Abstract
Chromodomain proteins (Chp1/Chp2/Swi6/Clr4) bind to methylated H3K9 (H3K9me) and regulate pericentric heterochromatin in fission yeast. Chp1 and Clr4 (H3K9-HMT), bind transcriptionally active heterochromatin, whereas Chp2/Swi6 (HP1 homologs) are recruited during the inactive state. We show that H3K4 acetylation (H3K4ac) plays a role in the transition of dimethylated H3K9 (H3K9me2) occupancy from Chp1/Clr4 to Chp2/Swi6. H3K4ac, mediated by Mst1, is enriched at pericentromeres concomitantly with heterochromatin reassembly. H3K4R (Lys → Arg) mutation increases Chp1 and decreases Chp2/Swi6 pericentric occupancy and exhibits centromeric desilencing. Consistent with structural data, H3K4ac specifically reduces Chp1/Clr4 affinity to H3K9me. We propose that H3K4ac mediates a chromodomain switch from Chp1/Clr4 to Swi6/Chp2 to allow heterochromatin reassembly.
Keywords: H3K4ac, HP1, RITS complex, chromodomain switch, heterochromatin
Schizosaccharomyces pombe is a key genetic model organism for studying heterochromatin establishment and maintenance (Grewal and Jia 2007). Its centromeres are large DNA structures composed of a central region that forms specialized CENP-A-dependent chromatin surrounded by outer repetitive regions (otr) corresponding to the pericentric heterochromatin (Fig. 1A; Clarke and Baum 1990). Similar to higher eukaryotes, S. pombe pericentric heterochromatin is dependent on HP1-like proteins—Swi6 and Chp2—that bind via their chromodomains to histone H3 methylated at K9 (H3K9me) (Bannister et al. 2001; Nakayama et al. 2001; Motamedi et al. 2008; Sadaie et al. 2008; Fischer et al. 2009). Given the importance of this interaction, faithful renewal of H3K9me is essential, and relies on an elaborate interplay between DNA replication, RNA Polymerase II-dependent transcription, the RNAi machinery, and chromatin-binding and chromatin-modifying enzymes. A challenging moment for heterochromatin is DNA duplication, when the incorporation of newly synthesized histones dilutes di- and trimethylated H3K9 (H3K9me2/3). Interestingly, the opening of chromatin during replication fork passage creates a window of opportunity for RNA Polymerase II to transcribe through the heterochromatic repeats (Chen et al. 2008). These transcripts provide both a source for the generation of siRNA and a template for the binding of the RITS (RNA-induced transcriptional silencing) complex. Indeed, Ago1 is loaded with siRNA and tethers RITS to the pericentric repeats through an interaction between the nascent transcripts and the matching siRNA (Reinhart and Bartel 2002; Volpe et al. 2002; Motamedi et al. 2004; Verdel et al. 2004). The RITS complex also contains Chp1, a chromodomain-containing protein whose high-affinity binding for H3K9me2/3 is also required for the binding of the complex onto the chromatin (Verdel et al. 2004; Schalch et al. 2009). The RITS complex then recruits the Clr4 methyltransferase-containing complex to methylate H3K9 on the newly deposited histones and allow the HP1-like proteins to mediate heterochromatin formation (Bernard et al. 2001; Nonaka et al. 2002; Fischer et al. 2009). Thus, the re-establishment of heterochromatin following DNA replication involves the sequential recruitment of the H3K9me-binding protein Chp1 and the HP1 homologs Swi6 and Chp2.
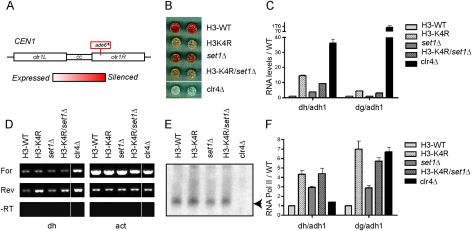
Figure 1.
H3K4R mutant cells display pericentric silencing defects. (A) Diagram illustrating the ade6+ gene insertion within the centromere 1 (otr1R∷Sph1) used for the colony color silencing assay. (B) Colony color silencing assay of the indicated strains containing ade6+ as shown in A. (C) Random primed RT followed by quantitative PCR (Q-PCR) using primers specific for the pericentric dh and dg repeats. The data are represented as mean ± SD, after normalization to the control adh1 euchromatic gene and to transcript levels in wild-type (WT) cells. (D) Strand-specific RT followed by semi-Q-PCR using primers specific for the pericentromeric dh repeat and the control act1 euchromatic gene. (E) Northern blot analysis of purified small RNA (<200 nucleotides), probed with a mix of oligos complementary to the centromeric siRNAs (Reinhart and Bartel 2002; Buhler et al. 2006). (F) ChIP with anti-RNA Polymerase II C-terminal domain repeat antibody analyzed by Q-PCR with primers specific for the pericentric dh and dg repeats. The data are represented as mean ± SD, after normalization to the control adh1 euchromatic gene and to ChIP levels in wild-type cells.
Results and Discussion
H3K4R (Lys → Arg), but not the H3K4 methyltransferase set1 mutant, is defective in centromeric silencing
While assaying a variety of mutants of histone H3 in fission yeast, we noticed that H3K4R mutants displayed a strong derepression of an ade6+ reporter gene inserted at the outer repeats of centromere 1 (otr1R∷Sph1) (Fig. 1A,B). The defect in pericentric silencing revealed by this colony color assay was confirmed at the molecular level by quantitative RT–PCR with primers specific for the dh and dg pericentric repeats (Fig. 1C). In S. pombe, the forward and reverse strands are differentially regulated: The forward strand is repressed at the transcriptional level, while the reverse strand is post-transcriptionally silenced by the RNAi machinery (Volpe et al. 2002). Strand-specific RT–PCR indicated that the reverse strand of the dh repeat was significantly more derepressed than the forward strand in the H3K4R mutant (Fig. 1D). Further analysis revealed that the increased levels of the reverse strand were not due to a defect in siRNA generation (Fig. 1E), but rather to increased presence of the RNA Polymerase II at the pericentric repeats (Fig. 1F). Importantly, the pericentric silencing defects were specific to the H3K4R mutant and were not shared by the set1Δ mutant (Fig. 1B–F). Since Set1 is the methyltransferase responsible for all detectable H3K4 methylation in fission yeast (Noma and Grewal 2002; Kanoh et al. 2003), this suggested the possibility that a modification on H3K4 other than methylation may be required for proper centromeric silencing.
H3K4 is modified by acetylation
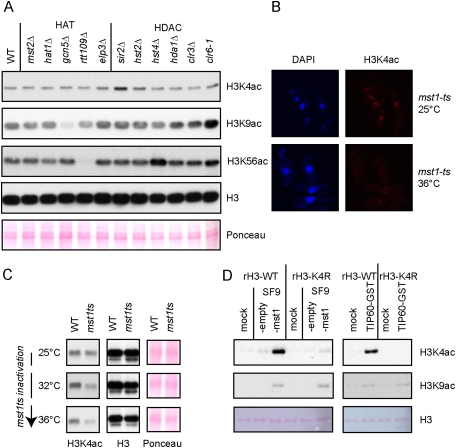
We hypothesized that this modification on H3K4 could be acetylation, since this mark was identified previously by mass spectrometry of histones purified from human cell lines (Garcia et al. 2007). We raised an antibody against the H3K4 acetylation (H3K4ac) modification that, after affinity purification, was highly reactive to the H3K4ac modification (Supplemental Fig. S3). When used in Western blotting with whole-cell extracts (WCEs) from S. pombe cells, our anti-H3K4ac antibody recognized a specific band at the level of histone H3 that was clearly reduced in the H3K4R mutant (Supplemental Fig. S4). Furthermore, immunofluorescence (IF) analysis with this antibody revealed a nuclear staining that disappeared in the H3K4R mutant (Supplemental Fig. S5). In order to identify the enzymes that catalyze acetylation and deacetylation of H3K4, we constructed a collection of fission yeast strains deleted or mutated for known or putative histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively. Western blot analysis of WCEs from this collection revealed that the H3K4ac signal was significantly increased in a sir2Δ mutant (Fig. 2A). Moreover, the H3K4R mutation in the sir2Δ background abolished the H3K4ac signal (Supplemental Fig. S4). In addition, a modest increase was observed in the hst2Δ and clr6-1 mutants, suggesting that several HDACs could participate in H3K4 deacetylation. Surprisingly, none of the HAT mutants initially tested affected the H3K4ac signal, while the screen readily worked for known HATs and HDACs regulating H3K9ac and H3K56ac (Fig. 2A). However, this initial collection did not contain Mst1, because this HAT is an essential protein. Therefore, we obtained a previously described thermosensitive (ts) mutant of mst1 (Gomez et al. 2008). IF analysis revealed that the nuclear signal obtained with the anti-H3K4ac antibody was greatly reduced when the mst1-ts mutant was switched to the nonpermissive temperature even for a very short period of time (2 h) (Fig. 2B). This result was confirmed by Western blot analysis of WCEs from mst1-ts cells grown at semi- or nonpermissive temperatures (Fig. 2C). Importantly, the Mst1 protein purified from Bacculovirus-infected insect cells—as well as its human homolog, TIP60—directly acetylated histone H3 at K4 in an in vitro HAT assay (Fig. 2D; Supplemental Fig. S6). Taken together, these results show that H3K4ac exists in S. pombe, and that Mst1 and the sirtuin Sir2 are the major HAT and HDAC, respectively, for H3K4.
Figure 2.
In vivo screen suggests that the essential mst1 HAT (TIP60 homolog) and the sirtuin sir2 HDAC are the major enzymes that catalyze H3K4ac and H3K4 deacetylation, respectively. (A) Western blot analysis of WCEs from cells of the indicated genotypes probed with a specific anti-H3K4ac (Supplemental Figs. S3–5) and control anti-H3K9ac and anti-H3K56ac antibodies. (B) IF with anti-H3K4ac antibody of mst1-ts cells exponentially grown at the permissive temperature (25°C) or shifted for 2 h at the nonpermissive temperature (36°C). (C) Western blot analysis with anti-H3K4ac antibody of wild-type (WT) or mst1-ts cells exponentially grown at the permissive temperature (25°C) or shifted for 4 h at the semipermissive (32°C) and nonpermissive (36°C) temperatures. (D) Western blot analysis with anti-H3K4ac and anti-H3K9ac antibodies of in vitro HAT assays with Mst1- and GST-tagged TIP60 using recombinant histone hH3.1 wild type or H3K4R as substrate. Mst1 was purified from SF9 insect cells infected with a bacculovirus containing mst1 ORF under a polyhedrin promoter.
H3K4ac occurs at the pericentric repeats and peaks after H3K9me2 at the cell cycle-regulated dh repeat
In order to test whether H3K4ac played a direct role at the pericentric heterochromatin, we performed chromatin immunoprecipitation (ChIP) experiments using our specific anti-H3K4ac antibody. As shown in Figure 3, A and B, H3K4ac was enriched specifically at the pericentric repeats compared with the euchromatic ura4-DS/E or adh1 gene. This enrichment was completely lost in the H3K4R mutant (Fig. 3B), suggesting that it was specific for H3K4ac and not another acetylation. Moreover, the acetyltransferase Mst1 was modestly enriched at the pericentric repeats (Supplemental Fig. S7), indicating that Mst1 might directly acetylate H3K4 on heterochromatin. Since S. pombe heterochromatin is cell cycle-regulated (Chen et al. 2008; Kloc et al. 2008), we tested whether H3K4ac occurred at a specific time frame at the pericentric repeats. For this, temperature-sensitive cdc10-V50 mutant cells were arrested at the G1/S boundary and released to grow synchronously (Xhemalce et al. 2007). ChIP analysis using the anti-H3K4ac and anti-H3K9me2 antibodies demonstrated that H3K4ac peaks at the dh repeat, and, more interestingly, it does so after H3K9me2 and at a time when heterochromatin reassembly takes place (Fig. 3C, graphs; Chen et al. 2008; Kloc et al. 2008). The bulk of H3K4ac is also cell cycle-regulated, and the cellular H3K4ac peaks in S phase in a similar fashion to H3K56ac (Fig. 3C, gels), providing a possible explanation for the sensitivity of H3K4R mutants to S-phase DNA damage (Supplemental Fig. S1).
Figure 3.
ChIP with specific anti-H3K4ac antibody shows that H3K4ac is enriched at pericentromeric repeats and peaks after H3K9me2 during cell cycle. (A) ChIP with anti H3K4ac antibody analyzed by multiplex PCR with ura4 primers recognizing simultaneously a ura4+ gene inserted at the centromere 1 (otr1R∷Sph1) and a truncated ura4-DSE gene at the endogenous ura4 euchromatic locus. (B) ChIP with anti-H3K4ac antibody analyzed by Q-PCR with primers specific for the pericentric dh and dg repeats and the control adh1 euchromatic gene. The data are represented as mean ± SD, after normalization to ChIP levels in H3K4R cells. (C, graphs) ChIP with anti-H3K9me2 and anti-H3K4ac antibodies analyzed by Q-PCR with dh and adh1 primers from cell cycle-synchronized cdc10-ts cells. (Gels) Western blot analysis with anti-H3K56ac and anti-H3K4ac antibodies of the ChIP inputs. H3K56ac was used to give the time frame of S phase as reported previously (Xhemalce et al. 2007).
H3K4R mutation does not affect H3K9 methylation, but rather the balance of the downstream chromodomain proteins at the pericentric repeats
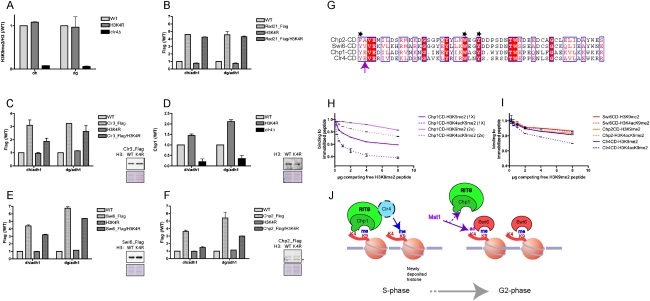
To gain insight into the mechanism by which H3K4ac may regulate pericentric heterochromatin, we analyzed by ChIP which steps of heterochromatin are disrupted in the H3K4R mutant: (1) H3K9 methylation, (2) cohesin recruitment, (3) histone deacetylation, and (4) H3K9me2/3-binding proteins. Consistent with H3K4ac occurring after H3K9 methylation at the pericentric repeats (Fig. 3C), the H3K4R mutation did not alter H3K9 methylation (Fig. 4A), suggesting that a step downstream from H3K9 methylation is perturbed in the H3K4R mutant. Furthermore, Rad21 presence (Fig. 4B) was not affected, indicating that H3K4R mutation does not affect the recruitment of cohesins at the pericentric repeats. However, the H3K4R mutation did have a clear effect on the binding of the downstream H3K9me2/3 chromodomain proteins, decreasing the binding of the HP1 homologs Swi6 and Chp2 and increasing Chp1 levels at the pericentric repeats (Fig. 4D–F; Supplemental Fig. S8). Importantly, the observed effects were not due to perturbed cellular levels of these proteins (Fig. 4D–F, Western blots). The use of the same Flag antibody to chromatin-immunoprecipitate Swi6 and Chp2 endogenously tagged with Flag allowed us to directly compare the effect of the H3K4R mutation on the recruitment of these proteins to the pericentric repeats. Our ChIP analysis revealed that Chp2 recruitment was more affected than that of Swi6 (Fig. 4E,F). Consistent with the role of Chp2 in the recruitment of Clr3 at the pericentric repeats (Motamedi et al. 2008; Sadaie et al. 2008; Fischer et al. 2009), we found that the levels of this HDAC were also decreased in the H3K4R mutant, albeit less severely than those of Chp2.
Figure 4.
H3K4R mutation does not affect H3K9 methylation, but rather the binding of the downstream chromodomain proteins, decreasing the HP1 homologs Swi6 and Chp2 and increasing Chp1 (RITS complex). (A) ChIP with anti-H3K9me2 antibody analyzed by Q-PCR with dh and dg primers from wild-type (WT), H3K4R, and clr4Δ cells. The data are represented as mean ± SD, after normalization to H3 and ChIP levels in wild-type cells. (B) ChIP with anti-Flag antibody analyzed by Q-PCR with dh and dg primers from untagged and rad21-Flag cells in wild-type or H3K4R background. The data are represented as mean ± SD, after normalization to the control adh1 euchromatic gene and ChIP levels in wild-type cells. (C) ChIP analysis as in B from clr3-Flag cells. (Gels) Western blot analysis with anti-Flag antibody of Clr3-Flag in wild-type and H3K4R background. (D) ChIP with anti-Chp1 antibody analyzed by Q-PCR with dh and dg primers from wild-type, H3K4R, and clr4Δ cells. The data are represented as mean ± SD, after normalization to the control adh1 euchromatic gene and ChIP levels in wild-type cells. (Gels) Western blot analysis with anti-Chp1 antibody of WCEs from wild-type and H3K4R cells. (E) ChIP analysis as in B from swi6-Flag cells. For the same analysis using a Swi6 antibody, see Supplemental Figure S8. (Gels)Western blot analysis with anti-Flag antibody of Swi6-Flag in wild-type and H3K4R background. (F) ChIP analysis as in B from chp2-Flag cells. (Gels) Western blot analysis with anti-Flag antibody of Chp2-Flag in wild-type and H3K4R background. The asterisk indicates a nonspecific cross-reacting band. (G) Alignment of the chromodomains of the two HP1 homologs, Swi6 and Chp2, as well as Chp1 and Clr4. The black asterisks indicate the aromatic cage residues that recognize H3K9me (Jacobs and Khorasanizadeh 2002). The purple arrow points to Chp1-E23 and its conservation in Clr4. (H) Peptide competition assays measuring the binding of the chromodomain of Chp1 to H3K9me2 and H3K4acK9me2 biotinylated peptides after addition of increasing amounts of competing free H3K9me2 peptide (Supplemental Material). Two different initial amounts (1× [50 μg] or 2× [100 μg]) of Chp1 were used (Supplemental Material). (I) Peptide competition assays measuring the binding of the chromodomains of Swi6, Chp2, and Clr4 to H3K9me2 and H3K4acK9me2 biotinylated peptides after addition of increasing amounts of competing free H3K9me2 peptide. An initial amount of 100 μg of the CD was used (Supplemental Material). (J) Model for how H3K4 acetylation may affect Chp1 binding to H3K9me at the S–G2-phase transition.
H3K4ac decreases the affinity of Chp1 for the H3 tail in vitro
Since Chp1 on one side and the HP1 proteins Swi6 and Chp2 on the other side share the same binding site, we hypothesized that H3K4ac may regulate the balance between the binding of these proteins to the H3K9me tail. The crystal structure of the Chp1 chromodomain in complex with a H3K9me3 peptide (Schalch et al. 2009) revealed that K4 of H3 engages in a salt bridge and van der Waals contacts with E23 of Chp1 (Fig. 4G). Importantly, a similar salt bridge is not observed between H3K4 and the chromodomain of the Swi6 and Chp2 homolog HP1 (Jacobs and Khorasanizadeh 2002). Finally, the mutation of E23 to V, an uncharged amino acid, significantly reduced the affinity of Chp1 for the H3K9me2/3 peptides. To determine if acetylation at H3K4 differentially affects the binding of the Chp1 chromodomain to the H3K9me tail, we performed peptide competition assays. In these assays, the Chp1 recombinant chromodomain was first bound in saturating conditions to beads coated with H3K9me2 or H3K4ac/K9me2 peptides. The bound Chp1-CD was then competed by the addition of increasing amounts of competing free H3K9me2 peptide (Supplemental Material). As shown in Figure 4H, the competing H3K9me2 peptide more effectively displaced Chp1 bound to the doubly modified H3K4ac/K9me2 peptide than to the singly modified H3K9me2 peptide. On the contrary, the binding of Swi6 and Chp2 to the H3K9me2 tail was not differentially modulated by H3K4 acetylation (Fig. 4I). Thus, consistent with the structural data (Jacobs and Khorasanizadeh 2002; Schalch et al. 2009), our assays show that H3K4ac has the ability to weaken the interaction between the chromodomain of Chp1 and the tail of H3, but has no effect on the binding of the HP1 homologs Swi6 and Chp2.
The fourth H3K9me-binding chromodomain protein present in S. pombe is the H3K9 methyltransferase Clr4 (Zhang et al. 2008). Since this enzyme is recruited to heterochromatin in the actively transcribed phase by the RITS complex (Chen et al. 2008), we asked if the binding of this protein was also affected by H3K4ac, as is the case for Chp1. Figure 4I shows that Clr4 is also sensitive to the presence of an acetylated H3K4. As highlighted on the protein sequence alignment in Figure 4G, the chromodomain of Clr4 also contains a glutamic acid at the same position as Chp1-E23, while the chromodomains of Swi6 and Chp2 contain an uncharged amino acid at this position. Since H3K4ac occurs after the peak of H3K9 methylation, these data suggest that H3K4ac could help to terminate the open state of chromatin (marked by pericentric transcription and the binding of Chp1 and Clr4) in order to allow for the recruitment of the HP1 homologs Chp2 and Swi6 to heterochromatin.
Conclusions
Our results identify H3K4ac as a novel modification involved in heterochromatin formation in S. pombe. Classically, acetylation has been defined as a modification involved in active transcription, so a role in heterochromatin was unexpected. However, H3K4ac has a role during S phase, when pericentric heterochromatin is transcribed (Chen et al. 2008; Kloc et al. 2008). During this time, DNA replication results in two newly formed DNA strands, where unmodified nucleosomes are deposited. The transmission of H3K9 methylation onto these nucleosomes involves the RITS complex and one of its members, Chp1 (Chen et al. 2008; Kloc et al. 2008). Chp1 anchors RITS to H3K9me nucleosomes and augments the recruitment of Clr4 and the H3K9 methylation of newly deposited nucleosomes (Fig. 4H; Partridge et al. 2002; Schalch et al. 2009). At the transition of S phase into G2, the HP1 proteins Swi6 and Chp2 need to be recruited to H3K9 methylated nucleosomes to enable heterochromatin reassembly. Our data suggest that it is at this juncture that H3K4ac has a role (Fig. 4J). We propose that H3K4ac deposition facilitates the binding of the HP1 proteins to H3K9me by decreasing the affinity of Chp1 for the K9 methylated H3 tail. It is interesting to note that this “chromodomain switch” is possible due to the distinct sensitivities of Chp1 and HP1 proteins to the presence of an acetyl group at H3K4. A further role for H3K4ac may be to compete directly with H3K4 methylation, an active transcription histone mark. It has been shown recently that H3K4 methylation is actively removed from heterochromatin in S. pombe. Indeed, mutation of the jumanji H3K4 demethylase Lid2 (Jarid1B homolog) is associated with increased levels of H3K4 methylation at the pericentric repeats and loss of centromeric silencing (Li et al. 2008). Finally, a conserved evolutionary role for H3K4ac is very likely, given the existence of human homologous enzymes TIP60 and Sirt1, which regulate this modification. It will be interesting to establish if, in humans, like in S. pombe, this acetylation plays an important role in the regulation of epigenetic events related to variegating loci, like heterochromatin or genes alternating between active and inactive states.
Materials and methods
All experiments were performed using the S. pombe strains described in Supplemental Table S1. More details about all of the strains and plasmids used are available on request. Importantly, the functionality of all tagged strains was checked by growth and centromeric silencing assays. The H3K4ac antibody used in this study was raised by Diagenode in collaboration with the EPITRON (EPIgenetic TReatment Of Neoplastic disease; http://www.epitron.eu) European Consortium and was affinity-purified. The details of all the other commercially available antibodies used in the study, including catalog and lot numbers, are incorporated in Supplemental Table S2. The mean and SD values were calculated from independent experiments, and the graphs were constructed with the GraphPad Prism software. Detailed Materials and Methods are available in the Supplemental Material.
Acknowledgments
We thank the anonymous reviewers of our manuscript for their constructive suggestions; Shiv Grewal for the clr6-1 mutant; Susan Forsburg for the mst1-ts mutant; Janet Partridge and Andrew Bannister for the Chp1-CD, the Chp2-CD, the Clr4-CD GST expression vector, and the full-length Swi6 GST expression vector (which served as a template for cloning the Swi6-CD); Marc Buehler for the siRNA purification protocol; Kyle Miller for help with the purification of the recombinant histones; Till Bartke for the recombinant histone hH3.1 expression vector and recombinant His6-TEV; and Andrew Bannister, Mark Dawson, and Kyle Miller for helpful discussions and critical comments on the manuscript. The T.K. laboratory is funded by grants from Cancer Research UK (CRUK) and the 6th Research Framework Program of the European Union (EPITRON, SMARTER, and HEROIC).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1881710.
Supplemental material is available at http://www.genesdev.org.
References
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC. Requirement of heterochromatin for cohesion at centromeres. Science. 2001;294:2539–2542. doi: 10.1126/science.1064027. [DOI] [PubMed] [Google Scholar]
- Buhler M, Verdel A, Moazed D. Tethering RITS to a nascent transcript initiates RNAi- and heterochromatin-dependent gene silencing. Cell. 2006;125:873–886. doi: 10.1016/j.cell.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Chen ES, Zhang K, Nicolas E, Cam HP, Zofall M, Grewal SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–737. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- Clarke L, Baum MP. Functional analysis of a centromere from fission yeast: A role for centromere-specific repeated DNA sequences. Mol Cell Biol. 1990;10:1863–1872. doi: 10.1128/mcb.10.5.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer T, Cui B, Dhakshnamoorthy J, Zhou M, Rubin C, Zofall M, Veenstra TD, Grewal SI. Diverse roles of HP1 proteins in heterochromatin assembly and functions in fission yeast. Proc Natl Acad Sci. 2009;106:8998–9003. doi: 10.1073/pnas.0813063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, et al. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Gomez EB, Nugent RL, Laria S, Forsburg SL. Schizosaccharomyces pombe histone acetyltransferase Mst1 (KAT5) is an essential protein required for damage response and chromosome segregation. Genetics. 2008;179:757–771. doi: 10.1534/genetics.107.085779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI, Jia S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- Kanoh J, Francesconi S, Collura A, Schramke V, Ishikawa F, Baldacci G, Geli V. The fission yeast spSet1p is a histone H3-K4 methyltransferase that functions in telomere maintenance and DNA repair in an ATM kinase Rad3-dependent pathway. J Mol Biol. 2003;326:1081–1094. doi: 10.1016/s0022-2836(03)00030-5. [DOI] [PubMed] [Google Scholar]
- Kloc A, Zaratiegui M, Nora E, Martienssen R. RNA interference guides histone modification during the S phase of chromosomal replication. Curr Biol. 2008;18:490–495. doi: 10.1016/j.cub.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Huarte M, Zaratiegui M, Vaughn MW, Shi Y, Martienssen R, Cande WZ. Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell. 2008;135:272–283. doi: 10.1016/j.cell.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–790. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Noma K, Grewal SI. Histone H3 lysine 4 methylation is mediated by Set1 and promotes maintenance of active chromatin states in fission yeast. Proc Natl Acad Sci. 2002;99:16438–16445. doi: 10.1073/pnas.182436399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y. Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol. 2002;4:89–93. doi: 10.1038/ncb739. [DOI] [PubMed] [Google Scholar]
- Partridge JF, Scott KS, Bannister AJ, Kouzarides T, Allshire RC. cis-acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr Biol. 2002;12:1652–1660. doi: 10.1016/s0960-9822(02)01177-6. [DOI] [PubMed] [Google Scholar]
- Reinhart BJ, Bartel DP. Small RNAs correspond to centromere heterochromatic repeats. Science. 2002;297:1831. doi: 10.1126/science.1077183. [DOI] [PubMed] [Google Scholar]
- Sadaie M, Kawaguchi R, Ohtani Y, Arisaka F, Tanaka K, Shirahige K, Nakayama J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–6988. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalch T, Job G, Noffsinger VJ, Shanker S, Kuscu C, Joshua-Tor L, Partridge JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Xhemalce B, Miller KM, Driscoll R, Masumoto H, Jackson SP, Kouzarides T, Verreault A, Arcangioli B. Regulation of histone H3 lysine 56 acetylation in Schizosaccharomyces pombe. J Biol Chem. 2007;282:15040–15047. doi: 10.1074/jbc.M701197200. [DOI] [PubMed] [Google Scholar]
- Zhang K, Mosch K, Fischle W, Grewal SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–388. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]