Abstract
In this study, a combination of active shape model (ASM) and active appearance model (AAM) was used to segment the left and right ventricles of normal and Tetralogy of Fallot (TOF) hearts on 4-D (3-D+time) MR images. For each ventricle, a 4-D model was first used to achieve robust preliminary segmentation on all cardiac phases simultaneously and a 3-D model was then applied to each phase to improve local accuracy while maintaining the overall robustness of the 4-D segmentation. On 25 normal and 25 TOF hearts, in comparison to the expert traced independent standard, our comprehensive performance assessment showed subvoxel segmentation accuracy, high overlap ratios, good ventricular volume correlations, and small percent volume differences. Following 4-D segmentation, novel quantitative shape and motion features were extracted using shape information, volume-time and dV/dt curves, analyzed and used for disease status classification. Automated discrimination between normal/TOF subjects achieved 90%–100% sensitivity and specificity. The features obtained from TOF hearts show higher variability compared to normal subjects, suggesting their potential use as disease progression indicators. The abnormal shape and motion variations of the TOF hearts were accurately captured by both the segmentation and feature characterization.
Index Terms: Active appearance model (AAM), active shape model (ASM), cardiac magnetic resonance imaging, disease characterization, shape and motion analysis, Tetralogy of Fallot (TOF)
I. INTRODUCTION
Tetralogy of Fallot (TOF) is a common congenital heart disease characterized by four typical features: right ventricular hypertrophy, right ventricular outflow tract (RVOT) obstruction, ventricular septal defect (VSD), and overriding aorta. Initial management of infants with TOF includes palliative surgery to augment the RVOT and close the VSD, but patients often suffer from pulmonary valve insufficiency after surgery. This can over time lead to right ventricular dilation and dysfunction eventually requiring pulmonary valve replacement (PVR) in the adult years [1]–[3]. Although TOF patients often have similar life expectancy as healthy people with an estimated prevalence of 80000 in the U.S. [4], they need to be monitored throughout their lives to identify whether further surgery is required. Cardiac magnetic resonance (MR) imaging has become essential for pre- and post-operative management of TOF patients to determine the necessity and optimal timing of PVR [5]–[8]. It has been shown that RV dilation, RV systolic dysfunction and the left ventricle (LV) systolic dysfunction are independent predictors of progressive LV dysfunction [9]. It has also been suggested that unfavorable LV and RV interaction may be an important indicator for poor post-operation outcome and for timing of PVR [9], [10].
The traditional functional indices measured in clinical analysis such as ventricular volumes and ejection fraction may not be sufficient to allow timely patient management and prevent long term sequelae [11], [12] because they do not fully utilize the rich information provided by the MR data to describe the complex ventricular remodeling process. In addition, due to the inevitable and large observer variability associated with the manual data analysis, it is difficult to utilize these traditional indices in long-term patient care to produce reliable prediction of disease progression. It has been found that the post-operative TOF RV with near-normal ejection fraction exhibits unique abnormal ventricular remodeling characteristics such as larger and rounder cross-sectional shape and more basal bulging [13], but the quantitative relationships between these shape features and clinical outcomes are still unknown.
To improve the quality of clinical analysis for TOF and other cardiac diseases, it is essential to have quantitative indices that are capable of describing and measuring shape and motion features. Furthermore, highly-automated and accurate methods must be developed to perform 4-D (3-D+time) segmentation and reduce observer variability.
The cardiac image segmentation problem has been intensively studied by many groups in the recent years. Although there are still some feature-based methods such as using threshold and watershed [14], most groups acknowledged the necessity of using deformable models or parametric models to incorporate anatomical knowledge into the segmentation.
The deformable model method often starts from an atlas (template mesh). The anatomic knowledge is included in the atlas and its feasible deformation is learned from training or clinical models. During segmentation, the atlas is deformed onto the target image using registration or transform guided by image features. Lötjönen et al. [15] segmented all four heart chambers in 3-D MR data using landmark probability distribution, probability atlas, and nonrigid registration. Ecabert et al. [16] proposed a shape-constrained deformable model for 3-D CT to combine the strengths of parametric and deformable models and to segment all four heart chambers using piecewise affine transform optimization. Bistoquet et al. [17] introduced an incompressible myocardium model to improve the clinical feasibility of LV and RV segmentations in 4-D MR data. Sermesant et al. [18] combined the intensity and deformable models to achieve 3-D biventricular segmentations in multiple modalities. Zheng et al. [19] constructed models of all four heart chambers with holes for inflow/outflow tracts and achieved segmentation initialization through marginal space learning and steerable features.
Active shape model (ASM) and active appearance model (AAM) are popular parametric models. The statistical property of the target shape or appearance (combination of shape and texture) is derived from a training set and denoted by compact modal indices. ASM and AAM have been used for various applications such as: 2-D+time LV segmentation of ultrasound data [20], 3-D LV or LV+RV segmentation of ultrasound and MR data [21], bi-temporal LV segmentation of MR data [22], 3-D LV segmentation of MR and CT data [23], 4-D LV shape fitting of MR and CT data [24], and modeling four heart chambers of 4-D CT data [25]. Andreopoulos et al. [26] achieved LV segmentation of 4-D MR images using 3-D AAM followed by 2-D+time ASM and found time-dependent segmentation errors—larger errors at end-systole than at enddiastole. Zhu et al. [27] performed LV segmentation of 4-D MR data in a phase-by-phase manner where the statistical and dynamic models were combined in 3-D segmentation. Several methods have been proposed to improve the quality and capability of parametric models using independent component analysis for modeling and segmentation [28], [29], enlarging ASM training sets [30], using sparse modeling with orthomax criterion [31], improving landmark correspondence [32], [33], and modifying ASM optimization [23], [34].
Most of the approaches to cardiac image processing, especially for MR, share several common limitations.
The manual tracing on short-axis MR data is often accepted as the independent standard without considering the side effects due to breathing motion, anisotropic voxel and inter-observer variability.
For parametric models, the curse of dimensionality due to a limited training set is widely known, but its effects on modeling and segmentation have not been fully evaluated.
The method has not been developed for and tested on diseased hearts with large shape and motion variations.
The segmentation error is often measured by the point-to-surface (or contour) distance between segmented mesh vertices and the independent standard. Such measurements may not fully describe the segmentation accuracy. How the segmentation error varies with respect to time in 4-D segmentation is often not fully measured.
Sometimes the traditional clinical indices are used as the main segmentation performance metrics, but such indices cannot measure the quality of shape and motion tracking.
How the segmentation accuracy affects the outcome of computer-aided diagnosis has not been discussed.
The potential for using the proposed method in disease characterization has been speculated on but not explored.
This study utilizes ASM and AAM to accomplish two main goals. 1) Develop an LV and RV segmentation method that is highly automated, applicable in clinical environment and capable of producing accurate 4-D segmentations of clinical-quality MR data. 2) Derive novel 4-D ventricular function indices and test their capabilities for distinguishing between normal and TOF hearts, and assessing post-operative status. A complete cardiac MR analysis pipeline was designed from defining an independent standard to disease characterization.
The unique contributions of this study are as follows.
The short-axis and long-axis MR images were corrected for breathing motion and fused into a single 4-D dataset that has complete ventricle coverage, improved quality and reduced ambiguity in base and apex locations.
Manual tracing was performed on the fused dataset using a custom-designed tracing application that provides better 4-D context to the user. In addition to smooth shape and motion, consistent LV mass and equal stroke volumes for normal hearts were utilized to produce an accurate independent standard.
A template-based method was implemented with efficient algorithms for automatic landmark generation.
New strategies for model construction, segmentation and disease characterization were designed to minimize the side effect of a limited training set. They include: separately modeled and segmented individual ventricles from normal and TOF hearts, preliminary segmentation followed by final segmentation, etc.
The performance of landmarking, modeling, and segmentation was evaluated with a large set of quantitative measures focusing on various aspects of accuracy.
The proposed functional indices were derived from independent standard and segmentation results to test their capabilities in disease characterization and the effects of imperfect segmentations on disease characterization.
The cardiac motion tracking capability of a 4-D hybrid ASM/AAM segmentation was improved by our reversed ASM tuning technique where new candidate shape is generated from shape modal indices and its quality tested by intensity-based criterion.
II. ACTIVE SHAPE AND APPEARANCE MODELS
The point distribution model (PDM) [35] describes the shape variation of a training set of objects. The coordinates of landmarks—a set of corresponding points representing the boundary—are concatenated to form a shape vector s, whose statistical properties are identified via principal component analysis (PCA) yielding
| (1) |
where s̄ is the mean shape and matrix Φ contains PCA modes (a.k.a. eigenvectors or components) ordered by their significance. After discarding less significant modes, the weight vector b (referred to as the modal indices in this paper) is used as compact alternative to approximate any shape s in the training set or synthesize a new feasible shape that is not in the training set but still within the allowed range of variations. The application of PDM in segmentation is ASM [36]. The local intensities along scan lines associated with landmarks are modeled from the training set. During ASM segmentation, the landmarks are iteratively updated based on image features and trained scan line models, then new modal indices are computed and restricted within the allowed range.
Similarly, the intensity pattern—the texture vector t—of the object can also be modeled by a statistical texture model once the correspondence of all interior voxels is established. Combining shape modal indices (bs) and texture modal indices (bt) with appropriate weighting Ws results in an appearance vector a that can be statistically modeled by AAM
| (2) |
The AAM segmentation [37] is an iterative process often referred to as the model matching. The appearance modal indices ba synthesize model texture and model shape. The model shape is mapped to the target image as the target shape using an affine transform. Another texture, the target texture, is sampled based on the target shape. The goal of the model matching is to find the optimal model parameters d (ba and the affine transform parameters) that minimize the differences between the model texture and the target texture. The classical AAM implementation minimizes the root-mean-square of the texture difference. The model parameters are iteratively updated using knowledge acquired from model training. The training typically employs reduced-rank multivariate linear regression on a set of known model parameters to find a Jacobian matrix R that is used to predict the model parameter displacement Δd based on the texture difference r(d) as
| (3) |
ASM and AAM have their strengths and weaknesses. AAM is good at object tracking but it may impose too stringent of constraints and the matching can be easily trapped at local minima. ASM is good at finding local features but it requires a good initialization and can be disturbed by neighboring nontarget objects. The strengths of AAM and ASM were combined in a multistage hybrid approach proposed by Mitchell et al. [38]. Its usage in MR ventricular segmentation yielded better results than AAM or ASM alone.
III. INDEPENDENT STANDARD CREATION
The short-axis (SA) MR images are typically traced in clinical analysis because they show clearly-identifiable cross-sectional views of the doughnut-shaped LV and the crescent-shaped RV depicted on 2-D slices. Due to the typically large slice thickness of 6–8 mm as opposed to the in-plane resolution of 1.5–2.5 mm, identifying the traceable borders in the last basal image is difficult and is the major source of volumetric measurement errors [39], [40]. However, the consistency of ventricular coverage is often given a higher priority in clinical practice. In the complementary set of standard MR images acquired from four chamber long-axis (LA) planes, the ventricular bases and apexes are easy to identify, but tracing LV endocardial borders is more troublesome due to difficulty of accurately identifying papillary muscles which are commonly included in the LV blood pool. Several proposed methods combined SA and LA tracings together to produce better LV shapes [41], [42] using simple geometric assumptions that are not feasible for the RV due to its complex shape. Cardiac MR images are also affected by the respiratory motion that can be simply modeled as misalignment of the ventricles between slices. The motion correction that uses only SA images [43] is feasible but not appropriate for shape analysis because it can potentially change the actual orientation and shape of the heart.
A. Image Fusion
The complementary information present in the SA and LA MR data is fully utilized by our image fusion technique. The motion correction method, although similar to Lötjönen’s approach [15], was developed independently. In addition, the motion-corrected data from both orientations are fused into a single 4-D image with isotropic voxels and improved quality as shown below.
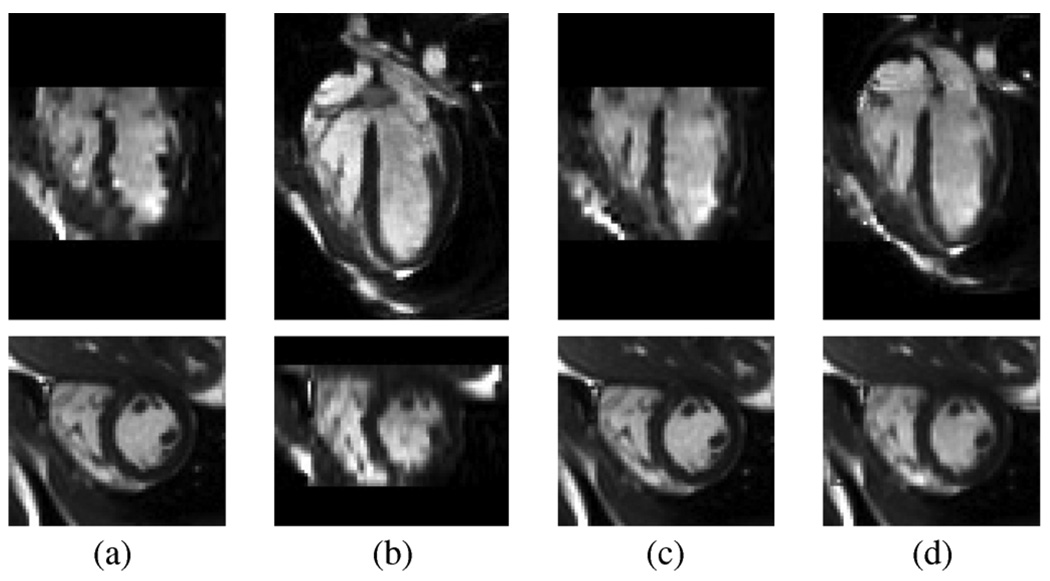
The number of phases (time frames) acquired per cardiac cycle in the MR scan is determined by the imaging protocol and the subject’s heart rate so it varies between 18 and 25. We first normalized our data to 16 phases per cycle using nearest neighbor (along time axis) interpolation that prevents the cardiac motion from introducing artifacts in a phase-by-phase fashion. The potential temporal motion change due to interpolation is very small in practice and is only limited to a very small portion of the cardiac cycle. Without motion correction, the end-diastolic isotropic 3-D images reconstructed by shifted linear interpolation [44] from SA and LA MR data are shown in Fig. 1(a) and (b).
Fig. 1.
Image fusion process illustrated by selected LA (top row) and SA (bottom row) slices of end-diastolic isotropic 3-D images reconstructed (a) from SA MR data before motion correction, (b) from LA MR data before motion correction, and (c) from motion-corrected SA MR data. (d) End-diastole of the final fused image; note the improved image consistency in both SA and LA panels.
After establishing correspondence between SA and LA MR data using imaging plane description in the DICOM file header, the motion in the SA MR data is corrected by registering each SA MR data [bottom row of Fig. 1(a)] to its counterpart in the 3-D image reconstructed from LA MR data [bottom row of Fig. 1(b)]. The registration is restricted to translation only and the criterion is minimal intensity difference. The result of motion correction is shown in Fig. 1(c). The motion correction of the LA MR data is then performed in the same way utilizing information from the motion-corrected SA MR data. The two images reconstructed from motion-corrected SA and LA MR data are fused into a single image as shown in Fig. 1(d), where the average intensity of the two reconstructed images was used in the overlapped region. The fused image has better coverage of the heart as well as improved image quality—note the difference in LA slices of Fig. 1(c) and 1(d). For SA MR data with d mm in-plane resolution, the isotropic voxel size of the fused image is d mm.
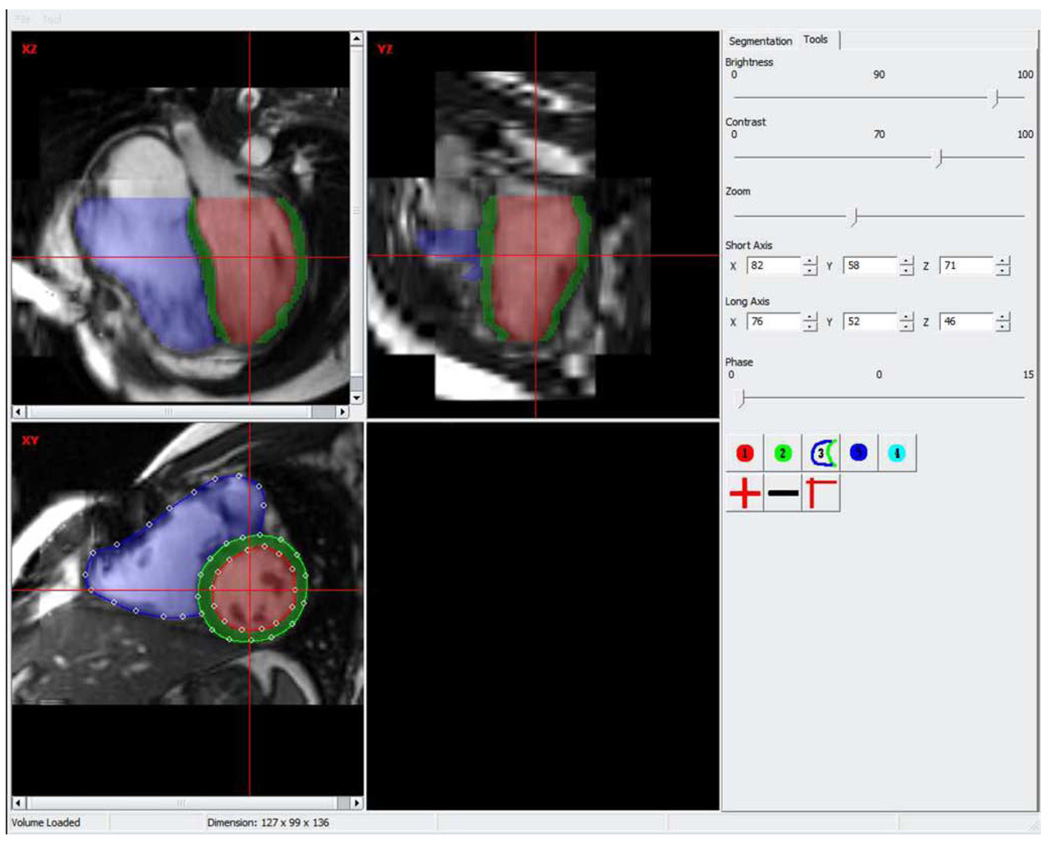
B. Manual Tracing
A custom-designed application is used for manual tracing of the fused image as shown in Fig. 2. This tracing environment provides sufficient 4-D context such that the ventricular apex and base locations, end-diastole and end-systole can be easily identified. The manual tracing is performed on user-selected short-axis slices as 2-D contours defined by spline control points. The control points are not used as landmarks and their number is determined by the user such that the resulting contour correctly describes the ventricular border. The contours are translated into 2-D distance maps. The 3-D shape on each phase is created by distance-based shape interpolation [45] and is used as the independent standard. The colored overlay shows the created shapes and provides the user with feedback regarding the tracing accuracy.
Fig. 2.
Screen shot of the manual tracing application.
IV. MODEL CONSTRUCTION
A. Landmark Generation
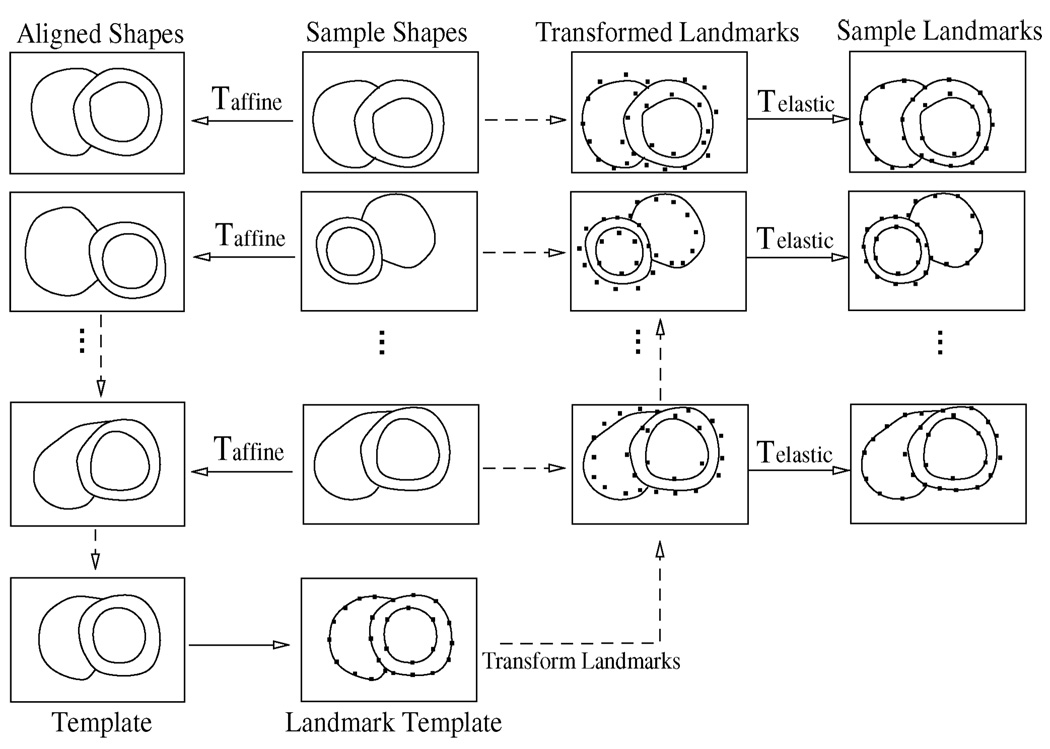
In order to automatically create landmarks on all sample shapes, a template-based general framework proposed by Frangi et al. [46] is adapted but implemented differently as illustrated in Fig. 3. The whole process is performed in a phase-by-phase manner treating a 4-D sample as 16 3-D shapes. It consists of the following steps.
Template creation: The sample shapes of the training set are aligned to a common reference frame using a set of affine transforms 𝒯affine that are restricted to rotation, translation, and scaling. The aligned shapes are blended to a template shape using a distance-based method [45].
Template landmarking: Create landmarks on the template shape. Apply individual inverse transform to map the landmarks back to the sample space as the transformed landmarks in Fig. 3 such that they are close to the original sample shapes.
Landmark propagation: Apply a set of elastic transforms 𝒯elastic to propagate the transformed landmarks onto the surfaces of the original sample shapes.
Fig. 3.
Flowchart of automated landmark generation.
1) Template Creation
A customized iterative closest point algorithm is used to find the optimal 𝒯affine with anisotropic scaling such that later on the transformed landmarks are brought as close to the sample shape as possible and the deformation associated with landmark propagation is reduced.
2) Template Landmarking
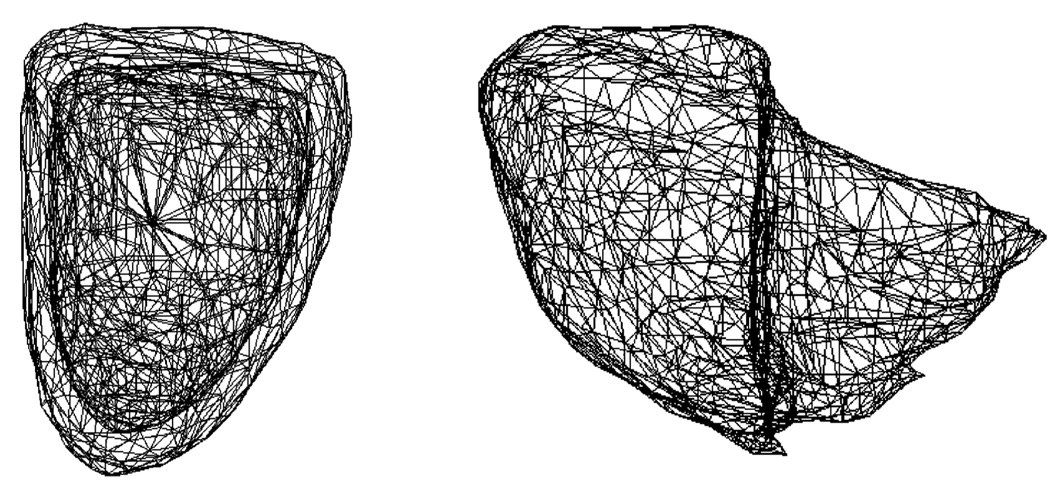
The landmarks on the template shape are created by three steps using VTK-designed filters: surface meshing to create dense triangular mesh on the template surface [47], surface smoothing to remove possible sharp edges [48], and triangle decimation to reduce the number of vertices and triangles [49]. The LV and RV are landmarked individually in this study using their own templates as shown in Fig. 4. For LV, its endocardial surface is landmarked first. To form landmarks for the epicardial surface, an exact copy of the landmarked endocardial surface is created and its vertices are moved along the surface normals to the epicardial surface. Both LV surfaces are therefore defined by the same number of vertices and triangles and exhibit the same topology.
Fig. 4.
Template landmark meshes of LV and RV.
3) Landmark Propagation
Because the transformed land-marks are reasonably close to the sample surface, an efficient method is designed. The landmarks, pi = (xi,yi,zi), are iteratively updated by an implicit function that minimizes the cost that consists of a similarity cost Cs and a regularization constraint Cr with appropriate weightings ws and wr.
| (4) |
The similarity cost Cs is defined as
| (5) |
where D is the distance map of the target sample shape L computed by distance transform D𝒯. It can be derived that iteratively minimizing Cs is equivalent to iteratively moving pi along the gradient direction of D(pi) as
| (6) |
Minimizing the similarity cost forces the landmarks onto the target surface. Because it is performed on each landmark without considering neighboring structure of the mesh, geometric and topological errors could be introduced. The regularization constraint Cr is used to preserve the spatial smoothness of the deformation. It is defined as the Dirichlet integral and can be calculated for the surface mesh by
| (7) |
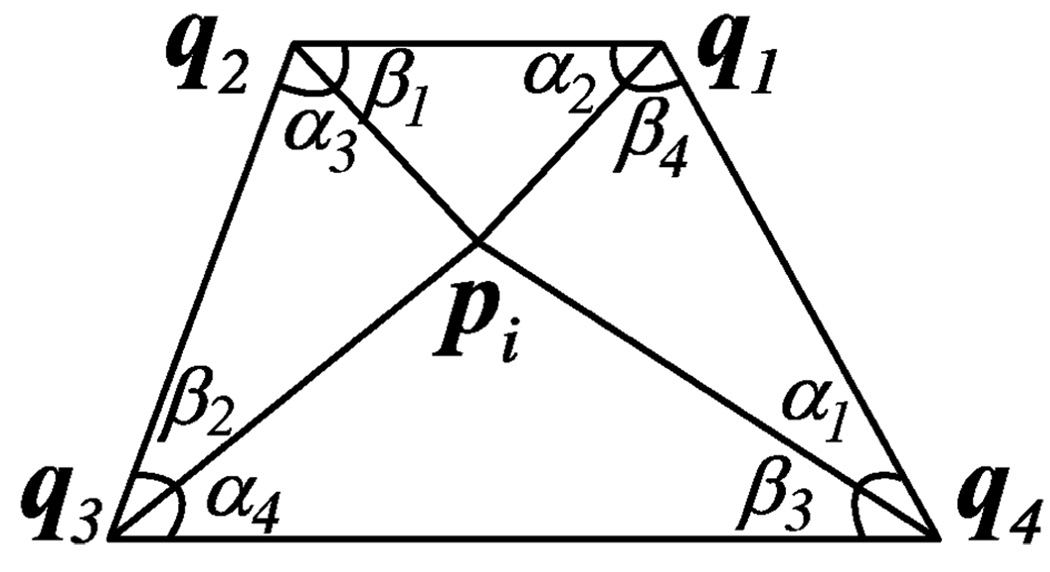
where |aj | is the length of the jth edge, αj and βj are two angles associated with the edge. Their definitions are shown in Fig. 5 where the jth edge is the edge between vertices (landmarks) pi and qj. It can be shown that when Cr is minimized, vertex pi must satisfy [50]
| (8) |
Fig. 5.
Surface vertex pi and its associated neighboring vertices qj.
Combining the optimization of Cs and Cr together, the landmark pi is iteratively updated by
| (9) |
In this study, the regularization constraint is not used to produce a discrete minimal surface as originally proposed [50], but instead to preserve the smoothness of the deformation. The iterative deformation defined by (9) therefore cannot introduce abrupt changes to the relative sizes and shapes of the triangles associated with any mesh vertex. The proper weightings, ws and wr are empirical values related to number of iterations. In this study, 200 iterations of landmark propagation are performed with ws = 0.03, wr = 0.01.
B. Texture Mapping
In this study, we use virtual surfaces to restrict the size of the texture vector such that it only contains intensity information that is useful for the segmentation. Two virtual surfaces are defined for each ventricle: one by enlarging the outermost surface (scaling it with respect to its centroid by 1.2), the other by shrinking the innermost surface (using a scale of 0.8). The vertices of the virtual surfaces are not used in the shape model as landmarks, but whenever the actual ventricular surface is changed during model matching, its associated virtual surfaces are recomputed and the corresponding texture vectors are re-sampled accordingly. In the 3-D space where the landmarking template is defined, uniformly spaced voxels between the two virtual surfaces are used to define texture mapping. The voxel correspondences are established using a barycentric coordinate system, where a voxel’s barycentric coordinates are defined by the vertices (landmarks) of the tetrahedron it resides in. Because all the virtual and actual surfaces of a ventricle are designed to have the same topology, building the tetrahedrons for texture mapping becomes trivial—connecting the vertices of the two corresponding triangles from the neighboring surfaces defines three tetrahedrons.
V. VENTRICULAR SEGMENTATION
The basic approach of the multistage hybrid ASM/AAM method proposed by Mitchell et al. [38] is adapted and implemented with additional new improvements. An AAM is first used to produce a shape sa. Then an ASM starts from sa and produces another shape ss. These two shapes are combined with an appropriate weight w as
| (10) |
The new combined shape s is then used to compute the new appearance modal indices using (2) to start a new AAM model matching until convergence. This hybrid approach provides extra momentum to bring model matching out of local minima and thus increases the chance of finding a global minimum. However, the appearance modal indices are associated with both shape and texture variations and the improvements in segmentation accuracy embedded in the combined shape s may not be fully incorporated into the resulting new appearance modal indices. In addition, an improved shape computed from updated appearance modal indices may be rejected as a better segmentation because the simultaneously updated texture modal indices may lead to increased matching error. To decouple this tie between shape and texture in the appearance model, a special segmentation fine tuning mechanism—brute-force reversed ASM—is designed as follows.
Starting from the current shape modal indices bo, compute shape and texture in the target image space and then compute the model matching error eo—root-mean-square error of texture difference r(d) in (3)—without changing texture modal indices. Set the number of shape modal indices to fine tune as J and i = 1.
Displace the ith shape modal index centered at bo within [−σi, σi] and each time by (1/K)σi, where σi = , and λi is the eigenvalue associated with the ith shape mode. The displaced shape modal indices are bn and the new matching error is computed as en. If en < eo, then b = bn and e = en.
If i < J, increment i and goto step 2 with bo = b and eo = e.
The new segmentation result is computed from b.
The traditional ASM first updates landmarks using image features and then applies the model constraints. Our method first creates a new shape by directly updating the shape modal indices and then tests whether the new shape fits the target image better based on the texture. Therefore, it is a reversed ASM method. Although it preforms a brute-force search in the model space and is more computationally expensive than traditional ASM, when used to fine tune the AAM segmentation in practice, it only uses 5%–10% of the total running time.
The full process of our hybrid ASM/AAM method is as follows.
Initializing the affine transform that fits the mean shape onto the target image.
Setting the total number of passes as P, setting p = 1.
Optimizing the affine transform parameters using Jacobian matrices provided by the model training as in (3).
Performing a hybrid ASM/AAM segmentation to compute the new appearance modal indices ba.
Optimizing the first M appearance modal indices individually, starting from the most significant one, using trained Jacobian matrices as in (3).
Optimizing the remaining less significant appearance modal indices simultaneously with the first M modal indices fixed.
If p < P, incrementing p and proceeding with step 3.
Performing another hybrid ASM/AAM segmentation.
Employing the reversed ASM fine tuning to optimize the first J shape modal indices.
VI. EXPERIMENTAL METHODS
A. Study Population and Independent Standard
Balanced steady-state free precession cardiac MR imaging was performed on 25 patients with repaired TOF and resultant pulmonic regurgitation (12 males, 13 females; ages: 18–60, median = 30). The study population also included 25 normal subjects (13 males, 12 females; ages: 22–38, median = 29). The normal subjects were scanned on a Siemens Avanto scanner (in-plane resolution 2.05 ±0.15 mm). TOF patients were scanned on a GE Signa scanner (in-plane resolution 1.48 ± 0.19 mm). The slice thickness was 6–8 mm and the slice gap was 0–2 mm. The acquired SA slices covered the whole heart and LA slices covered at least 70% of the heart. To prevent body fat from influencing motion correction, the MR data were cropped using two manually defined rectangular regions that enclosed the complete LV and RV on short-axis and long-axis views, respectively. The cropped MR data were motion corrected and fused into 4-D datasets as described in Section III-A.
The fused 4-D datasets were manually traced by a cardiologist on short-axis slices using our tracing application. Three ventricular borders were traced: LV endocardium, LV epicardium, and RV epicardium. The papillary muscles were included in the LV blood pool enclosed by the LV endocardial border. The manually traced slices were not limited to slices present in the original MR imaging planes and not all the interpolated slices were traced. When the ventricles exhibited small shape changes between slices, the distance between traced slices was about 6–8 mm. In the basal region, where the ventricles often show sudden 2-D shape changes, the traced slices were often 2–4 mm apart. After the initial pass of manual tracing, the tracings were repeatedly edited. The ventricular shapes were created and loaded into the tracing application as shown in Fig. 2, and the quantitative volume measurements of all cardiac phases were recorded. Working in consensus with a second cardiologist, the expert who performed the tracing then checked the volume measures and observed the resulting shapes to make appropriate small changes on the tracings until the ventricular shapes showed correctly identified end-diastole, end-systole, base and apex locations, and smooth 3-D shape and motion. Extra effort was made to guarantee correct depiction of the LV myocardial motion and the constancy of LV mass throughout the cardiac cycle. For normal subjects, stroke volumes of the LV and RV were expected to be approximately the same.
B. Two-Step Segmentation
To minimize the loss of segmentation accuracy due to a small training set and large shape variability, the normal and TOF hearts as well as the LV and RV were modeled and segmented separately. The LV or RV was segmented using a two-step approach. In the first step, the preliminary segmentation was performed using a 4-D model. It was manually initialized by fitting the 4-D mean shape onto the target image. The fitting was visually inspected and adjusted mainly on the first cardiac phase, which corresponds to the peak of R-wave in EKG-gated MR acquisition. The full hybrid ASM/AAM process listed in Section V was applied with M = 10 and J = 10. In the second step, the preliminary segmentation was used as initialization of the final segmentation which was performed in a phase-by-phase manner using a single 3-D model that was created from 3-D shape/texture samples of all cardiac phases. The reversed ASM fine tuning was not performed in the final segmentation and M = 20 was used. In both steps, P = 1 and w = 0.35 were used.
C. Performance Assessment
The segmentation performance was assessed using a hold-out validation strategy. All of the 25 4-D datasets in the normal or TOF population were randomly divided into five groups each containing five datasets. The datasets in any group were segmented using models created and trained from datasets in the other four groups. The sizes of the training sets were therefore 20 for 4-D models and 320 for 3-D models. This approach guarantees that a complete independence of the training and testing sets is maintained throughout the multistage process. The ventricle (VE, either LV or RV) produced by any stage was compared to the independent standard (IS) using labeled image—a voxel is labeled as background or one of the several objects—that has the same resolution as the 4-D fused image.
The following measurements were computed on individual cardiac phases of all datasets.
Average signed and absolute surface positioning errors in voxels and millimeters using 3-D distance transform [51]. For a voxel on the resulting ventricular surface, a negative signed error indicates it is inside the surface of the independent standard.
Milliliter volumes of LV blood pool, LV myocardium (LV mass), combined LV blood pool and mass, and RV.
- Overlap ratio and percent volume difference as
(11) (12)
The following metrics were derived to assess the performance.
Mean and standard deviation (σ) of average surface positioning errors, overlap ratios and percent volume differences.
Equation and correlation coefficient R of linear regressions of VE versus IS performed on all volumes, end-diastole volume (EDV) and end-systole volume (ESV) of LV and RV.
p-value of paired t-tests that compare the distributions of volumetric measurements computed from VE and IS.
To visualize the time-dependence of the performance, the following plots were used.
Normalized volume-time curves (nVTC) of all datasets. The LV and RV volumes were normalized by their EDVs, the LV masses were normalized by its average over the cardiac cycle.
Scatter plots of percent volume differences and temporal curves of mean and mean ± σ of percent volume differences of individual phases.
D. Disease Characterization
The objective of this task is to identify quantitative features that are unique to TOF and its progression. The shape and motion features were derived from the labeled images of the ventricles. The cardiac motion features were derived from the volumetric measurements. The independent standard and the final segmentation were analyzed to test whether they can extract the same or similar disease features.
The 4-D PDMs of the LV, RV, and LV+RV combined—by concatenating shape vectors of LV and RV—were constructed using all 50 datasets as the training set. The ventricular landmarks were generated again from the labeled volumes of the independent standard or the final segmentation. Having a single training set that contains an equal number of normal and TOF hearts is suitable to identify TOF features in the context of normal ventricles. The shape modal indices were computed and analyzed.
The nVTCs of the LV, RV, and LV+RV were used to represent the cardiac motion. Each nVTC was treated as a vector including ventricular volumes of all cardiac phases. The normalization by EDV removes the overall size variation and therefore performs a similar task to the Procrustes Analysis in PDM construction. In addition, dV/dt curves were also computed from the nVTCs and analyzed. The nVTCs and dV/dt curves do not include any shape knowledge. The nVTCs contain information about ejection fraction while dV/dt curves include no direct ejection fraction knowledge and focus on the rate of volume changes between phases. Extracting modal indices from nVTCs of all datasets was performed by PCA.
The modal indices extracted from the shape model or volume-based curve model were fed into a classifier using linear discriminative analysis to separate TOF patients from normal subjects. The classification performance was evaluated by leave-one-out validation and measured by specificity and sensitivity. In this binary classification of positive TOF and negative normal hearts, the results are represented by numbers of true positive (TP), false positive (FP), true negative (TN), and false negative (FN). The specificity and sensitivity were computed by
| (13) |
The classifications were performed using one to five strongest modal indices and the best combination of specificity and sensitivity achieved was reported as the final classification performance.
VII. RESULTS
A. Quality of the Independent Standard
The motion correction and image fusion were performed on all datasets and assessed by visual inspection. The resulting fused images had no remaining motion artifact and no image quality degradation. Due to the time-consuming nature of the manual tracing, observer variability was not assessed. Again, 50 fused 4-D images were traced by a cardiologist and further inspected and approved by a senior cardiology expert. The total time spent on tracing and editing on each 4-D dataset by the two experts was about 6–8 h and at least 50% of the time was spent on the initial contour drawing.
B. Quality of the Landmarking
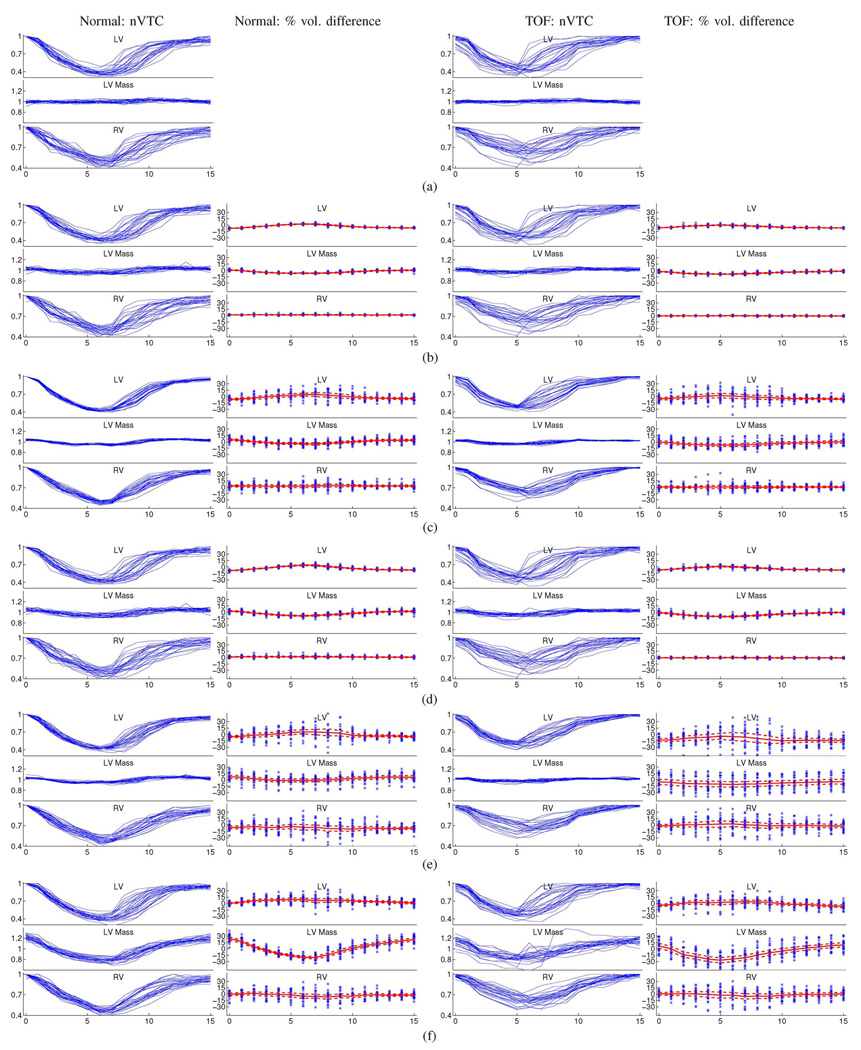
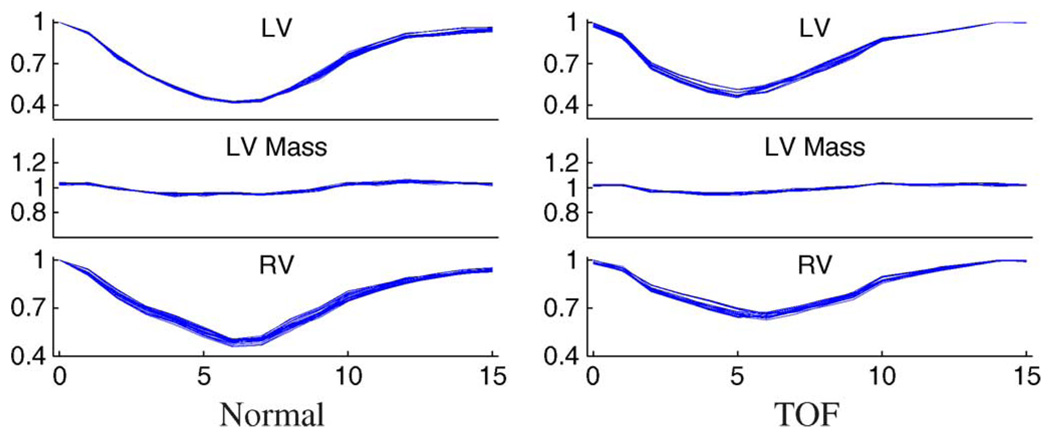
The numbers of landmarks used for 3-D ventricular surfaces were: 400 for LV endocardium, 400 for LV epicardium, and 800 for RV epicardium. The total running time of all automated land-marking steps performed on all 50 LVs and 50 RVs was approximately one hour on a PC with Intel Core 2 Duo E6400 processor and 2 GB of RAM. The landmarking error measured by average landmark-to-surface distance was 0.00 ± 0.02 voxels, but it does not necessarily mean that the landmark-defined surfaces accurately represent the actual ventricular shapes. The performance metrics defined in Section VI-C were used to assess the land-marking quality by comparing the landmark-defined ventricles and the independent standard. The results are shown in Fig. 6(b) and Table I and Table II. The average surface positioning errors show subvoxel accuracy in mean and standard deviation. The signed errors and volume-based metrics show a weak trend towards underestimation. The volume curves in Fig. 6(b) are almost the same as those of the independent standard in Fig. 6(a) to a naked eye, and there are slightly more pronounced fluctuations in the distribution of the LV mass curves. The mean percent volume differences of LV and LV mass begin to show a time-dependent pattern even though they are almost constant within the cardiac cycle and the standard deviations are very small.
Fig. 6.
Motion tracking capability of each process stage are visualized by nVTCs of normal (column 1) and TOF (column 3) hearts and percent volume differences of normal (column 2) and TOF (column 4) hearts. The mean and mean ± σ curves are shown in percent volume difference plots. In each plot, shown from top to bottom are measurements for LV, LV mass, and RV; the horizontal axis depicts cardiac phases 0–15 representing the entire cardiac cycle.
TABLE I.
Average Surface Positioning Errors and Overlap Ratios (mean ± σ)
| Normal | TOF | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Average Surface Positioning Error |
Overlap | Average Surface Positioning Error |
Overlap | |||||||
| sgn voxel | sgn mm | abs voxel | abs mm | % | sgn voxel | sgn mm | abs voxel | abs mm | % | |
| Landmarking | ||||||||||
| LV Endo | −0.12±0.18 | −0.24±0.36 | 0.37±0.06 | 0.76±0.11 | 95.4±0.6 | −0.18±0.19 | −0.28±0.29 | 0.49±0.08 | 0.76±0.11 | 95.4±0.7 |
| LV Epi | −0.19±0.07 | −0.40±0.14 | 0.37±0.04 | 0.75±0.09 | 96.7±0.4 | −0.33±0.09 | −0.50±0.12 | 0.50±0.07 | 0.78±0.09 | 96.3±0.5 |
| RV | 0.17±0.05 | 0.35±0.12 | 0.40±0.07 | 0.83±0.14 | 94.7±0.9 | 0.09±0.08 | 0.15±0.13 | 0.62±0.10 | 0.96±0.13 | 95.1±0.7 |
| 4D Modeling | ||||||||||
| LV Endo | 0.00±0.36 | 0.00±0.74 | 0.73±0.14 | 1.49±0.29 | 90.8±2.4 | −0.02±0.53 | −0.02±0.80 | 1.12±0.25 | 1.74±0.38 | 89.3±2.8 |
| LV Epi | −0.01±0.24 | −0.02±0.50 | 0.69±0.14 | 1.42±0.30 | 93.6±1.4 | −0.14±0.37 | −0.22±0.57 | 1.07±0.21 | 1.67±0.35 | 92.1±1.7 |
| RV | 0.26±0.26 | 0.54±0.53 | 0.84±0.19 | 1.71±0.39 | 88.7±2.5 | 0.25±0.38 | 0.40±0.60 | 1.40±0.29 | 2.17±0.44 | 88.6±2.2 |
| 3D Modeling | ||||||||||
| LV Endo | −0.09±0.21 | −0.18±0.44 | 0.55±0.09 | 1.13±0.16 | 93.1±1.3 | −0.14±0.23 | −0.22±0.35 | 0.66±0.11 | 1.03±0.15 | 93.7±1.1 |
| LV Epi | −0.15±0.09 | −0.32±0.19 | 0.50±0.09 | 1.02±0.19 | 95.4±0.9 | −0.29±0.09 | −0.44±0.13 | 0.67±0.11 | 1.04±0.16 | 95.1±0.8 |
| RV | 0.18±0.10 | 0.37±0.21 | 0.58±0.11 | 1.18±0.22 | 92.2±1.5 | 0.12±0.09 | 0.20±0.15 | 0.93±0.17 | 1.45±0.23 | 92.4±1.1 |
| Preliminary Segmentation | ||||||||||
| LV Endo | 0.11±0.48 | 0.22±0.92 | 0.97±0.24 | 1.97±0.48 | 87.5±3.6 | −0.36±0.88 | −0.54±1.34 | 1.55±0.48 | 2.41±0.73 | 84.4±5.3 |
| LV Epi | 0.14±0.39 | 0.27±0.81 | 0.98±0.24 | 2.01±0.48 | 90.8±2.3 | −0.46±0.86 | −0.72±1.32 | 1.63±0.43 | 2.53±0.68 | 87.5±3.8 |
| RV | 0.07±0.41 | 0.13±0.84 | 1.11±0.21 | 2.27±0.45 | 83.5±3.6 | 0.42±0.73 | 0.65±1.14 | 1.95±0.44 | 3.04±0.71 | 83.7±4.0 |
| Final Segmentation | ||||||||||
| LV Endo | 0.31±0.29 | 0.65±0.59 | 0.82±0.14 | 1.67±0.30 | 89.9±2.7 | 0.12±0.51 | 0.19±0.80 | 1.10±0.28 | 1.71±0.45 | 89.6±3.2 |
| LV Epi | 0.22±0.48 | 0.50±0.98 | 0.89±0.20 | 1.81±0.40 | 91.8±1.9 | −0.17±0.66 | −0.26±1.05 | 1.26±0.34 | 1.97±0.58 | 90.6±3.1 |
| RV | 0.12±0.32 | 0.23±0.65 | 1.04±0.19 | 2.13±0.39 | 85.2±2.9 | 0.32±0.70 | 0.50±1.08 | 1.88±0.48 | 2.92±0.73 | 84.2±4.2 |
TABLE II.
Results of Linear Regressions (Equation and R), Percent Volume Differences (mean ± σ), and Paired t Test (p Value)
| Normal | TOF | |||||||
|---|---|---|---|---|---|---|---|---|
| Linear Regression |
diff(%) | p | Linear Regression |
diff(%) | p | |||
| Eq. (ml) | R | Eq. (ml) | R | |||||
| Landmarking | ||||||||
| LV | 0.90x+5.8 | 1.00 | −3.3±3.7 | 0.90 | 0.90x+6.1 | 1.00 | −4.1±2.9 | 0.45 |
| RV | 1.01x+0.6 | 1.00 | 1.5±1.3 | 0.21 | 0.99x+1.7 | 1.00 | −0.7±1.1 | 0.70 |
| LVMass | 0.94x+2.9 | 0.99 | −3.4±3.6 | <0.05 | 0.93x+1.1 | 0.99 | −5.6±3.2 | <0.05 |
| LVEDV | 0.93x−0.7 | 1.00 | −7.9±1.3 | 0.57 | 0.93x+0.7 | 1.00 | −7.0±1.0 | 0.41 |
| LVESV | 0.98x+2.1 | 1.00 | 2.3±1.7 | 0.53 | 0.98x+1.7 | 1.00 | 0.5±1.5 | 0.63 |
| RVEDV | 1.01x+0.1 | 1.00 | 1.3±0.9 | 0.70 | 0.99x+2.1 | 1.00 | −0.8±1.1 | 0.94 |
| RVESV | 1.02x−0.0 | 1.00 | 1.7±1.6 | 0.67 | 0.99x+0.9 | 1.00 | −0.6±1.1 | 0.89 |
| 4D Modeling | ||||||||
| LV | 0.89x+7.6 | 0.98 | −1.6±9.1 | 0.27 | 0.87x+10.7 | 0.97 | −3.0±9.7 | 0.07 |
| RV | 1.00x+2.5 | 0.98 | 2.5±6.8 | 0.32 | 0.96x+9.5 | 0.99 | −0.2±6.3 | 0.84 |
| LVMass | 0.97x+3.4 | 0.96 | 0.0±6.9 | 0.97 | 0.93x+3.1 | 0.92 | −4.3±8.7 | <0.05 |
| LVEDV | 1.00x−9.4 | 0.98 | −7.2±5.1 | 0.28 | 0.94x−2.5 | 0.96 | −7.6±5.0 | 0.18 |
| LVESV | 0.83x+11.7 | 0.96 | 5.6±8.5 | 0.60 | 0.79x+16.2 | 0.94 | 4.9±11.4 | 0.63 |
| RVEDV | 1.11x−15.7 | 0.98 | 1.7±5.1 | 0.75 | 0.98x+1.8 | 0.99 | −1.7±4.2 | 0.89 |
| RVESV | 0.89x+12.5 | 0.98 | 4.1±7.9 | 0.70 | 0.94x+14.1 | 0.98 | 2.6±8.1 | 0.89 |
| 3D Modeling | ||||||||
| LV | 0.89x+6.9 | 1.00 | −3.2±4.8 | 0.09 | 0.89x+7.7 | 1.00 | −4.1±3.6 | <0.05 |
| RV | 1.01x+0.8 | 1.00 | 1.3±2.3 | 0.59 | 0.99x+0.4 | 1.00 | −0.9±1.4 | 0.74 |
| LVMass | 0.93x+4.5 | 0.97 | −2.7±5.3 | 0.09 | 0.93x+1.5 | 0.98 | −5.4±4.6 | <0.05 |
| LVEDV | 0.91x+0.3 | 1.00 | −8.7±1.8 | 0.15 | 0.91x+2.3 | 1.00 | −7.7±1.2 | 0.14 |
| LVESV | 0.97x+3.6 | 1.00 | 4.3±2.9 | 0.66 | 0.97x+2.9 | 1.00 | 2.0±2.2 | 0.84 |
| RVEDV | 1.02x−1.2 | 1.00 | 0.9±1.8 | 0.88 | 0.99x−0.3 | 1.00 | −1.0±l.5 | 0.91 |
| RVESV | 1.03x−0.5 | 1.00 | 1.8±2.9 | 0.82 | 0.99x+1.1 | 1.00 | −0.6±1.3 | 0.95 |
| Preliminary Segmentation | ||||||||
| LV | 0.85x+12.4 | 0.97 | −0.8±11.5 | 0.33 | 0.73x+17.8 | 0.91 | −11.0±16.0 | <0.05 |
| RV | 0.87x+8.5 | 0.94 | −7.0±11.6 | <0.05 | 0.90x+20.4 | 0.95 | −0.8±12.9 | 0.53 |
| LVMass | 0.86x+15.7 | 0.91 | 2.3±9.6 | 0.19 | 0.79x+12.8 | 0.78 | −8.1±15.0 | <0.05 |
| LVEDV | 0.92x+3.4 | 0.98 | −5.8±4.9 | 0.33 | 0.81x+6.18 | 0.88 | −16.0±10.2 | <0.05 |
| LVESV | 0.69x+19.2 | 0.92 | 6.5±12.2 | 0.54 | 0.68x+19.6 | 0.84 | −0.6±16.4 | 0.80 |
| RVEDV | 1.00x−11.1 | 0.95 | −6.6±7.7 | 0.36 | 0.96x+3.8 | 0.97 | −3.6±8.7 | 0.74 |
| RVESV | 0.76x+16.4 | 0.90 | −4.5±15.0 | 0.52 | 0.83x+37.1 | 0.90 | 4.4±17.2 | 0.84 |
| Final Segmentation | ||||||||
| LV | 0.97x+7.1 | 0.98 | 5.3±8.1 | 0.10 | 0.88x+12.2 | 0.97 | −0.7±9.7 | 0.47 |
| RV | 0.95x+3.0 | 0.96 | −2.8±10.0 | 0.29 | 0.94x+10.7 | 0.95 | −1.8±12.3 | 0.43 |
| LVMass | 0.88x+13.9 | 0.76 | 1.6±17.0 | 0.14 | 0.67x+24.2 | 0.64 | −8.4±18.6 | <0.05 |
| LVEDV | 0.98x+3.6 | 0.98 | 0.2±3.7 | 0.98 | 0.89x+7.8 | 0.94 | −6.8±7.2 | 0.22 |
| LVESV | 1.01x+4.1 | 0.96 | 8.7±10.3 | 0.33 | 0.93x+7.5 | 0.95 | 5.1±9.4 | 0.55 |
| RVEDV | 1.05x−11.0 | 0.96 | −1.3±6.9 | 0.90 | 0.97x+8.7 | 0.97 | −0.4±8.4 | 0.97 |
| RVESV | 0.84x+9.8 | 0.94 | −4.3±12.2 | 0.57 | 0.81x+29.8 | 0.89 | −1.8±16.3 | 0.74 |
C. Quality of the Appearance Modeling
The inherent consequence of using a small number of modal indices to represent complex shape and motion is the inevitable loss of detail. When the training set is small, such loss could affect the expected best segmentation accuracy. Using the same hold-out strategy as used in the segmentation, the appearance modal indices of all datasets were computed and used to synthesize the ventricular shapes approximated by the model. This procedure can be thought of as an ideal segmentation when all the model parameters that need to be optimized are known. The synthesized shapes were compared with the independent standard and the results are shown in Fig. 6(c) and (d) in Table I and Table II.
From 4-D to 3-D modeling, the standard deviations of average positioning errors decrease by 0.1–0.3 voxels. The means of absolute errors decrease by 0.3 voxels in normal hearts and 0.5 voxels in TOF hearts. Such improved accuracy of the 3-D modeling is also shown in the volumetric metrics where the mean overlaps increase by 2%–4% and the standard deviations of percent volume differences decrease by 4%–6%. The important improvements introduced by these relatively small numbers can be seen in Fig. 6(c) and (d). The 4-D modeling results show more clustered distribution of nVTCs and wider spread of percent volume differences than for the landmarking results, while the distributions of nVTCs and percent volume differences for the 3-D modeling results are very similar to those for the landmarking results.
D. Segmentation Performance
Training Jacobian Matrices for the 3-D or 4-D AAM of 20 hearts on the same PC used for the landmarking took approximately one hour for normal LV or RV and 90 min for TOF LV or RV. The manual segmentation initializations for LV and RV took less than 5 min, and consisted of the user adjusting the transform parameters to align the transformed mean shape onto the first phase of the target 4-D image such that the base and apex locations were correctly defined and the ventricular sizes were approximated. The preliminary LV or RV segmentation in 4-D took approximately 2–3 min. The final segmentation in 3-D took approximately 5 min for normal LV or RV and 10 min for TOF LV or RV. The total time used to segment a 4-D dataset was approximately 15 min for a normal heart and 30 min for a TOF heart. The TOF segmentation had longer running time mainly because the texture vector of the enlarged RV—even sampled at similar grid density as the normal RV—is substantially longer.
The performance assessments of preliminary and final segmentations are shown in Fig. 6(e) and (f) and Table I and Table II. Compared with their corresponding theoretical optima suggested by the modeling quality assessment, the means of the absolute positioning errors are off by 0.2–0.3 voxels for normal and TOF hearts, the standard deviations are off by 0.1–0.2 voxels for normal hearts and 0.2–0.3 voxels for TOF hearts.
The final segmentation produced better accuracy than the preliminary segmentation. The means of the absolute positioning errors are reduced by 0.1 voxels for normal hearts and 0.1–0.4 voxels for TOF hearts. The overlap ratios show increased mean and decreased standard deviation. In the volumetric metrics for LV and RV, better agreement with the independent standard is shown in increased slopes and decreased interceptions of linear regressions, increased correlation coefficients, and closer-to-zero means and decreased standard deviations of percent volume differences. The LV and RV nVTCs of the final segmentation also show improvements in motion tracking over the preliminary segmentation, especially in the rapid-fill diastole stage where the LV undergoes rapid expansion after reaching end-systole. However, the final segmentation show decreased LV mass accuracy.
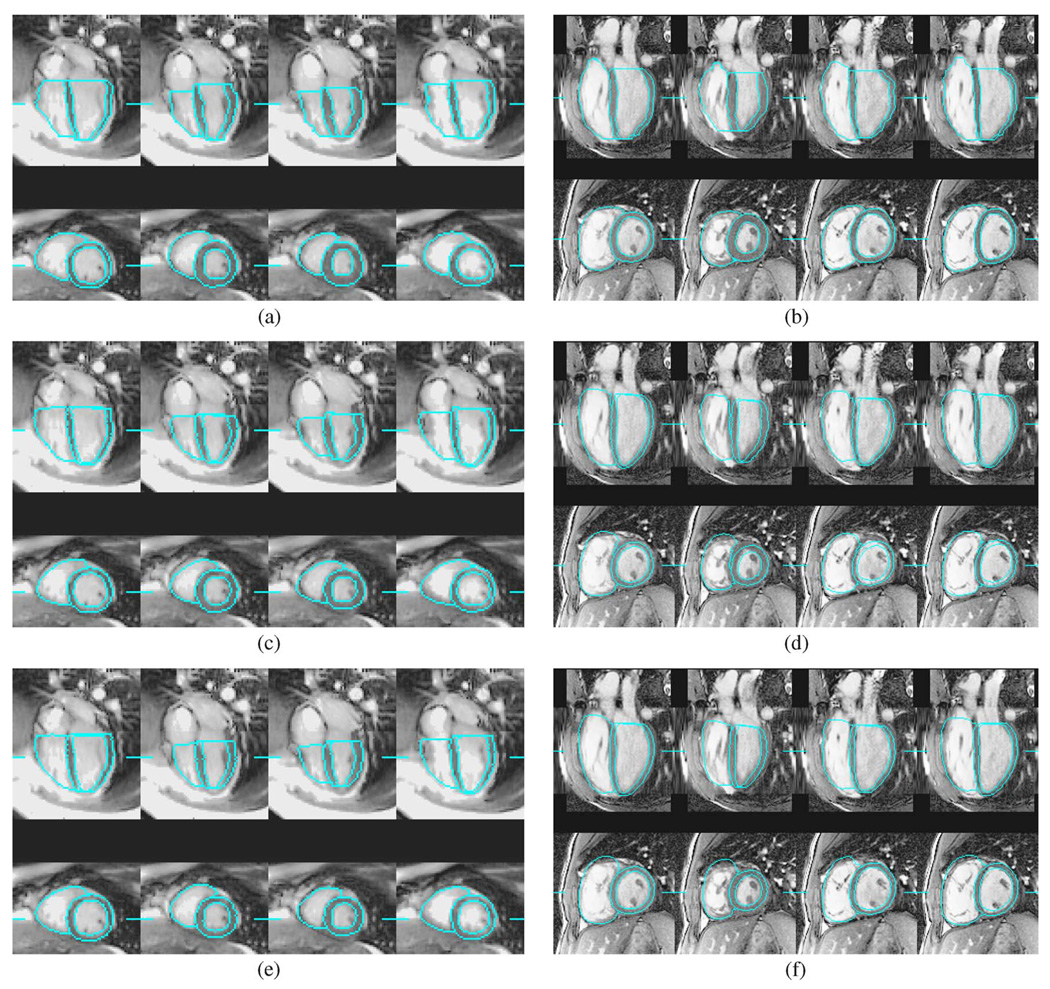
Fig. 7 shows the independent standard and segmentation result on a normal heart and a TOF heart at cardiac phases 0, 3, 7, and 11. Both preliminary and final segmentations produced smooth and accurate ventricular shapes for the normal heart. The final segmentation recovered some shape details that were not captured by the preliminary segmentation, and produced better RV short-axis borders. However, the final segmentation showed reduced accuracy in myocardial thickness on phases 3 and 7 due to inaccurate LV epicardial tracking.
Fig. 7.
Segmentation of a normal heart and a TOF heart at cardiac phases 0, 3, 7, and 11. The line segments mark the locations of slices. (a) Normal: independent standard. (b) TOF: independent standard; (c) normal: preliminary segmentation. (d) TOF: preliminary segmentation. (e) Normal: final segmentation. (f) TOF: final segmentation.
The TOF-affected heart in Fig. 7 has a unique motion pattern. It almost reaches end-systole early at phase 3, then expands quickly and reaches the end of rapid-filling diastole at phase 7. The preliminary segmentation could not fully capture this unique motion pattern as in Fig. 7(d). The final segmentation achieved correct cardiac motion tracking as in Fig. 7(f), where the LV endocardial surface is accurately identified. However, the final segmentation also introduced an underestimated LV myocardium at phase 3 of Fig. 7(f). Note that the user accidentally missed one apex SA slice that should have been traced on phase 3, but the AAM segmentations identified the correct apex location.
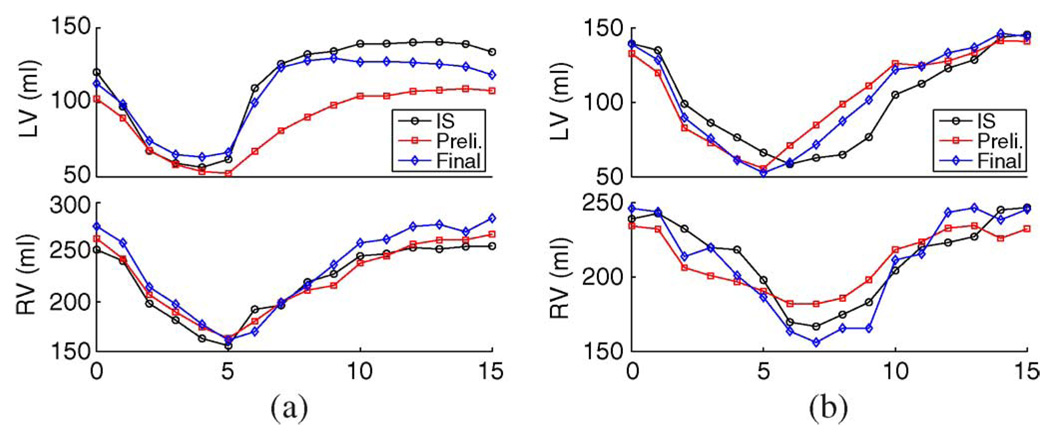
Fig. 8 shows the volume curves of the independent standard, preliminary segmentation, and final segmentation of two TOF hearts. The final segmentation’s capability of producing better cardiac motions is demonstrated in Fig. 8(a) for LV motion and Fig. 8(b) for RV motion.
Fig. 8.
Examples of improved motion tracking in the final segmentation shown by volume curves of LV (top row) and RV (bottom row) of two TOF hearts. (IS: independent standard, Preli: preliminary segmentation.)
To test the benefits of the reversed ASM fine tuning, the preliminary segmentation was performed without the reversed ASM step. The resulting nVTCs of the segmented volumes shown in Fig. 9 are significantly different from those in Fig. 6(c). Note that the segmentation tightly follows the mean motion pattern not allowing for subject-specific differences. Consequently, disease patterns may be masked by this strong influence of the underlying motion pattern.
Fig. 9.
Distributions of nVTCs of preliminary segmentations without reversed ASM. In each plot, shown from top to bottom are curves for LV, LV mass, and RV; the horizontal axis depicts cardiac phases 0–15 representing the entire cardiac cycle.
E. Disease Characterization
Table III lists the classification performance achieved using modal indices of 4-D shapes, nVTCs and dV/dt curves derived from ventricular volumes of different stages. The 4-D shape model analyzes both ventricular shape and motion variations and captures the difference between normal and TOF ventricles even for the LV whose shape is often not severely affected by TOF. The shape model analysis of the final segmentation achieved almost the same classification performance as that of the independent standard.
TABLE III.
Classification Sensitivity (Sens.) and Specificity (Spec.) Using Modal Indices of 4-D Shape nVTCs and dV/dt Curves
| LV | RV | LV+RV | ||||
|---|---|---|---|---|---|---|
| sens. | spec. | sens. | spec. | sens. | spec | |
| 4D Shape Model | ||||||
| IS | 1.00 | 1.00 | 1.00 | 0.96 | 0.96 | 1.00 |
| Preli. Seg. | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Final Seg. | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| nVTC Model | ||||||
| IS | 0.76 | 0.80 | 0.84 | 0.88 | 0.76 | 1.00 |
| Preli. Seg. | 0.92 | 0.92 | 1.00 | 1.00 | 0.96 | 1.00 |
| Final Seg. | 0.96 | 0.88 | 0.96 | 1.00 | 0.88 | 1.00 |
| dV/dt Model | ||||||
| IS | 0.92 | 0.80 | 0.92 | 0.88 | 0.92 | 0.96 |
| Preli. Seg. | 0.96 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| Final Seg. | 0.92 | 0.92 | 1.00 | 1.00 | 0.96 | 1.00 |
When the ventricular motion patterns are partially described by the nVTCs, the difference between normal and TOF hearts begins to diminish. Lower classification performance was achieved by using nVTCs of the independent standard. The dV/dt curves measure the rate of cardiac contraction and expansion but exclude the ejection fraction knowledge. Using the independent standard, the dV/dt model achieved better classification performance than the nVTC model.
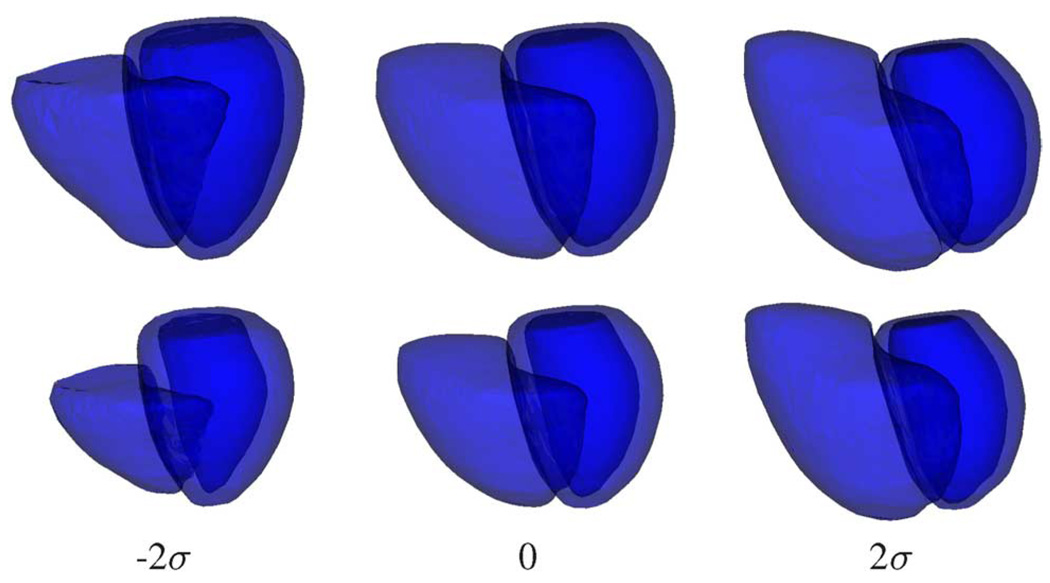
Fig. 10 shows the shape variations at phases 0 and 7 introduced by varying the value of the first modal index of the LV+RV 4-D shape model derived from the independent standard. The first shape mode is associated with variations in LV myocardium thickness, LV orientation, bulging of apex, ventricular height along the long axis, and severity of RV enlargement. In this example, increasing the value of the first modal index introduces more TOF-related shape features to the heart. Very similar shape variation patterns were observed in the shape model derived from the final segmentation.
Fig. 10.
Shape variations at phases 0 (top row) and 7 (bottom row) introduced by varying the first modal index of the LV+RV 4-D shape model derived from the independent standard. The values of the first modal index are: − 2σ, 0 (mean shape), and 2σ.
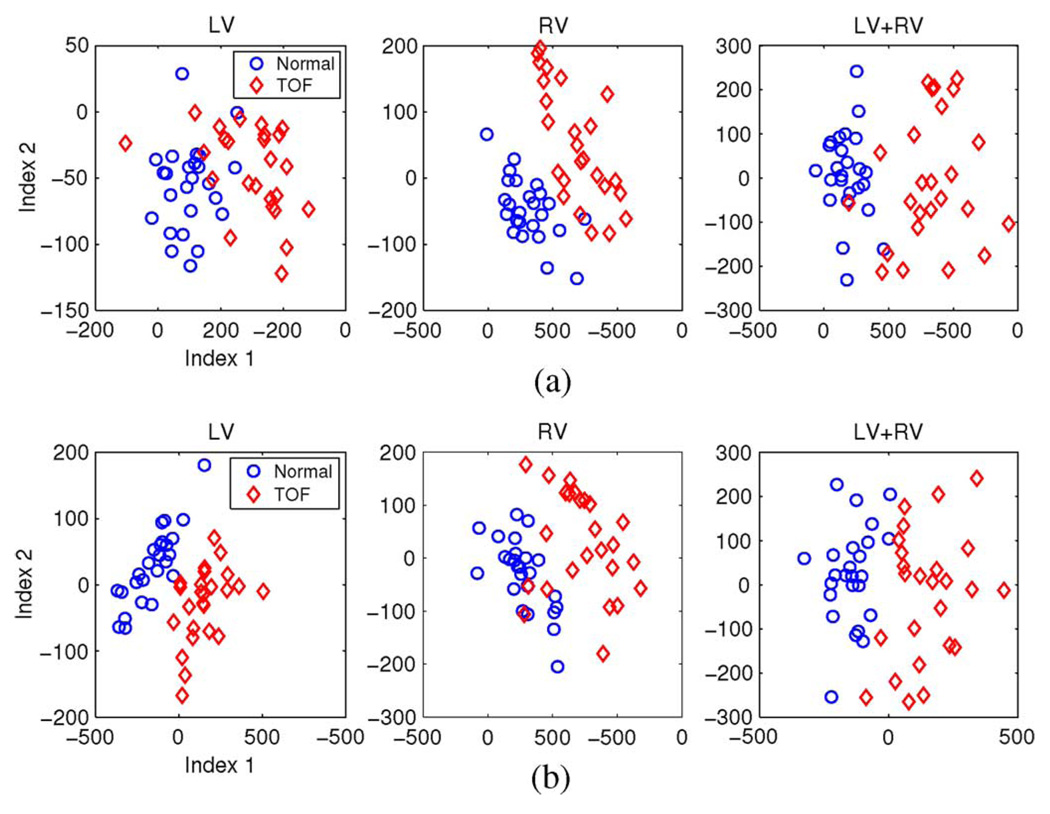
Fig. 11 shows the distributions of the first two modal indices of 4-D shape models of LV, RV, and LV+RV derived from the independent standard and the final segmentation. The strongest mode captures the main difference between normal and TOF hearts, which also agrees with the visualization in Fig. 10. In the shape model space, the normal and TOF hearts have clear differences in clustering of modal index distribution. Although the normal and TOF clusters have slightly different distribution patterns using models derived from the independent standard and the final computerized segmentation, such small differences have almost no effect on the classification performances listed in Table III.
Fig. 11.
Distributions of the two strongest modal indices of 4-D shape models of (from left to right) LV, RV, and LV+RV combined derived from (a) the independent standard and (b) the final segmentation.
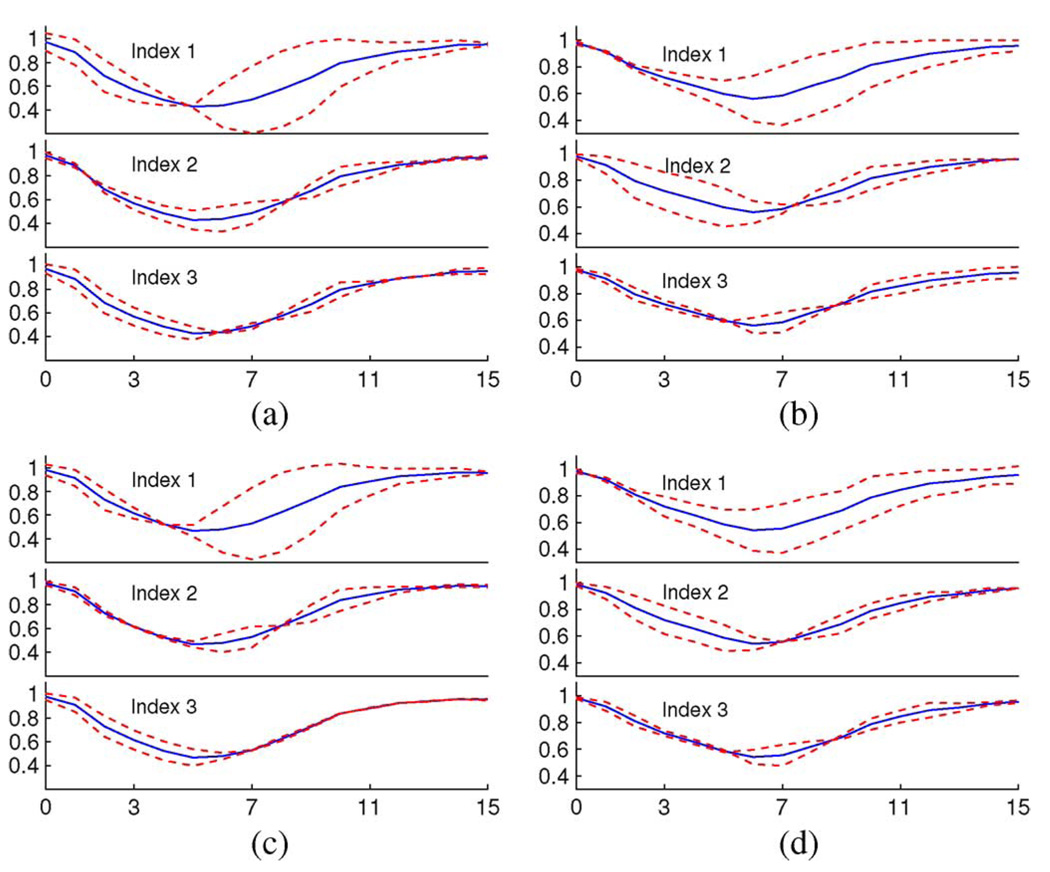
Fig. 12 shows the nVTC variations associated with varying the first three modal indices within [−2σ, 2σ] together with the mean nVTCs. In spite of the difference in classification performance, the patterns of nVTC variations captured by the models derived from the independent standard and the final segmentation are almost the same. The first mode is mainly associated with different diastolic motion and the second mode with systolic motion. Detailed observations showed that the ranges of variations embedded in the final segmentations were slightly smaller than those in the independent standard.
Fig. 12.
The nVTC variations associated with varying first three modal indices (showing from top to bottom) by ±2σ (dashed lines) and the mean nVTCs (solid lines) of models derived from the independent standard (IS) and final segmentation. (a) IS LV nVTC. (b) IS RV nVTC. (c) final seg. LV nVTC. (d) final seg. RV nVTC.
VIII. DISCUSSION
Although our image fusion and customized tracing application reduced observer variability, the manual tracings are still subject to inevitable uncertainties and inconsistencies. The results produced by varying the base and apex locations identified on LA views by one or even two voxels can still be deemed as clinically plausible due to lack of certainty about location of the atrio-ventricular valves, but the resulting fluctuation of the ventricular volume is large. The ventricular border tracings on the SA slices are affected by inconsistent criteria used across different hearts and cardiac phases. Such observer variability also exists when comparing the computer segmentation with the independent standard and contributes to imperfect accuracy indicated by validation metrics such as the overlap ratio.
Representing 3-D shapes by landmarks always introduces loss of detail. Our quality testing showed that such a loss yields only very small landmarking errors and the important ventricular shape features are intact. Therefore, the landmarked surfaces provide sufficiently accurate representations of the ventricular shapes. An alternative landmark generation approach is to generate landmarks on the first phase of the 4-D datasets and then propagate the landmarks throughout the cardiac cycle. We did not use this approach because small geometric errors introduced by one propagation will be accumulated and potentially amplified in other cardiac phases.
One consequence of a small training set is that the model-synthesized shape could be significantly different from the independent standard because the model cannot fully capture the variability of the training set and cannot approximate data that are not in the training set. Our intensive quality testing revealed that the 3-D modeling has only slightly worse accuracy than the landmarking, while it achieves better accuracy, especially in motion tracking, than 4-D modeling. Because 4-D modeling has a limited motion tracking capability as shown in Fig. 6(c), using 3-D models for final segmentation seems preferable. Fig. 6(d) also shows the potential error patterns of the segmentations such as underestimation of LV EDV, overestimation of LV ESV and reduced LV mass at end-systole.
Another consequence of a small training set is the under-trained model matching that produces imperfect segmentation. The training set contains insufficient information to produce Jacobian matrices that can generate parameter displacement with correct sign and enough magnitude to guide the model matching. The model matching therefore can be easily trapped at local minima and lead to imperfect segmentation. This under-training problem is more serious in the preliminary segmentation as shown in Fig. 9. The preliminary segmentation benefits from the reversed ASM fine tuning that provides some extra momentum during the model matching, but it still cannot achieve good cardiac motion tracking since the full motion variation is not captured by the 4-D modeling. Although it has been shown that the ASM training can be improved by an artificially enlarged training set [30], applying a similar technique to our approach is not appropriate because the clinical validity of the training set is critical.
To achieve accurate segmentation and minimize side effects of a small training set by reducing shape variations to be included in the models, the LV and RV as well as normal and TOF hearts are modeled with anisotropic scaling and segmented separately. Manual initialization was chosen due to its robustness and efficient utilization of expert knowledge. The combination of manual initialization and preliminary segmentation is necessary in our approach because it utilizes learned motion patterns and reduces human effort in manual initialization, which is also made more intuitive when anisotropic scaling is used. Without preliminary segmentation, the subsequent final segmentation using 3-D phase-by-phase approach either requires more human interaction or does not offer a comparable performance. When constructing the shape model used for disease characterization, isotropic scaling is used to preserve all potential disease features. The possible very small intersections and holes in the septal region—because the septum is included in both LV and RV—were corrected during segmentation and we found that they have no influence on our disease characterization.
Our final segmentation uses the same single 3-D model of all cardiac phases. Using multiple 3-D models—one model for one or several phases—is possible in principle, but such approach uses a smaller training set than the single-model approach and tracking severe irregular ventricular motion [Fig. 8(a)] and its associated appearance variations [Fig. 7(b)] becomes more difficult. The single-model final segmentation achieved improved accuracy and motion tracking compared to the preliminary segmentation. However, the segmentation performance is still bounded by the inherent limitations of the classical model matching approach because it cannot guarantee that 1) the accurate segmentation corresponds to the minimum intensity-based matching error, 2) the simplified multi-variable optimization using Jacobian matrices will find the global minimum, and 3) different but equally plausible initializations will converge to the same solution. The final LV endocardium and RV segmentations are improved because temporal motion constraints that may be too stringent are excluded from the 3-D model. The segmentation of LV epicardial surface with weak intensity evidence suffers from losing constraints on myocardium thickness and results in temporal fluctuation of LV mass. Due to this problem with the epicardium, the reversed ASM fine tuning was not performed in the final segmentation.
The 4-D ventricular shape and motion are complicated and so is the validation, where a single metric only partially describes the accuracy. For example, a segmentation that is a slightly shifted version of the independent standard but accurately captures the actual shape and motion features will correspond to reduced overlap. Our validation was performed on a large set of metrics to provide a comprehensive interpretation of segmentation accuracy. We believe that the ultimate segmentation performance assessment is reflected by the performance of disease characterization—whether the segmentation captures the same disease features as the independent standard. Our results show that the reported segmentation errors have almost no effect on the distribution and classification of the 4-D shape modal indices. The volume-based models built from segmentation produce slightly different classification results than those from the independent standard, but Fig. 12 shows that they contain the same types of motion variation with only slightly smaller ranges of modal indices.
The current running time of a full segmentation process, 15–30 min, is comparable to that of a reported 3-D ASM using fuzzy interface [23], if it is used for full cardiac cycle segmentation. The segmentation can be further optimized with a multi-resolution approach and the final segmentation can benefit from parallel computing. With the latest development in hardware, the human interaction can further be streamlined by utilizing virtual reality [52]. It is expected that the fully optimized and integrated application will need less than 10 min to produce comprehensive quantitative morphology and function results for clinical analysis.
IX. CONCLUSION
In this paper, a complete cardiac MR image processing pipeline was designed and implemented, from creating the independent standard to disease characterization. The pipeline was carefully designed to meet the requirement of clinical applicability where many practical limitations must be overcome.
Based on knowledge learned from quality assessment of modeling and model training, the segmentation was designed as a two-stage process with goal-oriented modeling and segmentation strategies. The preliminary segmentation utilizes a 4-D hybrid ASM/AAM algorithm with special reversed-ASM fine tuning. The final segmentation step was designed to achieve better cardiac motion tracking as well as improved shape details. The human intervention was minimized to manual initialization on only one cardiac phase. The comprehensively assessed segmentation accuracy showed that even when trained on a small population, the AAM-based segmentation method achieved very good performance.
In the final stage, a set of novel shape and motion features were designed using 4-D shape model and volume-based curves. The normal/TOF classification of these features achieved 90%–100% sensitivity and specificity using both segmetation result and independent standard, which further proves the high quality of the segmentation. We also showed that the features extracted from the independent standard and from the segmentation result represent very similar disease characteristics in shape and motion. In addition, the TOF hearts exhibit less clustered distributions in the feature space, suggesting these features can potentially be used as disease status and/or disease progression indicators.
In conclusion, the left and right ventricles of normal and TOF hearts can be accurately segmented by our AAM-based method, and the extracted shape model and volume-based features can correctly identify the unique characteristics of TOF and potentially its progression.
ACKNOWLEDGMENT
The contribution of M. Thomas during preparation of the independent standard is gratefully acknowledged.
This work was supported in part by NIH NHLBI and NIH NIBIB grants R01-HL071809 and R01-EB004640.
Footnotes
Color versions of one or more of the figures in this paper are available online at http://ieeexplore.ieee.org.
Contributor Information
Honghai Zhang, Department of Electrical and Computer Engineering, The University of Iowa, Iowa City, IA 52242 USA (honghai-zhang@uiowa.edu).
Andreas Wahle, Department of Electrical and Computer Engineering, The University of Iowa, Iowa City, IA 52242 USA (andreas-wahle@uiowa.edu).
Ryan K. Johnson, Division of Pediatric Cardiology, Department of Pediatrics, The University of Iowa, Iowa City, IA 52242 USA ryan-johnson@uiowa.edu
Thomas D. Scholz, Division of Pediatric Cardiology, Department of Pediatrics, The University of Iowa, Iowa City, IA 52242 USA thomas-scholz@uiowa.edu
Milan Sonka, Department of Electrical and Computer Engineering, the Department of Ophthalmology and Visual Sciences, and the Department of Radiation Oncology, The University of Iowa, Iowa City, IA, 52242 USA (milan-sonka@uiowa.edu).
REFERENCES
- 1.Anderson RH, Tynan M. Tetralogy of Fallot—A centennial review. Int. J. Cardiol. 1988 Dec;vol. 21(no. 3):219–232. doi: 10.1016/0167-5273(88)90100-3. [DOI] [PubMed] [Google Scholar]
- 2.Gatzoulis MA, Webb GD, Daubeney PEF. Diagnosis and Management of Adult Congenital Heart Disease. New York: Churchill Livingstone; 2003. [Google Scholar]
- 3.Anderson RH, Weinberg PM. The clinical anatomy of tetralogy of Fallot. Cardiol. Young. 2005 Feb;vol. 15(no. 1):38–47. doi: 10.1017/s1047951105001010. [DOI] [PubMed] [Google Scholar]
- 4.Hoffman JIE, Kaplan S, Liberthson RR. Prevalence of congenital heart disease. Am. Heart J. 2004 Mar;vol. 147(no. 3):425–439. doi: 10.1016/j.ahj.2003.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Helbing WA, de Roos A. Clinical applications of cardiac magnetic resonance imaging after repair of tetralogy of Fallot. Pediatr. Cardiol. 2000;vol. 21(no. 1):70–79. doi: 10.1007/s002469910009. [DOI] [PubMed] [Google Scholar]
- 6.Schwerzmann M, Samman AM, Salehian O, Holm J, Provost Y, Webb GD, Therrien J, Siu SC, Silversides CK. Comparison of echocardiographic and cardiac magnetic resonance imaging for assessing right ventricular function in adults with repaired tetralogy of Fallot. Am. J. Cardiol. 2007 Jun;vol. 99(no. 11):1593–1597. doi: 10.1016/j.amjcard.2007.01.035. [DOI] [PubMed] [Google Scholar]
- 7.Lai WW, Gauvreau K, Rivera ES, Saleeb S, Powell AJ, Geva T. Accuracy of guideline recommendations for two-dimensional quantification of the right ventricle by echocardiography. Int. J. Car-diovasc. Imag. 2008 Oct;vol. 24(no. 7):691–698. doi: 10.1007/s10554-008-9314-4. [DOI] [PubMed] [Google Scholar]
- 8.Baur LHB. Magnetic resonance imaging: The preferred imaging method for evaluation of the right ventricle. Int. J. Cardiovasc. Imag. 2008 Oct;vol. 24(no. 7):699–700. doi: 10.1007/s10554-008-9321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knauth AL, Gauvreau k, Powell AJ, Landzberg MJ, Walsh EP, Lock JE, del Nido PJ, Geva T. Ventricular size and function assessed by cardiac MRI predict major adverse clinical outcomes late after tetralogy of Fallot repair. Heart. 2008 Feb;vol. 94(no. 2):211–216. doi: 10.1136/hrt.2006.104745. [DOI] [PubMed] [Google Scholar]
- 10.Geva T, Sandweiss BM, Gauvreau K, Lock JE, Powell AJ. Factors associated with impaired clinical status in long-term survivors of tetralogy of Fallot repair evaluated by magnetic resonance imaging. J. Am. Coll. Cardiol. 2004 Mar;vol. 43(no. 6):1068–1074. doi: 10.1016/j.jacc.2003.10.045. [DOI] [PubMed] [Google Scholar]
- 11.Therrien J, Siu SC, McLaughlin PR, Liu PP, Williams WG, Webb GD. Pulmonary valve replacement in adults late after repair of tetralogy of Fallot: Are we operating too late? J. Am. Coll. Cardiol. 2000 Nov;vol. 36(no. 5):1670–1675. doi: 10.1016/s0735-1097(00)00930-x. [DOI] [PubMed] [Google Scholar]
- 12.Therrien J, Siu SC, Harris L, Dore A, Niwa K, Janousek J, Williams WG, Webb G, Gatzoulis MA. Impact of pulmonary valve replacement on arrhythmia propensity late after repair of tetralogy of Fallot. Circulation. 2001 May;vol. 103(no. 20):2489–2494. doi: 10.1161/01.cir.103.20.2489. [DOI] [PubMed] [Google Scholar]
- 13.Sheehan FH, Ge S, Vick GW, Urnes K, Kerwin WS, Bolson EL, Chung T, Kovalchin JP, Sahn DJ, Jerosch-Herold M, Stolpen AH. Three-dimensional shape analysis of right ventricular remodeling in repaired tetralogy of Fallot. Am. J. Cardiol. 2008 Jan;vol. 101(no. 1):107–113. doi: 10.1016/j.amjcard.2007.07.080. [DOI] [PubMed] [Google Scholar]
- 14.Cousty J, Najman L, Couprie M, Clément-Guinaudeau S, Goissen T, Garot J. Automated, accurate and fast segmentation of 4-D cardiac MR images. Functional Imaging and Modeling of the Heart. 2007;vol. 4466:474–483. [Google Scholar]
- 15.Lötjönen J, Kivistö S, Koikkalainen J, Smutek D, Lauerma K. Statistical shape model of atria, ventricles and epicardium from short- and long-axis Mr images. Med. Imag. Anal. 2004 Sep;vol. 8(no. 3):371–386. doi: 10.1016/j.media.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 16.Ecabert O, Peters J, Schramm H, Lorenz C, von Berg J, Walker MJ, Vembar M, Olszewski ME, Subramanyan K, Lavi G, Weese J. Automatic model-based segmentation of the heart in CT images. IEEE Trans. Med. Imag. 2008 Sep;vol. 27(no. 9):1189–1201. doi: 10.1109/TMI.2008.918330. [DOI] [PubMed] [Google Scholar]
- 17.Bistoquet A, Oshinski J, Škrinjar O. Left ventricular deformation recovery from cine MRI using an incompressible model. IEEE Trans. Med. Imag. 2007 Sep;vol. 26(no. 9):1136–1153. doi: 10.1109/TMI.2007.903693. [DOI] [PubMed] [Google Scholar]
- 18.Sermesant M, Peyrat J-M, Chinchapatnam P, Billet F, Mansi T, Rhode K, Delingette H, Razavi R, Ayache N. Toward patient-specific myocardial models of the heart. Heart Fail. Clin. 2008 Jul;vol. 4(no. 3):289–301. doi: 10.1016/j.hfc.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 19.Zheng Y, Barbu A, Georgescu B, Scheuering M, Comaniciu D. Four-chamber heart modeling and automatic segmentation for 3-D cardiac CT volumes using marginal space learning and steerable features. IEEE Trans. Med. Imag. 2008 Nov;vol. 27(no. 11):1668–1681. doi: 10.1109/TMI.2008.2004421. [DOI] [PubMed] [Google Scholar]
- 20.Bosch JG, Mitchell SC, Lelieveldt BPF, Nijland F, Kamp O, Sonka M, Reiber JHC. Automatic segmentation of echocardio-graphic sequences by active appearance motion models. IEEE Trans. Med. Imag. 2002 Nov;vol. 21(no. 11):1374–1383. doi: 10.1109/TMI.2002.806427. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell SC, Bosch JG, Lelieveldt BPF, van der Geest RJ, Reiber JHC, Sonka M. 3-D active appearance models: Segmentation of cardiac MR and ultrasound images. IEEE Trans. Med. Imag. 2002 Sep;vol. 21(no. 9):1167–1178. doi: 10.1109/TMI.2002.804425. [DOI] [PubMed] [Google Scholar]
- 22.Stegmann MB, Pedersen D. Bi-temporal 3-D active appearance models with applications to unsupervised ejection fraction estimation. Proc. SPIE Med. Imag. 2005 Apr;vol. 5747:336–350. [Google Scholar]
- 23.van Assen HC, Danilouchkine MG, Dirksen MS, Reiber JHC, Lelieveldt BPF. A 3-D active shape model driven by fuzzy inference: Application to cardiac CT and MR. IEEE Trans. Inf. Technol. Biomed. 2008 Sep;vol. 12(no. 5):595–605. doi: 10.1109/TITB.2008.926477. [DOI] [PubMed] [Google Scholar]
- 24.Perperidis D, Mohiaddin RH, Edwards PJ, Rueckert D. Segmentation of cardiac MR and CT image sequences using model-based registration of a 4-D statistical model. Proc. SPIE Med. Imag. 2007;vol 6512 [Google Scholar]
- 25.Ordas S, Oubel E, Leta R, Carreras F, Frangi AF. A statistical shape model of the heart and its application to model-based segmentation; Proc. SPIE Med. Imag.; 2007. p. 65111K. [Google Scholar]
- 26.Andreopoulos A, Tsotsos JK. Efficient and generalizable statistical models of shape and appearance for analysis of cardiac MRI. Med. Imag. Anal. 2008 Jun;vol. 12(no. 3):335–357. doi: 10.1016/j.media.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Y, Papademetris X, Sinusas AJ, Duncan JS. Bidirectional segmentation of three-dimensional cardiac MR images using a subject-specific dynamical model; Proc. MICCAI 2008; Berlin, Germany: 2008. pp. 450–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Üzümcü M, Frangi AF, Sonka M, Reiber JHC, Lelieveldt BPF. ICA vs. PCA active appearance models: Application to cardiac MR segmentation; Proc. MICCAI 2003; 2003. pp. 451–458. [Google Scholar]
- 29.Suinesiaputra A, Frangi AF, Lamb HJ, Reiber JHC, Lelieveldt BPF. Automatic prediction of myocardial contractility improvement in stress mri using shape morphometrics with independent component analysis; Proc. IPMI 2005; 2005. pp. 321–332. [DOI] [PubMed] [Google Scholar]
- 30.Tölli T, Koikkalainen J, Lauerma K, Lötjönen J. Artificially enlarged training set in image segmentation; Proc. MICCAI 2006; 2006. pp. 75–82. [DOI] [PubMed] [Google Scholar]
- 31.Stegmann MB, Sjöstrand K, Larsen R. Sparse modeling of landmark and texture variability using the orthomax criterion; Proc. SPIE Med. Imag. 2006; 2006. pp. 485–496. [Google Scholar]
- 32.Marsland S, Twining CJ, Taylor CJ. A minimum description length objective function for groupwise nonrigid image registration. Proc Image Vis. Comput. 2008;vol. 26(no. 3):336–346. [Google Scholar]
- 33.Davies RH, Twining CJ, Taylor C. Groupwise surface correspondence by optimization: Representation and regularization. Med. Imag. Anal. 2008;vol. 12(no. 6):787–796. doi: 10.1016/j.media.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 34.Liu J, Udupa JK. Oriented active shape models. IEEE Trans. Med. Imag. 2009 Apr;vol. 28(no. 4):571–584. doi: 10.1109/TMI.2008.2007820. [DOI] [PubMed] [Google Scholar]
- 35.Cootes TF, Cooper D, Taylor CJ, Graham J. A trainable method of parametric shape description. Proc. Br. Machine Vis. Conf. 1991:54–61. [Google Scholar]
- 36.Cootes TF, Taylor CJ. Active shape models—“Smart snakes. Proc. Br. Machine Vis. Conf. 1992:266–275. [Google Scholar]
- 37.Cootes TF, Edwards GJ, Taylor CJ. Active appearance models; Proc. Eur. Conf. Comput. Vis; 1998. pp. 484–498. [Google Scholar]
- 38.Mitchell SC, Lelieveldt BPF, van der Geest RJ, Bosch HG, Reiber JHC, Sonka M. Multistage hybrid active appearance model matching: Segmentation of left and right ventricles in cardiac MR images. IEEE Trans. Med. Imag. 2001 May;vol. 20(no. 5):415–423. doi: 10.1109/42.925294. [DOI] [PubMed] [Google Scholar]
- 39.Debatin JF, Nadel SN, Sostman HD, Spritzer CE, Evans AJ, Grist TM. Magnetic resonance imaging-cardiac ejection fraction measurements. phantom study comparing four different methods. Invest. Radiol. 1992 Mar;vol. 27(no. 3):198–204. [PubMed] [Google Scholar]
- 40.Pattynama PM, Doornbos J, Hermans J, van der Wall EE, de Roos A. Magnetic resonance evaluation of regional left ventricular function. effect of through-plane motion. Invest. Radiol. 1992 Sep;vol. 27(no. 9):681–685. doi: 10.1097/00004424-199209000-00004. [DOI] [PubMed] [Google Scholar]
- 41.Bloomer TN, Plein S, Radjenovic A, Higgins DM, Jones TR, Ridgway JP, Sivananthan MU. Cine MRI using steady state free precession in the radial long axis orientation is a fast accurate method for obtaining volumetric data of the left ventricle. J. Magn. Reson. Imag. 2001 Dec;vol. 14(no. 6):685–692. doi: 10.1002/jmri.10019. [DOI] [PubMed] [Google Scholar]
- 42.Swingen CM, Seethamraju RT, Jerosch-Herold M. Feedback-assisted three-dimensional reconstruction of the left ventricle with MRI. J. Magn. Reson. Imag. 2003 May;vol. 17(no. 5):528–537. doi: 10.1002/jmri.10290. [DOI] [PubMed] [Google Scholar]
- 43.Stegmann MB, Olafsdottir H, Larsson HBW. Unsupervised motion-compensation of multi-slice cardiac perfusion MRI. Med. Imag. Anal. 2005;vol. 9(no. 4):394–410. doi: 10.1016/j.media.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 44.Blu T, Thévenaz P, Unser M. Linear interpolation revitalized. IEEE Trans. Image Process. 2004 May;vol. 13(no. 5):710–719. doi: 10.1109/tip.2004.826093. [DOI] [PubMed] [Google Scholar]
- 45.Raya SP, Udupa JK. Shape-based interpolation of multidimensional objects. IEEE Trans. Med. Imag. 1990 Mar;vol. 9(no. 1):32–42. doi: 10.1109/42.52980. [DOI] [PubMed] [Google Scholar]
- 46.Frangi A, Rueckert D, Schnabel J, Niessen W. Automatic construction of multiple-object three-dimensional statistical shape models: Application to cardiac modeling. IEEE Trans. Med. Imag. 2002 Sep;vol. 21(no. 9):1151–1166. doi: 10.1109/TMI.2002.804426. [DOI] [PubMed] [Google Scholar]
- 47.Lorensen WE, Cline HE. Marching cubes: A high resolution 3-D surface construction algorithm. Comput. Graph. 1987;vol. 21:163–169. [Google Scholar]
- 48.Taubin G, Zhang T, Golub G. Optimal surface smoothing as filter design IBM, Tech. Rep. RC-20404. 1996
- 49.Hoppe H. New quadric metric for simplifying meshes with appearance attributes; Proc. IEEE Visualizat; 1999. Oct, pp. 59–66. [Google Scholar]
- 50.Pinkall U, Polthier K. Computing discrete minimal surfaces and their conjugates. Exper. Math. 1993;vol. 2(no. 1):15–36. [Google Scholar]
- 51.Maurer CR, Qi R, Raghavan V. A linear time algorithm for computing exact Euclidean distance transforms of binary images in arbitrary dimensions. IEEE Trans. Pattern Anal. Mach. Intell. 2003 Feb;vol. 25(no. 2):265–270. [Google Scholar]
- 52.Sonka M, Beichel R, Bornik A, Reitinger B, Sorantin E, Werkgartner G. Computer-aided liver surgery planning. In: Armato S, Brown M, editors. Multidimensional Image Processing, Analysis, and Display. Oak Brook, IL: Radiological Soc. North America; 2005. pp. 227–236. [Google Scholar]