Abstract
Over the last thirteen years, the field of optical imaging has expanded from in vitro fluorescence microscopy of cells to in vivo imaging of living animals. Recent advances in optical imaging of bacterial infection have been propelled by the invention of genetic methods that produce fluorescent and bioluminescent bacteria, and also the discovery of synthetic fluorescent probes that selectively target bacterial cell surfaces. Optical imaging is an effective method of conducting longitudinal studies of bacterial infection in small animals such as nude mice. It can be used to address questions in medical microbiology concerning migration and colonization and it is an attractive method for determining the efficacy of antibiotic therapies.
Introduction
In contrast to conventional imaging methods (X-ray, MRI, ultrasound), which display differences in anatomical features or physicochemical responses, molecular imaging employs a molecular probe that emits a signal only from the site of probe localization or activation. Recent advances in molecular imaging provide new opportunities for the study of disease models, especially in the preclinical phase of the drug discovery process where imaging can quickly determine efficacy endpoints in living subjects and readily allow longitudinal studies [1]. An important point with the latter application is the expectation that molecular imaging will lead to a decrease in the number of laboratory animals that must be sacrificed [2]. Typically, in vivo imaging employs “smart” molecular probes that can selectively target specific types of cells and report their location. Alternatively, the target cells are genetically modified to express products that act as signaling beacons. Until recently, in vivo imaging of disease and disease models has been dominated by radioactive probes, and to a lesser extent, MRI contrast agents. In comparison to these relatively mature imaging modalities, in vivo optical imaging is much less developed, but it has several attractive features that make it a promising technology for laboratories that study disease models. Compared to MRI and nuclear imaging, optical imaging is relatively cheap, safe, and operationally simple. Furthermore, the time-frame for signal acquisition is quite short such that optical imaging can be used to collect kinetic data on dynamic biochemical processes such as enzyme action and related gene expression.
Optical imaging detects emitted light, in the form of bioluminescence or fluorescence, and creates a contrast image that locates a targeted molecule, cell, or tissue. A growing number of vendors sell in vivo whole-body optical imaging stations for small animals at prices that start around US$65,000. The specific focus of this article is optical imaging of bacterial infection. Although relatively new, bacterial imaging is a rapidly improving technology, in part because it is increasingly easy to genetically modify pathogenic bacteria so that they are bioluminescent or fluorescent. Bioluminescent bacteria are engineered to express luciferase enzymes that catalyze the light-generating oxidation of substrates such as luciferin in the presence of oxygen and ATP [3]. The luminescence is detected using a charge coupled device (CCD) and the signal intensity is proportional to the number of viable bacterial cells. A major advantage with bioluminescent bacteria is the inherently low background signal that is emitted by the host animal. However, the emission spectrum is generally quite broad and only a small percentage extends beyond 600 nm, which means that tissue penetration is limited (perhaps 2-3 cm) [4,5]. A related problem is that light scattering is wavelength dependent, and signal intensity changes quickly with depth of sample (usually there is a ten-fold loss of signal intensity with each centimeter of tissue depth), which means that it is quite challenging to make accurate quantitative comparisons between different imaging sites.
In addition to bioluminescence, bacteria can also be engineered to express fluorescent proteins. Many different mutants of green fluorescent protein (GFP) have been developed via directed evolution, and there are now examples that emit at all colors in the visible spectrum including red [6,7]. Since the fluorescent bacteria have to be irradiated, there are several factors that can diminish signal sensitivity such as undesired absorption of light with wavelengths below 600 nm by other biomolecules in the sample, increased background signal due to autofluorescence, and non-optimal quantum yields.
A current limitation with optical imaging using genetically modified bacteria is restricted tissue penetration of the light. Imaging probes that emit near-IR radiation, with wavelengths in the region of 650-900 nm, have much greater penetration depths; however, there are presently no optimal genetic reporter groups that emit with appreciable brightness in the near-IR. An alternative approach is to develop exogenous synthetic probes that are highly stable and have extremely bright near-IR fluorescence. The development of synthetic fluorescent probes is advancing rapidly with the discovery of new and improved organic dyes and luminescent nanoparticles. Hybrid systems are also being reported such as fusions of bioluminescent proteins with inorganic nanoparticles like quantum dots [8]. The following section provides a concise review of recent efforts to image in vitro and in vivo models of bacterial infection. Most of these studies have been conducted for one of two purposes. One major goal is to explore questions in medical microbiology concerning the migration and colonization properties of different bacterial strains in living animals. The alternative goal is to develop rapid and reliable methods of determining the efficacy of candidate antibiotic therapies.
Imaging In Vitro Models of Bacterial Infection
The growth of bacteria in culture is considered the most straightforward in vitro model of infection. Measurements of optical density are often used to assess antibiotic efficacy and pharmacokinetics prior to testing in mouse models. Bacteria expressing bioluminescent [9,10], or fluorescent [11,12] genetic reporters have been incorporated in these basic in vitro systems as a way of enhancing the sensitivity of the toxicity assays. More sophisticated in vitro models of infection employ a mixed culture of mammalian and bacteria cells. For example, Garcia-Medina and co-workers incubated Pseudomonas aeruginosa expressing GFP with primary murine epithelial cells [13]. They used fluorescence microscopy to demonstrate bacterial invasion and biofilm formation within the epithelial cells. As these cells invaded healthy tissue, they formed pod-like structures that were insensitive to antibiotic treatment. Similar studies have been performed to demonstrate liver uptake of GFP expressing Salmonella enterica [14]. In this case, the liver cells displayed specific antigens on their surface to facilitate uptake of the bacteria, followed by proinflammatory cytokine production. It is apparent that in vitro models of infection are useful as simple early screens for identifying antibiotic candidates and they may also provide tractable methods for studying the infection process under controlled conditions. Nevertheless, a more relevant picture of bacterial pathogenesis, as well as a more realistic assessment of antibiotic efficacy, is gained from in vivo infection models.
Imaging In Vivo Models of Bacterial Infection
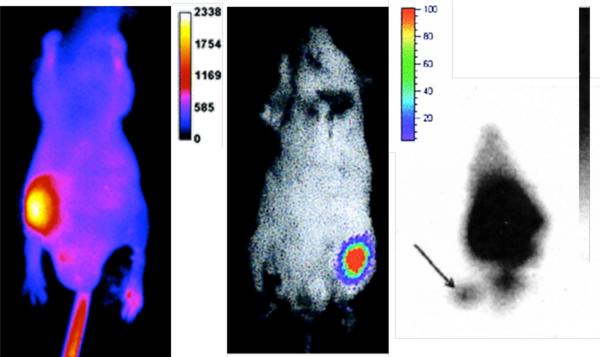
The most basic and commonly used murine infection model utilizes a rear thigh muscle for bacterial injection. The rear flank is clearly separated from all major organs, and bacterial migration away from the thigh is usually very slow, which reduces any confusion as to the origin of the light signal. Furthermore, the uninfected contralateral thigh provides a convenient control with which to compare signal intensity. The thigh infections are often employed as the starting point to validate a new imaging method. Shown in Figure 1 are the results from three different studies of mice with Staphylococcus aureus thigh infections. The left frame shows the infection, as detected using a synthetic near-IR fluorescent probe (ex. 794 nm, em. 810 nm) with affinity for the anionic surfaces that are ubiquitously present on all bacterial cells [15]. After intravenous injection of the probe, the site of infection was clearly marked by a fluorescence signal that was 4-fold greater than the rest of the body. E. coli infections were also readily detected using this probe. The center frame of Figure 1 shows the light output from a bioluminescent strain of S. aureus [16,17]. The site of infection emits blue/green light (em. 490 nm) that could be observed for days, although the light emission faded away after antibiotic treatment. Repeating the experiments with a bioluminescent E. coli strain gave similar results [18]. As a comparison to nuclear imaging, the right frame of Figure 1 shows an image of a S. aureus thigh infection targeted with radiolabeled antibodies (see arrow) and imaged with a gamma ray detector [19]. Although the infection site can be identified, the signal for target thigh compared to non-target is relatively poor. While radioimaging is presently superior for deep tissue imaging, optical methods appear to be sufficient for detecting S. aureus infection in the thigh of a nude mouse.
Figure 1.
S. aureus thigh infections as detected using fluorescence (left), bioluminescence (middle) and radioimaging (right). Images reprinted with permission from the Journal of the American Chemical Society (left), American Society for Microbiology (center), and Journal of Nuclear Medicine (right).
The development of a new in vivo infection model, beyond the thigh model, is often as straightforward as injecting bacteria in a new body part or organ of the mouse. For example, GFP expressing E. coli were administered orally to mice, and their progress through the gastrointestinal tract was monitored by fluorescence [20]. In another study, the lungs of a mouse were infected with bioluminescent Streptococcus pneumonia by introducing the cells through the nose into the airway (Figure 2, center) [21,22]. When these same cells are introduced via an intracranial injection, they infect the brain and spine (Figure 2, right) [23,24]. In both of these cases, antibiotic treatment mitigated light output from the invading cells. A urinary tract infection model was developed using bioluminescent Pseudomonas aeruginosa injected into the bladder via the urethra (Figure 2, left) [25]. In this example, researchers were able to follow the infection to the kidneys and monitor antibiotic treatment efficacy by measuring light output. When bacterial cells self-aggregate into tight clusters, they form biofilms which can shield the encapsulated bacteria from antibiotic action. Researchers at Caliper Life Sciences Corporation (formerly Xenogen Corporation) have shown that optical imaging is an effective way to study the protective properties of biofilms in vivo. For example, bioluminescent strains of S. aureus and P. aeruginosa were used to form biofilms on catheters, which were then implanted subcutaneously in living mice. For each of these biofilms, the light signal from the bacteria was monitored for days, and upon drug treatment. In one study, with bioluminescent S. aureus biofilms, the antibiotics tobramycin and ciprofloxacin were found to be ineffective, but rifampin treatment produced a dose-dependent decline in bioluminescence which correlated with decreased colony counts [26]. After treatment, cell numbers rebounded, signaling the formation of an antibiotic resistance sub-population of bacteria. A second imaging study monitored urinary tract infection with bioluminescent Pseudomonas aeruginosa and Proteus mirabilis and could follow the entire disease process, including ascending urinary tract infection, treatment efficacy, and population rebound [27].
Figure 2.
Infections of the bladder (left), lungs (middle) and brain/spine (right) by bioluminescent bacteria. Images reprinted with permission from the American Society of Microbiolgy.
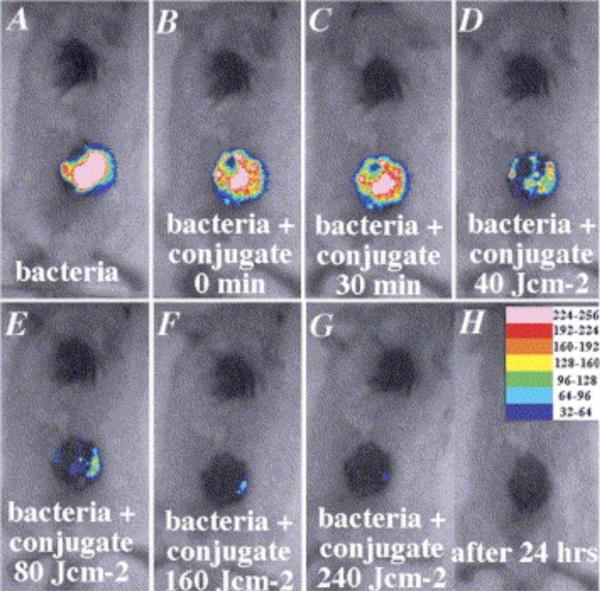
In vivo imaging with bioluminescent bacteria is an effective way of determining the efficacy of light-based therapies for open wound infections. This disease model is quite compatible with the low tissue penetration of the emitted bioluminescence. A recent evaluation was made of the bactericidal action of an 810 nm diode laser in a cutaneous wound infection. Open circular wounds were induced on the backs of rats, then infected with bioluminescent E. coli and irradiated at a fluence of 260 J/cm2 which increased the temperature to 45° C. A complete loss of bacterial bioluminescence from the wound was noted after 48 h. The antibacterial effect was attributed to dessication of the wound rather than a direct photodynamic effect via excitation of bacterial chromophores [28]. A related study employed laser irradiation to induce Photodynamic Therapy (PDT) of an open wound infection [29]. In this case, the wound was pretreated with a photosensitizer, a non-toxic dye that can be excited by red light to produce reactive oxygen species that kill the neighboring cells. PDT was recently demonstrated to cure an otherwise lethal P. aeruginosa wound infection in mice. Mice with wounds infected with bioluminescent P. aeruginosa quickly developed an illness consistent with systemic sepsis. The inoculum gave a sufficiently bright signal to allow two logs of signal reduction to be accurately measured. The infected wounds were treated with the polycationic conjugate, Pl-ce6, a photosenstizer which selectively targets the bacteria and absorbs 660 nm irradiation. Laser irradiation produced a fluence-dependent loss of luminescence over 40 min with complete loss of bacteria viability when the mice were imaged the next day (Figure 3). Control wounds that were untreated with the Pl-ce6 conjugate or were not irradiated showed a two-fold increase in luminescence signal due to growth in the nutrient rich medium of the wound. Further experiments explored the utility of this PDT approach in a deeper infection model, namely bioluminescent S. aureus in the mouse thigh. Upon direct injection of the photosensitizing conjugate into the infected thigh, and subsequent irradiation (160 J/cm2), the bioluminence signal decreased by >99%. However, in 4 out of the 5 mice, the treated thighs showed a reoccurrence of the bioluminescence on succeeding days. This study nicely illustrates how optical imaging maximizes the information gained from longitudinal studies, and demonstrates how the required number of laboratory animals can be minimized.
Figure 3.
Response of bioluminescent P. aeruginosa infection in a murine open wound model to treatment with conjugate photosensitizer followed by laser irradiation. Images reprinted with permission from Elsevier.
Important insights and discoveries in medical microbiology have been realized using bioluminescent bacteria in murine model systems of infection. Contag and co-workers made the surprising discovery that bioluminescent Listeria monocytogenes, a life threatening pathogen, will colonize the gall-bladder after systemic infection [30]. The gall-bladder acts as a reservoir for the bacteria to reproduce extracellularly, and then re-invade the host via access to the digestive tract through the bile ducts. A more recent study of Brucella infection in mice employed bioluminescent Brucella melitensis [31]. Bioluminescent imaging of infected interferon regulatory factor-1 knockout mice identified acute infection in many tissues, including the salivary glands suggesting a previously unknown tissue preference. These examples demonstrate how optical imaging greatly facilitates the study of sub-lethal bacterial infections in small animals, which are difficult to analyze using classical methods. The technology helps indentify virulence determinants that may control tissue specific replication.
Recent studies have demonstrated that bacteria will localize within tumors [32]. This has sparked an interest in using bioluminescent microbes to image tumors and metastases [33]. The bacteria can be delivered systemically and will localize selectively within tumors to provide an imaging signal for many days. While typically a safe haven for anaerobic microbes, aerobic bacteria like E.coli and S. typhimurium have also demonstrated tumor affinity as they evade the immune system [34]. The left panel of Figure 4 shows colonization of a breast cancer tumor (4T7 cells) with bioluminescent E. coli DH5α [35]. An interesting alternative tumor infection system was reported by Min and colleagues, who utilized the unique near-IR fluorescent properties of the purple bacterium Rhodobacter sphaeroides to image cancer [36]. These bacteria are photosynthetic, and absorb light at 850 nm with emission at 900 nm. The fluorescence emission for these cells was maximized by genetically removing the electron acceptors in the photosynthetic reaction center, such that more energy is dissipated as near-IR light emission. Four days after injecting these bacterial cells into the blood stream of tumor bearing mice, near-IR fluorescence was detected from the tumor (Figure 3, right). The infection of tumors by luminescent and fluorescent bacteria represents a promising emerging strategy to image in vivo models of cancer and its metasteses.
Figure 4.
Cancer imaging using bioluminescent (left) or NIR fluorescent (right) bacteria. Images reprinted with permission from BC Decker Publishing.
Method Comparison
At present, the most common method to optically image bacteria in small animals is bioluminescence which has the potential for high signal to noise ratio in shallow tissue since mammalian tissue essentially does not emit photons. The bioluminescence signal can persist for many days, and even reoccur if a diminished population recovers after incomplete antibiotic therapy, but signal dependence on the growth phase and microenvironment of the bacteria can cause undesired variability during longitudinal studies. For example, the requirement for oxygen may limit the use of luciferase to aerobic environments. In comparison, genetically encoded fluorescent proteins provide a signal source that may be less sensitive to environment and bacterial cell health. Both bioluminescence and fluorescent bacteria have limited detection depths due to attenuation of the light intensity at wavelengths between 300 and 600 nm by blood, biomolecules, and skin pigments. In addition, autofluorescence with visible light below 600 nm is a major background problem that can be edited, however, with new spectral imaging techniques [37]. The major advantage with synthetic near-IR probes is the radiation can potentially penetrate up to 15-20 cm of tissue, although this imaging outcome has yet to be demonstrated in a compelling way [38]. High intensity near-IR fluorescent probes will improve the feasibility of imaging deep tissue locations and will facilitate attempts to improve tomographic 3D reconstruction [39]. Disadvantages with synthetic exogenous probes include potential toxicity, and the loss of signal due to probe clearance or decomposition, which is not desired in longitudinal studies. A major attraction with synthetic probes is their potential to detect pathogenic bacteria in the clinic.
Model Translation to Humans
In general, animal infection models mirror those in humans because, in both cases, the bacteria have the same genetic blue-print. Indeed, clinical isolates are often used in murine models of infection, and bioluminescent versions of these clinical isolates are expected to exhibit the same host/pathogen interactions. However, if the goal is to locate an acquired bacterial infection in a human patient, then exogenous probes will likely have to be used, since these invasive bacteria will not express genetic reporters. As optical imaging technology evolves to include instrumentation that is suited for humans, the need for strategies that target bacteria with extremely bright near-IR probes will increase.
Conclusions
The concurrent appearance of genetic methods to prepare bioluminescent and fluorescent bacteria and the associated technology for their detection in vitro and in vivo has spurred the innovative improvement of several mouse models of infection. With classical infection models, bacteria are injected at various sites in mice, and large numbers of animals were required for dissection at each time point to follow disease progression. In contrast, optical imaging greatly increases the information gained from longitudinal studies, and in principle, the technology should reduce the number of animal that must be sacrificed. In the near-future it is likely that optical imaging of infection sites will evolve from the simple planar imaging method that is commonly used to-day, to more sophisticated techniques such as high-resolution time-resolved imaging and 3-D tomographic reconstruction. There will also be increased development of multicomponent reporter systems that will provide a simultaneous readout of separate biological events occurring during the infection process. Optical imaging is already a proven method for studying host/pathogen interactions in disease models, and in time, it will likely become a clinical method for detecting and identifying infection sites in humans.
Table I.
Comparison of Three Optical Methods for In Vivo Imaging of Bacterial Infection Models
| Bioluminescence | Fluorescent Proteins | Targeted Fluorescent Probes | |
|---|---|---|---|
| Advantages | • Inherently low background. • Amenable to longitudinal studies • Close mimics of natural bacterial strains • Ability to detect infection recovery |
• Amenable to longitudinal studies • Near-IR versions under development • No substrate needed • Ability to detect infection recovery |
• Bright near-IR probes are best for deep tissue • Activatable by enzymes • Possible translation to clinic • Multimodal probes under development |
| Disadvantages | • Limited tissue penetration of visible luminescence • Bacteria must be encoded with genetic reporter • Signal can vary • Substrate is often needed |
• Limited tissue penetration of visible emission • Autofluorescence • Bacteria must be encoded with genetic reporter |
• Probe preparation • Probe may affect bacteria function • Probes may be toxic or decompose • Not ideal for longitudinal studies. |
| Best Use of Method | • Study of bacterial pathogenesis • Antibiotic screening |
• Study of bacterial pathogenesis • Antibiotic screening |
• Detection of bacteria which do not express genetic reporters. • Clinical potential |
| How to Get Access to the Method | • Academic literature • Instrumentation vendors |
• Academic literature • Instrumentation vendors |
• Academic literature • Instrumentation vendors |
| References | [3] | [7] | [15] |
| Relevent Patents | [40] | [41] | [N/A] |
References
- 1.Jaffer FA, et al. Molecular Imaging in the Clinical Arena. JAMA. 2005;293:855–862. doi: 10.1001/jama.293.7.855. [DOI] [PubMed] [Google Scholar]
- 2.Hudson M. The Welfare and Scientific Advantages of Non-invasive Imaging of Animals Used in Biomedical Research. Anim. Welfare. 2005;14:303–317. [Google Scholar]
- 3.Doyle TC, et al. In Vivo Bioluminescence Imaging for Integrated Studies of Infection. Cellular Microbiol. 2004;6:303–317. doi: 10.1111/j.1462-5822.2004.00378.x. [DOI] [PubMed] [Google Scholar]
- 4.Bashkatov AN, et al. Optical Properties of Human Skin, Subcutaneous and Mucous Tissues in the Wavelength Range from 400 to 2000 nm. J. Phys. D: Appl. Phys. 2005;38:2543–2555. [Google Scholar]
- 5.Shah K, et al. Molecular Optical Imaging: Applications Leading to the Development of Present Day Therapeutics. NeuroRx®: J. Am. Soc. Exp. NeuroTherap. 2005;2:215–225. doi: 10.1602/neurorx.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiong T, et al. In vivo Optical Imaging of Bacterial Infection and Antibiotic Response in Intact Nude Mice. Proc. of SPIE. 2005;5699:19–25. [Google Scholar]
- 7.Shaner NC, et al. A Guide to Choosing Fluorescent Proteins. Nat. Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 8.So MK, et al. Self-illuminating Quantum Dot Conjugates for In Vivo Imaging. Nat. Biotechnol. 2006;24:339–343. doi: 10.1038/nbt1188. [DOI] [PubMed] [Google Scholar]
- 9.Salisbury V, et al. Use of a Clinical Escherichia coli Isolate Expressing Lux Genes to Study the Antimicrobial Pharmacodynamics of Moxifloxacin. J. Antimicrob. Chemother. 1999;43:829–832. doi: 10.1093/jac/43.6.829. [DOI] [PubMed] [Google Scholar]
- 10.Tenhami M, et al. Measurement of Effects of Antibiotics in Bioluminescent Staphylococcus aureus RN4220. Antimicrob. Agents Chemotherap. 2001;45:3456–3461. doi: 10.1128/AAC.45.12.3456-3461.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehtinen J, et al. Real-Time Monitoring of Antimicrobial Activity with the Multiparameter Microplate Assay. J. Microbiol. Meth. 2006:381–389. doi: 10.1016/j.mimet.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Gogoi SK, et al. Green Fluorescent Protein-Expressing Escherichia coli as a Model System for Investigating the Antimicrobial Activities of Silver Nanoparticles. Langmuir. 2006;22:9322–9328. doi: 10.1021/la060661v. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Medina R, et al. Pseudomonas aeruginosa Acquires Biofilm-Like Properties within Airway Epithelial Cells. Infect. Immun. 2005;73:8298–8305. doi: 10.1128/IAI.73.12.8298-8305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson C, et al. Liver Dendritic Cells Present Bacterial Antigens and Produce Cytokines upon Salmonella Encounter. J. Immunol. 2004;172:2496–2503. doi: 10.4049/jimmunol.172.4.2496. [DOI] [PubMed] [Google Scholar]
- 15.Leevy WM, et al. Optical Imaging of Bacterial Infection in Living Mice Using a Fluorescent Near-Infrared Molecular Probe. J. Am. Chem. Soc. 2006;128:16476–16477. doi: 10.1021/ja0665592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis KP, et al. Monitoring Bioluminescent Staphylococcus aureus Infections in Living Mice Using a Novel luxABCDE Construct. Infect. Immun. 2000;68:3594–3600. doi: 10.1128/iai.68.6.3594-3600.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuklin NA, et al. Real-Time Monitoring of Bacterial Infection In Vivo: Development of Bioluminescent Staphylococcal Foreign-Body and Deep-Thigh-Wound Mouse Infection Models. Antimicrob. Agents Chemother. 47:2740–2748. doi: 10.1128/AAC.47.9.2740-2748.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rocchetta HL, et al. Validation of a Noninvasive, Real-Time Imaging Technology Using Bioluminescent Escherichia coli in the Neutropenic Mouse Thigh Model of Infection. Antimicrob. Agents Chemother. 2001;45:129–137. doi: 10.1128/AAC.45.1.129-137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welling MM, et al. 99mTc-Labeled Antimicrobial Peptides for Detection of Bacterial and Candida albicans Infections. J. Nuc. Med. 2001;42:788–794. [PubMed] [Google Scholar]
- 20.Zhao M, et al. Spatial-Temporal Imaging of Bacterial Infection and Antibiotic Response in Intact Animals. Proc. Nat. Acad. Sci. 2001;98:9814–9818. doi: 10.1073/pnas.161275798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Francis KP. Visualizing Pneumococcal Infections in the Lungs of Live Mice Using Bioluminescent Streptococcus pneumoniae Transformed with a Novel Gram-positive lux Transposon. Infect. Immun. 2001;69:3350–3358. doi: 10.1128/IAI.69.5.3350-3358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orihuela CJ, et al. Tissue-Specific Contributions of Pneumococcal Virulence Factors to Pathogenesis. J. Inf. Dis. 2004;190:1661–1669. doi: 10.1086/424596. [DOI] [PubMed] [Google Scholar]
- 23.Kadurugamuwa JL. Reduction of Astriogliosis by Early Treatment of Pneumococcal Meningitis Measured by Simultaneous Imaging, In Vivo of the Pathogen and Host Response. Infect. Immun. 2005;73:7836–7843. doi: 10.1128/IAI.73.12.7836-7843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro MA, et al. Comparative Therapeutic Efficacy of Clinafloxacin in a Pneumococcal Meningitis Mouse Model. J. Antimicrob. Chemo. 2000;45:489–492. doi: 10.1093/jac/45.4.489. [DOI] [PubMed] [Google Scholar]
- 25.Kadurugamuwa JL, et al. Noninvasive Biophotonic Imaging for Monitoring of Catheter-Associated Urinary Tract Infections and Therapy in Mice. Infect. Immun. 2005;73:3878–3887. doi: 10.1128/IAI.73.7.3878-3887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kadurugamuwa JL, et al. Direct Continuous Method for Monitoring Biofilm Infection in a Mouse Model. Infect. Immun. 2003;71:882–890. doi: 10.1128/IAI.71.2.882-890.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadurugamuwa JL, et al. Noninvasive Optical Imaging Method to Evaluate Postantibiotic Effects of Biofilm Infection In Vivo. Antimicrob. Agents Chemother. 2004;48:2283–2287. doi: 10.1128/AAC.48.6.2283-2287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jawhara S, et al. Monitoring of Bactericidal Action of Laser by In Vivo Imaging of Bioluminescent E. Coli in a Cutaneous Wound Infection. Lasers Med. Sci. 2006;21:153–159. doi: 10.1007/s10103-006-0388-8. [DOI] [PubMed] [Google Scholar]
- 29.Demidova TN, et al. Monitoring Photodynamic Therapy of Localized Infections By Bioluminescence Imaging of Genetically Engineered Bacteria. J. Photochem. Photobiol. B: Biology. 2005;81:15–25. doi: 10.1016/j.jphotobiol.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardy J, et al. Extracellular Replication of Listeria moncytogenes in the Murine Gall Bladder. Science. 2004;303:851–853. doi: 10.1126/science.1092712. [DOI] [PubMed] [Google Scholar]
- 31.Rajashekara G, et al. Temporal Analysis of Pathogenic Events in Virulent and Avirulent Brucella Melitensis Infections. Cell. Microbiol. 2005;7:1459–1473. doi: 10.1111/j.1462-5822.2005.00570.x. [DOI] [PubMed] [Google Scholar]
- 32.Ryan RM, et al. Use of Bacteria in Anti-Cancer Therapies. BioEssays. 2005;28:84–94. doi: 10.1002/bies.20336. [DOI] [PubMed] [Google Scholar]
- 33.Yu YA, et al. Optical Imaging: Bacteria, Viruses, and Mammalian Cells Encoding Light-Emitting Proteins Reveal the Locations of Primary Tumors and Metastases in Animals. Anal. Bioanal. Chem. 2003;377:964–972. doi: 10.1007/s00216-003-2065-0. [DOI] [PubMed] [Google Scholar]
- 34.Yu YA, et al. Visualization of Tumors and Metastases in Live Animals with Bacteria and Vaccinia Virus Encoding Light-Emitting Proteins. Nat. Biotech. 2004;22:313–320. doi: 10.1038/nbt937. [DOI] [PubMed] [Google Scholar]
- 35.Min J-J, et al. Specific Targeting Breast Cancer and Its Metastatic Lesions Using Light-Emitting Bacteria. Mol. Imaging. 2006;5:321. [Google Scholar]
- 36.Min J-J, et al. Targeting Solid Tumors Using Near-Infrared Fluorescence Emitting Purple Bacteria. Mol. Imaging. 2006;5:320. [Google Scholar]
- 37.Tam JM, et al. Improved In Vivo Whole-Animal Detection Limits of Green Fluorescent Proetin-Expressing Tumor Lines by Spectral Fluorescence Imaging. Molec. Imag. 2007 Doi: 10.2310/7290.2007.00023. [PubMed] [Google Scholar]
- 38.Tung C, et al. A Receptor-Targeted Near-Infrared Fluorescence Probe for In Vivo Tumor Imaging. ChemBiochem. 2002;8:784–786. doi: 10.1002/1439-7633(20020802)3:8<784::AID-CBIC784>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 39.Zacharakis G, et al. Volumetric Tomography of Fluorescent Proteins Through Small Animals In Vivo. Proc. Natl. Acad. Sci., USA. 2005;102:18252–18257. doi: 10.1073/pnas.0504628102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Contag PR, et al. Noninvasive Localization of a Light-Emitting Conjugate in a Mammal. 2005 US Patent 6,916,462.
- 41.Tsien RY, et al. Modified Green Fluorescent Proteins. 2004 U. S. Patent 6,800,733.