Summary
Polycomb group (PcG) proteins are conserved epigenetic transcriptional repressors that control numerous developmental gene expression programs and have recently been implicated in modulating embryonic stem cell (ESC) fate. We identified the PcG protein PCL2 (polycomb-like 2) in a genome-wide screen for regulators of self-renewal and pluripotency and predicted that it would play an important role in mouse ESC fate determination. Using multiple biochemical strategies, we provide evidence that PCL2 is a Polycomb Repressive Complex 2 (PRC2)-associated protein in mouse ESCs. Knockdown of Pcl2 in ESCs resulted in heightened self-renewal characteristics, defects in differentiation and altered patterns of histone methylation. Integration of global gene expression and promoter occupancy analyses allowed us to identify PCL2 and PRC2 transcriptional targets and draft regulatory networks. We describe the role of PCL2 in both modulating transcription of ESC self-renewal genes in undifferentiated ESCs as well as developmental regulators during early commitment and differentiation.
Introduction
Control over ESC fate decisions is accomplished through a variety of molecular, genetic and epigenetic events. Exogenous control of cell fate can be achieved by a limited number of factors. When grown in fetal bovine serum (FBS)-containing medium and in the presence of murine embryonic fibroblast feeder cells (Evans and Kaufman, 1981; Martin, 1981) or the cytokine leukemia inhibitory factor (LIF) (Smith et al., 1988; Smith and Hooper, 1987; Williams et al., 1988), mouse ESCs remain undifferentiated. BMP4, provided by the serum, functions in the presence of LIF to maintain pluripotency by inducing phosphorylation and nuclear localization of Smad1, followed by up-regulation of Id proteins that block neural differentiation (Ying et al., 2003a).
Three transcription factors are known to be critical in the establishment and/or maintenance of ESC pluripotency. OCT4 (Pou5f1) has a highly conserved role in maintaining pluripotent cell populations (Morrison and Brickman, 2006; Nichols et al., 1998) and its expression level dictates ESC fate (Niwa et al., 2000). SOX2 forms a complex with OCT4 and is necessary to cooperatively activate target genes (Ambrosetti et al., 1997; Yuan et al., 1995). NANOG is critical for initiating pluripotency and maintaining OCT4 levels, even in the absence of LIF (Chambers et al., 2003; Chambers et al., 2007; Mitsui et al., 2003) while it is itself regulated by OCT4 and SOX2 (Rodda et al., 2005).
The polycomb group (PcG) proteins, first described in D. melanogaster, regulate epigenetic states and are required for proper repression of homeotic genes during development (Schuettengruber et al., 2007; Schwartz and Pirrotta, 2007). PcG proteins have been identified in either the PRC1 or the PRC2 multi-protein complex (Lund and van Lohuizen, 2004). The core components of PRC2 in Drosophila are Extra Sex Combs, E(Z) and SU(Z)12. These proteins are found in distinct complexes with additional accessory proteins that undergo dynamic changes during development (Furuyama et al., 2003; Kuzmichev et al., 2005). Core PRC2 members are highly conserved between species (Schuettengruber et al., 2007) and their mouse orthologs are EED, EZH2 and SUZ12, respectively (Cao et al., 2002; Czermin et al., 2002). Through the methyltransferase activity of EZH2, PRC2 induces gene repression by trimethylating lysine 27 on histone 3 (3meH3K27) (Cao et al., 2002; Pasini et al., 2004; Silva et al., 2003).
In Drosophila, PCL (polycomb-like) is present in a subset of PRC2 complexes (O’Connell et al., 2001; Tie et al., 2003). Evidence suggests that PCL is required to generate high levels of 3meH3K27 at some target genes. However, gene repression and 3meH3K27 of many known PRC2 targets is not abolished in the absence of PCL, suggesting that PRC2 can function independently of PCL at many target genes (Nekrasov et al., 2007). In contrast to other PRC2 members, the lack of PCL produces a mild homeotic phenotype, specifically affecting target gene expression in central nervous system and mesoderm tissues (Duncan, 1982; O’Connell et al., 2001). PCL2 is a mammalian ortholog of PCL and defects reported in animal models of Pcl2 deficiency are consistent with lack of PCL in Drosophila. In G. gallus (Wang et al., 2004), Pcl2 regulates left-right axis specification and in X. laevis (Kitaguchi et al., 2001), central nervous system gene expression is disrupted by the lack of Pcl2. Finally, hypomorphic Pcl2 gene trap mice have pleiotropic defects including posterior transformation of axial skeleton, stunted growth, hydrocephaly, hunchback and incisor abnormalities (Wang et al., 2007).
Here, we investigated the role of PCL2 in mouse ESCs. Reduction of PCL2 resulted in heightened self-renewal characteristics and inefficient differentiation to the three germ layers. PCL2 was found to associate with the core PRC2 complex and using chromatin immunoprecipitation (ChIP) coupled with massively parallel DNA sequencing (ChIP-seq) we found that PCL2 is highly enriched at many, but not all, locations of PRC2 enrichment. Loss of PCL2 did not abolish global levels of 3meH3K27 but did result in decreased 3meH3K27 at specific targets and altered patterns of 3meH3K27 during early commitment. Integrating PCL2-PRC2 targets with the ESC self-renewal circuitry revealed a key role for PCL2-PRC2 in limiting transcription of ESC self-renewal genes in undifferentiated ESCs as well as controlling developmental regulators during commitment and early differentiation.
Results
Identification of Mammalian Pcl2
In a screen for regulators of ESC fate, we hypothesized that networks critical for the stability of the ESC state would be differentially regulated at the initiation of commitment (Walker et al., 2007). Transcript levels of Mtf2 (metal response element binding transcription factor 2) decreased during differentiation (Figure 1A) and promoter occupancy data indicated that Mtf2 is bound by both OCT4 and NANOG (Loh et al., 2006). Further inspection revealed that the protein product of Mtf2 is orthologous to Drosophila PCL. Both proteins contain a TUDOR domain and two PHD zinc finger domains (Figure 1B) and MTF2 shares 73% and 88% similarity to the PCL ortholog, PCL2, studied in X. laevis (Kitaguchi et al., 2001) and G. gallus (Wang et al., 2004), respectively. The mouse and human MTF2 protein sequences share 96% identity. Thus, we will refer to the mammalian Mtf2 transcript as Pcl2. Mammals have two other PCL orthologs: PHF1 and PHF19 which have 63% homology with PCL2 amongst the conserved domains. Interestingly, expression analysis of Pcl2 in mice show that it is uniquely expressed in undifferentiated ESCs and during early embryonic development (Figure S1).
Figure 1. Down-regulation of Pcl2 leads to enhanced self-renewal and impaired differentiation capacity.

A) Microarray expression profile of Pcl2 throughout both −LIF and +RA monolayer differentiation of OCT4:eGFP R1 ESCs. B) PCL2 has three predicted isoforms. C) Relative expression of Pcl2 mRNA in control and shRNA knockdown clones. D) Relative expression of Oct4, Nanog, and Sox2 in three different Pcl2 shRNA knockdown clones compared to wild-type R1 and two mismatch controls as determined by qPCR. Data are presented as mean ± standard deviation of triplicate experiments. E&F) OCT4 protein was measured by quantitative immunofluorescence in 10,000 individual cells of three different Pcl2 shRNA knockdown clones over 72 hr in both +LIF and -LIF conditions. In E) the distribution of OCT4 expression is illustrated in a histogram for one representative Pcl2 shRNA knockdown clone and a mismatch control. The distribution is best modeled as a double-Gaussian (red curve) which is a compilation of the OCT4-negative population (green curve) and the OCT4-positive population (blue curve). The vertical red line represents the threshold between OCT4 +ve and −ve populations and the percentage of cells above and below this threshold is reported. In F) the percentage of cells that had lost OCT4 expression at each time point is reported. Error bars represent the standard deviation of triplicate experiments. G&H) Three different Pcl2 shRNA knockdown clones, wild-type R1 and two mismatch controls were plated in +LIF and the distribution of colonies on each plate that were undifferentiated or fully differentiated were determined by morphology and alkaline phosphatase (ALP) expression. The results are an average of three separate experiments, each performed in triplicate. I) Fluorescent images depict Pcl2 shRNA knockdown and mismatch control cells stained for smooth muscle actin, Nestin, alpha fetoprotein or OCT4 following EB differentiation. Differentiation experiment was performed in duplicate. J) EBs were formed in suspension and cultured for 25 days (left panel). After 25 days they were trypsinized, cultured in +LIF for 3 days and stained for ALP (right panel).
Down-regulation of Pcl2 results in enhanced self-renewal of ESCs
To examine the function of Pcl2 in ESCs, endogenous Pcl2 expression was stably knocked down using short hairpin RNA (shRNA) (Kunath et al., 2003). We designed three shRNA sequences targeting different regions of the Pcl2 mRNA as well as a mismatch control sequence to rule out indirect effects. One shRNA sequence resulted in the most significant reduction in Pcl2. Three of these clones, each exhibiting at least 70% knockdown in Pcl2 transcript compared to the mismatch control and wild-type R1 ESCs, were chosen for further analysis (Figure 1C) and used in all subsequent experiments. However, stable clones expressing the additional two shRNA sequences were analyzed in many of the assays and showed similar results (data not shown). We also confirmed that knockdown in Pcl2 mRNA levels resulted in reduced protein levels (Figure S2B,C).
To test the effect of Pcl2 depletion on ESC self-renewal, we measured transcript levels of Oct4, Nanog and Sox2 and found that they were increased in Pcl2 knockdowns (Figure 1D) over both wild type and mismatch controls. Next, we performed a high-content fluorescence imaging assay to examine OCT4 protein levels in single cells (Davey and Zandstra, 2006; Walker et al., 2007). The OCT4 distribution is best estimated by a double Gaussian curve and a threshold delineating OCT4 +ve and -ve cells was calculated as the intersection of these two curves (Davey and Zandstra, 2006). Results are reported as the percentage of cells above and below this threshold. At three hours after plating, the cells were viable and contained over 90% OCT4 positive cells (Figure 1E). Cells were cultured under multiple differentiative conditions to study the effects of known self-renewal mediators LIF and BMP4. In all cases, Pcl2 knockdowns maintained a larger OCT4 +ve population (Figure 1E, 1F; BMP4 data not shown). However, the intensity of the OCT4 +ve peak in knockdown cells was not increased compared to the control (Figure 1E). Thus, individual cells did not over-express OCT4. Instead, a greater percentage of the population was OCT4 +ve. This resulted in the appearance of over-expression in bulk analyses (Figure 1D and 4A) and highlights the importance of single-cell analyses in distinguishing distinct cell states within a heterogeneous population.
Figure 4. PCL2 knockdown disrupts 3meH3K27 during differentiation.
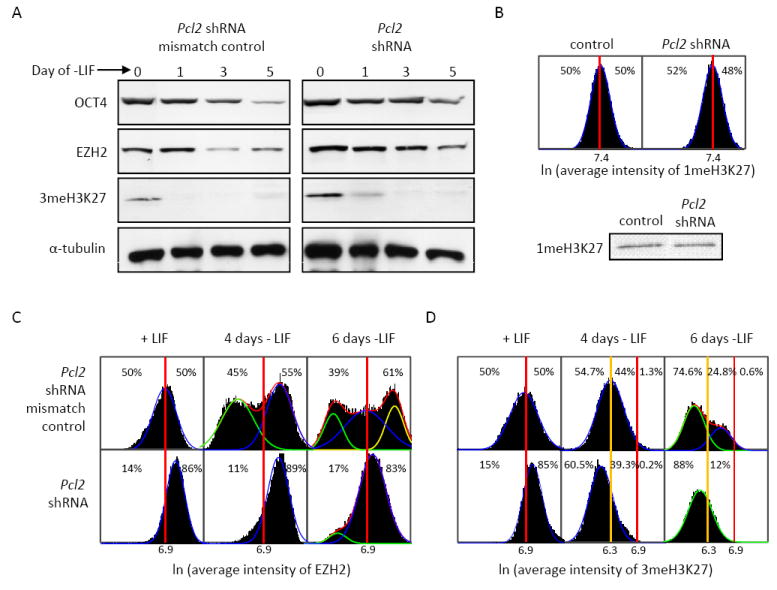
A) Immunoblot analysis demonstrates levels of OCT4, EZH2 and 3meH3K27 in Pcl2 shRNA knockdown and mismatch control cells. α-tubulin was used as a loading control. B) H3K27 monomethylation levels in control vs. knockdown. C&D) EZH2 (C) and 3me-H3K27 (D) protein was measured in 10,000 individual cells over six days in -LIF conditions. The distributions are best modeled as multi-peak Gaussians (red curve) that is a compilation of the low population (green curve), mid population (blue curve) and high population (yellow curve).
In clonogenic assays (Peerani et al., 2009; Zhang et al., 2006), cultures were stained for alkaline phosphatase (ALP) activity and colonies were stringently scored, with only colonies resembling those in Figure 1G being counted as undifferentiated. Pcl2 knockdown cells formed undifferentiated colonies with much greater efficiency than controls (Figure 1H). Collectively, these results suggest that knockdown of Pcl2 promoted self-renewal and maintained the undifferentiated state of ESCs during the initial time points following the cue to differentiate.
Down-regulation of Pcl2 results in impaired differentiation capacity
To test the effect of Pcl2 on differentiation capacity, we performed directed neuroectoderm differentiation (Ying et al., 2003b) assays and found Pcl2 knockdown cells maintained high levels of OCT4 with no observable expression of Nestin within a five day period (data not shown). Next, we used embryoid body (EB) formation to determine how the cells would respond when stimulated to differentiate for a prolonged period. Compared to controls, knockdown EBs remained rounded and in general, cells did not migrate outwards, nor did they acquire a differentiated morphology. Based on immunostaining, control EBs formed large areas of both smooth muscle actin (SMA, mesoderm) and Nestin (neuroectoderm)-positive cells while knockdown EBs formed very few differentiated cells (Figure 1I). Furthermore, knockdown EBs did not form α-fetoprotein (endoderm)-positive cells and maintained high levels of OCT4 expression throughout the cultures (Figure 1I). We also tested whether expression of lineage specific genes was disrupted and found that although transcripts of some markers of mesoderm and ectoderm were eventually expressed, their expression was delayed (Figure S1B). We also observed that endoderm markers were expressed to different extents in knockdowns compared to controls (Figure S1B).
To test whether knockdown EBs would eventually differentiate after extended culture, EBs were grown in suspension for 25 days (Kaji et al., 2006). After 25 days, control cultures consisted of cystic EBs (Figure 1J, top left) which, when plated on gelatin, developed into cells of differentiated morphology that did not exhibit ALP activity (Figure 1J, top right). In contrast, knockdown EBs remained as small, tightly compacted groups of cells (Figure 1J, bottom left) and retained the capacity to form undifferentiated ALP +ve colonies (Figure 1J, bottom right).
These data suggest that Pcl2 is critically involved in early commitment and differentiation. Knockdowns appeared unable to progress to terminal differentiation of some cell types and retained a population of self-renewing ESC-like cells.
Ectopic expression of PCL2 partially rescues knockdown phenotype
To verify that the phenotype observed was specific to the depletion of Pcl2, we performed rescue experiments by ectopically expressing Pcl2-IRES-βgeo with a doxycycline-inducible transposon expression system (Woltjen et al., 2009). Clones expressing the transgene encoding the predominate ESC isoforms (Figure S2A,B) were selected based on β-galactosidase expression (Figure 2A), PCL2 protein levels (Figure 2B) and Pcl2 transcript (Figure 2C) following 72 hours of treatment with doxycycline. In colony forming assays, over-expression of PCL2 was not sufficient to return the normal ratio of differentiated to undifferentiated colonies in +LIF (Figure 2D,E). However, under −LIF conditions, rescue clones regained the ability to differentiate (Figure 2E) and contained a normal percentage of OCT4 negative cells (Figure 2F). Finally, after long-term culture in doxycycline, expression of ESC-specific genes Oct4, Sox2, Nanog, Esrrb and Tcl1 were decreased (Figure 2G). These data demonstrate that following over-expression of Pcl2 in the knockdown clones, cells recover the ability to down-regulate the pluripotency network and differentiate.
Figure 2. Induced expression of PCL2 partially rescues knockdown phenotype.

A) Two Pcl2 shRNA clones containing the doxycycline inducible Pcl2-IRES-βgeo expression cassette were stained for β-galactosidase expression before and 72 hours after addition of doxycycline to the media. B) By Western analysis, PCL2 protein was increased in the two rescue clones following doxycycline induction. C) By qPCR analysis, Pcl2 transcript was increased following doxycycline induction. D&E) The rescue clones were plated in +LIF or -LIF, with or without doxycycline and the distribution of colonies on each plate that were undifferentiated or fully differentiated were determined by morphology and alkaline phosphatase (ALP) expression. F) The percentage of cells in the rescue clones that had lost OCT4 expression by 72 hours in both +LIF and −LIF, with or without doxycycline. G) Relative transcript levels of ESC specific, self-renewal genes before and after long term culture of rescue clones in doxycycline. Expression levels are reported as a percentage of expression in wild-type R1 cells.
PCL2 is a PRC2-associated protein
In Drosophila, PCL associates with PRC2 (O’Connell et al., 2001; Savla et al., 2008); therefore, we examined whether murine PCL2 associated with the PRC2 complex. Gel filtration analysis indicated that PCL2 is found in two chromatographically distinct peaks, a large molecular mass peak that co-eluted with SUZ12, EZH2 and EED, and a smaller peak, which may represent the monomeric form of PCL2 or smaller subcomplexes. SUZ12 and EED were also detected in fractions corresponding to the smaller PCL2 peak (Figure 3A). These associations were confirmed by co-immunoprecipitation experiments (Figure 3B). Previous reports have suggested that PCL2 does not directly interact with EZH2 (O’Connell et al., 2001), which prompted us to further substantiate its association with SUZ12 using a non-biased epitope-tagged SUZ12 system coupled to mass spectrometry.
Figure 3. PCL2 is a PRC2 associated protein.

A) Gel filtration chromatography of ESC nuclear proteins indicating that PCL2, EED, EZH2 co-elute in the same fractions. B) Immunoblot analysis demonstrates that EZH2 and SUZ12 protein are detected following IP with a PCL2-specific antibody. In addition, PCL2 protein is present following IP with an EZH2 antibody. C) Coomassie blue stained gel of affinity purified SUZ12 complex. D) Mass spectrometry results showing interaction of PCL2 with Suz12-HF.
We purified SUZ12 associated proteins from a mouse ESC line containing a 6xHis-3xFLAG tag (HF) targeted to the C-terminus of one allele of the Suz12 gene, which resulted in expression of a full-length tagged SUZ12 protein (Singla et al., 2010). SUZ12 associated proteins were then separated by SDS-PAGE and stained with Coomassie blue to identify interacting proteins (Figure 3C). Mass spectrometric analysis indicated that the two prominent bands at approximately 100 and 55kDa contained proteins SUZ12, EZH1, EZH2, and RBBP4, RBBP7, AEBP2, EED, and PCL2, respectively (Figure 3D). In addition to a number of previously known interactors of the PRC2 complex including RBBP4, RBBP7, and AEBP2 (Figure 3D) (Kim et al., 2009; Kuzmichev et al., 2002), we identified fifteen peptides of PCL2 (Table S1).
Taken together, these data demonstrate that PCL2 associates with the PRC2 complex in mouse ESCs.
PCL2 knockdown disrupts global 3meH3K27 during differentiation
The PRC2 complex is responsible for mediating gene repression by trimethylating H3K27. Loss of the core members of PRC2 results in disruption of the complex and decreased global 3meH3K27. Reduction of PCL2 using shRNA led to moderately increased levels of EZH2 (Figure 4A and 4C) and global 3meH3K27 (Figure 4A and 4D) in the undifferentiated state, while levels of 1meH3K27 were unchanged (Figure 4B). Following LIF withdrawal, EZH2 initially declined, and by day six the control cells comprised three distinct cell populations expressing different levels of EZH2 (Figure 4C). In the knockdown cells, EZH2 levels did not decrease during the first four days of LIF withdrawal, and a small portion of cells displayed decreased EZH2 at day six (Figure 4C). 3meH3K27 levels decreased in both control and knockdown cells immediately following differentiation. Consistent with their undifferentiated state, the knockdown cells had slightly more 3meH3K27 during the first day of differentiation but contained similar levels to the control cells by day four of differentiation. At day six, control cells consisted of two different populations of cells displaying two low but distinct levels of 3meH3K27, whereas knockdown cells comprised a single population (Figure 4D). These data reveal Pcl2 is dispensable for EZH2 expression and global 3meH3K27 in undifferentiated cells. However, upon the withdrawal of LIF, levels of both EZH2 and 3meH3K27 decline below the levels of the control.
Genome-wide assessment of PCL2 targets
To determine which genes were deregulated as a result of Pcl2 knockdown, we performed expression microarray analysis. 2585 probes (1152 genes) (Table S2) were down-regulated and 1715 probes (770 genes) (Table S2) were up-regulated (Figure 5A) based on a fold-change cutoff of 1.5 and p-value less than 0.05. Gene ontology (GO) analysis (Beissbarth and Speed, 2004) indicated that up-regulated genes are involved in transcription, chromatin remodeling, cell cycle and DNA repair (Figure 5B) and included several key markers of undifferentiated ESCs. The majority of down-regulated genes are involved in development and differentiation (Figure 5C).
Figure 5. Genome-wide assessment of PCL2 targets.

A) Volcano plot of differentially expressed genes between Pcl2 shRNA knockdown cells and mismatch control cells cultured in +LIF. Vertical red line represents a fold change of ±1.5. Microarrays were performed in triplicate and the horizontal red line represents a cut-off p-value of 0.05. B & C) GO analysis of genes up-regulated and down-regulated in knockdown cells. D) Location of enriched DNA sequence bound by PCL2 as determined by ChIP-seq. E) Histogram illustrating the distribution of enriched sequences in relation to known TSS. F) GO analysis illustrating the biological function GO terms over-represented amongst the genes that were enriched in the ChIP-seq experiment. Statistically over-represented GO terms were determined by comparing the incidence of a GO term within the input gene list (observed, red bar) to the incidence of that GO term among the entire mouse genome recorded in the MGI database (expected, yellow bar). Fisher’s Exact Test was used to determine a p-value for each term.
To identify direct targets of PCL2, we performed ChIP-seq, using a commercial antibody directed towards PCL2. The antibody was first tested for specificity by over-expressing Pcl2 in 293T cells (Figure S2A) as well as in the knockdown studies (Figure S2B, S2C). ChIP DNA was sequenced using the Solexa 1G platform, aligned to the mouse genome and enrichment analysis was performed as described previously (Johnson et al., 2007), resulting in 3,847 enriched regions with a FDR of 0.3% (Table S3). Our analyses revealed that 47% of targets were within 100 kb of a known transcriptional start site (TSS) (Figure 5D) and 34%, or 1597 targets, were within 2 kb of a known TSS (Figure 5E). Further, GO analysis found that targets were involved in processes such as development, pattern specification and differentiation (Figure 5F).
PCL2 binds a subset of PRC2 targets and is associated with H3K27 trimethylation at those targets
We have shown that PCL2 associates with PRC2; however, it is not clear whether PCL2 is present at all PRC2 target locations. To identify specific targets regulated by the PCL2-PRC2 complex, we compared our ChIP-seq data to published ChIP-seq datasets for PRC2 components EZH2 and SUZ12 in mouse ESCs (Ku et al., 2008), as well as sites containing the histone modifications 3meH3K27 (catalyzed by PRC2) and 3meH3K4. This bivalent epigenetic mark occupies the promoters of developmental regulators in mouse ESCs (Bernstein et al., 2006; Ku et al., 2008). Finally, we incorporated the ChIP-seq dataset for PRC1 component RING1B. Targets positive for both PRC2 and PRC1 binding are evolutionarily conserved and present on the majority of developmentally associated PRC2 targets (Ku et al., 2008).
By integrating these ChIP-seq datasets, we established that 953 were highly enriched for PCL2, EZH2 and SUZ12 and 96% of these targets were enriched for the bivalent domain (3meH3K27+3meH3K4) (Figure S3A, Table S3) and 65% were bound by RING1B. A second class of 772 targets was bound by EZH2 and SUZ12, but enrichment of PCL2 was below the three-fold cutoff threshold. Comparable to the PCL2-PRC2 targets, 94% of these targets were enriched for the bivalent domain and 55% were bound by RING1B. The remaining subsets consisted of fewer targets and were not as likely to contain the bivalent domain or RING1B. Thus, PCL2 is highly enriched in a subset of PRC2 targets and these targets are likely to be enriched for the bivalent domain as well as RING1B.
Figure 6A shows the aligned occupancy results of fourteen genes whose expression was deregulated in the Pcl2 knockdowns. Oct4, Nanog and Tcl1 were not bound by PCL2, or any of the PRC2 members but were up-regulated in the knockdowns. These genes are highly expressed in ESCs, consistent with exhibiting 3meH3K4, a well-validated transcriptional activation mark, but not 3meH3K27. Thus, these genes are likely to be indirectly inhibited by PCL2.
Figure 6. PCL2 binds a subset of PRC2 targets and is associated with 3meH3K27.

A) Binding profiles of 14 PCL2 and PRC2 targets. Genomic range in each panel extends from -10kb from the TSS to the end of the gene. B-F) ChIP-qPCR validation of 14 selected promoter regions in both wild type and knockdown cells for PCL2, EZH2, EED, SUZ12 and 3meH3K27. Values are calculated as the fold enrichment above a control without antibody. Wild-type vs. knockdown samples were normalized to a fraction of the Input material for each primer set tested to adjust for differences in starting amounts.
Tbx3 is also highly expressed in ESCs and was highly over-expressed in Pcl2 knockdowns. Consistent with this, Tbx3 exhibits the activating 3meH3K4 modification. Interestingly, it is also bound by PCL2, EZH2 and SUZ12 and has low levels of 3meH3K27. This suggests that Tbx3 is controlled by PCL2-PRC2 through the modulation of 3meH3K27.
The remaining ten targets are developmental genes that were regulated in the Pcl2 knockdowns. Overall, the binding profile of PCL2 shows remarkable overlap with the profiles of the other PRC2 members, strongly suggesting that they are binding to the same region of DNA.
ChIP-qPCR with antibodies directed against each PRC2 component was performed on fourteen targets. Additionally, ChIP-qPCR was done on the knockdown cells to determine the effect of PCL2 depletion on PRC2 occupancy and 3meH3K27 at specific target promoters (Figure 6B-F). 3meH3K27 as well as all PRC2 members were enriched at these eleven targets. SUZ12 and EED enrichment were greatly reduced in the knockdown cells (Figure 6C and 6D), and EZH2 appeared less affected by loss of PCL2 (Figure 6E). Reduction in PCL2 expression does not disrupt the interactions of the core PRC2 components (Figure S3B). Finally, despite our observation that global 3meH3K27 was not depleted by loss of PCL2, we observed that 3meH3K27 enrichment was reduced by 2 to 3 fold in the Pcl2 knockdown cells at these specific targets of PCL2 (Figure 6F). Collectively these data are consistent with findings in Drosophila (Nekrasov et al., 2007) and suggests that PCL2 promotes PRC2 function at specific targets and, despite being dispensable when assayed for global levels of 3meH3K27, PCL2 is required at specific PCL2-PRC2 targets to achieve proper 3meH3K27.
Our analysis also showed that PCL2 binds to a subset of 483 targets independently of PRC2. There is evidence that PCL2 can act as an activating transcription factor in mouse L-cells (Remondelli and Leone, 1997; Remondelli et al., 1997) and thus may function independently of PRC2 at some targets. Upon further analysis, we found that most of these targets showed weak PCL2 enrichment and were not regulated following knockdown of Pcl2. Of the remaining 152 genes that were both bound and regulated, we then carefully analyzed the binding profiles of the other PRC2 members and found that, despite being below the cutoff threshold in the EZH2 and SUZ12 experiments, there was binding above background for nearly all. Ultimately we identified a subset of 23 targets that appeared to be bound by PCL2 and not by other PRC2 proteins and that were also regulated following Pcl2 knockdown. ChIP-qPCR at 11 of these targets confirmed that PCL2 was weakly bound in the wild-type cells and depleted in the knockdown cells (Figure S3C). EZH2 and 3meH3K27 were absent at these targets and were not altered following knockdown of Pcl2 (Figure S3D-F). However, binding patterns of SUZ12 were remarkably similar to those of PCL2 and SUZ12 was displaced from these targets following knockdown of Pcl2 (Figure S3D). Enrichment of SUZ12 at some of these targets (Figure S3D) suggests that they were false negatives in the SUZ12 ChIP-seq experiment. Together these data suggest that if PCL2 regulates these targets, it may do so in cooperation with SUZ12 through a mechanism independent of 3meH3K27. However, with the exception of Inhb1, regulation following knockdown or over expression is not significant (Figure S3H and S3I) and these targets are not regulated following the withdrawal of LIF (Figure S3H). It is possible that regulation of these targets is not critical in undifferentiated ESCs. It remains possible that PRC2 may play a role in regulating these targets in alternate cell types.
Predicting and testing PCL2 dependent ESC regulatory networks
To draft potential PCL2-PRC2 dependent ESC regulatory networks, we combined our ChIP-seq and microarray expression data with published ChIP-PET experiments for OCT4 and NANOG (Loh et al., 2006). The proximal promoter of Pcl2 is bound by both OCT4 and NANOG and since Pcl2 is down-regulated concomitantly with Oct4 and Nanog, we reason that OCT4 and NANOG activate Pcl2. To extend the network, we incorporated our previously published time course microarray data of differentiating ESCs (Walker et al., 2007). The complete network of 953 PCL2-PRC2 targets is summarized in Figure S4 and reveals that there is substantial overlap between the targets of OCT4, NANOG and PCL2-PRC2. We found two distinct classes of targets. First, in the case of ESC-specific genes, OCT4 and NANOG exert an activating pressure that is opposed by the repressive effect of PCL2-PRC2. The interaction of three genes known to be important for ESC self-renewal (Tbx3, Klf4 and Foxd3) are highlighted in Figure 7A. The second class of OCT4, NANOG and PCL2-PRC2 targets comprise developmental regulators that are directly repressed by PCL2-PRC2 (Figure 7B) but also repressed indirectly by OCT4 and NANOG through the activation of the extended pluripotency network (Figure S4).
Figure 7. Predicting PCL2 dependent regulatory networks.

A) Identification of one network motif that functions to modulate ESC pluripotency and self-renewal. Genes connected by an arrow were activated by the initiating gene, whereas genes connected by a T-bar were repressed. Activation and repression is supported by promoter occupancy studies or microarray expression changes following knockdown. B) Summary of the categories of developmental genes found to be targeted by the PCL2-PRC2 complex. C) A pool of Dharmacon siRNAs targeting the Pcl2 transcript were used to acutely knock down Pcl2. Cells were re-transfected 48 hours after the first transfection to maintain depletion of Pcl2. Relative expression of Pcl2 as well as a set of potential targets were measured 24 and 72 hours following transfection, in both +LIF and −LIF. D) Pcl2 expression was induced in the rescue clones and transcript levels of the potential targets were measured 72 hours after doxycycline induction. Transcript levels are reported relative to their expression level in wild type ESCs. E) Microarray results from Pcl2 knockdowns and Suz12 and Ezh2 null cells.
To test the response of target genes immediately following perturbation of Pcl2, we used both transient siRNA and inducible over-expression. Pcl2 transcript was depleted by 90% at 24 hours post siRNA transfection and target expression was elevated compared to the siRNA control by 72 hours (Figure 7C). Conversely, we used the aforementioned rescue clones to induce expression of Pcl2 and observed that targets were decreased 72 hours after doxycycline treatment (Figure 7D).
In the case of the developmental genes, increased expression following transient siRNA (Figure 7C) is in contrast to the reduced expression of those same targets in the long-term culture of the Pcl2 shRNA knockdown cells. We propose that this effect is due to increased repressive pressure on developmental targets exerted by the pluripotency network, which is over-expressed in the knockdown cells. Gene expression resulting from the long-term depletion of Pcl2 is consistent with expression patterns of developmental targets in Suz12 and Ezh2 null ESCs (Pasini et al., 2007; Shen et al., 2008) (Figure 7E), although it is most similar to Suz12 null cells.
Discussion
We predict that PCL2-PRC2 represses key members of the pluripotency network (Figure 7A, 7C-E). Cells of the inner cell mass and ESCs, their in vitro derivatives, must undergo self-renewal while remaining poised to differentiate in response to extrinsic cues. Accumulated evidence suggests that ESCs maintain self-renewal through the autoregulation of the pluripotency TFs Oct4, Nanog and Sox2. In addition, some of the primary targets of these TFs, such as Tbx3, Klf4 and Foxd3, support the pluripotency phenotype in part by stabilizing Oct4, Nanog and Sox2 through feedback mechanisms (Hanna et al., 2002; Ivanova et al., 2006; Kim et al., 2008). It has recently been shown that over-expression of either Tbx3 or Klf4 can support LIF independent self-renewal (Niwa et al., 2009). Additionally, over-expression of Klf4 can be used to revert adult fibroblasts to a pluripotent state and convert EpiSCs to ESCs (Guo et al., 2009; Takahashi and Yamanaka, 2006). Thus, it is likely that over-expression of Tbx3 and Klf4 due to depletion of PCL2 is a major determinant of the Pcl2 knockdown phenotype, as predicted in our draft network (Figure 7A). Such a self-reinforcing network should result in a stable, self-renewing population of pluripotent stem cells.
However, experience reveals that even when grown under self-renewal culture conditions, a small but noticeable percentage of ESCs commits and undergoes morphologically distinguishable differentiation. Furthermore, upon removal of LIF or induction of differentiation by addition of external signals such as retinoic acid, wild type ESCs are able to rapidly down-regulate these genes in response to external signals. We propose that PCL2 is critical for maintaining the capacity of ESCs to commit rather than self-renew by exerting repressive pressure on Tbx3, Foxd3 and Klf4 through a feed-forward mechanism, thus dampening the autoregulatory network driving self-renewal to the point the cells can respond to external inputs (Figure 7A). Feed-forward motifs have been shown in bacteria to accelerate target response time and maintain steady-state levels of the target (Alon, 2007) and stabilization of transcript levels of Tbx3, Foxd3 and Klf4 and indirectly of Oct4, Nanog, and Sox2 through PCL2-PRC2 repression may be further beneficial to ESCs because it allows activation by a strong promoter, without risk of over-shooting the optimal pluripotency TF concentration. Thus, one potential function of PCL2-PRC2 repression of ESC specific genes is to allow rapid activation of the extended pluripotency network once the initiating TFs have been activated. Another potential function of PCL2-PRC2 repression is to regulate ESC fate by interrupting the self-reinforcing network that maintains pluripotency once the ESC is exposed to a differentiative environment.
Genome-wide promoter occupancy studies in ESCs have shown that EZH2, SUZ12 and EED also bind a large number of TFs required for development, leading to the hypothesis that PcG proteins maintain the ESC state by repressing developmental regulators that would otherwise cause the cells to differentiate (Boyer et al., 2006; Lee et al., 2006). However, knockdown and knockout studies of PcG proteins in ESCs have not supported this hypothesis. In the mouse, ablation of any of the three core PRC2 components does not result in embryonic lethality until the time of gastrulation (Faust et al., 1995; O’Carroll et al., 2001; Pasini et al., 2004) and both Suz12 and Eed null ESCs can be derived. Suz12 null ESCs maintain an undifferentiated morphology and high levels of ESC markers even after withdrawal of self-renewal signals and are unable to differentiate into mature cell types (Pasini et al., 2007), which is similar to our findings for Pcl2 knockdowns in ESCs. Additionally, Eed null ESCs express heightened levels of differentiation genes but also maintain high levels of ESC markers and can be taken to high passage without losing their undifferentiated morphology (Chamberlain et al., 2008). This suggests that PCL2-PRC2 and in general PRC2 target repression is not required to maintain the undifferentiated state. However, consistent with the established role of PRC2 in Drosophila gastrulation (Schuettengruber et al., 2007), and recent work implicating PcG in pattern formation (Oktaba et al., 2008), PCL2 depletion most dramatically affected PCL2-PRC2 target gene expression profiles during the early stages of ESC commitment to differentiation.
Despite the observed effect of PCL2 in modulating 3meH3K27 and PRC2 levels at its target promoters, we found that the integrity of the interactions between the core PRC2 components could still be observed in cell lysates from Pcl2 knockdown cell lines. This could suggest that PCL2 is involved in the recruitment of the PRC2 complex but not in its formation or stability.
It is counter-intuitive to consider that Pcl2 is both highly expressed in ESCs but required for repression of the self-renewal circuitry. The emerging landscape of the self-renewal regulatory network is of a dense and interconnected system of auto-regulatory and feed-back loops (Kim et al., 2008). We propose that one function of PCL2 is to provide the ESC with a mechanism to hold this system in check. Repression of self-renewal genes by Tcf3 represents one other such ESC regulatory mechanism that has recently been identified (Yi et al., 2008). It will also be interesting to determine whether depletion of PCL2 will enhance the ability of adult cells to be reprogrammed into induced pluripotent stem (iPS) cells. It is possible that the forced down-regulation of PCL2 could result in hyper-activation of the self-renewal regulatory network, providing a selective advantage for reprogrammed cells.
Experimental Procedures
ES cell culture
R1 ESCs were cultured as described previously (Walker et al., 2007).
shRNA vector design, construction and electroporation
Three independent shRNA sequences were designed, cloned, electroporated into R1 ESCs, and clones derived as previously described (Walker et al., 2007). Mismatch controls were created by altering five of the 21 bases in the chosen Pcl2 shRNA sequence. Pcl2 target and mismatch sequences are provided in Supplementary Methods.
Immunoprecipitation
R1 ESCs were pelleted and resuspended in co-IP lysis buffer. 2 ug of antibody and 1 mg of total protein were combined followed by immobilization with protein A/G beads (Pierce). Sample was separated from beads by centrifugation and run on a 15% SDS-PAGE for standard Western Blot analysis. Details of buffer solutions, antibody specifications and Western blot procedure provided in Supplementary Methods.
Protein Quantification in Single Cells
Cells were plated, cultured, stained and imaged as described previously (Walker et al., 2007). Ten thousand individual cells were imaged and Gaussian curves and statistics were generated using Origin 6.1 software. Additional details are provided in Supplementary Methods.
Quantitative Real-Time PCR
Total RNA extraction and reverse transcription were performed as described (Walker et al., 2007). Serial dilutions of mouse genomic DNA were used to generate standard curves to adjust for the efficiency of each primer (Yun et al., 2006). Mouse genomic DNA standards or the cDNA template were added to the qPCR reaction in a final volume of 10μL containing: 2x SYBR master mix (Roche) and 0.5 μM primers. Standard Roche PCR protocol was performed on the Roche Light Cycler 480. qPCR primers are provided in Supplementary Methods.
Clonogenic assay and ALP staining
Single cell suspensions were plated in a 12-well dish at a density of 500 cells/well. Colonies were grown in +LIF for 5 days with media changed daily. ALP staining was performed as described previously (Walker et al., 2007).
Microarray hybridizations
Total RNA was extracted from Pcl2 mismatch controls and one Pcl2 shRNA clone with RNeasy columns (Qiagen). RNA quality was tested using an Agilent Bioanalyzer before performing standard cDNA synthesis (Invitrogen Superscript) and in vitro transcription (IVT) (Enzo IVT kit). 10 ug of RNA was used for IVT and 15 ug of cRNA was used for hybridization (EukGE-WS2v4 kit) to the Mouse Genome 430 2.0 GeneChip. Scanning was performed using the Affymetrix GeneChip Scanner 3000 and analysis done using GCOS1.4 to obtain signal-log-ratios of the control to the sample. Hybridizations of three biological replicates for both the control and Pcl2 shRNA clone were performed.
ChIP-seq and ChIP-qPCR
DNA sonicated to approximately 1kb lengths was supplied to The Centre for Applied Genomics (TCAG) for high-throughput sequencing or used for qPCR. Details of buffer solutions and qPCR primers provided in Supplementary Methods.
Generation of the HF-tagged allele of Suz12
An allele of Suz12 encoding a full-length protein with a carboxy-terminal 3xFLAG-TEV-6xHis tag was created using a Cre recombinase-mediated modification of a gene trap allele. Briefly, we obtained an ES cell line with a gene trap insertion in the seventh intron of the Suz12 locus (XG122, Bay Genomics). Transient expression of Cre recombined Lox sites flanking the splice acceptor, and Cre-mediated recombination was also used to subsequently insert the coding sequence of the nine 3’ exons and tags.
Purification of Suz12-HF and Identification of Suz12 interacting proteins by mass spectrometry
Suz12-HF was purified according to the protocol of Andrew Krutchinsky (Deng et al., 2009). Modifications are noted in the Supplementary Methods. After being separated by SDS-PAGE and stained with Coomassie blue (GelCode Blue, Pierce), protein bands were excised and processed for digestion with trypsin (Promega) as described (Shevchenko et al., 1996) and analyzed by LCMS.
Gel filtration Chromatography
Nuclear protein extract was prepared from undifferentiated mouse ESCs as described by (Dignam et al., 1983). Proteins (5 mg/ml) were separated by AKTA FPLC using a Superose 6 HR 10/30 gel filtration column (GE Healthcare). Approximately 1 mg of protein was loaded onto the column in a 50 mM Phosphate buffer containing .015 M NaCl (pH 7.0). Proteins were eluted at a flow rate of 0.4 ml/min in the same buffer and 250 μl fractions were collected and analyzed by Western blotting.
Database Submissions
Microarray data have been deposited at NCBI GEO: accession number GSE16364.
Raw ChIP-seq data has been deposited at NCBI GEO: accession number GSE16526.
Supplementary Material
Acknowledgments
We thank Hamid Bolouri and Joanna Wysocka for critical reading of the manuscript; Barbara Panning for critical reading of the manuscript and the generous gift of the Eed antibody; Diego Pasini and Kristian Helin for providing the Suz12 microarray datasets; Tammy Reid, Qing Lan, Christine To, Paul Cassar, Anna Omelyanenko and Janet Manias of the Stanford lab; and Junjun Zhang, Xiaolin Wang, Zhizhou Hu and Chao Lu at TCAG and Jenny Gu for technical expertise. The mass spectrometry analysis was carried out in the UCSF mass spectrometry facility, which is headed by Alma Burlingame. This work was supported by operating grants from CIHR (MOP-74528) and Canadian Cancer Society (19122) and infrastructure from Genome Canada, CFI and Ontario (MRI) to WLS. The Suz12 affinity tag targeting, mass spec analysis, and confirmation studies were supported by NIH (RO1AR054396), CIRM (RN2-00919), the Burroughs Wellcome Fund, the Packard Foundation, and the Sandler Family Supporting Foundation grants to JFR and NIH, Keck Foundation, and Searle Foundation grants to NJK. EW was supported by from Ontario Graduate Studentship in Science & Technology and a CIHR Banting and Best CGS Doctoral Research Award; WYC was supported by a MRI Postdoctoral Fellowship; and WLS was supported by a Canada Research Chair. JT is supported by operating grant from the CIHR and NCIC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alon U. Network motifs: theory and experimental approaches. Nature reviews. 2007;8:450–461. doi: 10.1038/nrg2102. [DOI] [PubMed] [Google Scholar]
- Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Molecular and cellular biology. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissbarth T, Speed TP. GOstat: find statistically overrepresented Gene Ontologies within a group of genes. Bioinformatics (Oxford, England) 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem cells (Dayton, Ohio) 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Davey RE, Zandstra PW. Spatial organization of embryonic stem cell responsiveness to autocrine gp130 ligands reveals an autoregulatory stem cell niche. Stem cells (Dayton, Ohio) 2006;24:2538–2548. doi: 10.1634/stemcells.2006-0216. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic acids research. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan IM. Polycomblike: a gene that appears to be required for the normal expression of the bithorax and antennapedia gene complexes of Drosophila melanogaster. Genetics. 1982;102:49–70. doi: 10.1093/genetics/102.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- Faust C, Schumacher A, Holdener B, Magnuson T. The eed mutation disrupts anterior mesoderm production in mice. Development (Cambridge, England) 1995;121:273–285. doi: 10.1242/dev.121.2.273. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Tie F, Harte PJ. Polycomb group proteins ESC and E(Z) are present in multiple distinct complexes that undergo dynamic changes during development. Genesis. 2003;35:114–124. doi: 10.1002/gene.10173. [DOI] [PubMed] [Google Scholar]
- Guo G, Yang J, Nichols J, Hall JS, Eyres I, Mansfield W, Smith A. Klf4 reverts developmentally programmed restriction of ground state pluripotency. Development (Cambridge, England) 2009;136:1063–1069. doi: 10.1242/dev.030957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna LA, Foreman RK, Tarasenko IA, Kessler DS, Labosky PA. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes & development. 2002;16:2650–2661. doi: 10.1101/gad.1020502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science (New York, NY) 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Kaji K, Caballero IM, MacLeod R, Nichols J, Wilson VA, Hendrich B. The NuRD component Mbd3 is required for pluripotency of embryonic stem cells. Nature cell biology. 2006;8:285–292. doi: 10.1038/ncb1372. [DOI] [PubMed] [Google Scholar]
- Kim H, Kang K, Kim J. AEBP2 as a potential targeting protein for Polycomb Repression Complex PRC2. Nucleic acids research. 2009 doi: 10.1093/nar/gkp149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaguchi T, Nakata K, Nagai T, Aruga J, Mikoshiba K. Xenopus Polycomblike 2 (XPcl2) controls anterior to posterior patterning of the neural tissue. Development genes and evolution. 2001;211:309–314. doi: 10.1007/s004270100157. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat Biotechnol. 2003;21:559–561. doi: 10.1038/nbt813. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes & development. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- Lund AH, van Lohuizen M. Polycomb complexes and silencing mechanisms. Curr Opin Cell Biol. 2004;16:239–246. doi: 10.1016/j.ceb.2004.03.010. [DOI] [PubMed] [Google Scholar]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Morrison GM, Brickman JM. Conserved roles for Oct4 homologues in maintaining multipotency during early vertebrate development. Development (Cambridge, England) 2006;133:2011–2022. doi: 10.1242/dev.02362. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nature genetics. 2000;24:372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Molecular and cellular biology. 2001;21:4330–4336. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell S, Wang L, Robert S, Jones CA, Saint R, Jones RS. Polycomblike PHD fingers mediate conserved interaction with enhancer of zeste protein. The Journal of biological chemistry. 2001;276:43065–43073. doi: 10.1074/jbc.M104294200. [DOI] [PubMed] [Google Scholar]
- Oktaba K, Gutierrez L, Gagneur J, Girardot C, Sengupta AK, Furlong EE, Muller J. Dynamic Regulation by Polycomb Group Protein Complexes Controls Pattern Formation and the Cell Cycle in Drosophila. Developmental cell. 2008 doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Lazzerini Denchi E, Helin K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. Embo J. 2004;23:4061–4071. doi: 10.1038/sj.emboj.7600402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peerani R, Bauwens C, Kumacheva E, Zandstra PW. Patterning mouse and human embryonic stem cells using micro-contact printing. Methods in molecular biology (Clifton, NJ) 2009;482:21–33. doi: 10.1007/978-1-59745-060-7_2. [DOI] [PubMed] [Google Scholar]
- Remondelli P, Leone A. Interactions of the zinc-regulated factor (ZiRF1) with the mouse metallothionein Ia promoter. The Biochemical journal. 1997;323(Pt 1):79–85. doi: 10.1042/bj3230079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remondelli P, Moltedo O, Leone A. Regulation of ZiRF1 and basal SP1 transcription factor MRE-binding activity by transition metals. FEBS letters. 1997;416:254–258. doi: 10.1016/s0014-5793(97)01212-x. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. The Journal of biological chemistry. 2005;280:24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development (Cambridge, England) 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nature reviews. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Molecular cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone h3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Singla V, Hunkapiller J, Santos N, Seol AD, Norman AR, Wakenight P, Skarnes WC, Reiter JF. Floxin, a resource for genetically engineering mouse ESCs. Nature methods. 2010 doi: 10.1038/nmeth.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Heath JK, Donaldson DD, Wong GG, Moreau J, Stahl M, Rogers D. Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature. 1988;336:688–690. doi: 10.1038/336688a0. [DOI] [PubMed] [Google Scholar]
- Smith AG, Hooper ML. Buffalo rat liver cells produce a diffusible activity which inhibits the differentiation of murine embryonal carcinoma and embryonic stem cells. Developmental biology. 1987;121:1–9. doi: 10.1016/0012-1606(87)90132-1. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Molecular and cellular biology. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, Der SD, Morris Q, Hughes TR, Zandstra PW, et al. Prediction and Testing of Novel Transcriptional Networks Regulating Embryonic Stem Cell Self-Renewal and Commitment. Cell Stem Cell. 2007;1:16. doi: 10.1016/j.stem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wang S, He F, Xiong W, Gu S, Liu H, Zhang T, Yu X, Chen Y. Polycomblike-2-deficient mice exhibit normal left-right asymmetry. Dev Dyn. 2007;236:853–861. doi: 10.1002/dvdy.21070. [DOI] [PubMed] [Google Scholar]
- Wang S, Yu X, Zhang T, Zhang X, Zhang Z, Chen Y. Chick Pcl2 regulates the left-right asymmetry by repressing Shh expression in Hensen’s node. Development (Cambridge, England) 2004;131:4381–4391. doi: 10.1242/dev.01269. [DOI] [PubMed] [Google Scholar]
- Williams RL, Hilton DJ, Pease S, Willson TA, Stewart CL, Gearing DP, Wagner EF, Metcalf D, Nicola NA, Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, et al. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Pereira L, Merrill BJ. Tcf3 functions as a steady-state limiter of transcriptional programs of mouse embryonic stem cell self-renewal. Stem cells (Dayton, Ohio) 2008;26:1951–1960. doi: 10.1634/stemcells.2008-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying QL, Nichols J, Chambers I, Smith A. BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell. 2003a;115:281–292. doi: 10.1016/s0092-8674(03)00847-x. [DOI] [PubMed] [Google Scholar]
- Ying QL, Stavridis M, Griffiths D, Li M, Smith A. Conversion of embryonic stem cells into neuroectodermal precursors in adherent monoculture. Nat Biotechnol. 2003b;21:183–186. doi: 10.1038/nbt780. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes & development. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- Yun JJ, Heisler LE, Hwang II, Wilkins O, Lau SK, Hyrcza M, Jayabalasingham B, Jin J, McLaurin J, Tsao MS, et al. Genomic DNA functions as a universal external standard in quantitative real-time PCR. Nucleic acids research. 2006;34:e85. doi: 10.1093/nar/gkl400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Walker E, Tamplin OJ, Rossant J, Stanford WL, Hughes TR. Zfp206 regulates ES cell gene expression and differentiation. Nucleic acids research. 2006;34:4780–4790. doi: 10.1093/nar/gkl631. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


