhibitory of theCentromere-Associated Protein E (CENP-E), GSK923295A Inhibits Cell Growth in Breast Cancer Cells. American Association of Cancer Research. 2009 Denver, CO; 2009. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.