Abstract
The CD22 antigen is a viable target for therapeutic intervention for B-cell lymphomas. Several therapeutic anti-CD22 antibodies as well as an anti-CD22-based immunotoxin (HA22) are currently under investigation in clinical settings. Coupling of anti-CD22 reagents with a nano-drug delivery vehicle is projected to significantly improve treatment efficacies. Therefore, we generated a mutant of the targeting segment of HA22 (a CD22 scFv) to increase its soluble expression (mut-HA22), and conjugated it to the surface of sonicated liposomes to generate immunoliposomes (mut-HA22-liposomes). We examined liposome binding and uptake by CD22+ B-lymphocytes (BJAB) by using calcein and/or rhodamine PE-labeled liposomes. We also tested the effect of targeting on cellular toxicity with doxorubicin-loaded liposomes. We report that: (i) Binding of mut-HA22-liposomes to BJAB cells was significantly greater than liposomes not conjugated with mut-HA22 (control liposomes), and mut-HA22-liposomes bind to and are taken in by BJAB cells in a dose and temperature-dependent manner, respectively; (ii) This binding occurred via the interaction with the cellular CD22 as pre-incubation of the cells with mut-HA22 blocked subsequent liposome binding; (iii) Intracellular localization of mut-HA22-liposomes at 37°C but not at 4°C indicated that our targeted liposomes were taken up through an energy dependent process via receptor-mediated endocytosis; and (iv) Mut-HA22-liposomes loaded with doxorubicin exhibited at least 2-3 fold more accumulation of doxorubicin in BJAB cells as compared to control liposomes. Moreover, these liposomes showed at least a 2-4 fold enhanced killing of BJAB or Raji cells (CD22+), but not SUP-T1 cells (CD22-). Taken together these data suggest that these 2nd-generation liposomes may serve as promising carriers for targeted drug delivery to treat patients suffering from B-cell lymphoma.
Keywords: Liposomes, targeting, B-cell Lymphoma, anti-CD22 ScFv, drug delivery
Introduction
CD22, a B-cell specific surface molecule is a member of the Ig super-family (1-3), and is known to be upregulated in non-Hodgkin's lymphoma (NHL) and other types of B-cell lymphomas (4;5). The transmembrane protein CD22 is a well-characterized molecule that rapidly internalizes upon antibody binding; the structure-function relationship of CD22 with the BCR complex has been investigated in detail (2;6). To treat B-cell lymphomas/malignancies, multiple approaches including CD22-specific antibodies alone (7-14), or as bioconjugates (15;16), as well as combination-immunotherapy have been developed (17-19). Among these, recombinant humanized anti-CD22 monoclonal antibodies, epratuzumab, inotuzumab and ozogamicin (18) have been extensively studied, and epratuzumab is currently under clinical trials for NHL. Another promising treatment approach includes use of a recombinant immunotoxin containing an anti-CD22 (Fv) fused to truncated Pseudomonas exotoxin A (HA22) (20-23). We aimed at improving the efficacy of an anti-CD22 targeted therapy by conjugating a novel CD22 specific scFv to a nano-drug delivery carrier.
Antibody-coated liposomes (immunoliposomes) have been explored for site-specific targeting of drugs and therapeutics for cancer treatment (24;25). However, in vivo success of immunoliposomes is subject to the availability of suitable targeting antibody molecules (that will trigger rapid receptor internalization) as well as formulations amenable to tunable drug release for cytosolic or intratumoral delivery. Various antibodies (mainly mAb-IgG) and antibody fragments (including F(ab’)2, Fab or scFvs) have been investigated for immunoliposome studies (for a detailed review, see (24)). For example, liposomes conjugated with CD19 and CD20 mAbs have been examined for B-cell targeting (26;27). Among antibody fragments, scFvs (being small in size) bear promise as targeting ligands for developing 2nd generation immunoliposomes. To date, HER2 scFv (28;29)-conjugated liposomes for drug delivery, and anti-TfR scFv-lipoplexes for gene delivery have been successfully developed (30;31).
Besides the use of antibodies and/or their fragments, selected cytokines (e.g. the B cell activating factor belonging to the TNF family, mBAFF) have also been used as ligands for delivery of liposomal drugs to B-cell lymphomas (32). A recent study by O'Donnell and colleagues demonstrated that immunoliposomes bearing an anti-CD22 mAb (HB22.7) resulted in significantly enhanced cytotoxicity of CD22+ cells by liposome-entrapped doxorubicin, an anti-cancer drug (33). Although, mAb-coated immunoliposomes may serve as suitable vehicles for delivery of cancer therapeutics, the Fc domain-mediated immune responses may limit their future clinical applications (34;35). Therefore, immunoliposomes coated with anti-HER2 scFvs have been developed as 2nd-generation immunoliposomes (28;29;36). Previous studies by de,Kruiff and colleagues have reported immunoliposomes to target B-cell lymphoma (37), and the biosynthetically lipid-tagged CD22 ScFv was generated using a semi-synthetic human library with potentially high affinity (38). This lipidated ScFv was incorporated into the liposomes by a detergent solubilization protocol. Since detergent solubilization method poses limitations to encapsulate payload of anticancer agents in the aqueous compartment of liposomes, alternate approaches are warranted to develop immunoliposomes for B-cell targeting.
This study was designed to generate immunoliposomes bearing high affinity anti-CD22 scFv for targeted drug delivery to B-cell lymphomas. An improved anti-CD22 scFv molecule (mut-HA22, M.W. 34 kDa) was developed and cysteine residues were introduced at the C-terminus for coupling to liposomes. Mut-HA22 bears the advantage as its parental form (HA22) has been well-studied and has been examined in clinical trials in the format of immunotoxin, where no inhibitory immune responses were reported (8). Mut-HA22 conjugation to the surface of preformed liposomes (loaded with calcein (as a model solute), or an anticancer drug, doxorubicin (DOX) in the aqueous compartment) is attained by malemide-cysteine chemistry, rendering such formulations suitable for future in vivo applications.
The observation that mut-HA22 specifically binds to cell-surface expressed CD22 renders it a suitable targeting ligand. Mut-HA22-liposomes bound to CD22+ BJAB cells in a dose-dependent manner, and were accumulated in these cells at 37°C. Our results on target-specific delivery of DOX by mut-HA22 liposomes, and their improved cytotoxicity further provide evidence for their projected application as drug carriers.
Methods
Reagents and Cell Lines
Tributylphosphine and Biogel A-0.5m were from Bio-Rad (Hercules, CA); all other reagents were from Sigma-Aldrich Inc. (St. Louis, MO) and of analytical grade. The CD22+ human B-lymphocyte cell line, BJAB (ATCC# HB-136) and CD22- human T-lymphocyte cell lines Jurkat (ATCC# CRL-10915) and SUP-T1, (ATCC# CRL-1942) were maintained in RPMI 1640 media supplemented with 10% heat-inactivated fetal bovine serum (FBS) and antibiotics (Invitrogen, Carlsbad, CA). Doxorubicin-hydrochloride (DOX-HCl) (Bedford Laboratories, Bedford, OH) and Doxil (Ben Venue Laboratories, Bedford, OH) were obtained through the NIH Pharmacy, Clinical Center, Bethesda, MD.
Lipids
1,2-Dipalmitoyl-sn-Glycero-3-Phosphocholine (DPPC), 1,2-Distearoyl-sn-Glycero-3-Phosphoethanolamine-N-[Maleimide(Polyethylene Glycol)2000] (DSPE-PEG2000-Maleimide), and 1,2-Dioleoyl-sn-Glycero-3-Phosphoethanolamine-N-(Lissamine Rhodamine B Sulfonyl) (Lissamine Rhodamine PE) (ex/em 557/571) were purchased from Avanti Polar Lipids, Inc. (Alabaster, AL). Lipid purity was routinely verified by thin layer chromatography with a silica gel stationary phase and a 65:25:4 chloroform:methanol:water (by volume) mobile phase (39). Molybdenum Blue Spray was used to visualize phospholipids (40) and other possible organic contaminants were visualized by placing developed TLC plates in an iodine chamber.
Mutated HA22 scFv (mut-HA22)
HA22 is an improved version of the immunotoxin BL22 with an improved affinity of the CD22 targeting moiety, the scFv portion (Xiao and Dimitrov, unpublished). In an attempt to further increase the affinity, we generated a mutational phage library based on HA22 scFv. In this library, each clone carried totally random residues on two adjacent positions in the CDR3 region. This phage library was used for panning against purified CD22-Fc (kindly provided by Ira Pastan). For conjugation purposes, site-directed mutagenesis was performed using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) to introduce a cysteine residue immediately before the stop codon of the selected HA22 scFv mutant.
Mut-HA22 binding to CD22+ cells by FACS Analysis
CD22 expression was confirmed by indirect immunostaining using the mut-HA22 and a commercially available anti-CD22 mouse IgG (MYG13 (raised against Raji cells of human origin, Santa Cruz Biotechnology, Inc, Santa Cruz, CA). BJAB (CD22+), SUP-T1 (CD22-), or Jurkat (CD22-) cells were incubated for 15 minutes at room temperature in PBS (containing 5% each of mouse serum, fetal bovine serum, goat serum, and human serum) to block nonspecific binding sites on the cell surface. The cells were resuspended to 107 cells/mL PBS supplemented with 5% FBS (PBS-FBS).
To stain CD22 using mut-HA22, 2 μg of the scFv was added to cells and incubated for 30 minutes in the dark at 4°C. The cells were then washed twice with cold PBS supplemented with 1% BSA (PBS-BSA), resuspended to 107 cells/mL PBS-FBS, and incubated with 1 μg mouse anti-penta-histidine IgG (QIAGEN, Valencia, CA) for 30 minutes in the dark at 4°C. The cells were then washed twice with cold PBS-BSA, resuspended to 107 cells/mL PBS-FBS, and incubated with a 1:32 dilution of FITC-anti-mouse IgG (Sigma, Saint Louis, MO) for 30 minutes in the dark at 4°C.
To stain using MYG13, 2 μg of the IgG was added to the cells and incubated for 30 minutes in the dark at 4°C. The cells were then washed twice with cold PBS-BSA, resuspended to 107cells/mL PBS-FBS, and incubated with a 1:32 dilution of the FITC-anti-mouse IgG for 30 minutes in the dark at 4°C. All cells were washed twice with cold PBS-BSA. Fluorescence of the samples was analyzed using a FACS caliber flow cytometer (Becton Dickinson, San Jose, CA), analyzing 10,000 viable cells per sample.
Preparation of Sonicated Unilamellar Liposomes
Liposomes were prepared by probe sonication as described (41). Lipids (in chloroform) were mixed at desired molar ratios according to Table 1 in a glass tube, dried under streaming nitrogen to create a film, then placed in a vacuum desiccator overnight. In some preparations, rhodamine-PE (0.1mol %) was also included to monitor binding to the cells (see below).
Table 1.
Liposome formulations used in this study.
| Liposomes | Lipids | mut-HA22 molecules/liposome (average) | Fluorescent Markers*/Drug ** | Size Analysis (nm) | |
|---|---|---|---|---|---|
| Fluorescently Labeled | |||||
| mut-HA22 Liposomes (Formulation I) | DPPC (96 mol%) DSPE-PEG2000-Maleimide (4 mol%) | 90 | Calcein/ Rhodamine-PE | See Fig. 2B | |
| 1 mol% DSPE-PEG2000-Maleimide mut-HA22 Liposomes (Formulation II) | DPPC (96 mol%) DSPE-PEG2000-Maleimide (1 mol%) DSPE-PEG2000 (3 mol%) | 47 | Calcein/ Rhodamine-PE | See Fig. 2B. | |
| Control Liposomes (Formulation III) | DPPC (96 mol%) DSPE-PEG2000 (4 mol%) | 0 | Calcein/ Rhodamine-PE | See Fig. 2B | |
|
Drug Loaded | |||||
| mut-HA22 Liposomes (Formulation IV) | DPPC (96 mol%) DSPE-PEG2000-Maleimide (4 mol%) | 90 | Doxorubicin | 123.0 | 7.9 |
| Control Liposomes (Formulation V) | DPPC (96 mol%) DSPE-PEG2000-Maleimide (4 mol%) | 0 | Doxorubicin | 110.5 | 6.6 |
| Empty Liposomes (Formulation VI) | DPPC (96 mol%) DSPE-PEG2000-Maleimide (4 mol%) | 0 | N/A | 86.9 | 4.0 |
Fluorescent liposomes were prepared with 50 mM Calcein and/or 0.1 mol% Rhodamine-PE.
Doxorubicin loading was typically 150-200 ng dox/nmol Pi.
For calcein loaded liposomes, lipid films were reconstituted in HBSE buffer (10mM HEPES, 150mM NaCl, 9.1 mM EDTA, pH 7.4) containing 50 mM calcein by vortexing and heating at 42°C. Multilamellar vesicles (MLVs) were sonicated in a room temperature water bath with a probe sonicator (W-373 Heat Systems-Ultrasonics, NY) for 6 cycles of 1 minute of sonication followed by 1 minute of rest. Titanium particles were removed by centrifugation (2000 xg, 5 minutes). Un-encapsulated calcein was removed from liposome-entrapped calcein on Sepharose CL-6B column (41). Liposome-rich fractions were pooled together and used for conjugation with mut-HA22. Inorganic phosphorus determination was used to determine the concentration of phospholipids in the liposomes (42).
For DOX-loaded liposomes, the ammonium sulfate gradient loading method was used, as previously described (43). Briefly, lipid films were reconstituted in 300 mM ammonium sulfate buffer at pH 7.4 with alternating periods of vortexing and incubation at 41°C. Sonicated liposomes were prepared as above, and were run on a PD-10 column, (GE Healthcare Life Sciences, Piscataway, NJ), pre-equilibrated with HBS pH 7.4, to exchange the external buffer. Liposome-rich fractions were pooled, immediately incubated with DOX at the ratio of 20mg liposomal lipids to 0.5 mg DOX, and incubated for 12-16 hours at room temperature to allow for DOX encapsulation. The DOX-loaded liposomes were separated from un-encapsulated drug by elution on a PD-10 column equilibrated with HBSE (pH 7.4). Fractions containing liposomes were pooled together and used for conjugation with the mut-HA22 scFv. Lipid content was determined as above. Extent of DOX loading was assayed by monitoring DOX fluorescence (in the presence of 0.5% Triton X-100 w/v) at 485/585 nm (ex/em) using a 96-well plate reader (SpectraMax M2e, Molecular Devices, Sunnyvale, CA). Known concentrations of DOX or Doxil (in a range of 0.2-2.0 μg per well) using 0.2 mL total volume were used to generate a calibration curve using the PBS+Triton X-100. Liposomal DOX was quantitated using the DOXIL standard curve.
Conjugation of mut-HA22 to the liposome surface (mut-HA22-liposomes)
The mut-HA22 scFv was reduced by incubation with tributylphosphine (2mM in HBSE buffer) under flowing argon for 4 hours. The reduced antibody was immediately added to either calcein or DOX-loaded sonicated liposomes from above. Typically, we used a ratio of 200 μg scFv for 10-20mg of phospholipids. The scFv-liposome mixture was incubated in the dark for approximately 16 hours at room temperature, sealed under argon. To block free maleimide groups, liposomes were then incubated with L-cysteine (1 mM final concentration) for 15 minutes. The resulting mut-HA22 liposomes were separated from any unconjugated antibody and L-cysteine using size exclusion chromatography (Sepharose CL-6B, GE Healthcare Life Sciences, Piscataway, NJ ), pre-equilibrated with HBSE buffer (pH 7.4). Liposome-rich fractions, detected continuously by a UV detector, were pooled, and concentrated with Centriplus Centrifugal Filter Devices (regenerated cellulose filter, MWCO 30 kDa, Millipore, Bedford, MA). Antibody conjugation to the liposomes was confirmed by gel electrophoresis. Samples were loaded in a 4-12% Bis-Tris gel (NuPage) and allowed to run in MES/SDS running buffer at 200V for 35 minutes under reducing conditions. To visualize the DSPE-PEG2000-maleimide-mut-HA22 conjugate, the gel was stained with Microwave Blue (Protiga, Frederick, MD). To quantify conjugated mut-HA22, known concentrations of reduced free mut-HA22 were run on the same gel and stained to create a calibration curve. Odyssey infrared imaging system software (LI-COR Biosciences, Lincoln, NE) was used to calculate the amount of protein based on infrared intensity using a reciprocal fit curve. The distribution of liposomal hydrodynamic size was determined by dynamic light scattering measurements (Zetasizer, MALVERN Instruments, Worcestershire, UK) (41).
Calcein leakage from liposomes was determined by incubation of liposomes in PBS supplemented with 0, 10 or 50% heat-inactivated FBS as described (41). Briefly, liposomes were diluted in the buffer at 1:10 ratios and incubated at 37°C for 0-24 hours. Calcein leakage was determined as described (41).
Analysis of the number of mut-HA22 Molecules bound to Liposomes
The following parameters were used to calculate the average number of mut-HA22 molecules per liposome (equation below): (a) mut-HA22/mL liposome solution (14.67 μg CD22/mL); (b) Phospholipids/ mL liposome solution (0.317 mg phospholipids/mL); (c) Liposome hydrodynamic diameter (110 nm) and (d) Number of phospholipids/liposome for approximately 110 nm liposomes (9×104) (44).
(*The mut-HA22 molecules per liposome were calculated based on 100% DPPC). Therefore, the average number of mut-HA22 molecules per liposome was 90. For 1 mole% DSPE-PEG2000-MaL in the liposomes, the average number of mut-HA22 molecules per liposome was 47.
Mut-HA22-Liposome/Cell Interaction Assays
Binding of fluorescently labeled mut-HA22 liposomes to BJAB or SUP-T1 cells was done as follows: In a typical experiment, 107 cells/mL PBS-FBS were incubated with 0-2 μg of liposome-conjugated mut-HA22 (or the equivalent amount of liposomal phospholipids for control liposomes) for 20-40 minutes at 4°C or 37°C. The exact conditions for individual experiments are described in figure legends. Cells were washed twice with cold PBS in 1% bovine serum albumin (PBS-BSA) and were analyzed for immunofluorescence either by microscopy or FACS analyzing 10,000 viable cells per sample.
Fluorescence Microscopy
To observe binding/uptake of liposomes, liposome-bound cells were placed in microwells and images were captured using a NIKON Eclipse TE200 (Melville, NJ) inverted microscope supplied with a 40X oil lens (N.A. 1.30) and processed as described (45). To further assess the uptake of liposomes, samples were analyzed using confocal microscopy. Z-stack images were acquired with Olympus FV1000, IX81 inverted laser scanning confocal microscope (Olympus America Inc., Center Valley, PA) equipped with a motorized XYZ stage (Olympus), multi Argon laser (488 nm) for visualizing calcein loaded liposomes. High-resolution 12-bit images (512 X 512 pixels) were acquired using a 60X oil immersion objective lens (Olympus PLAPON NA 1.42) coupled with 4-5 optical zoom, resulting in a lateral resolution of 0.082 microns. We captured 14-16 optical slices at 1.7 micron intervals. All optical sections were captured using line-based Kalman filter available on the FV1000 microscope. Post acquisitions, the Z-stacks were analyzed using Olympus Fluoview Viewer, version 1.6.a (Olympus America Inc.).
Cell Viability Assays
Cells (5 × 105/mL) were incubated in RPMI1640 phenol-red free media with various concentrations of DOX-loaded liposomes for one hour at 37°C. Following incubation, cells were washed with RPMI1640 and plated in triplicates in a 96 well, flat bottom polystyrene plate in a total volume of 100 μL at 5 × 104 cells per well. The incubations were continued for an additional 72 hours at 37°C, 5% CO2. In some samples, cells were incubated with equivalent concentrations of free DOX or liposomes and the samples were incubated for 72 hours at 37°C without removal of the drug. At the end of incubations, cytotoxicity was determined using a cell viability kit (Cell Titer Blue assay kit, Promega, Madison, WI). Cell viability was calculated as percentage of control cells without DOX or liposomal treatment. Empty liposomes (DPPC:DSPE-PEG2000-Maleimide, 96:4) were also used as controls to determine lipid-mediated non-specific toxicity. Using the previously determined phospholipid concentrations of liposomal samples, equivalent amounts of empty liposomes were incubated with cells in the same manner as described above for DOX-loaded liposomes.
We also determined accumulation of liposomal DOX in the cells following a published protocol (46). Cells (105 per sample) were incubated with liposomes (0- 20 μg liposomal DOX per mL) in eppendorf tubes for 1 hour at 37°C. The cells were washed with culture medium without phenol red (0.5 mL×3). The pellets were resuspended in PBS (0.1 mL per sample), and transferred to 96-well plates. DOX accumulation was determined as above, except the standard curve was generated in the presence of untreated cells.
Results and Discussion
(i) Preparation and Characterization of mut-HA22
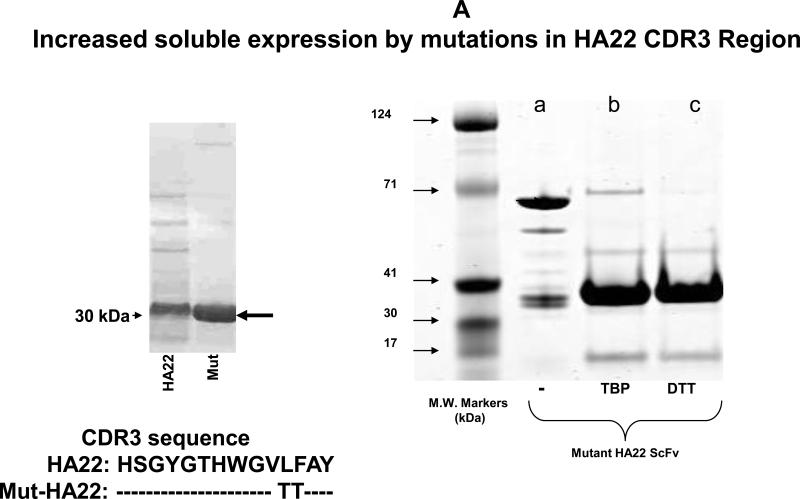
Previous studies have described immunoliposomes using a biosynthetically lipid-tagged CD22 ScFv, though further characterization of this ScFv (such as ligand-affinity and internalization properties) has not been clearly documented. We aimed at developing a CD22 scFv mutant from a well-characterized CD22 ScFv (HA22). From the mutational library based on HA22, a mutant with two amino acid changes in the HA22 CDR3 was selected from phage panning that showed slightly increased binding affinity but significantly increased soluble expression (Xiao and Dimitrov, unpublished data and Figure 1A). A cysteine residue was introduced in front of its stop codon and the resulting clone was named mut-HA22.
Figure 1. Characterization of mut-HA22.
Figure 1A: Analysis of mut-HA22. Mut-HA22 was analyzed by gel electrophoresis. (Left Panel), mutations to the CDR3 region of HA22 increased soluble expression of the scFv. (Right Panel), verification of mut-HA22 reduction by gel electrophoresis: (a) mut-HA22, shown as a monomer and dimer, (b) mut-HA22 reduced with tributylphosphine (TBP), and (c) mut-HA22 reduced with dithiothreitol (DTT).
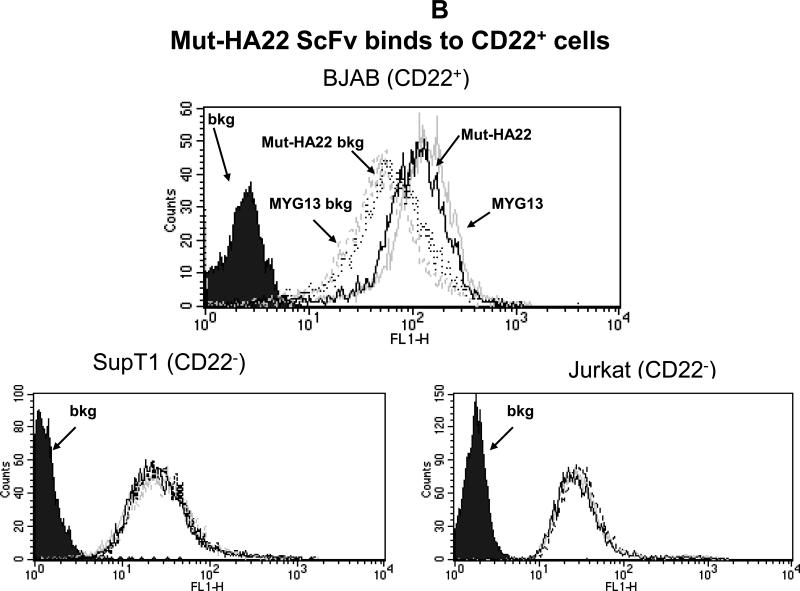
Figure 1B: Mut-HA22 binds to CD22-expressing cells. To analyze specific binding of mut-HA22 with CD22, BJAB cells were incubated with 2μg mut-HA22 followed by sequential incubations with mouse anti-histidine IgG, and FITC-conjugated goat anti-mouse IgG. A commercial anti-CD22 IgG MYG13 was used as positive control, and was stained with FITC-conjugated goat anti-mouse IgG. SUP-T1 and Jurkat cells were used as negative controls. Graphs representing corresponding cell types are indicated. Fluorescence distributions of stained cells are: solid black curves, cells alone; black lines, immunostaining of mut-HA22; grey lines, immunostaining of MYG13; black dotted lines, mut-HA22's secondary Abs (mouse anti-his IgG and FITC conjugated goat anti-mouse IgG; dotted grey lines, MYG13's secondary Abs (FITC-conjugated goat anti-mouse IgG). These results were reproducible from at least three independent experiments. Bkg, background binding.
We have demonstrated that treatment of mut-HA22 dimers with TBP (a mild reducing agent) or dithiothreitol (DTT) resulted in nearly complete reduction to monomers (see Figure 1A, right panel). TBP-reduced mut-HA22 was used for liposome conjugation (41;47). We also examined the effect of mutations in the CDR3 region of HA22 on its binding to cell-surface expressed CD22. Since mut-HA22 contains a His-tag, an anti-His mouse antibody (+FITC-conjugated mouse Ab) was used to detect binding of mut-HA22 to cells. We show that incubation of CD22-expressing BJAB cells with mut-HA22 resulted in CD22-specific binding (Figure 1B, top panel, solid grey lines) as compared to incubations without mut-HA22 (i.e. cells incubated only with mouse anti-His+FITC conjugated mouse Ab, Figure 1B, top panel, broken grey lines). We also tested cell-surface expression of CD22 on BJAB cells by using a commercial anti-CD22 mAb, MYG13. The results shown in Figure 1B indicate that mut-HA22 bound to CD22 with similar affinity to that of MYG13 (solid black lines MYG13; broken black lines, FITC-conjugated anti-mouse Ab alone). The background binding with secondary antibodies to various cells shown in Figure 1B is similar to the data supplied by the manufacturer. Since we did not observe an increase in background fluorescence in CD22- cells (SUP-T1 or Jurkat T lymphocytes) after incubations with mut-HA22 or MYG13, we conclude that mut-HA22 binding to BJAB cells is CD22 antigen specific. Hence, mut-HA22 is likely to serve as a suitable ligand for CD22 targeting.
(ii) Mut-HA22- Conjugated Liposomes
Various liposome formulations were prepared according to Table 1. DPPC was used as the matrix lipid and a pegylated lipid (4 mol %) was included for future in vivo applications. DPPC liposomes have been previously demonstrated to release their contents at ≈41°C due to the phase-transition and therefore, are known to have thermosensitive properties (48). Calcein (λ ex/em 494 nm/517 nm) was encapsulated in the liposomes (50 mM) and some preparations also included a fluorescent lipid, rhodamine PE (λ ex/em 557nm/571nm). DSPE-PEG2000-MaL was included to conjugate mut-HA22 via malemide-cysteine chemistry on the surface of liposomes (mut-HA22-liposomes). Control liposomes (without mut-HA22 conjugation) were prepared under identical conditions except DSPE-PEG2000 (instead of DSPE- PEG2000-MaL) was included in the liposomes. The antibody conjugation to the liposome surface was based on a chemical reaction between maleimide of the lipid with the thiol group on the C-terminus of mut-HA22 (28;49). Conjugation of mut-HA22 to preformed DPPC/DSPE-PEG2000-maleimide liposomes yielded immunoliposomes with 90 mut-HA22 per liposome (41). Initially we examined antibody conjugation with liposomes containing either 1 mol % or 4 mol % DSPE-PEG2000-MaL. The liposomes containing 1 mol% DSPE-PEG2000-MaL also contained 3 mol% DSPE-PEG2000 (Table 1). We used a molar ratio of 1:41 mut-HA22-PEG2000-MaL for conjugation, considering the number of lipids only in the outer monolayer of liposomes (see Methods section). Initial analysis of mut-HA22-liposome conjugation was done by separation of unconjugated antibody from the mut-HA22 conjugated liposomes on a size exclusion column, essentially as described (41).
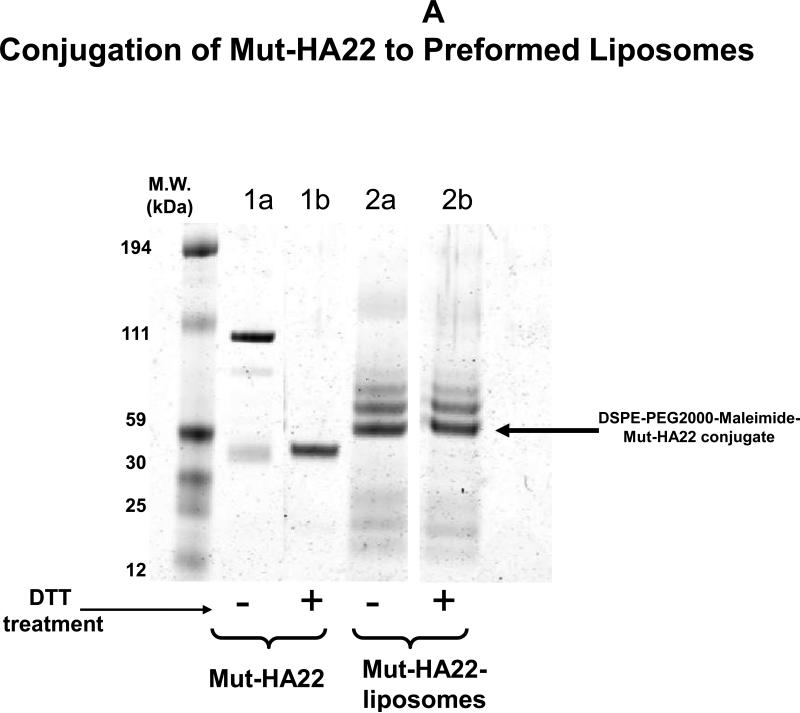
To confirm mut-HA22 coupling to the liposomes, we examined a molecular weight shift of mut-HA22 following conjugation with DSPE-PEG2000-MaL by gel electrophoresis. The data are shown in Figure 2A. We observed a shift in molecular weight of mut-HA22 monomers (Figure 2A, lane 1b) upon conjugation with liposomes (Fig. 2A, lane 2a). DTT pre-treatment of the conjugated liposomes did not result in any change in the electrophoresis pattern (Fig. 2A, lane 2b), confirming that mut-HA22 was covalently coupled to the PEG lipid. We also observed additional bands in the lanes 2a/b. Although the exact nature of additional bands requires further characterization, it is likely that availability of additional SH- groups on the mut-HA22 may have resulted in coupling of more than one PEG lipid. Control liposomes (lipid compositions without maleimide functional group) did not show any antibody binding above background levels under identical conditions confirming the specificity of this reaction (data not shown).
Figure 2. Characterization of mut-HA22-liposomes.
Figure 2A: Determination of mut-HA22 conjugation to liposomes. Concentrated liposomes were prepared in sample buffer with or without DTT, and were loaded on a 4-12% Bis-Tris gel (under non-reducing conditions). The gel was run in MES-SDS running buffer at 200V for 35 minutes and proteins were stained with Microwave Blue. Conjugation of mut-HA22 to DSPE-PEG2000-Maleimide is verified by the molecular weight increase of free mut-HA22 to liposomal mut-HA22 (indicated by the arrow).
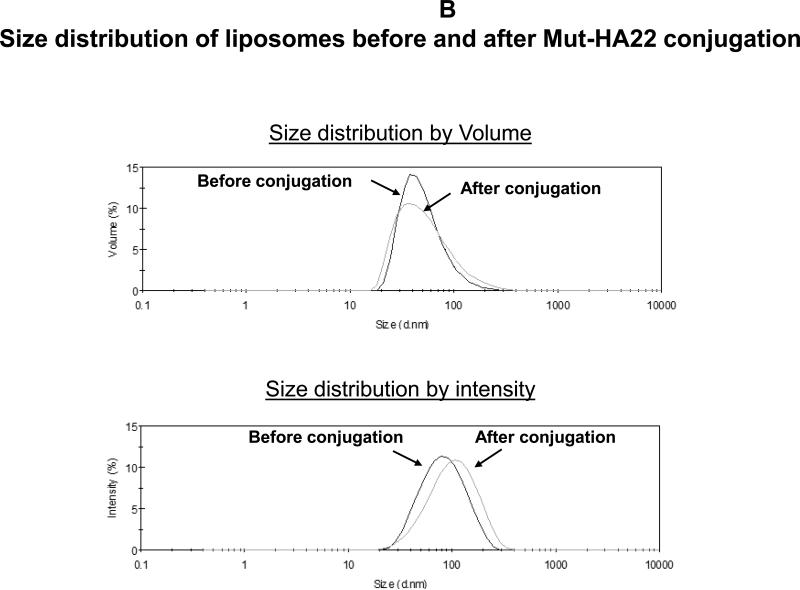
Figure 2B: Hydrodynamic size distribution of liposomes before and after conjugation with mut-HA22. Hydrodynamic size was measured by backlight scattering in phosphate buffered saline with a Malvern Zetasizer Nano ZS instrument. The data are plotted as volume (top panel) and intensity (bottom panel) weighted distributions.
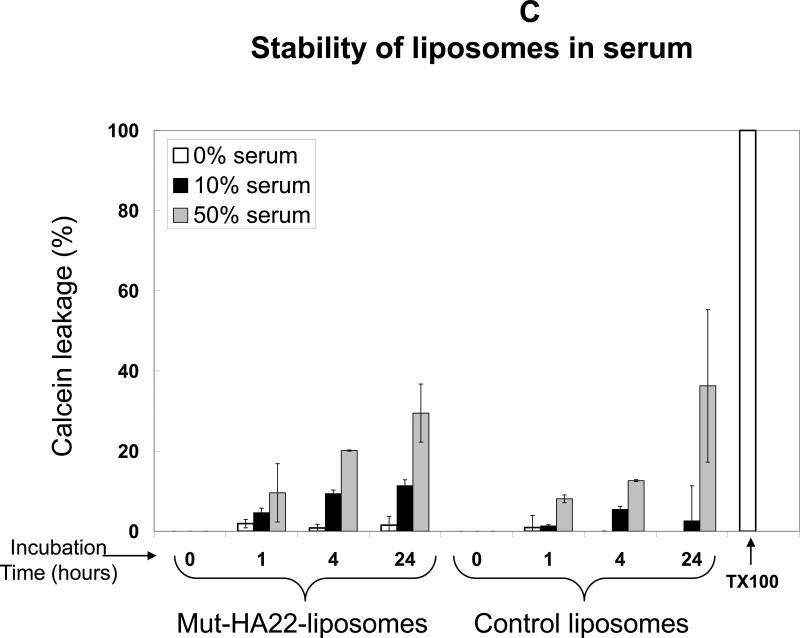
Figure 2C: Stability of mut-HA22-liposomes in the presence of serum at 37°C. Calcein release from liposomes was measured after liposome incubation at 37°C in PBS with 0, 10, or 50% FBS for 0, 1, 4, or 24 hours before and after addition of Triton X-100 (TX100). Leakage is expressed as a percent of total, where 100% calcein leakage was obtained by addition of 20 μl Triton-X100 (10 % w/v) and 0% calcein leakage was the background fluorescence value before liposome incubation at 37°C. Error bars represent ± SD from at least three samples within a single experiment.
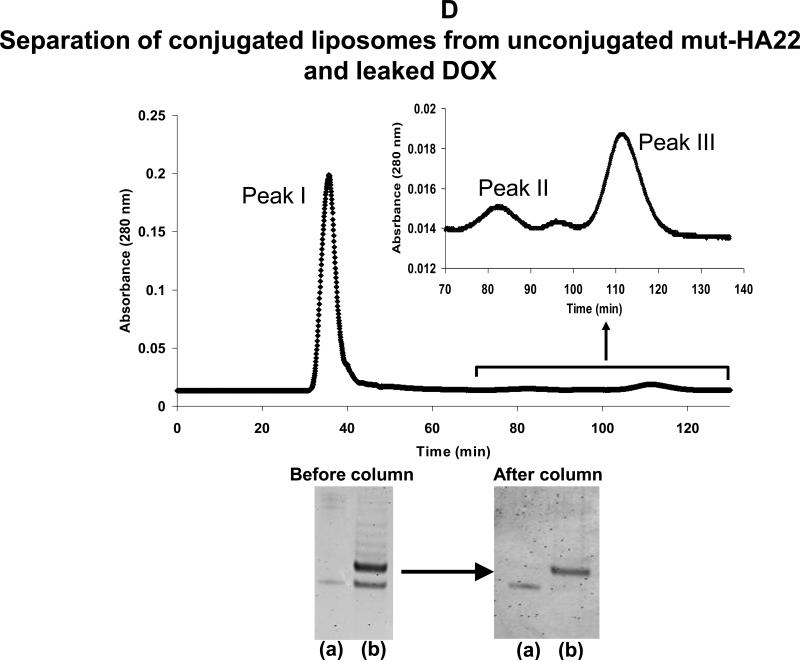
Figure 2D: Elution profile of DOX-loaded, mut-HA22 liposomes on Sepharose CL-6B column
Top panel: mut-HA22 conjugated liposomes were separated from free, unconjugated mut-HA22 scFv through size exclusion chromatography on a Sepharose CL-6B column (1x40cm). 1 mL fractions were collected at an elution rate of 0.37 mL/min. Absorbance at 280nm (as measured by UV detector) is shown. Peak I: DOX-loaded, mut-HA22 conjugated liposomes; Peak II: unconjugated mut-HA22 scFv; Peak III: Free DOX.
Bottom panel: Samples in peak I were analyzed (see Fig 2A) to confirm mut-HA22 conjugation to liposomes. (a): mut-HA22 scFv standard; (b): DOX-loaded, mut-HA22 conjugated liposomes before and after Sepharose CL-6B column. The fractions in peak II were analyzed for the presence of mut-HA22 by gel electrophoresis as described (see Methods section, data not shown). Similarly, all fractions were also analyzed for DOX and peak III was assigned accordingly (data not shown).
To examine the effect of mut-HA22 conjugation on the physical properties of liposomes, samples were characterized for their size distribution by dynamic light scattering measurements. Data presented in Figure 2B demonstrate that mut-HA22 conjugation to the liposome surface did not have any significant effect on their size distribution. Our liposomes exhibited an average size distribution of 80-100 nm, with an average 90 mut-HA22 molecules per liposome.
(iii)Liposome Stability studies in Serum
For future medical applications, it is critical that physical properties of mut-HA22-liposomes are not altered upon storage or upon interactions with plasma components prior to targeting to permissive cells and/or tissues. Therefore, we examined stability of liposomes in the presence of serum. Liposomes were incubated in PBS supplemented with 0-50% heat-inactivated serum at 37°C for various time periods, and extent of calcein release was measured. Results are shown in Figure 2C. An enhancement of calcein release from liposomes was observed with an increase in incubation time and percent serum in the incubation medium. Typically, 5-10% calcein leaked from mut-HA22-liposomes upon incubation in PBS containing 10% FBS for 1-24 hours. Similarly, incubations in the presence of PBS containing 50% serum resulted in 10-30% calcein leakage from mut-HA22 liposomes after 1-24 hour incubations (Figure 2C). Control liposomes showed similar leakage data (Figure 2C). The results are representative of two independent measurements form the same liposome batch. Liposomes could be stored at room temperature in the dark for up to 5 weeks without any appreciable calcein release or change in their size distribution (data not shown).
(iv) Interaction of mut-HA22-liposomes with Receptor Expressing Cells
The preceding section demonstrates that mut-HA22 conjugation to the liposomes occurred via the chemical reaction between cysteines on the antibody and malemide groups on the PEG lipid of the liposomes. The antibody conjugation had no significant effect on the physical properties of liposomes. Therefore, our next set of experiments were designed to evaluate the binding (and uptake) of these liposomes via mut-HA22 and CD22 interactions. We have used BJAB (CD22+) and SUP-T1 (CD22-) cells for these studies. Liposome binding and/or internalization via the CD22 was examined by fluorescence microscopy and by FACS analysis.
(a) Cellular association of mut-HA22-liposomes occurs via the interaction of cell-surface expressed CD22
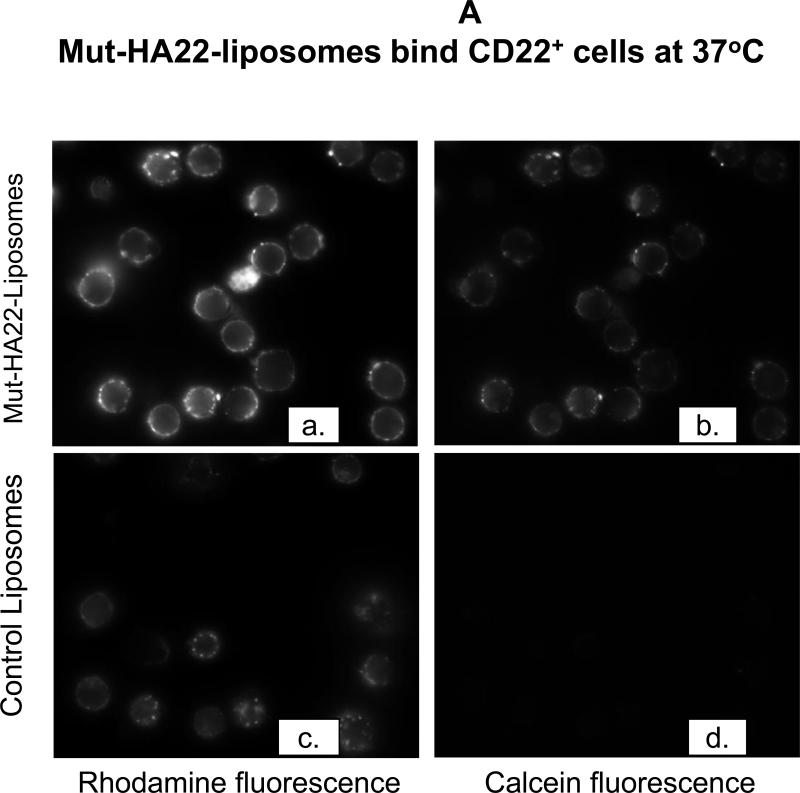
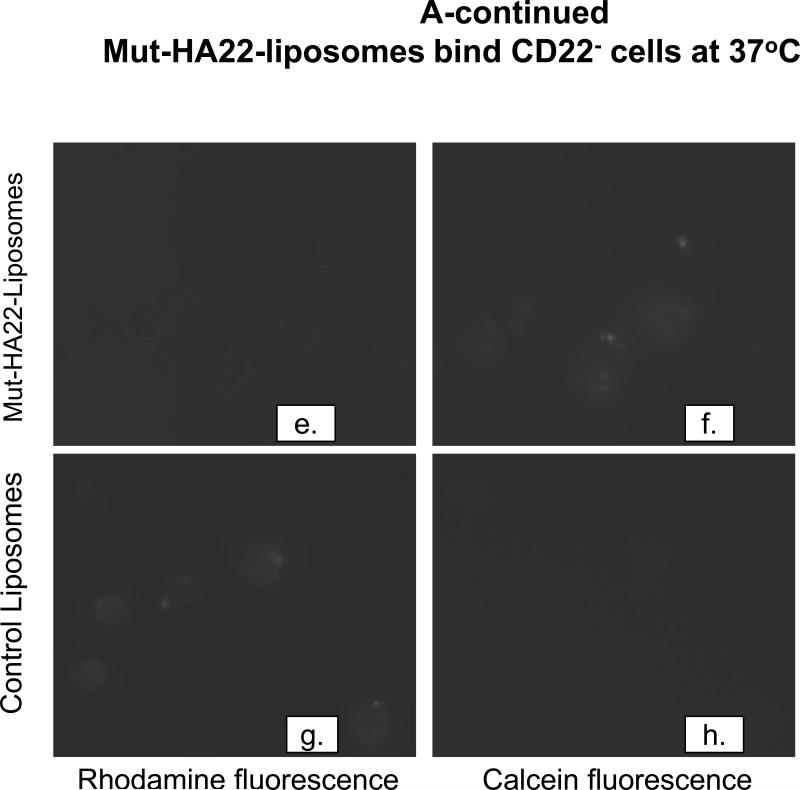
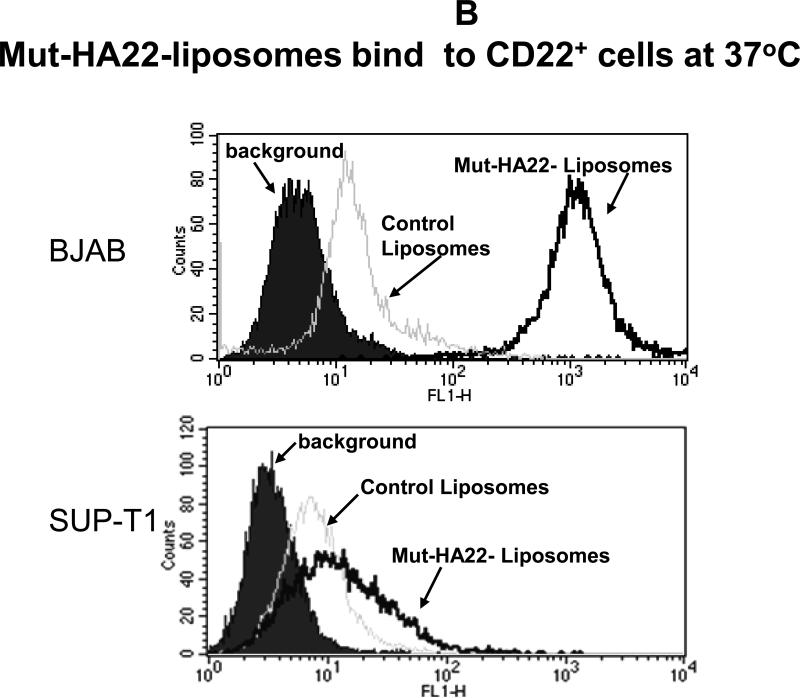
To investigate CD22-mediated binding of liposomes, BJAB or SUP-T1 cells were incubated with 2 μg liposome-conjugated mut-HA22/106 cells at 37°C for 30 minutes, and liposome binding was determined by fluorescence measurements either by (i) microscopy or (ii) FACS. The results are presented in Figure 3A (fluorescence microscopy images) and Figure 3B-D (FACS). Mut-HA22-liposomes showed significant binding to BJAB cells (Figure 3A, a-d) based on calcein and rhodamine fluorescence. On the other hand, control liposomes showed only a low level of liposome binding. Interestingly, we did not see any calcein with control liposomes, whereas there was background binding based on rho-PE fluorescence. We attribute this discrepancy to calcein quenching within the liposomes. Binding of liposomes with CD22- SUP-T1 cells is shown in Figure 3A (e-h). It is clear that mut-HA22-liposomes did not show enhanced binding compared to control liposomes, corroborating our interpretation that the mut-HA22 binds specifically to the CD22 on the cell surface. Moreover, in our initial experiments, we also tested liposomes using the lipid composition used for mut-HA22-liposomes but without any mut-HA22 conjugation. These liposomes did not show any CD22-specific binding above background, confirming CD22-specific interactions of mut-HA22-liposomes (data not shown). Next, we analyzed liposome binding to cells by FACS analysis. The data presented in Figure 3B, clearly show that mut-HA22-liposomes bind to BJAB cells with high affinity, as compared to that of control liposomes (Fig. 3B, top panel). In contrast to the microscopy data, we observed a slight increase in binding of mut-HA22-liposomes to SUP-T1 cells as compared to control liposomes (Figure 3B, lower panel). Nevertheless, mut-HA22-liposome binding to BJAB cells was significantly greater than that for SUP-T1 cells.
Figure 3. Binding of mut-HA22-liposomes to CD22-expressing cells.
Cells (107/mL in RPMI-1640, 10% serum) were incubated with fluorescently labeled mut-HA22-liposomes (2 μg of mut-HA22/46μg of phospholipids) or control liposomes (0 μg of mut-HA22/46 μg of phospholipids) for 40 minutes at 37°C. At the end of incubations, cells were centrifuged and washed twice with PBS-BSA to remove unbound liposomes; cell-bound fluorescence was examined by microscopy and FACS.
Figure 3A: Binding of mut-HA22-liposomes to CD22-receptor expressing cells.
Fluorescence images were captured using the Nikon Eclipse TE200 inverted microscope with a 40X oil objective (N.A. 1.30). Images shown are for calcein and rhodamine fluorescence as indicated ((a-d), BJAB cells, (e-h) SUP-T1 cells).
Figure 3B: Flow cytometry analysis of incubated cells. The data shown are for calcein fluorescence.
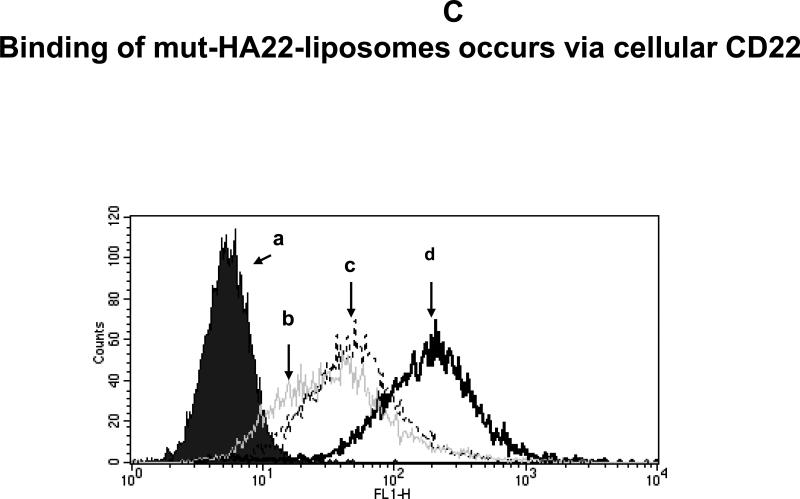
Figure 3C: Pre-incubation of cells with free mut-HA22 inhibits subsequent binding of liposomes.
BJAB cells were incubated with (b) 8, (c) 2, or (d) 0 μg of free mut-HA22 at 4°C for 30 minutes in the dark, washed twice with cold PBS-BSA, and then incubated with fluorescently labeled mut-HA22-liposomes (1μg/106 cells) at 4°C for 30 minutes ((a) cells only)).
(b) Binding of mut-HA22-liposomes to BJAB cells is inhibited by free mut-HA22
The results presented in preceding section(s) demonstrate that mut-HA22-liposomes interact with the CD22-expressing cells. To further establish dependence of the liposome binding via the CD22, we pre-incubated BJAB cells with varying concentrations of unconjugated mut-HA22 (0-8 μg mut-HA22/106 cells) for 30 minutes. The incubations were performed at 4°C to prevent internalization of mut-HA22. The unbound antibody was removed and the samples were incubated with calcein-loaded mut-HA22-liposomes (1 μg/106 cells) for additional 30 minutes at 4°C. The liposome-associated cells were quantified by FACS. The results presented in Figure 3C clearly demonstrate that pre-incubation of BJAB cells with free antibody blocked the association with the liposomes. We conclude that binding of the mut-HA22 conjugated liposomes occurs by specific interactions with cellular CD22.
(v)Effect of Liposomal mut-HA22 Density and Dose-dependence on mut-HA22-liposome-cell binding
It has been shown earlier that immunoliposomes bear the advantage to present multivalent ligand-receptor interactions, as compared to their monovalent counter-parts (50). Furthermore, the average number of antibody molecules per liposome is likely to have a significant effect on liposome binding and/or internalization by receptor-expressing cells (50). As demonstrated in our previous sections, mut-HA22-liposomes interact with CD22-receptor expressing cells. Our next experiments were designed to evaluate the optimal number of mut-HA22 molecules per liposome and liposome to cell ratios for CD22-specific interactions.
(a) Liposome-cell association increases with an increase in mut-HA22 scFv molecules per Liposome
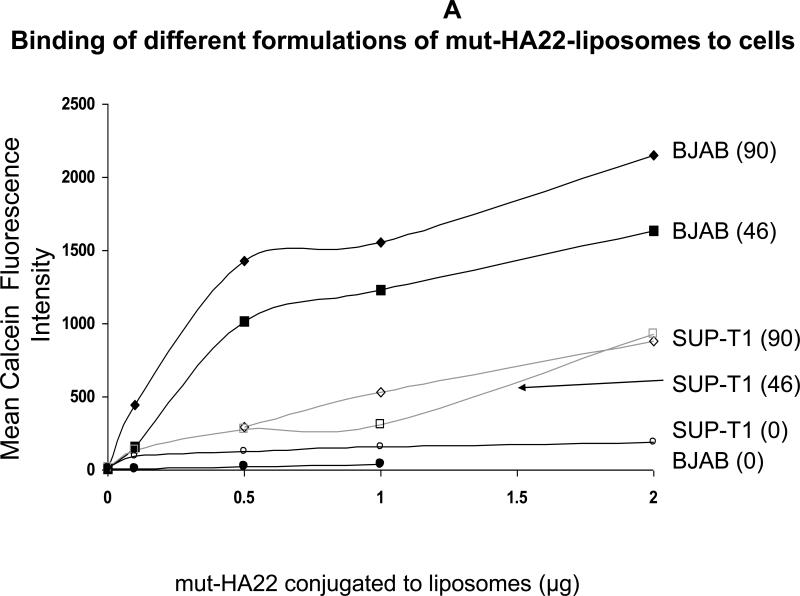
In our initial experiments, we investigated the relationship between amount of mut-HA22 conjugation to liposomes and the liposome binding to the target receptor. We generated mut-HA22 scFv conjugated liposomes using either 1 mol% (Table 1, Formulation II) or 4 mol% DSPE-PEG2000-MaL (Table 1, Formulation I). The conjugation yields in the two formulations were 47 and 90 scFv molecules/liposome respectively. Liposome-cell suspensions were incubated at 37°C for 30 minutes, and liposome binding was observed by monitoring calcein fluorescence by FACS. Data are summarized in Figure 4A, and the values are presented as mean fluorescence intensities. We observed an increase in cell-associated fluorescence with an increasing dosage of liposomes (Formulations I, II) for CD22-expressing BJAB cells. By comparison, a small increase in binding was observed above background when SUP-T1 cells were used for incubations. It is also evident from the data presented in Figure 4A that liposomes prepared using the 4 mol% DSPE-PEG2000-MaL lipid (Table 1, Formulation I) exhibited an increase in CD22-dependent binding as compared to the liposomes prepared using only 1 mol% DSPE-PEG2000-MaL lipid (Figure 4A). Since our future objective is to develop these liposomes for treatment of B-cell lymphoma, we used Formulation I (containing 90 scFv molecules per liposome) for further analysis.
Figure 4.
A: Binding of mut-HA22--liposomes with 1% or 4% DSPE-PEG2000-Maleimide to CD22- expressing cells.
Cells (107/mL in RPMI-1640, 10% serum) were incubated with various concentrations of calcein-loaded mut-HA22-liposomes for 40 minutes at 37°C. At the end of incubations, cells were centrifuged and washed twice with PBS-BSA to remove unbound liposomes; cell-bound fluorescence was examined by FACS. The graph represents mean fluorescence intensity values (y-axis). Three liposome preparations (see Table 1) corresponding to either 0 mol% (=0 mut-HA22/liposome), 1 mol% (=47 mut-HA22/liposome) or 4 mol% (=90 mut-HA22/liposome) were used for incubations with either BJAB or SUP-T1 cells. The amounts of liposomal mut-HA22 added are given in x-axis. Cell types are indicated at the end of each curve. The numbers in parentheses indicate the average number of mut-HA22/liposome.
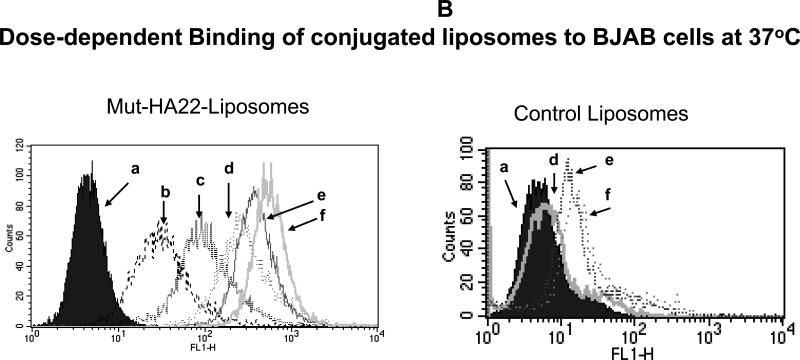
Figure 4B: Dose-dependent binding of mut-HA22-liposomes to BJAB cells.
BJAB cells were incubated with various concentrations of calcein-loaded liposomes for 40 minutes at 37°C. At the end of incubations, cells were washed and analyzed for calcein fluorescence by FACS. Concentrations of liposome-conjugated mut-HA22 added to cells: (a) cells alone, (b) 0.01 μg mut-HA22, (c) 0.05 μg mut-HA22 (d), 0.2 μg mut-HA22, (e) 0.5 μg mut-HA22, and (f) 2 μg mut-HA22. Control liposomes contained the corresponding phospholipid quantities without any mut-HA22.
(b) Dose-dependent binding of mut-HA22 liposomes
Since the degree of association of mut-HA22-liposomes to BJAB cells was dependent both on the initial input and the total (average) number of antibody molecules per liposome (Figure 4A), our next experiments were designed to evaluate the amount of liposomes needed for optimal binding to cellular CD22. Mut-HA22-liposomes (0-2 μg liposomal protein) were incubated with BJAB cells at 37°C and the extent of liposome binding was monitored by FACS analysis of increased cell fluorescence (Figure 4B). We observed that binding of liposomes increased up to a ratio of 0-2 μg liposomal protein/106 cells Control liposomes on the other hand did not show any significant increase in liposome binding (a. 0 μg, b. 0.1 μg, c. 0.5 μg, d. 1 μg, e. 2μg).
(v) Intracellular Uptake of mut-HA22-liposomes
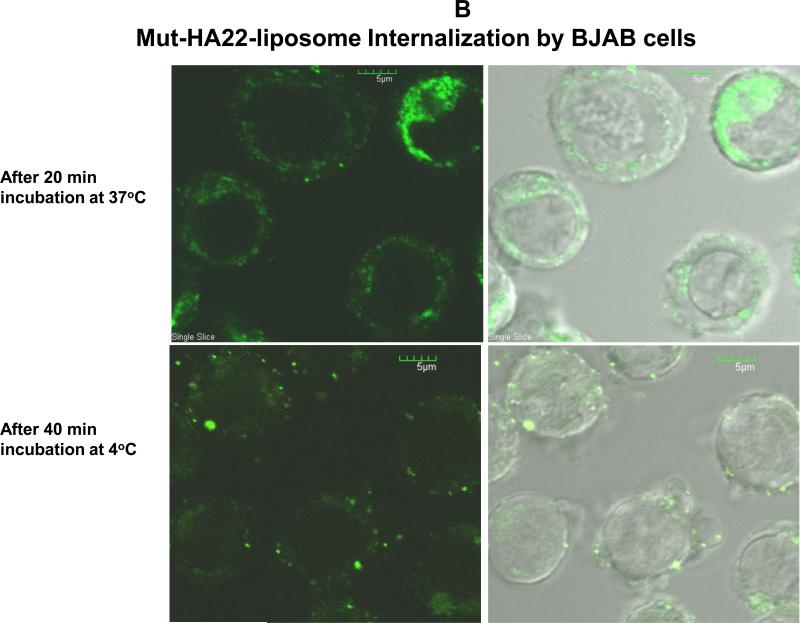
It has been proposed that internalization of targeted liposomes by the tumor or diseased cells is important to achieve effective therapeutic index and delivery of payload of drugs (51). Therefore, we examined interactions of liposome-BJAB cells, (i) by comparing liposome-cell association at 4°C versus 37°C, and (b) by visualizing intracellular accumulation of calcein loaded liposomes at 37°C by confocal microscopy. The results are presented in Figure 5.
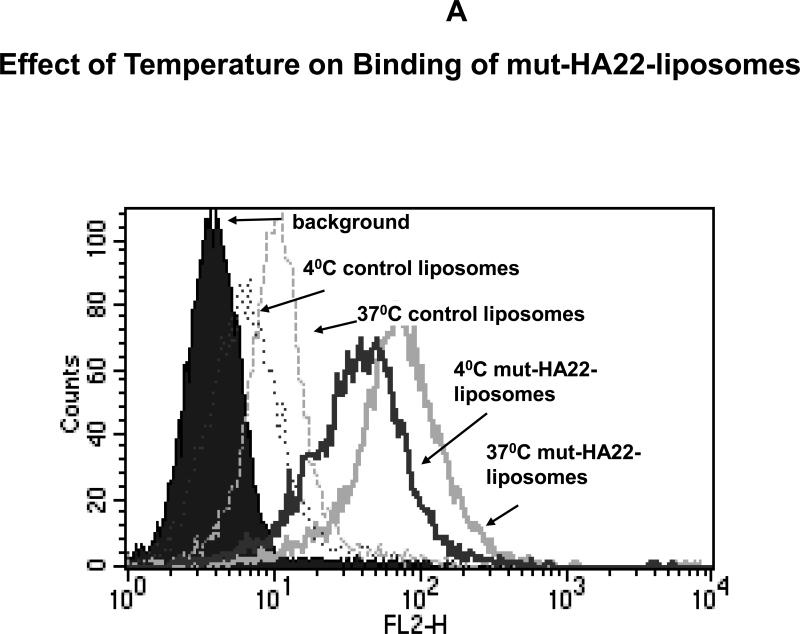
Figure 5. Uptake of mut-HA22-liposomes by BJAB cells at 37°C.
BJAB cells (107/mL RPMI-1640) were incubated with liposomes at 37°C or 4°C. Cells were then washed twice with cold PBS-BSA and kept on ice until analyzed.
Figure 5A: Temperature Dependent Binding/Uptake of mut-HA22-liposomes by BJAB cells. Cells were incubated with 4 μg of liposome-conjugated mut-HA22 (or equivalent amount of phospholipids for control liposomes) for 40 minutes at 37°C or 4°C (see methods section). Cells were then washed and rhodamine fluorescence of cells was analyzed by FACS.
Figure 5B: Intracellular Localization of mut-HA22-scFv liposomes by BJAB Cells. Cells were incubated with 0.2 μg of liposome-conjugated mut-HA22 (or equivalent amount of phospholipids for control liposomes) for 20 or 40 minutes at 37 or 4°C. Z-stacks of 14-16 optical slices at 1.7 micron intervals were acquired. Calcein fluorescence of cells is shown in a single slice of the acquired images.
Figure 5A shows that the incubation temperature affected binding of liposomes to the cells. It is clear from the results that liposome binding was increased at 37°C (solid grey line, Fig. 5A) as compared to that at 4°C (solid black line, Fig. 5A). We also observed similar increase at 37°C when non-targeted (control) liposomes were used (dotted lines, black, 4°C, grey, 37°C). However, the extent of mut-HA22-liposome binding to BJAB cells was significantly greater than control liposomes at both temperatures.
Next, we examined intracellular uptake by confocal microscopy. The results presented in Figure 5B clearly show that a major population of mut-HA22-liposomes was accumulated intracellularly following incubations at 37°C for 20 minutes (Fig 5B, a. calcein, b. calcein/phase overlay). In contrast, incubations of liposomes at 4°C up to 40 minutes showed majority of liposomes only on the surface of cells (Fig 5B, c, calcein; d, calcein/phase overlay). To obtain three-dimensional information on the cellular location of the fluorescence, a series of optical sections were made through infected BJAB cells. The results further confirmed that in BJAB cells incubated at 37°C with mut-HA22-liposomes, there were both surface and intracellular foci of fluorescence corresponding to intracellular and extracellular liposomes (data not shown).
(vi) DOX accumulation and DOX-mediated cytotoxicity
Immunoliposomes have long been studied as vehicles for site-specifc delivery of drugs and pharmaceutics (24). Recently, anti-HER2 scFv-DOXIL (29) and anti-CD22 mAb-DOXIL (33) have demonstrated improved DOX-mediated cell killing in vitro in comparison to DOXIL alone (33), as well as in animal studies (29). As shown in preceding sections, we have demonstrated that mut-HA22 liposomes are internalized by CD22+ cells via CD22-mut-HA22 interactions. To further confirm the potential of these formulations in the clinic, we encapsulated DOX using the well-established ammonium sulfate remote loading protocol (43), followed by conjugation of mut-HA22 on the surface of DOX-loaded liposomes (Table I, formulations IV, DOX- mut-HA22 liposomes & V, DOX-control liposomes). DOX loading was quantitated by measuring DOX in the samples in the presence of Triton X-100 (Figure 6A, left panel). Our liposomes typically had 150-200 ng DOX/nmol Pi. Unconjugated mut-HA22 was separated from mut-HA22 liposomes by gel permeation column chromatography on a Sepharose CL-6B column. DOX-mut-HA22 liposomes (peak I, Figure 2D) were eluted in the void volume whereas unconjugated mut-HA22 (peak II, Figure 2D) was eluted in the included volume. We did not observe any appreciable leakage of DOX from liposomes during the conjugation step (Figure 2D, peak III). The ratios of DOX to the phospholipid lipid were 180 ng DOX/nmol inorganic phosphate (Pi) and 157 ng DOX /nmol Pi for control liposomes (Formulation V, Table I) and mut-HA2 liposomes (Formulation IV, table I) respectively. The effect of DOX loading and mut-HA22 conjugation on the average size distribution is shown in Table I. A slight increase in average diameter upon DOX loading (110.5 ± 6.6 nm) and mut-HA22 conjugation (123.0 ± 7.9 nm) was observed when compared with liposomes before DOX loading (86.9 ± 4.0 nm).
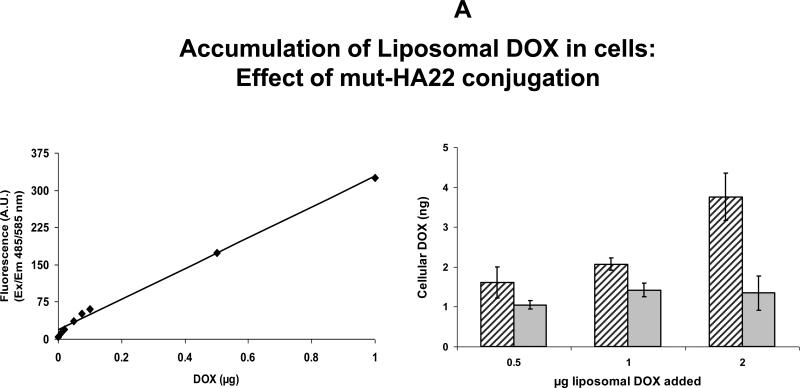
Figure 6.
A: Cellular accumulation of liposomal DOX by Raji cells
Cells (3×105 cells per sample) were incubated with 0.5 – 2 ug of liposomal DOX for one hour at 37°C. The cells were then washed with phenol-red free culture medium (0.5mL×3) and resuspended in 0.3 mL of PBS and transferred to a 96-well plate (0.1mL×3). Cellular accumulation of DOX was determined using a standard curve of known DOX concentrations in the presence of cell lysates without incubation with liposomes (left panel). Right panel: accumulation of DOX in nanograms. Solid gray bars, control liposomes; black diagonal line bars, mut-HA22 liposomes. Error bars represent ± SD of triplicate samples within a single experiment.
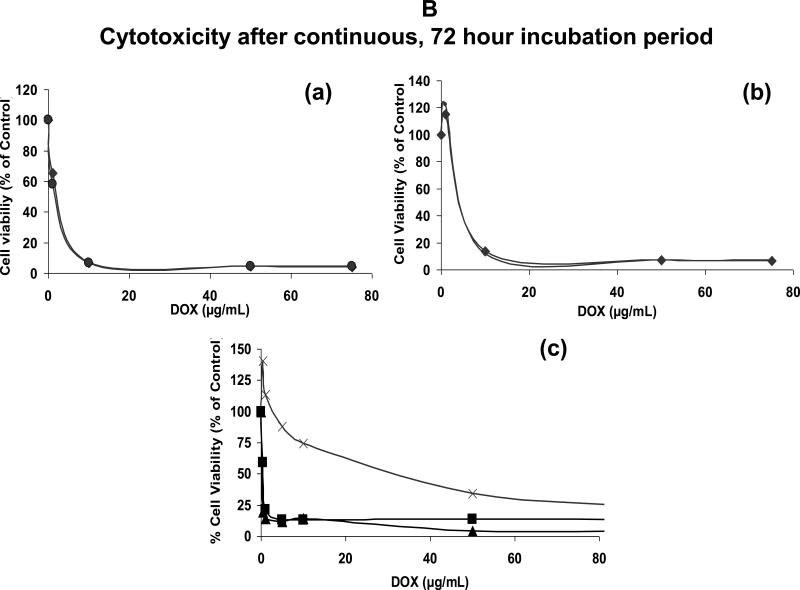
Figure 6B: Cell viability in the continuous presence of liposomal DOX
Cells were plated in duplicates in a 96-well plate (105/0.1mL per well) and incubated with 0-7.5 ug of liposomal or free DOX for 72 hours at 37°C. Cell viability was calculated taking control sample (without DOX) as 100%. Values represent an average of duplicate samples.
(a) and (b) Cytotoxicity by liposomal DOX: (a) BJAB (CD22+), (b) SUP-T1 (CD22-) mut-HA22 liposomes (◆) and control liposomes (●).
(c) Cytotoxicity by free DOX: SUP-T1 (X), BJAB (▲), and Raji (■).
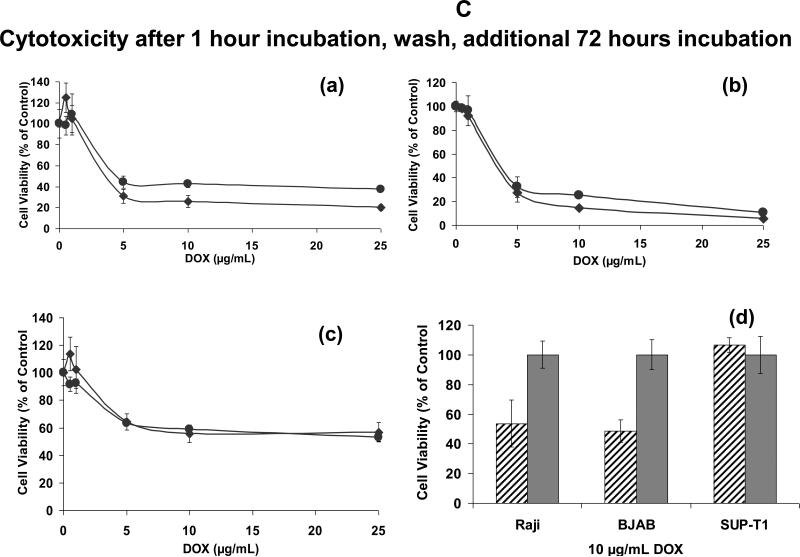
Figure 6C: Cell viability following pre-incubation with liposomes
Cells were incubated in triplicates with liposomal DOX for 1 hour, and any unbound liposomes were washed off. Following an additional 72-hour incubation, cell viability was determined (see Figure 6B).
(a) Raji (CD22+), (b) BJAB (CD22+), and (c) SUP-T1 (CD22-) mut-HA22 liposomes (◆) and control liposomes (●)
(d) A snapshot of cell viability at a concentration of 10 ug/mL liposomal DOX is shown (Data from the respective curves in Figure 6C (a-c)). In this graph, the total number of cells remaining viable in the presence of control liposomes were taken as 100%. mut-HA22 liposomes (diagonal line bars), control liposomes (gray bars)
Results on cellular interaction of DOX-loaded liposomes are shown in Figure 6. Our initial experiments were aimed at determining effect of mut-HA22 conjugation on the uptake of liposomes by CD22+ cells. Increasing amounts of DOX-loaded liposomes were incubated with Raji cells for 1 hour at 37°C. Subsequently, unbound liposomes were removed and cellular DOX was quantitated in cell lysates (see Methods section). Data presented in Figure 6A (right panel) clearly show that conjugation of mut-HA22 to the liposomes resulted in at least a 2 fold increase in accumulation of DOX in the cells. Moreover, we observed a dose-dependent DOX accumulation in these samples. In contrast, control liposomes did not show an increase in DOX accumulation with increasing dose of the liposomes (Figure 6A). A standard curve for DOX quantitation is also shown in Figure 6A (left panel).
Next, we examined the effect of liposomal DOX on cellular viability (Figure 6B&C). First, DOX-mediated cell killing was determined in continuous presence of DOX-loaded liposomes or free DOX. When DOX was encapsulated in liposomes, > 90 % cell killing was observed at 10 μg DOX/mL for all the cell lines tested (Figure 6B (a, BJAB, b, SUP-T1), circles, control liposomes; diamonds, mut-HA22 liposomes). On the other hand, >90% cell killing was observed at relatively lower DOX concentrations (1-2 μg free DOX /mL) for BJAB (solid triangles, Figure 6B,) and Raji cells (squares, Figure 6B,c), consistent with previous studies (33). The only exception was SUP-T1 cells, as those required very high concentrations of free DOX to achieve maximum cytotoxicity (crosses, Figure 6B, c). It is likely that intracellular DOX from SUP-T1 cells was pumped out of the cells by some membrane transport mechanisms. We also examined non-specific effects mediated by liposomal lipids (unrelated to DOX-mediated cytotoxicity) by incubating empty liposomes (not loaded with DOX) with cells. We did not observe any cell killing at concentrations equivalent to the amounts of liposomes used in our cytotoxicity experiments.
Although our initial results clearly demonstrated CD22-specific binding and uptake of mut-HA22 liposomes by fluorescence (Figures 3-5) and liposomal DOX accumulation (Figure 6A), the cell viability results (Figure 6B(a)) did not exhibit any selectivity by mut-HA22 liposomes. We surmise that continuous incubations of liposomes in cell cultures for 72 hours superseded the kinetics of mut-HA22 liposomes internalization. To mimic in vivo conditions of liposome uptake by tumors and clearance by reticuloendothelial systems, we measured cell viability for 72 hours following removal of unbound liposomes after the first 60 minutes of incubation (29;33). In the previous cell viability experiment (Figure 6B), we observed maximum cell killing by liposomal DOX at a concentration of 10 μg DOX/mL. Therefore we tested concentrations of DOX up to 25 μg/mL in these experiments. Enhanced cytotoxicity by mut-HA22 liposomes (diamonds) versus control liposomes (circles) was observed for CD22+ cells (Figure 6C(a), Raji and (b), BJAB), but not CD22- cells (Figure 6C(c), SUP-T1). The relative increase in cellular toxicity (~50%) by mut-HA22 liposomes in comparison with control liposomes is shown in Figure 6C(d) (diagonal bars, mut-HA22 liposomes; solid bars, control liposomes). Our observations on DOX-mediated cytotoxicity of mut-HA22 liposomes to CD22+ cells are consistent with previous reports on immunoliposomes (29;33). Taken together, our fluorescence-based binding/uptake, DOX accumulation, and cytotoxicity data show that mut-HA22 liposomes may serve as promising candidates for treatment of B-cell lymphoma.
Conclusions
Generation of high-affinity antibodies and antibody fragments is an important parameter for their in vivo applications (52). The therapeutic index of these biologicals can be further enhanced by conjugation to nanoparticles (such as immunoliposomes) to promote multivalent interactions. In addition, these targeted nanoparticles have the potential to deliver a payload of chemotherapeutics to the tumor site. We have developed immunoliposomes by conjugation to a high affinity anti-CD22 scFv. The newly designed mut-HA22-liposomes described herein may serve as viable carriers for delivery of chemotherapeutic agents to treat B cell lymphoma. However, further experiments are needed to demonstrate in vivo drug delivery potential of these targeted particles.
ACKNOWLEDGEMENTS
We wish to thank Ira Pastan and Mitchel Ho, Laboratory of Molecular Biology, NCI, NIH for providing reagents. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We thank Dr. Prabhakar Gudla and Ms. Kim Peifley (Image Analysis Laboratory, ATP, SAIC-Frederick} for help with the confocal microscopy. We thank Julie Belanger and Eli Heldman for critical reading of the manuscript.
Abbreviations
- Mut-HA22
modified anti-CD22 scFv
- DPPC
1,2-dipalmitoyl-sn-glycero-3-phosphatidylcholine (16:0 PC)
- Egg PC
L-α-phosphatidyl choline
- DSPE-PEG2000
1, 2-disteaoryl-sn-glycero-3-phosphatidylethanolamine-N-[Methoxy(Polyethylene glycol)-2000](Ammonium Salt)
- DSPE-PEG2000 MaL
1,2-disteaoryl-sn-glycero-3-phosphatidylethanolamine-N-[Maleimide-(Polyethylene glycol)-2000](Ammonium Salt)
- HBSE (10mM HEPES
150mM NaCl, 9.1mM EDTA, pH 7.5)
- PBS
phosphate buffered saline
- PBS-BSA
phosphate buffered saline supplemented with 1% bovine serum albumin
- PBS-FBS
phosphate buffered saline supplemented with 5% fetal bovine serum
- DOX
doxorubicin-hydrochloride
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tedder TF, Tuscano J, Sato S, Kehrl JH. CD22, a B lymphocyte-specific adhesion molecule that regulates antigen receptor signaling. Annu Rev Immunol. 1997;15:481–504. doi: 10.1146/annurev.immunol.15.1.481. [DOI] [PubMed] [Google Scholar]
- 2.Shan D, Press OW. Constitutive endocytosis and degradation of CD22 by human B cells. J Immunol. 1995;154:4466–75. [PubMed] [Google Scholar]
- 3.Sato S, Tuscano JM, Inaoki M, Tedder TF. CD22 negatively and positively regulates signal transduction through the B lymphocyte antigen receptor. Semin Immunol. 1998;10:287–97. doi: 10.1006/smim.1998.0121. [DOI] [PubMed] [Google Scholar]
- 4.Dorken B, Moldenhauer G, Pezzutto A, et al. HD39 (B3), a B lineage-restricted antigen whose cell surface expression is limited to resting and activated human B lymphocytes. J Immunol. 1986;136:4470–9. [PubMed] [Google Scholar]
- 5.Schwarting R, Stein H, Wang CY. The monoclonal antibodies alpha S-HCL 1 (alpha Leu-14) and alpha S-HCL 3 (alpha Leu-M5) allow the diagnosis of hairy cell leukemia. Blood. 1985;65:974–83. [PubMed] [Google Scholar]
- 6.Sliedregt LA, van Rossenberg SM, Autar R, et al. Design and synthesis of a multivalent homing device for targeting to murine CD22. Bioorg Med Chem. 2001;9:85–97. doi: 10.1016/s0968-0896(00)00224-8. [DOI] [PubMed] [Google Scholar]
- 7.Siegel AB, Goldenberg DM, Cesano A, Coleman M, Leonard JP. CD22-directed monoclonal antibody therapy for lymphoma. Semin Oncol. 2003;30:457–64. doi: 10.1016/s0093-7754(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 8.Leonard JP, Coleman M, Ketas JC, et al. Phase I/II trial of epratuzumab (humanized anti-CD22 antibody) in indolent non-Hodgkin's lymphoma. J Clin Oncol. 2003;21:3051–9. doi: 10.1200/JCO.2003.01.082. [DOI] [PubMed] [Google Scholar]
- 9.Leonard JP, Coleman M, Ketas JC, et al. Epratuzumab, a humanized anti-CD22 antibody, in aggressive non-Hodgkin's lymphoma: phase I/II clinical trial results. Clin Cancer Res. 2004;10:5327–34. doi: 10.1158/1078-0432.CCR-04-0294. [DOI] [PubMed] [Google Scholar]
- 10.Leonard JP, Coleman M, Ketas J, et al. Combination antibody therapy with epratuzumab and rituximab in relapsed or refractory non-Hodgkin's lymphoma. J Clin Oncol. 2005;23:5044–51. doi: 10.1200/JCO.2005.13.821. [DOI] [PubMed] [Google Scholar]
- 11.Furman RR, Coleman M, Leonard JP. Epratuzumab in non-Hodgkin's lymphomas. Curr Treat Options Oncol. 2004;5:283–8. doi: 10.1007/s11864-004-0019-1. [DOI] [PubMed] [Google Scholar]
- 12.Coleman M, Goldenberg DM, Siegel AB, et al. Epratuzumab: targeting B-cell malignancies through CD22. Clin Cancer Res. 2003;9:3991S–4S. [PubMed] [Google Scholar]
- 13.Carnahan J, Wang P, Kendall R, et al. Epratuzumab, a humanized monoclonal antibody targeting CD22: characterization of in vitro properties. Clin Cancer Res. 2003;9:3982S–90S. [PubMed] [Google Scholar]
- 14.Carnahan J, Stein R, Qu Z, et al. Epratuzumab, a CD22-targeting recombinant humanized antibody with a different mode of action from rituximab. Mol Immunol. 2007;44:1331–41. doi: 10.1016/j.molimm.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Krauss J, Arndt MA, Vu BK, Newton DL, Rybak SM. Targeting malignant B-cell lymphoma with a humanized anti-CD22 scFv-angiogenin immunoenzymedouble dagger. Br J Haematol. 2005;128:602–9. doi: 10.1111/j.1365-2141.2005.05356.x. [DOI] [PubMed] [Google Scholar]
- 16.Ho M, Kreitman RJ, Onda M, Pastan I. In vitro antibody evolution targeting germline hot spots to increase activity of an anti-CD22 immunotoxin. J Biol Chem. 2005;280:607–17. doi: 10.1074/jbc.M409783200. [DOI] [PubMed] [Google Scholar]
- 17.DiJoseph JF, Armellino DC, Boghaert ER, et al. Antibody-targeted chemotherapy with CMC-544: a CD22-targeted immunoconjugate of calicheamicin for the treatment of B-lymphoid malignancies. Blood. 2004;103:1807–14. doi: 10.1182/blood-2003-07-2466. [DOI] [PubMed] [Google Scholar]
- 18.DiJoseph JF, Popplewell A, Tickle S, et al. Antibody-targeted chemotherapy of B-cell lymphoma using calicheamicin conjugated to murine or humanized antibody against CD22. Cancer Immunol Immunother. 2005;54:11–24. doi: 10.1007/s00262-004-0572-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DiJoseph JF, Dougher MM, Kalyandrug LB, et al. Antitumor efficacy of a combination of CMC-544 (inotuzumab ozogamicin), a CD22-targeted cytotoxic immunoconjugate of calicheamicin, and rituximab against non-Hodgkin's B-cell lymphoma. Clin Cancer Res. 2006;12:242–9. doi: 10.1158/1078-0432.CCR-05-1905. [DOI] [PubMed] [Google Scholar]
- 20.Weldon JE, Xiang L, Chertov O, et al. A protease-resistant immunotoxin against CD22 with greatly increased activity against CLL and diminished animal toxicity. Blood. 2008 doi: 10.1182/blood-2008-08-173195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onda M, Beers R, Xiang L, Nagata S, Wang QC, Pastan I. An immunotoxin with greatly reduced immunogenicity by identification and removal of B cell epitopes. Proc Natl Acad Sci U S A. 2008;105:11311–6. doi: 10.1073/pnas.0804851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Z, Mahesh SP, Shen dF, et al. Eradication of tumor colonization and invasion by a B cell-specific immunotoxin in a murine model for human primary intraocular lymphoma. Cancer Res. 2006;66:10586–93. doi: 10.1158/0008-5472.CAN-06-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bang S, Nagata S, Onda M, Kreitman RJ, Pastan I. HA22 (R490A) is a recombinant immunotoxin with increased antitumor activity without an increase in animal toxicity. Clin Cancer Res. 2005;11:1545–50. doi: 10.1158/1078-0432.CCR-04-1939. [DOI] [PubMed] [Google Scholar]
- 24.Allen TM. Ligand-targeted therapeutics in anticancer therapy. Nat Rev Cancer. 2002;2:750–63. doi: 10.1038/nrc903. [DOI] [PubMed] [Google Scholar]
- 25.Torchilin VP. Targeted pharmaceutical nanocarriers for cancer therapy and imaging. AAPS J. 2007;9:E128–E147. doi: 10.1208/aapsj0902015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sapra P, Allen TM. Improved outcome when B-cell lymphoma is treated with combinations of immunoliposomal anticancer drugs targeted to both the CD19 and CD20 epitopes. Clin Cancer Res. 2004;10:2530–7. doi: 10.1158/1078-0432.ccr-03-0376. [DOI] [PubMed] [Google Scholar]
- 27.Sapra P, Moase EH, Ma J, Allen TM. Improved therapeutic responses in a xenograft model of human B lymphoma (Namalwa) for liposomal vincristine versus liposomal doxorubicin targeted via anti-CD19 IgG2a or Fab’ fragments. Clin Cancer Res. 2004;10:1100–11. doi: 10.1158/1078-0432.ccr-03-0041. [DOI] [PubMed] [Google Scholar]
- 28.Nellis DF, Giardina SL, Janini GM, et al. Preclinical manufacture of anti-HER2 liposome-inserting, scFv-PEG-lipid conjugate. 2. Conjugate micelle identity, purity, stability, and potency analysis. Biotechnol Prog. 2005;21:221–32. doi: 10.1021/bp049839z. [DOI] [PubMed] [Google Scholar]
- 29.Laginha KM, Moase EH, Yu N, Huang A, Allen TM. Bioavailability and therapeutic efficacy of HER2 scFv-targeted liposomal doxorubicin in a murine model of HER2-overexpressing breast cancer. J Drug Target. 2008;16:605–10. doi: 10.1080/10611860802229978. [DOI] [PubMed] [Google Scholar]
- 30.Xu L, Tang WH, Huang CC, et al. Systemic p53 gene therapy of cancer with immunolipoplexes targeted by anti-transferrin receptor scFv. Mol Med. 2001;7:723–34. [PMC free article] [PubMed] [Google Scholar]
- 31.Xu L, Huang CC, Huang W, et al. Systemic tumor-targeted gene delivery by anti-transferrin receptor scFv-immunoliposomes. Mol Cancer Ther. 2002;1:337–46. [PubMed] [Google Scholar]
- 32.Zhang L, Gao H, Chen L, et al. Tumor targeting of vincristine by mBAFF-modified PEG liposomes in B lymphoma cells. Cancer Lett. 2008;269:26–36. doi: 10.1016/j.canlet.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell RT, Martin SM, Ma Y, Zamboni WC, Tuscano JM. Development and characterization of CD22-targeted pegylated-liposomal doxorubicin (IL-PLD). Invest New Drugs. 2009 doi: 10.1007/s10637-009-9243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sapra P, Allen TM. Ligand-targeted liposomal anticancer drugs. Prog Lipid Res. 2003;42:439–62. doi: 10.1016/s0163-7827(03)00032-8. [DOI] [PubMed] [Google Scholar]
- 35.Lian T, Ho RJ. Trends and developments in liposome drug delivery systems. J Pharm Sci. 2001;90:667–80. doi: 10.1002/jps.1023. [DOI] [PubMed] [Google Scholar]
- 36.Kirpotin DB, Drummond DC, Shao Y, et al. Antibody targeting of long-circulating lipidic nanoparticles does not increase tumor localization but does increase internalization in animal models. Cancer Res. 2006;66:6732–40. doi: 10.1158/0008-5472.CAN-05-4199. [DOI] [PubMed] [Google Scholar]
- 37.de KJ, Storm G, van BL, Logtenberg T. Biosynthetically lipid-modified human scFv fragments from phage display libraries as targeting molecules for immunoliposomes. FEBS Lett. 1996;399:232–6. doi: 10.1016/s0014-5793(96)01335-x. [DOI] [PubMed] [Google Scholar]
- 38.Dekruif J, Terstappen L, Boel E, Logtenberg T. Rapid Selection of Cell Subpopulation-Specific Human Monoclonal-Antibodies from A Synthetic Phage Antibody Library. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:3938–42. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta CM, Bali A. Carbamyl analogs of phosphatidylcholines. Synthesis, interaction with phospholipases and permeability behavior of their liposomes. Biochim Biophys Acta. 1981;663:506–15. doi: 10.1016/0005-2760(81)90178-8. [DOI] [PubMed] [Google Scholar]
- 40.Goswami SK, Frey CF. Spray detection of phospholipids on thin-layer chromatograms. J Lipid Res. 1971;12:509–10. [PubMed] [Google Scholar]
- 41.Puri A, Kramer-Marek G, Campbell-Massa R, et al. HER2-specific affibodyconjugated thermosensitive liposomes (Affisomes) for improved delivery of anticancer agents. J Liposome Res. 2008;18:293–307. doi: 10.1080/08982100802457377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.AMES BN, DUBIN DT. The role of polyamines in the neutralization of bacteriophage deoxyribonucleic acid. J Biol Chem. 1960;235:769–75. [PubMed] [Google Scholar]
- 43.Haran G, Cohen R, Bar LK, Barenholz Y. Transmembrane ammonium sulfate gradients in liposomes produce efficient and stable entrapment of amphipathic weak bases. Biochim Biophys Acta. 1993;1151:201–15. doi: 10.1016/0005-2736(93)90105-9. [DOI] [PubMed] [Google Scholar]
- 44.Zhu J, Yan F, Guo Z, Marchant RE. Surface modification of liposomes by saccharides: vesicle size and stability of lactosyl liposomes studied by photon correlation spectroscopy. J Colloid Interface Sci. 2005;289:542–50. doi: 10.1016/j.jcis.2005.03.088. [DOI] [PubMed] [Google Scholar]
- 45.Rawat SS, Eaton J, Gallo SA, et al. Functional expression of CD4, CXCR4, and CCR5 in glycosphingolipid-deficient mouse melanoma GM95 cells and susceptibility to HIV-1 envelope glycoprotein-triggered membrane fusion. Virology. 2004;318:55–65. doi: 10.1016/j.virol.2003.08.042. [DOI] [PubMed] [Google Scholar]
- 46.Xiong XB, Ma Z, Lai R, Lavasanifar A. The therapeutic response to multifunctional polymeric nano-conjugates in the targeted cellular and subcellular delivery of doxorubicin. Biomaterials. 2010;31:757–68. doi: 10.1016/j.biomaterials.2009.09.080. [DOI] [PubMed] [Google Scholar]
- 47.Marty C, Schwendener RA. Cytotoxic tumor targeting with scFv antibody-modified liposomes. Methods Mol Med. 2005;109:389–402. doi: 10.1385/1-59259-862-5:389. [DOI] [PubMed] [Google Scholar]
- 48.Yatvin MB, Weinstein JN, Dennis WH, Blumenthal R. Design of liposomes for enhanced local release of drugs by hyperthermia. Science. 1978;202:1290–3. doi: 10.1126/science.364652. [DOI] [PubMed] [Google Scholar]
- 49.Nellis DF, Ekstrom DL, Kirpotin DB, et al. Preclinical manufacture of an anti-HER2 scFv-PEG-DSPE, liposome-inserting conjugate. 1. Gram-scale production and purification. Biotechnol Prog. 2005;21:205–20. doi: 10.1021/bp049840y. [DOI] [PubMed] [Google Scholar]
- 50.Houck KS, Huang L. The role of multivalency in antibody mediated liposome targeting. Biochem Biophys Res Commun. 1987;145:1205–10. doi: 10.1016/0006-291x(87)91565-8. [DOI] [PubMed] [Google Scholar]
- 51.Park JW, Benz CC, Martin FJ. Future directions of liposome- and immunoliposome-based cancer therapeutics. Semin Oncol. 2004;31:196–205. doi: 10.1053/j.seminoncol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 52.Dimitrov DS, Marks JD. Therapeutic antibodies: current state and future trends--is a paradigm change coming soon? Methods Mol Biol. 2009;525:1–27. xiii. doi: 10.1007/978-1-59745-554-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]