Abstract
The effect of macromolecular crowding on the binding of ligands to a receptor near membranes is studied using Brownian dynamics simulations. The receptor is modeled as a reactive patch on a hard surface and the ligands and crowding agents are modeled as spheres that interact via a steep repulsive interaction potential. When a ligand collides with the patch, it reacts with probability prxn. The association rate constant (k∞) can be decomposed into contributions from diffusion-limited (kD) and reaction-limited (kR) rates, i.e., 1/k∞ = 1/kD + 1/kR. The simulations show that kD is a nonmonotonic function of the volume fraction of crowding agents for receptors of small sizes. kR is always an increasing function of the volume fraction of crowding agents, and the association rate constant k∞ determined from both contributions has a qualitatively different dependence on the macromolecular crowding for high and low values of the reaction probability prxn. The simulation results are used to predict the velocity of the membrane protrusion driven by actin filament elongation. Based on the simple model where the protrusive force on the membrane is generated by the intercalation of actin monomers between the membrane and actin filament ends, we predict that crowding increases the local concentration of actin monomers near the filament ends and hence accelerates the membrane protrusion.
Introduction
The importance of macromolecular crowding on biochemical reaction kinetics is now widely recognized (1–4). There have been many studies, for example, on crowding effects on protein association in solution (5–7). In this work, we focus on protein association reactions occurring at the membrane, a problem that has received scant attention from theory or computation. Association reactions at the membrane are relevant to many biological processes, such as the binding of a signaling protein to its cognate receptor, or the intercalation of actin monomers between the membrane and actin filament ends (8,9). An understanding of crowding effects on these reactions is therefore of interest.
Much of our understanding of crowding effects comes from simple hard sphere models for crowding agents. From a dynamical perspective, the presence of crowding agents has two opposing effects that affect the reaction rates. As the volume fraction of crowding agents is increased, the diffusion coefficient of the reactants decreases, but the probability of a recollision after an initial unreactive collision increases. The net reaction rate reflects a balance between these effects (7,10,11). The Smoluchowski theory was in quantitative agreement with computer simulation results for hard sphere reactants with hard sphere crowding agents (7).
In this work, we perform the computer simulations for the association reaction between ligands and a receptor on a membrane. For this problem, the Smoluchowski theory cannot be solved analytically and we look to numerical simulations for insight. The ligands are modeled as spherical particles and the receptor is a square reactive patch on a hard surface. The size of the reactive patch on the surface is much smaller than that of the entire surface and the rest of the surface is inert to the reaction. The crowding agents are modeled as spheres (identical to the ligands). The degree of crowding is increased by increasing the volume fraction, ϕ, of the crowding agents from 0 to 0.4. When a ligand collides with the patch, it reacts with a probability, prxn. The reaction probability prxn is introduced to mimic the effect of the orientation-dependent anisotropic reactivity.
We calculate the long-time reaction rate constant k∞ and show that it can be decomposed into a diffusive (kD) and a reactive (kR) contribution, i.e., 1/k∞ = 1/kD + 1/kR. By fitting the simulation results, we determine the effect of crowding on kD and kR separately.
The most interesting result compared with the previous study of the crowding effect on the protein association reactions in bulk solution is that for some cases the diffusion-controlled rate of contact between ligands and a receptor, kD, is a nonmonotonic function of the volume fraction of crowding agents: kD increases with increasing ϕ for low volume fractions and then decreases as ϕ is increased further. This is in contrast to the behavior in bulk solution where kD always decreases as ϕ is increased. The nonmonotonic behavior in kD of this work arises from two competing effects. The presence of crowding agents hinders the approach of ligands to the surface, but once they are at the surface it also hinders the motion of the ligands away from the surface and thus the diffusive search for the receptor is confined to the surface (12) and becomes more efficient (13). This latter effect becomes unimportant when the size of the patch becomes large, and in that limit, kD decreases monotonically as ϕ is increased.
As in the study of protein association in solution, kR increases with crowding due to the increased density of ligands near a membrane surface as ϕ is increased. We derive an analytical expression for kR based on the ligand density at the wall, which is in excellent agreement with the simulation results. We also interpret this increase in terms of the increased probability of recollision of ligands with the receptor after an initial unreactive collision. The association rate constant k∞ determined from both contributions has a qualitatively different dependence on the macromolecular crowding for high and low values of prxn because different values of prxn only change kR, whereas kD is kept the same.
We study the velocity of membrane protrusion driven by actin filament elongation (4,8,9,14–17) by considering a simple model where the intercalation of actin monomers between the membrane and the growing filaments (14–17) is approximated as an association reaction between a monomer and a reactive patch. We estimate that the process is in the reaction-controlled regime with small values of reaction probability, prxn ∼10−5. Our analysis suggests that crowding effects increase the velocity of membrane protrusion by a factor of 4–6 under typical physiological conditions.
The rest of this article is organized as follows: The model and simulations are described in the next section. Simulation results are then discussed, and summary and conclusions are presented at the end.
Model and Methods
The system consists of NL ligands and Nc crowding agents confined between hard surfaces. On one of the surfaces is a receptor, modeled as a square reactive patch. The size of the reactive patch is much smaller than that of the entire surface and the rest of the surface is inert to the reaction. Any two particles of the ligands or crowding agents interact via a smooth repulsive interaction potential given by
| (1) |
where ɛ is the Lennard-Jones well-depth and σ is the sphere diameter; σ is used as the unit of length in this work. We set ɛ = kBT where kB is the Boltzmann constant and T is the temperature. The reactive patch has a side length of b, which varies from 0.25σ and 4σ, although most results are presented for b = σ. A schematic presentation of the simulation box is given in Fig. 1. The dimension of simulation box is Lx × Ly × Lz = 12σ × 12σ × 16σ. In all simulations, NL = 9, which corresponds to a ligand concentration of 100 μM for σ = 4 nm. The volume fraction of crowding agents, ϕ = Ncπσ3/6LxLyLz, is varied between 0 and 0.4 by changing the number of crowding agents, Nc, from 0 at ϕ = 0 to 1751 at ϕ = 0.4. Although we limit our study to a specific concentration of ligands to focus on the effect of macromolecular crowding, the effect of ligand concentration has been studied by Brownian dynamics simulations and the generalized Smoluchowski equation including the osmotic pressure and the distance-dependent diffusion coefficient (18,19).
Figure 1.
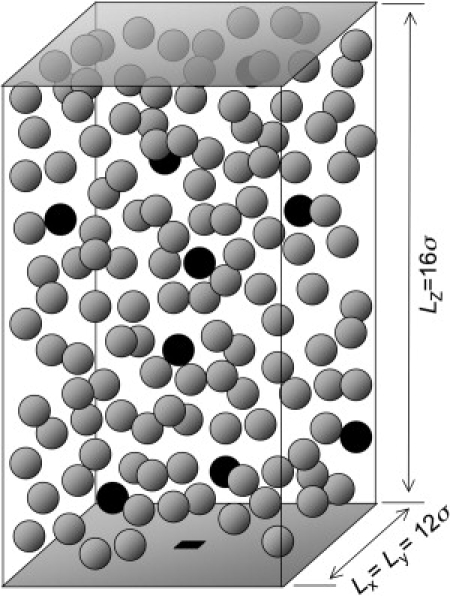
Simulation box with dimension of Lx × Ly × Lz = 12σ × 12σ × 16σ. A receptor modeled as a square reactive patch and ligands as spherical particles are colored in black. The rest of spherical particles colored in gray are crowding agents.
The system is evolved using conventional Brownian dynamics (BD) (20) without hydrodynamic interactions. At each time step Δt, the position, ri(t) of particle i is updated via
| (2) |
where Fi(t) is the total force acting on the bead i and Ri(Δt) is a random displacement with a Gaussian distribution function with zero mean and variance-covariance 〈Ri(Δt)Rj (Δt)〉 = 2D0Δtδij. D0 is the diffusion coefficient of the spheres in pure solvent. D0 sets the timescale and τBD = σ2/D0 is used as the unit of time in this work. A time step Δt = 10−4 τBD is used for all the simulations. This ensures that a particle does not move more than its radius in one time step whereas the number of time steps for the calculation of the rate constant is minimized. When the time steps Δt = 5 × 10−5 τBD and Δt = 10−5 τBD are used, the differences in the diffusion coefficients calculated in bulk solution are negligible, implying that the time step employed in this work is small enough. The total force Fi(t) is given by the gradient of U(r) where U(r) is the repulsive interactions between particles given in Eq. 1. For this study, the specific values of simulation parameters are considered: σ = 4 nm, D0 = 71.5 μm2/s, and then τBD = 0.22 μs.
The only difference from the standard BD algorithm is the treatment of the surfaces. If a sphere goes beyond the surface in a given time-step, the time-step is reduced by the amount required for that sphere to barely touch the surface. The velocity of the colliding sphere is then reset to mimic an elastic collision between the sphere and the surface. The coordinates on the surface at the moment of collision are recorded to determine whether the ligand collides with a receptor.
The reaction is described by L + R → LR where L, R, and LR stand for the ligand, the receptor, and their complex, respectively. The reaction rate constant is obtained as described in the previous work (7). When a ligand L collides with a reactive patch R, the reaction is assumed to occur with a reaction probability on collision, prxn. The ligands are not uniformly reactive spherical particles and their collisions with the surface reactive patch do not always lead to the reaction. The reaction probability on collision prxn is introduced to mimic the effect of such orientation-dependent anisotropic reactivity, and the effect of crowding is investigated at a range of prxn between 0.001 and 1.
The quantity directly obtained from the simulation trajectories is the survival probability, SR(t), which is the probability that the receptor R remains unreacted after time t. The time-dependent reaction rate constant k(t) is related to the survival probability via (7,11,18,19,21,22),
| (3) |
or
| (4) |
where CL is the concentration of ligands L which, in this work, is assumed to be constant. We focus on the long time-limit of the time-dependent rate constant, i.e., k∞ ≡ k(t → ∞), which we obtain from a linear least-squares fit of ln SR(t) at long times; the slope of this curve divided by CL gives k∞. In this work, ln SR(t) is fitted to Eq. 4 in the time intervals during which SR(t) is between 0.001 and 0.10 for statistically reliable data.
The survival probability is calculated using the method similar to the one suggested by Dong et al. (21) and Zhou and Szabo (22), which has also been adopted in a recent article (7). For each volume fraction a number, Ntrj, of independent trajectories are obtained. The surface on the bottom is divided into Npatch identical reactive patches, each of which is labeled R in turn and, for every choice of R, the spherical particles are grouped into Ngrp groups with NL number of L particles. Each group of L particles participates in the reaction independently with the reactive patch R, and hence in each trajectory there are 2NpatchNgrp reactive sets of ligands and a receptor, where a factor of 2 is included as either the top or the bottom surface can be considered to contain the receptor. The positions of ligands in each reactive set are kept track of until any ligand in this reactive set associate with the receptor. Each trajectory is stopped once all the reactive sets are consumed. Because a reaction is considered to occur only once for each reactive set, the concentration of ligands is always constant CL = 100 μM in each reactive set and the competition between receptors on the membrane is not taken into account. In total, there are 2NpatchNgrpNtrj configurations participating in the reactive events.
This greatly amplifies the volume of data collected. For example, for the total volume fraction of 0.10 and b = σ, there is a total of 288 reactive patches on the top and the bottom surfaces which can be labeled as a reactant R in turn, resulting in the 288 different configurations for the reaction. Once a patch is labeled as R, 440 particles are divided to 48 groups of nine ligands, only one group of which is considered to be reactive in turn. One single trajectory gives 13,824 (= 288 × 48) reactive events, and with only 39 trajectories a total of 539,136 reactive events is recorded. In the simulations reported, the number of independent trajectories Ntrj is adjusted such that the number of reactive events is >500,000.
Results and Discussion
The reaction rate constant k∞
The rate constant, k∞, is a nonmonotonic function of the volume fraction of crowding agents for large values of prxn but becomes a monotonically increasing function of ϕ for small values of prxn. This can be seen in Fig. 2 A, which depicts k∞ as a function of ϕ for various values of prxn. This implies that the ligand binding to the receptor on the membrane can be either accelerated or decelerated by crowding depending on the value of prxn. Note that even at prxn = 1 the rate constant is an increasing function of ϕ at small volume fractions. This is in contrast to the effect of crowding on the reaction between spherical particles, where for prxn = 1 crowding always decreased the rate of reaction.
Figure 2.
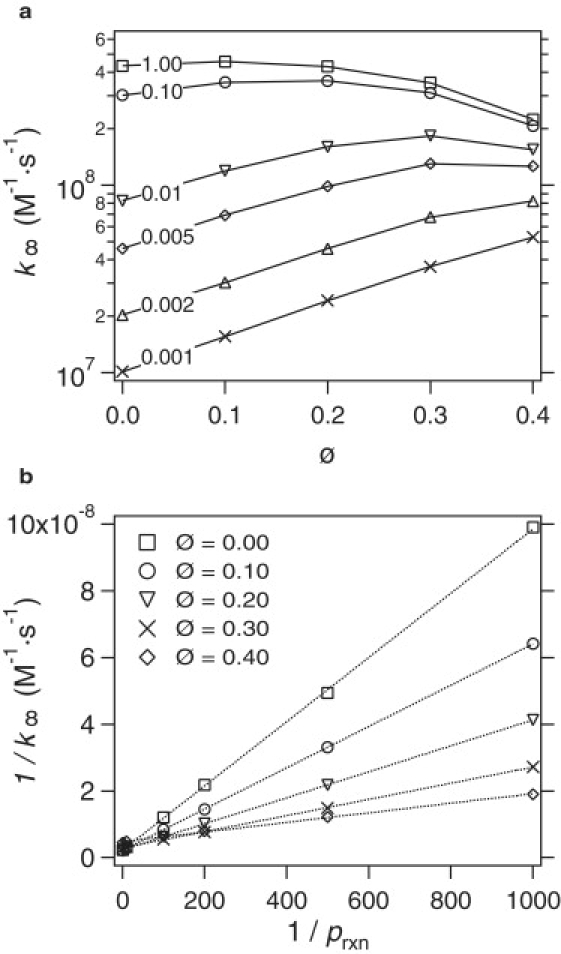
Simulations results for the rate constant (k∞). (a) The absolute rate constant (in units of M−1 s−1) as a function of total volume fraction for various values of the reaction probability, prxn. Numbers on each plot are the reaction probabilities prxn. (b) 1/k∞ as a function of 1/prxn for different volume fractions of crowding agents. Symbols are simulation results and the line is a linear fit to Eq. 6.
By analogy with the solution of the Smoluchowski theory for the spherically symmetric system we suggest that the rate constant can be decomposed into two terms based on the rate constant for an encounter, kD, and the rate constant for reaction, kR, once an encounter has occurred. This results in a simple Ansatz
| (5) |
which can be written as
| (6) |
where k′R is the rate constant of reaction between a encounter pair of a uniformly reactive spherical particle and a reactive patch on the surface.
The simulation results are consistent with the Ansatz in Eq. 6. Fig. 2 B shows that 1/k∞ is proportional to 1/prxn for all values of ϕ and by fitting the data to a straight line we obtain kD and k′R at each ϕ.
The encounter rate constant kD
kD is the diffusion-controlled rate of the first contact between ligands and a square reactive patch on the surface, and in the absence of crowding agents the simulation results for kD are in good agreement with theory. For the diffusion-controlled reaction of point particles with a circular patch on an infinite reflecting plane, kD = 4Da, where D is the diffusion coefficient of ligands and a is the radius of the circular receptor on the membrane (23,24). If we assume the expression is valid for the square patch with the same area as the circle, i.e., b2 = πa2, . For prxn = 1.00, M−1 s−1, which is in good agreement (within 20%) with the simulation result of kD = 4.71 × 108b/σ M−1 s−1, where we have used D = 71.5 μm2/s and σ = 4 nm.
One expects crowding to monotonically decrease kD but this only happens when the size of the patch is large. Fig. 3 depicts as a function of ϕ for various values of b. For large patches, kD decreases as ϕ is increased. For smaller patches, i.e., b ≤ 2σ, kD is a nonmonotonic function of ϕ. This is a surprising result because one expects crowding to significantly slow down diffusion. The ratio of the diffusion coefficient in the crowded conditions to that with no crowding, D/D0, measured from simulations in bulk solution is also presented in Fig. 3. Although the theory predicts the reduced kD in accord with the reduced diffusion D, even for the largest patch the theory significantly underestimates kD.
Figure 3.
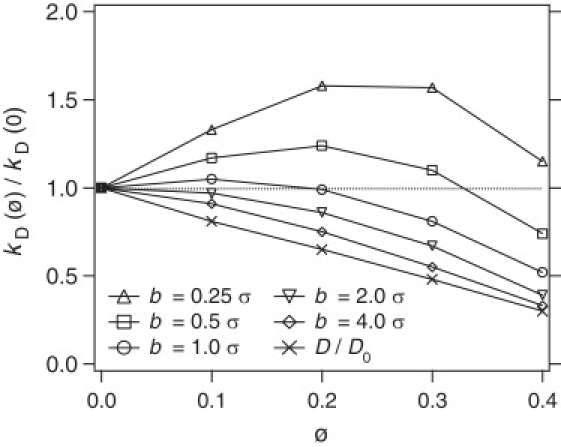
The encounter rate constant kD(ϕ), divided by the value in the absence of crowding kD(0), as a function of crowding volume fraction, for various values of b. D/D0 is the ratio of the diffusion coefficient in the crowded conditions to that with no crowding.
The nonmonotonic behavior of kD can be understood by noting that the diffusive contact of ligands with a small reactive patch on the large surface is determined by two diffusive processes of importance: the diffusion of ligands to the surface and, when they hit inert surface other than the reactive patch, the diffusive search along the surface. In the absence of crowding agents, once a ligand reaches the surface other than the reactive patch, the reflecting boundary condition causes it to diffuse away and the latter diffusive process is of little significance. In the presence of crowding agents, however, the high density of crowding agents near the surface confines the ligand to the vicinity of the surface for a period of time to facilitate the diffusive search along the surface. For small patches, the probability that a ligand finds the reactive patch on the surface upon first approach is low, but the presence of crowding agents confines the ligand to the vicinity of the surface (12) and therefore increases the probability that the ligand can find the reactive patch by diffusing parallel to the surface after hitting the inert surface (13). Crowding therefore has two effects on kD: The initial approach of the ligands to the surface is hindered by crowding, but the diffusive search along the surface is facilitated by crowding. The resulting kD is a balance between these two effects and consequently kD can be a nonmonotonic function of ϕ.
The above picture can be quantified by considering the number of surface sites visited by a ligand when it is in the vicinity of the surface. We define the vicinity of the surface by the region between the surface and the second peak in the density profile of particles (see inset of Fig. 4). We divide the surface into a grid with spacing 0.25σ and count the number of the square sites visited by the ligand before it leaves the vicinity of the surface, i.e., goes a distance beyond z∗. Fig. 4 depicts the probability distribution function of the number of sites visited for ϕ = 0 and 0.3 (the second peak at ϕ = 0.3 is chosen to define the vicinity of the surface for both cases). For ϕ = 0, the distribution is sharply peaked and goes rapidly to zero before 25 sites are visited, but for ϕ = 0.3 the distribution function has a long tail and the ligand often visits more than 80 sites before it leaves the vicinity of the surface. The presence of crowding therefore greatly increases the probability of finding a reactive patch after an initial collision with the inert surface. This more than compensates for the decrease in D with increasing ϕ.
Figure 4.
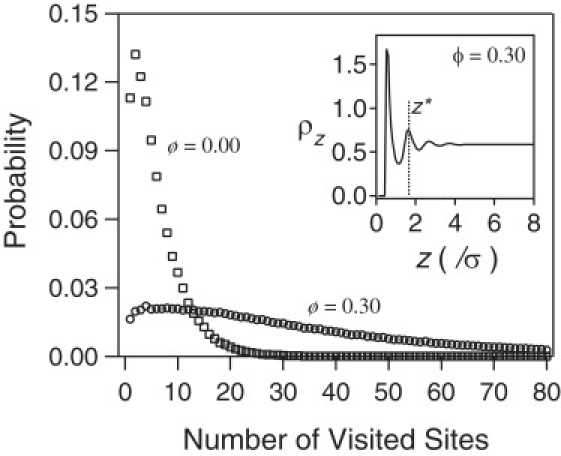
Probability distribution of the number of surface sites visited by a single ligand after the first collision with the wall and when it is within a distance z∗ from the wall. (Inset) Density profile of particles for ϕ = 0.30; z∗ is the position of the second peak in the density profile.
This nonmonotonic dependence of kD for small reactive patches is not observed for the protein association reaction in the bulk solution studied in the previous work (7). It is because the diffusive contact of proteins is determined only by the mutual approach by diffusion and no other diffusive process near the protein surface is involved, due to the isotropic reactivity on the protein surface assumed in our model.
The intrinsic rate constant, kR
The simulations show that crowding always increases the intrinsic reaction rate constant, kR. Fig. 5 depicts the ratio kR(ϕ)/kR(0) as a function of ϕ for b = σ. According to Minton's formulation (5), kR = Γ where is the limiting value of kR in the absence of crowding and Γ is the nonideality factor given by Γ = γLγR/γLR, where γX is the activity coefficient of the species X. Crowding always increases Γ because the configuration with the ligand at the surface is more favorable than the ligand in the bulk.
Figure 5.
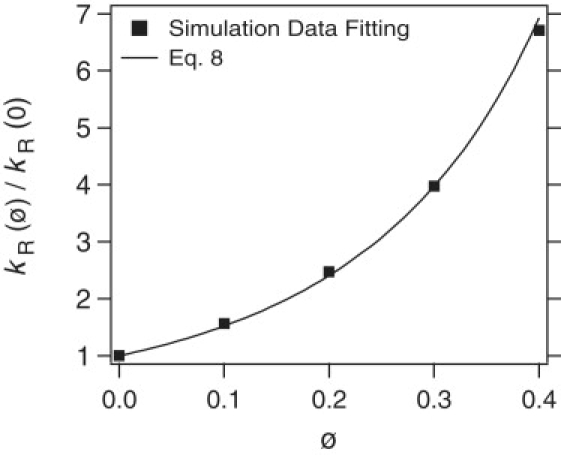
Comparison of simulation results (squares) for the ratio of kR to that at ϕ = 0 to theoretical predictions (line) from Eq. 8 for b = σ.
We can obtain an analytical expression for Γ for hard sphere crowding agents. Because the receptor R is a reactive patch on the surface, it is not affected by crowding and therefore γR = 1. For the ligand and the complex, the activity coefficient can be obtained from the excess chemical potential, μexX, via μexX = kBT ln γX. For hard sphere ligands, μexX = −kBT ln PX, where PX is the probability of inserting species X, i.e., PL is the probability of inserting a ligand sphere in bulk solution and PLR is the probability of inserting a ligand sphere at the surface. This gives Γ = γL/γLR = PLR/PL, and therefore
| (7) |
If we assume that the probability of inserting a component is proportional to the density of that component at that position, then PLR/PL ≈ ρw/ρb, where ρw and ρb are the densities at the surface wall and in the bulk, respectively. For hard spheres, the wall sum rule states that kBTρw = p, where p is the pressure (25). For a fluid at a single surface, such that there is a bulklike region far away, p is the bulk pressure. We assume that the surfaces are far enough apart that the fluid in the middle is bulklike, and use the bulk hard sphere Carnahan-Starling equation of state (26) for the pressure. The final result is
| (8) |
The crowding dependence of kR predicted from Eq. 8 is in excellent agreement with simulation data, as seen in Fig. 5. This implies that the increase of kR is caused by the increased density of ligands near the membrane due to the presence of crowding agents. In many applications prxn is quite small, and the effect of crowding on the reaction rates come primarily from kR. In this event, Eq. 8 provides a simple analytic form to estimate the increase in reaction rates from crowding effects.
The increase of kR with increasing ϕ can also be understood by considering the recollision probability α(t), which is the probability that a ligand-receptor pair recollide at least once in a time duration t given that they collided but did not react at time t = 0. The presence of crowding agents makes an escape of the ligand less likely, and therefore increases the recollision probability. The increased probability is depicted in Fig. 6. The difference does not seem large but it makes a huge difference when a great number of collisions are involved for the association in case of low value of prxn. It is very interesting that the same effect that facilitates the diffusive search along the surface when the ligands collided with the inert surface (increasing kD) also increases the frequency of recollision when the ligands collided with the reactive patch, but failed to react (increasing kR).
Figure 6.
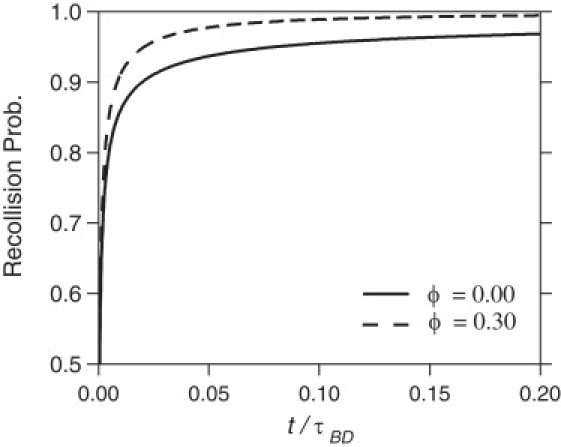
Recollision probability, α(t), as a function of time for volume fractions ϕ = 0 and 0.30.
Application to the membrane protrusion driven by actin filament elongation
We use our model to study the velocity of membrane protrusion driven by actin filament elongation. It has been widely accepted that the membrane protrusion occurs (see Fig. 7) via the intercalation of actin monomers into a gap between the membrane and the actin filament due to the undulations of both the membrane and the filament (14–17). Once a monomer enters the gap, an association reaction occurs with a probability related to its anisotropic reactivity. In our model, the intercalation of actin monomers is modeled as an association of actin monomers with a reactive patch on the surface below which a filament end is assumed to be located. The probability of a gap opening due to thermal undulations of the membrane and the filaments is incorporated in the calculation of the rate constant in terms of the reaction probability, i.e., the reaction probability prxn is intended to mimic the effect of a gap opening as well as the anisotropic reactivity between the actin monomer and the filament end.
Figure 7.
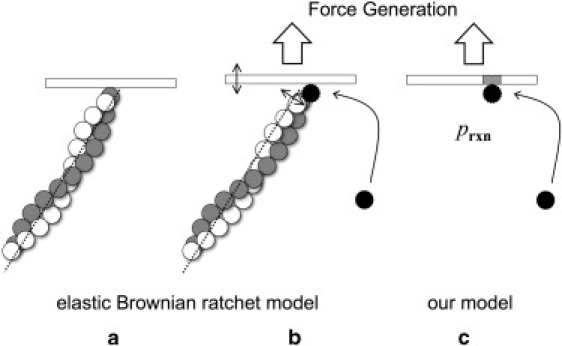
Model of membrane protrusion by actin polymerization. The elastic Brownian ratchet model is depicted in panels a and b where one end of actin filaments buttresses the membrane, fluctuations open a gap between the membrane and the growing filament, and an actin monomer intercalates into the gap to create a protrusive force. In our model, the intercalation of actin monomers into a gap is considered as an association of actin monomers with a reactive patch on the surface, with the gap opening probability and anisotropic reactivity taken into account via the reaction probability prxn.
We estimate the reaction probability to be very small, prxn ∼10−5, which makes membrane protrusion a reaction-controlled process. The reaction probability prxn can be divided into two contributions arising from the probability, pgap, of gap opening, and the probability, panr, of anisotropic reactivity. The value pgap can be estimated from the force, f, by the membrane and the distance, δ; the membrane advances due to the addition of a monomer: pgap ∼ (14–17). With δ = 2.2 nm, kBT = 4.1 pN · nm, and f ≈ 2 – 10 pN (16), we get pgap ≈ 5 × 10−3 ∼0.3. On the other hand, panr can be estimated from the rate constant measured from experiments in solution. Experimentally, the ratio of the observed rate constant to the estimated rate constant in the diffusion-controlled limit (kexp/kD) is 0.02 (27), and it corresponds to k∞/kD in our previous simulation results when panr = 0.001 in the absence of crowding agents (7). We therefore estimate that prxn = pgap · panr ≈ 5 × 10−6 − 3 × 10−4, i.e., roughly prxn ∼10−5. This is a very small reaction probability which suggests that the membrane protrusion process cannot be diffusion-controlled.
We estimate the velocity of membrane protrusion from
| (9) |
where CL is the concentration of monomers, and k∞ is determined from the estimated value of prxn. In the above, k∞ takes into account not only the monomer association rate but also the effect of the opposing force by the membrane () in terms of prxn. In principle one could calculate k∞ from simulations, but for low values of prxn this is computationally very intensive. Instead we use Eq. 6 to deduce the long-time rate constant, with kD and k′R obtained from simulations at higher values of prxn ≥ 0.001. This is a very accurate approximation over the range of prxn for which we have simulation results, and is expected to become more accurate for lower values of prxn, where k∞ ≈ kR = k′Rprxn.
The velocity of membrane protrusion, V, is presented in Fig. 8 for various choices of the parameter prxn. In the absence of crowding, the values of V are consistent with the predictions of the elastic Brownian ratchet model (16). Crowding increases the velocity of membrane protrusion by a factor of ∼4 (ϕ = 0.3) to ∼6 (ϕ = 0.4). This acceleration of the membrane protrusion arises from an increase in kR, which in turn originates from the increased local concentration of monomers at the surface.
Figure 8.

The velocity of membrane protrusion predicted based on Eq. 9 for prxn = 0.001, 0.0005, 0.0002, and 0.0001. Numbers on each plot are the reaction probabilities prxn.
Conclusions
We study the effect of macromolecular crowding on the ligand binding to a receptor on a membrane using Brownian dynamics simulations, and apply the results to the study of membrane protrusion by actin polymerization. Ligands and crowding agents are all modeled as spherical particles of the same diameter, and a receptor is modeled as a square reactive patch on the membrane surface. When a ligand encounters a receptor, via a collision, they react with a probability prxn, which mimics the fact that the ligand and the receptor must orient properly for a reaction to occur.
The qualitative impact of crowding agents on the association rate constant depends strongly on the value of prxn. For prxn ∼1, the rate constant of the ligand-receptor association k∞ depends largely on the rate of encounter between ligands and a receptor kD, whereas for smaller prxn the association rate constant is determined by the rate of reaction between a encounter pair of a ligand and a receptor kR. Consequently the ligand-receptor association can be either accelerated or decelerated by crowding.
The most interesting result compared with the previous study of the crowding effect on protein association reactions in solution is that the encounter rate constant kD is a nonmonotonic function of the volume fraction of crowding agents. For protein association reactions in solution, kD always decreases with increasing crowding. The nonmonotonic behavior is only observed for small sizes of receptor; for large receptors the association rate constant decreases monotonically with crowding as in solution. The rate constant of reaction between a encounter pair kR increases with crowding due to the increased probability of finding ligands near the membrane. Simulation data are in a good agreement with a theoretical prediction based on the density at a membrane. We predict that for low values of prxn, k∞ can be accelerated at most by a factor of 6.9 even for a crowding agent volume fraction of 0.4.
When the model is applied to an example of membrane protrusion driven by actin polymerization, it is found that the crowding accelerates the membrane protrusion by increasing the local concentration of actin monomers in the vicinity of the actin filament ends. Although based on a simplified model ignoring the complications of more recent models (28,29), this study suggests that crowding can be an important factor in accelerating the association of actin monomers in membrane protrusion.
It is important to note, however, that this work focuses on excluded volume effects on the reaction rates. Other interactions, such as van der Waals and electrostatic interactions, between ligands and a receptor on the membrane, could play an important role in realistic situations. In addition, the nonspecific interactions between ligands and the membrane could also affect the reaction kinetics by increasing the probability of the ligands around the receptor (30). Among other considerations of interest in realistic situations is the size effect of crowding agents. For smaller crowding agents than the ligands, the depletion effect may enhance the association of ligands with the receptor, whereas the recollision frequency of ligands with the surface due to larger crowding agents may not be as great as that due to crowding agents of the same size. These represent possible future directions of this work.
Acknowledgments
This article is based upon work supported by the National Science Foundation under grant No. CHE-0717569.
References
- 1.Zimmerman S.B., Minton A.P. Macromolecular crowding: biochemical, biophysical, and physiological consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 2.Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- 3.Ellis R.J., Minton A.P. Cell biology: join the crowd. Nature. 2003;425:27–28. doi: 10.1038/425027a. [DOI] [PubMed] [Google Scholar]
- 4.Rafelski S.M., Theriot J.A. Crawling toward a unified model of cell mobility: spatial and temporal regulation of actin dynamics. Annu. Rev. Biochem. 2004;73:209–239. doi: 10.1146/annurev.biochem.73.011303.073844. [DOI] [PubMed] [Google Scholar]
- 5.Minton A.P. Molecular crowding: analysis of effects of high concentrations of inert cosolutes on biochemical equilibria and rates in terms of volume exclusion. Methods Enzymol. 1998;295:127–149. doi: 10.1016/s0076-6879(98)95038-8. [DOI] [PubMed] [Google Scholar]
- 6.Wieczorek G., Zielenkiewicz P. Influence of macromolecular crowding on protein-protein association rates—a Brownian dynamics study. Biophys. J. 2008;95:5030–5036. doi: 10.1529/biophysj.108.136291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J.S., Yethiraj A. Effect of macromolecular crowding on reaction rates: a computational and theoretical study. Biophys. J. 2009;96:1333–1340. doi: 10.1016/j.bpj.2008.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pollard T.D., Blanchoin L., Mullins R.D. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- 9.Pollard T.D., Borisy G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 10.Rice S.A. Diffusion-Limited Reactions. Elsevier; Amsterdam, The Netherlands: 1985. Comprehensive chemical kinetics. [Google Scholar]
- 11.Szabo A. Theory of diffusion-influenced fluorescence quenching. J. Phys. Chem. 1989;93:6929. [Google Scholar]
- 12.Snook I.K., Henderson D. Monte Carlo study of a hard-sphere fluid near a hard wall. J. Chem. Phys. 1978;68:2134–2139. [Google Scholar]
- 13.Berg O.G., von Hippel P.H. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 14.Peskin C.S., Odell G.M., Oster G.F. Cellular motions and thermal fluctuations: the Brownian ratchet. Biophys. J. 1993;65:316–324. doi: 10.1016/S0006-3495(93)81035-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mogilner A., Oster G. Cell motility driven by actin polymerization. Biophys. J. 1996;71:3030–3045. doi: 10.1016/S0006-3495(96)79496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mogilner A., Edelstein-Keshet L. Regulation of actin dynamics in rapidly moving cells: a quantitative analysis. Biophys. J. 2002;83:1237–1258. doi: 10.1016/S0006-3495(02)73897-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mogilner A. Mathematics of cell motility: have we got its number? J. Math. Biol. 2009;58:105–134. doi: 10.1007/s00285-008-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senapati S., Wong C.F., McCammon J.A. Finite concentration effects on diffusion-controlled reactions. J. Chem. Phys. 2004;121:7896–7900. doi: 10.1063/1.1795132. [DOI] [PubMed] [Google Scholar]
- 19.Dzubiella J., McCammon J.A. Substrate concentration dependence of the diffusion-controlled steady-state rate constant. J. Chem. Phys. 2005;122:184902. doi: 10.1063/1.1887165. [DOI] [PubMed] [Google Scholar]
- 20.Ermak D.L., McCammon J.A. Brownian dynamics with hydrodynamic interactions. J. Chem. Phys. 1978;69:1352–1360. [Google Scholar]
- 21.Dong W., Baros F., Andre J.C. Diffusion-controlled reactions. I. Molecular dynamics simulation of a noncontinuum model. J. Chem. Phys. 1989;91:4643–4650. [Google Scholar]
- 22.Zhou H.X., Szabo A. Comparison between molecular dynamics simulations and the Smoluchowski theory of reactions in a hard-sphere liquid. J. Chem. Phys. 1991;95:5948–5952. [Google Scholar]
- 23.Hill T.L. Effect of rotation on the diffusion-controlled rate of ligand-protein association. Proc. Natl. Acad. Sci. USA. 1975;72:4918–4922. doi: 10.1073/pnas.72.12.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shoup D., Lipari G., Szabo A. Diffusion-controlled bimolecular reaction rates. The effect of rotational diffusion and orientation constraints. Biophys. J. 1981;36:697–714. doi: 10.1016/S0006-3495(81)84759-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rickayzen G., Augousti A. Integral equations and the pressure at the liquid-solid interface. Mol. Phys. 1984;52:1355–1366. [Google Scholar]
- 26.Carnahan N.F., Starling K.E. Equation of state for nonattracting rigid spheres. J. Chem. Phys. 1969;51:635–636. [Google Scholar]
- 27.Drenckhahn D., Pollard T.D. Elongation of actin filaments is a diffusion-limited reaction at the barbed end and is accelerated by inert macromolecules. J. Biol. Chem. 1986;261:12754–12758. [PubMed] [Google Scholar]
- 28.Dickinson R.B., Caro L., Purich D.L. Force generation by cytoskeletal filament end-tracking proteins. Biophys. J. 2004;87:2838–2854. doi: 10.1529/biophysj.104.045211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vavylonis D., Kovar D.R., Pollard T.D. Model of formin-associated actin filament elongation. Mol. Cell. 2006;21:455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou H.X., Szabo A. Enhancement of association rates by nonspecific binding to DNA and cell membranes. Phys. Rev. Lett. 2004;93:178101. doi: 10.1103/PhysRevLett.93.178101. [DOI] [PubMed] [Google Scholar]


