Abstract
We report a significant methodological advance in the application of double electron-electron resonance (DEER) spectroscopy to measure long-range distances in spin-labeled membrane proteins. In the pseudo two-dimensional environment of proteoliposomes, a steep intermolecular background shapes DEER signals leading to long accumulation times, complicating data analysis and reducing the maximal measurable distances from 70 Å down to ∼40–50 Å. To eliminate these limitations, we took advantage of the homogeneity and monodispersity of a class of discoidal nanoscale phospholipid bilayers in conjunction with the micromolar DEER sensitivity at Q-band (34 GHz) microwave frequency. Spin-labeled mutants of the ABC transporter MsbA were functionally reconstituted at a ratio of one functional dimer per nanoscale apolipoprotein-bound bilayer (NABB). DEER echo intensities from NABB-reconstituted MsbA have linear baselines reflecting a three-dimensional spatial distribution. This results in an order-of-magnitude higher sensitivity at Q-band relative to proteoliposomes and restores the maximal observable distance effectively increasing experimental throughput. The advances described here set the stage for the use of DEER spectroscopy to analyze conformational dynamics of sample-limited eukaryotic membrane proteins.
Main Text
Double electron-electron resonance (DEER) is an unparalleled tool in structural biology enabling the measurements of long-range distances (20–70 Å) between pairs of spin labels in biomolecules (1–3). These distances provide powerful restraints in modeling protein folds (4–6) and defining protein conformational changes (4,7–10). Electron paramagnetic resonance (EPR) parameters are obtained in nativelike environments regardless of the protein molecular mass under well-defined biochemical conditions in the absence of conformationally selective crystal lattice forces. Furthermore, identical spin labels are attached at introduced cysteines simplifying the labeling procedure compared to optical resonance energy transfer, which normally requires differential labeling with a donor-acceptor pair.
Application of DEER spectroscopy to membrane proteins is hindered by significant reduction in the accessible distance range, loss of sensitivity and compromised throughput. The DEER time domain signal is a product of an intramolecular term arising from dipolar coupling between spin labels within the same protein and an intermolecular term, also referred to as a background or form factor, describing the bulk spatial distribution of proteins (2). The relative contribution of the latter can dominate the signal if multiple protein molecules are incorporated in the two-dimensional environment of a liposome severely affecting interpretation. Whereas intramolecular distances can be obtained in detergent micelles across the entire theoretical range, this large background contribution in liposomes reduces practical distances measured to an upper limit of ∼50 Å (11). In some instances, the DEER signals are effectively uninterpretable, thereby reducing experimental throughput. Approaches to minimize the background contribution include the use of large lipid/protein molar ratios and/or reconstitution in the presence of unlabeled protein (9,12). The concomitant loss of sensitivity and increased sample requirements severely limit the number of distance restraints that can be obtained. DEER signals are screened in detergent micelles and a subset of sites is selected for distance measurements in liposomes. Alternatively, protein solubilization in bicelles removes the two-dimensional background without compromising sensitivity (11). Although representing a more nativelike environment, bicelles require detergents and restrict the type of phospholipids that can be used.
In this report we demonstrate the synergistic convergence of two technologies—discoidal nanoscale lipoprotein-bound bilayers (13–15) and Q-band (34 GHz microwave frequency) pulsed EPR—to circumvent the factors limiting DEER sensitivity and distance range in lipid bilayers, thereby effecting a significant increase in experimental throughput. Nanolipoprotein phospholipid bilayers, most commonly referred to as Nanodiscs (Sligar Lab, University of Illinois at Urbana-Champaign, Urbana, IL), are a class of soluble nanoscale assemblies of lipids surrounded by an annulus of amphipathic protein, i.e., an engineered fragment of apolipoprotein A-1, also referred to the membrane scaffold protein (MSP) (13,14). By forming a uniform belt around the bilayers, MSP constrains their dimensions and ensures relative monodispersity without restrictions on the nature of lipid headgroups or acyl chain lengths. These two characteristics facilitate the reconstitution of a single protein functional unit per bilayer disk through careful manipulation of the molar ratios of the constituents (16).
A number of shortcomings balance the expected sensitivity gains from the removal of the two-dimensional background. Sample amounts are limited by the requirement to recombinantly express the MSP. Furthermore, these samples do not concentrate readily, resulting in a working concentration ranging from 10 to 50 μM. Superior sensitivity for limited sample volumes is theoretically predicted and experimentally observed at higher microwave frequencies than the X-band frequency typically used for DEER measurements. Specifically, commercially available pulsed EPR at Q-band affords a remarkable increase in absolute DEER sensitivity relative to X-band. Initially demonstrated in model biradicals (17), the advantages of Q-band DEER were recently described and extended to a water-soluble peptide (18).
We found the increased sensitivity at Q-band necessary and sufficient to implement nanoscale apolipoprotein-bound bilayer (NABB) technology for DEER distance measurements. Distances and distance changes were determined in a set of mutants of the ABC transporter MsbA selected to fingerprint the ATP-induced conformational rearrangements previously demonstrated in proteoliposomes (9). Spin-label mobility and distance to a symmetry-related label were measured in NABB-reconstituted MsbA at sites in the nucleotide-binding domain (NBD 561), intracellular helix 1 (IH1 115), and transmembrane helix 3 (TM3 143, 158) (Fig. 1 A).
Figure 1.
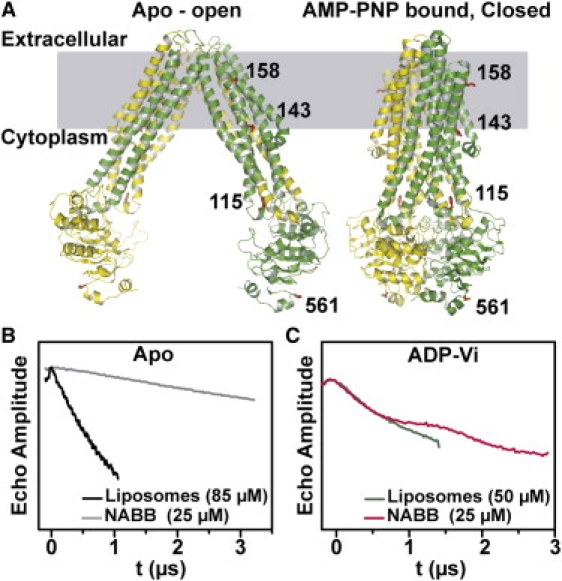
(A) Ribbon representation of MsbA open and closed structures highlighting sites for distance measurements. (B and C) Q-band raw DEER data for site 561.
Expression, purification, and functional analysis of MsbA were carried out after previously published procedures (19). In this work, the MSP is Zebrafish apolipoprotein A-I, which forms NABB particles of equivalent properties to Nanodiscs (15). NABB were assembled from solutions of Escherichia coli lipids mixed with detergent micelles, MSP, and detergent-solubilized MsbA (13), as described in the Supporting Material. MsbA-containing NABBs were then purified by size-exclusion chromatography. MsbA/NABB elutes as a single symmetric peak before and resolvable from the empty NABB on the size-exclusion column providing a convenient means for separation (13). Starting from molar ratios of excess MSP and lipids, a single MsbA dimer is incorporated per NABB, as verified by SDS-PAGE analysis demonstrating approximately one MSP per MsbA monomer (data not shown). MsbA is functional in NABB with specific ATP turnover rates similar to those measured in liposomes (Table S1 in the Supporting Material). Furthermore, EPR lineshapes are similar in NABB and unilamellar liposomes suggesting local dynamics are unchanged (Fig. S1 in the Supporting Material).
As predicted, the reconstitution of a single transporter per NABB dramatically improves DEER signal quality by reducing the large intermolecular component. This was observed in all samples and highlighted in Fig. 1 for site 561. Particularly instructive is the DEER signal obtained from the apo intermediate where the distance between the spin labels are outside the accessible range (Cα-Cα distance of 86 Å) (20). In this case, the echo decay consists primarily of the background arising from dipolar coupling between MsbA dimers. The slower decay observed in NABB demonstrates the more disperse distribution of MsbA relative to unilamellar proteoliposomes. Trapping of MsbA in the high energy posthydrolysis intermediate, by addition of ATP and vanadate (Vi) (10), reduces the distance between the symmetry-related spin labels at site 561. This is reflected in the rapid decay of the DEER signal and the appearance of dipolar oscillations, which is masked in the liposome sample by the background component. Superposition of the Q-band DEER raw data (Fig. 1 C) from proteoliposomes and NABB reveals the improvement in signal/noise of the latter. MsbA was twofold more concentrated (50 μM) in the liposome samples and required twofold longer collection time for half the echo evolution time. Thus, a further 5–10-fold increase in effective sensitivity is achieved by using NABB at Q-band.
Fig. 2 illustrates a case where no meaningful distances could be extracted from proteoliposome samples using X-band DEER. In contrast, the NABB data for MsbA spin-labeled at site 143 have well-defined baselines amenable to analysis. The sensitivity improvements are remarkable: the NABB ADP-Vi sample (Fig. 2 B) was sevenfold less concentrated and required 30% less collection time. In general, the shapes of the distance distributions are improved greatly by the superior signal/noise ratio (Fig. 2 C). Table 1 shows that the pattern of distance and distance changes are almost identical to those obtained in polar asolectin liposomes (9), suggesting little effects of lipid composition on the outline of MsbA conformational cycle. This pattern was also unchanged in NABB made of polar asolectin lipids (data not shown).
Figure 2.
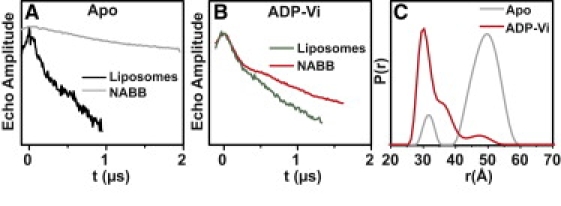
(A and B) Raw DEER data for site 143. The liposome data were obtained at X-band. (C) Distance distributions, P(r), for the two MsbA intermediates in NABB determined by Tikhonov regularization.
Table 1.
ATP-induced distance changes between symmetry-related spin labels in NABB-reconstituted MsbA
| Experimental distance Rav (Å), NABB |
Calculated distance ΔRCα- Cα (Å) |
||||||
|---|---|---|---|---|---|---|---|
| Mutant | Location | Apo | ADP-Vi | ΔRav | Open | AMPPNP | ΔRCα- Cα |
| 158 | TM3 | 39.5 ± 3 | 45.0 ± 6 | −5.5 | 41 | 46 | −5 |
| 143 | 49.5 ± 4 | 30.0 ± 2 | 19.5 | 47 | 28 | 19 | |
| 115 | IH1 | 54.0 ± 5 | 32.0 ± 1 | 22 | 62 | 31 | 31 |
| 561 | NBD | >70 | 42.0 ± 2 | >28 | 86 | 36 | 50 |
In summary, we developed an improved biochemical/spectroscopic approach to significantly increase sensitivity and range of distance measurements in spin-labeled membrane proteins. The quality of the raw DEER data at Q-band is vastly superior, despite a fivefold reduction in both typical sample concentration and volume relative to X-band. Improvement of the first generation Q-band pulsed instrumentation will additionally enhance the concentration sensitivity. Limited microwave power leads to long pulses (shortest π/2 pulse is 16 ns on our system), which restricts the fraction of excited spins, thereby reducing the amplitude of the spin echo. More importantly, power limitation compromises the depth of modulation of the DEER signal, which contains the dipolar evolution function. Current resonator design limits sample volumes in favor of high quality factors to compensate for the low power. Newly introduced microwave amplifiers promise to substantially increase the available power abrogating these problems. As these limitations are overcome, we expect to achieve submicromolar sensitivity, which will be critical for application of DEER to eukaryotic membrane proteins.
Acknowledgments
We thank Dr. Thomas Sakmar for providing the Zebrafish apolipoprotein expression system, Smriti for contributing the data for MsbA 561 in liposomes, and Jared Godar and Derek P. Claxton for critical reading of the manuscript.
This work was supported by National Institute of General Medical Sciences grant No. R01-077659.
Supporting Material
References and Footnotes
- 1.Borbat P.P., McHaourab H.S., Freed J.H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. J. Am. Chem. Soc. 2002;124:5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 2.Jeschke G. Distance measurements in the nanometer range by pulse EPR. ChemPhysChem. 2002;3:927–932. doi: 10.1002/1439-7641(20021115)3:11<927::AID-CPHC927>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 3.Jeschke G., Polyhach Y. Distance measurements on spin-labeled biomacromolecules by pulsed electron paramagnetic resonance. Phys. Chem. Chem. Phys. 2007;9:1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 4.Alexander N., Bortolus M., Meiler J. De novo high-resolution protein structure determination from sparse spin-labeling EPR data. Structure. 2008;16:181–195. doi: 10.1016/j.str.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borbat P.P., Freed J.H. Measuring distances by pulsed dipolar ESR spectroscopy: spin-labeled histidine kinases. Methods Enzymol. 2007;423:52–116. doi: 10.1016/S0076-6879(07)23003-4. [DOI] [PubMed] [Google Scholar]
- 6.Hilger D., Polyhach Y., Jeschke G. Backbone structure of transmembrane domain IX of the Na+/proline transporter PutP of Escherichia coli. Biophys. J. 2009;96:217–225. doi: 10.1016/j.bpj.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borbat P.P., Surendhran K., Mchaourab H.S. Conformational motion of the ABC transporter MsbA induced by ATP hydrolysis. PLoS Biol. 2007;5:2211–2219. doi: 10.1371/journal.pbio.0050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smirnova I., Kasho V., Kaback H.R. Sugar binding induces an outward facing conformation of LacY. Proc. Natl. Acad. Sci. USA. 2007;104:16504–16509. doi: 10.1073/pnas.0708258104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou P., Bortolus M., McHaourab H.S. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J. Mol. Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou P., McHaourab H.S. Alternating access of the putative substrate-binding chamber in the ABC transporter MsbA. J. Mol. Biol. 2009;393:574–585. doi: 10.1016/j.jmb.2009.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Georgieva E.R., Ramlall T.F., Eliezer D. Membrane-bound α-synuclein forms an extended helix: long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. J. Am. Chem. Soc. 2008;130:12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Q., Kim M., Cafiso D.S. Membrane hydrocarbon thickness modulates the dynamics of a membrane transport protein. Biophys. J. 2008;95:2849–2858. doi: 10.1529/biophysj.108.133629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boldog T., Li M., Hazelbauer G.L. Using Nanodiscs to create water-soluble transmembrane chemoreceptors inserted in lipid bilayers. Methods Enzymol. 2007;423:317–335. doi: 10.1016/S0076-6879(07)23014-9. [DOI] [PubMed] [Google Scholar]
- 14.Nath A., Atkins W.M., Sligar S.G. Applications of phospholipid bilayer Nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 15.Banerjee S., Huber T., Sakmar T.P. Rapid incorporation of functional rhodopsin into nanoscale apolipoprotein bound bilayer (NABB) particles. J. Mol. Biol. 2008;377:1067–1081. doi: 10.1016/j.jmb.2008.01.066. [DOI] [PubMed] [Google Scholar]
- 16.Boldog T., Grimme S., Hazelbauer G.L. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc. Natl. Acad. Sci. USA. 2006;103:11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Höfer P., Heilig R., Schmalbein D. The superQ-FT accessory for pulsed EPR, ENDOR and ELDOR at 34 GHz. Bruker SpinReport. 2003;152/153:37–43. [Google Scholar]
- 18.Ghimire H., McCarrick R.M., Lorigan G.A. Significantly improved sensitivity of Q-band PELDOR/DEER experiments relative to X-band is observed in measuring the intercoil distance of a leucine zipper motif peptide (GCN4-LZ) Biochemistry. 2009;48:5782–5784. doi: 10.1021/bi900781u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong J., Yang G., McHaourab H.S. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–1028. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- 20.Ward A., Reyes C.L., Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc. Natl. Acad. Sci. USA. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


