Abstract
Nonhomologous end-joining (NHEJ) inhibition at telomeres ensures that native chromosome ends do not fuse together. But the occurrence and consequences of rare telomere fusions are not well understood. It is notably unclear whether a telomere fusion could be processed to restore telomere ends. Here we address the behavior of individual dicentrics formed by telomere fusion in the yeast Saccharomyces cerevisiae. Our approach was to first stabilize and amplify fusions between two chromosomes by temporarily inactivating one centromere. Next we analyzed dicentric breakage following centromere reactivation. Unexpectedly, dicentrics often break at the telomere fusions during progression through mitosis, a process that restores the parental chromosomes. This unforeseen result suggests a rescue pathway able to process telomere fusions and to back up NHEJ inhibition at telomeres.
Keywords: NHEJ, Rap1, Telomere, centromere, dicentric, karyotype stability
Telomeres are the DNA–protein complexes at the ends of linear chromosomes. They solve the end replication problem resulting from semiconservative DNA replication (Walmsley et al. 1983; Greider and Blackburn 1985; Shore and Bianchi 2009). They also protect the native chromosome ends from the DNA damage repair and checkpoint pathways that act on ends generated by double-strand breaks (de Lange 2009). Evolution solved these problems differently in prokaryotes and in eukaryotes. In several bacteria and viruses, chromosomes are linear and have covalently closed hairpin telomere ends. Replication produces inverted repeats that are processed into two covalently closed hairpins by a specialized resolvase (Aihara et al. 2007). In other words, in these prokaryotic organisms, breakage of the fused telomeres is a normal part of the chromosome replication and segregation cycle. In eukaryotes, telomeres are stable, open double-strand ends. In most cases they display a short 3′ single-strand overhang, and their sequences are made of an oriented short G-rich repeated motif (e.g., TG1–3 in the yeast Saccharomyces cerevisiae and TTAGGG in vertebrates). The repeated motifs allow the concentration of specialized proteins that recognize them and establish telomere functions.
One function of telomeres in eukaryotes is to prevent fusions by the nonhomologous end-joining (NHEJ) double-strand break repair pathway (Riha et al. 2006). The normal role of NHEJ is to fuse the two ends of a double-strand break. NHEJ requires three conserved factors: the KU DNA end-binding protein; the Rad50–Mre11–Xrs2NBS1 complex; and Ligase 4 with its cofactors, Lif1XRCC4 and Nej1CERNU/XLF (Daley et al. 2005; Callebaut et al. 2006; Deng et al. 2009; Rass et al. 2009; Xie et al. 2009). NHEJ inhibition at telomeres is established by proteins bound to the double-stranded telomeric repeats. In the fission yeast Schizosaccharomyces pombe, the Taz1 protein binds telomeric DNA and recruits the Rap1 protein. Both Taz1 and Rap1 are required for NHEJ inhibition (Ferreira and Cooper 2001; Miller et al. 2005). In mammals, TRF2, a Taz1 ortholog, recruits RAP1 to telomeres, and both TRF2 and RAP1 establish NHEJ inhibition, likely through more than one pathway (van Steensel et al. 1998; Smogorzewska et al. 2002; Celli and de Lange 2005; Bae and Baumann 2007; Sarthy et al. 2009). In S. cerevisiae, the Rap1 protein directly binds the telomere sequences and establishes several parallel pathways to inhibit NHEJ (Pardo and Marcand 2005; Marcand et al. 2008). The synergy between different pathways ensures that fusions remain rare. In the absence of telomerase-mediated telomere elongation, fusions occur mostly between extremely short telomeres, as observed in yeast (Chan and Blackburn 2003; Mieczkowski et al. 2003), plants (Heacock et al. 2004), worms (Lowden et al. 2008), and human cells (Capper et al. 2007). However, telomere fusion frequencies in wild-type physiological contexts are yet unknown.
Telomere fusions between chromosomes lead to dicentric chromosomes, whose expected detrimental consequences are observed in several experimental situations. For instance, TRF2 inhibition in human cells results in anaphase bridges (van Steensel et al. 1998). In mice, liver cells with extensive telomere fusions induced by TRF2 loss fail to proceed normally through anaphase (Denchi et al. 2006). In fission yeast lacking Taz1 and in budding yeast with a conditional Rap1 mutant, telomere fusions cause some cell death (Ferreira and Cooper 2001; Pardo and Marcand 2005). Most commonly studied dicentrics are created by double-strand break-induced rearrangements and were first described by Barbara McClintock in the 1940s (McClintock 1941, 1942). They form anaphase bridges that break somewhere between the two centromeres, causing further rearrangements and genome instability, which eventually lead to cell death (Bajer 1964; Mann and Davis 1983; Haber et al. 1984; Surosky and Tye 1985; Koshland et al. 1987; Hill and Bloom 1989; Kramer et al. 1994; Jannink et al. 1996; Lo et al. 2002; Han et al. 2009; Paek et al. 2009; Pennaneach and Kolodner 2009). The mechanism of dicentric breakage is unknown. The high elasticity of mitotic chromosomes does not suggest a simple mechanical shearing by spindle forces (Marko 2008).
Interestingly, telomere fusions that occur spontaneously in the germline can sometimes get stabilized. An example in human evolution is chromosome 2, which comes from a fusion between two chromosomes that are distinct in apes (Lejeune et al. 1973). The head-to-head telomeric repeats are still present at the fusion point (IJdo et al. 1991), and one centromere is inactivated (Avarello et al. 1992). Recently, individual chromosome fusions were selected from fission yeast cells with an irreversible centromere deletion (Ishii et al. 2008). Here we address the behavior of individual dicentrics formed by telomere fusion in S. cerevisiae. Our approach was to first stabilize and amplify individual fusions between two chromosomes by temporarily inactivating one centromere in cells with frequent telomere–telomere fusions. Next, we analyzed dicentric breakage following centromere reactivation. Unexpectedly, dicentrics often break at the telomere fusions.
Results
Selection of individual telomere fusions
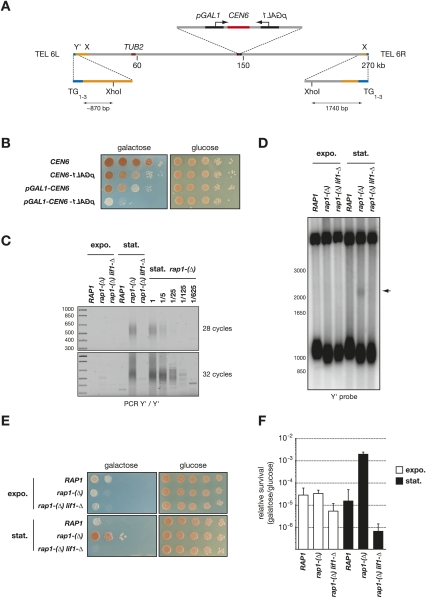
First, we needed a new tool to stabilize and select individual telomere fusions. In S. cerevisiae, a centromere can be partially inactivated by transcription from a galactose-inducible GAL1 promoter directed toward the centromere (Hill and Bloom 1987). The inactivated centromere is detached from the spindle. When transcription is turned off, the centromere is rapidly reattached to the spindle, and its function is restored (Tanaka et al. 2005). Centromere inactivation will stabilize chromosome fusions. In addition, it will create a positive selection for the fusions, since the missegregation of an unfused acentric chromosome will have a detrimental effect on cell growth, and chromosome fusion will rescue normal segregation and viability (Ishii et al. 2008). To increase the stringency of this selection, we chose to inactivate the centromere of chromosome 6. Increased copy number of this chromosome kills the cells, likely because it carries the TUB2 gene encoding the β subunit of tubulin (Katz et al. 1990; Torres et al. 2007). In a haploid context, a missegregation of chromosome 6 will lead to one cell missing the chromosome and one cell with two copies, both being unviable. We inserted a GAL1 promoter on each side of chromosome 6 centromere (CEN6) (Fig. 1A). As shown in Figure 1B, growth on galactose-containing medium is partially impaired when one promoter is present on one side of CEN6, and is further inhibited when one promoter is present on each side of CEN6. On glucose, transcription is repressed and cell growth is normal. In this study, we chose to use the construct with two GAL1 promoters because it appeared more effective at inactivating CEN6.
Figure 1.
Selection of individual telomere fusions. (A) Schematic representation of S. cerevisiae chromosome 6. Y′ are 5- to 7-kb subtelomeric elements present in one to four copies of about half of the chromosome ends. The left end of chromosome 6 (TEL 6L) contains at least one Y′ and an ∼500-bp X subtelomeric element followed by an ∼10-kb stretch with homology with other subtelomeric regions. The right end of chromosome 6 (TEL 6R) contains an X element followed by a unique sequence with no homology with the rest of the genome. (B) Loss of cell viability in response to CEN6 inactivation by transcription. Strains Lev554 (CEN6), Lev1064 (CEN6-pGAL1), Lev1211 (pGAL1-CEN6), and Lev1212 (pGAL1-CEN6-pGAL1) were grown overnight in glucose-containing rich medium, diluted by 10-fold serial dilutions, plated on synthetic media, and incubated for 3 d (glucose) or 4 d (galactose) at 30°C . The ade2-1 mutation present in this background causes the cells to accumulate purine precursors when they start to exhaust the adenine from the medium. These precursors give a red color to the colonies. Colonies with a large fraction of sick or dead cells remain white. (C) Amplification of fusions between Y′ telomeres. Strains Lev1212 (RAP1), Lev728 [rap1-(Δ)], and Lev730 [rap1-(Δ) lif1-Δ] were grown exponentially in glucose-containing rich medium (expo.) and allowed to reach stationary phase in 6 d (stat.). Fusions were amplified by PCR of 28 and 32 cycles with two primers annealing to Y′ ends. Genomic DNA from rap1-(Δ) cells in stationary phase was diluted serially to provide a semiquantitative estimation of the relative fusion frequency and of the method sensitivity. (D) Detection by Southern blot of fusions between Y′ telomeres in rap1-(Δ) cells in stationary phase. Genomic DNA was digested with XhoI, separated in a 0.9% agarose gel, and probed with a Y′ distal fragment. The size marker on the left is in base pairs. The mean telomere length in rap1-(Δ) cells is ∼240 bp; i.e., ∼60 bp shorter that in wild-type cells. An arrow marks the position of the Y′–Y′ telomere fusions. (E) Selection of survivors to CEN6 inactivation in rap1-(Δ) cells in stationary phase (see the Materials and Methods). (F) Quantification of the relative frequency of survivors to CEN6 inactivation from three independent experiments.
Next, we introduced this conditional centromere in cells where telomere fusions by NHEJ are frequent, using a previously described RAP1 conditional degron allele called rap1-(Δ). In rap1-(Δ) cells, rap1 protein level decreases, and fusions accumulate when cells reach stationary phase (Pardo and Marcand 2005). Figure 1C shows the amplification of telomere fusions by PCR with two primers annealing at the end of the Y′ sequences, the subtelomeric elements present at the tip of about half of the chromosome ends in S. cerevisiae (Louis and Haber 1992). Fusions are easily amplified from rap1-(Δ) cells that have reached stationary phase, but not from RAP1 cells or from rap1-(Δ) lif1-Δ cells defective for NHEJ. In rap1-(Δ) cells growing exponentially, fusions appear at least 100 times less frequently than in stationary phase. Fusions between telomeres should also be detectable by Southern blot if their frequency is high enough, a method first used in human cells (van Steensel et al. 1998). In yeast, Y′ telomeres display a conserved XhoI restriction site at an 850- to 900-base-pair (bp) distance from the beginning of the TG1–3 telomeric repeats (Fig. 1A). Figure 1D shows that, in rap1-(Δ) cells that have reached stationary phase, a new smear is detectable at about twice the average size of the XhoI telomeric restriction fragments in these cells. This smear is not detected in rap1-(Δ) lif1-Δ cells defective for NHEJ. The intensity of the smear is ∼3% of the signal from the Y′ ends, suggesting an average of one fusion between two Y′ telomeres in a quarter or a third of the rap1-(Δ) cells in stationary phase, assuming a random distribution in the population.
When stationary rap1-(Δ) cells with a conditional CEN6 were spread on galactose-containing plates, survivors to CEN6 inactivation appear at a frequency of ∼0.2% (Fig. 1E,F). Survivors are at least 50 times less frequent in rap1-(Δ) cells growing exponentially and in rap1-(Δ) lif1-Δ cells in stationary phase, indicating an overall correlation between survivors of CEN6 inactivation and the presence of telomere fusions in the cells prior to CEN6 inactivation. Since S. cerevisiae has 16 chromosomes, our previous estimation from Figure 1D that there is at least one fusion in a quarter of the stationary rap1-(Δ) cells predicts that ∼3% of the cells should have chromosome 6 fused to another chromosome, supposing the odds are equal among all telomeres. The discrepancy between this simple prediction and the observed 0.2% frequency of survivors suggests that, in stationary rap1-(Δ) cells, some telomeres may be more exposed to NHEJ than chromosome 6 telomeres, or that fusions between the two ends of chromosome 6, unselected in the assay, may be more frequent than fusions between chromosome 6 and another chromosome. In addition, fusions may be distributed unequally among the cells.
Characterization of individual telomere fusions
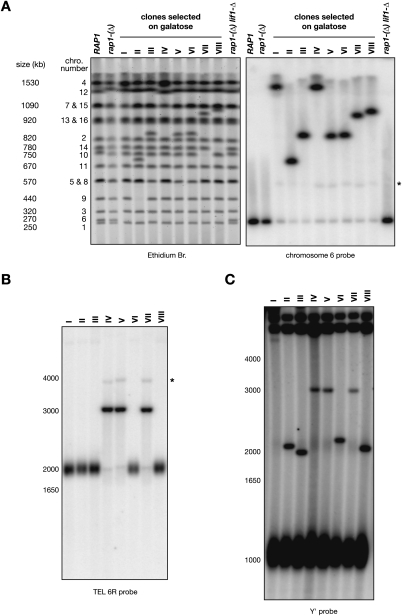
The karyotype of eight individual clones selected on galactose from stationary rap1-(Δ) cells was analyzed by pulse-field electrophoresis (Fig. 2A, left panel). In all clones, chromosome 6 as well as another chromosome are missing at their native position, and a single new chromosome appears at a size close to the sum of the two missing chromosomes. For instance, in clone II, chromosomes 6 (∼270 kb) and 9 (∼440 kb) are missing, and a new chromosome appears between 670 and 750 kb, indicating that chromosomes 6 and 9 are fused together. Hybridization with a probe from chromosome 6 shows for each clone the main signal at the position of the new chromosome (Fig. 2A, right panel). Weaker signals are present at the native position of chromosome 6 and at ∼550 kb, about twice the size of chromosome 6 (see below).
Figure 2.
Identification of eight individual telomere fusions. (A) Strains Lev1212 (RAP1), Lev728 [rap1-(Δ)], and Lev730 [rap1-(Δ) lif1-Δ] were grown in glucose-containing synthetic medium, and clones I–VIII selected on galactose from Lev728 were grown in galactose-containing synthetic medium. Chromosomes were separated by PFGE, labeled with ethidium bromide (left panel), and probed with a fragment from chromosome 6 (right panel). The size and position of the native chromosomes are labeled on the left. An asterisk marks the position of the weak ∼550-kb band. (B) Detection of the fusions with TEL 6R by Southern blot. Genomic DNA was digested with XhoI, separated in a 0.9% agarose gel, and probed with a TEL 6R-specific fragment. The size marker on the left is in base pairs. An asterisk marks the position of the weak ∼4-kb band. (C) Detection of the fusions with Y′ telomeres by Southern blot. Genomic DNA was digested with XhoI, separated in a 0.9% agarose gel, and probed with a Y′ distal fragment. The size marker on the left is in base pairs. The mean telomere length in rap1-(Δ) cells grown in galactose-containing medium is ∼180 bp, ∼60 bp shorter than in the same cells grown in glucose-containing medium (data not shown).
To further characterize the individual fusions, we first used the XhoI restriction site present at 1740 bp from the TG1–3 telomeric repeats at the right end of chromosome 6 (TEL 6R) (Fig. 1A). In a Southern blot with a TEL 6R probe (Fig. 2B), several clones display a discrete band at ∼3 kb instead of the telomeric smear at ∼2 kb, indicating that the right telomere of chromosome 6 is fused in these clones. The size of the bands suggests fusions with Y′ telomeres. A faint smear is also visible around 2 kb as well as two weak discrete bands at ∼2 kb and ∼4 kb (see below). The same samples were further analyzed by Southern blot with XhoI and a Y′ probe (Fig. 2C). The clones with a fusion involving TEL 6R display the same ∼3-kb band seen with a TEL 6R probe, confirming a fusion between TEL 6R and a Y′ telomere in these clones. Other clones display a new discrete band between 2 and 2.2 kb, suggesting a fusion between the left end of chromosome 6 (TEL 6L) and another Y′ telomere. Clone I displays a band above 4 kb, suggesting a fusion between TEL 6L and an X telomere. The subtelomeric sequences adjacent to the fusions were confirmed by PCRs with an X and a Y′ primer (clone I), with two Y′ primers (clones II, III, VI, and VIII), and with a TEL 6R and a Y′ primer (clones IV, V, and VII) (data not shown). The fusions of clones III and VIII display a restriction site at the junction of the two telomeres, allowing the cloning and sequencing of each telomere involved in the fusion. Clone III had fused two telomeres of 147 and 95 bp of TG1–3, and clone VIII had fused two telomeres of 159 and 152 bp (their sequences are shown in the Material and Methods). Thus, inactivation of one centromere allows the selection and propagation of individual telomere fusions with head-to-head telomeric repeats at the junction. The length of the repeats in the cloned fusions is shorter than the mean telomere length in rap1-(Δ) cells (∼240 bp). One interpretation is that telomere fusion may be favored by cooperation between the reduction of Rap1-binding sites in the shortest telomeres and the partial loss of function of the rap1-(Δ) mutant (Pardo and Marcand 2005). Another bias that we observed among a larger pool of selected fusions is that TEL 6L is fused more frequently than TEL 6R, and that some chromosomes are fused more frequently than others to chromosome 6 (e.g., chromosome 5) (data not shown). Again, it might be that some telomeres are relatively more exposed to NHEJ than others in stationary rap1-(Δ) cells. The constraints of chromosomes organization in the nucleus might also favor fusions between telomeres that are more likely to interact transiently (Therizols et al. 2010).
Breakage of dicentrics formed by telomere fusion
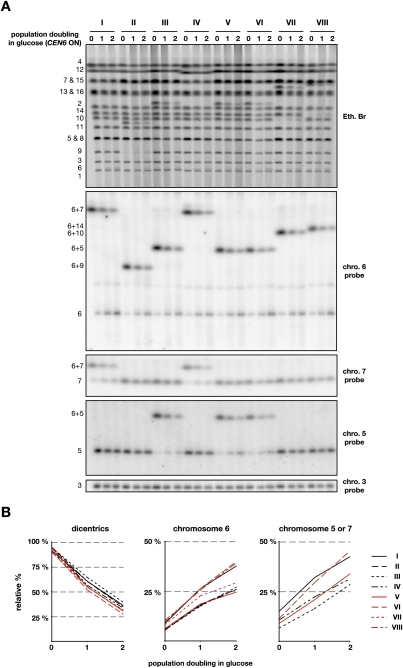
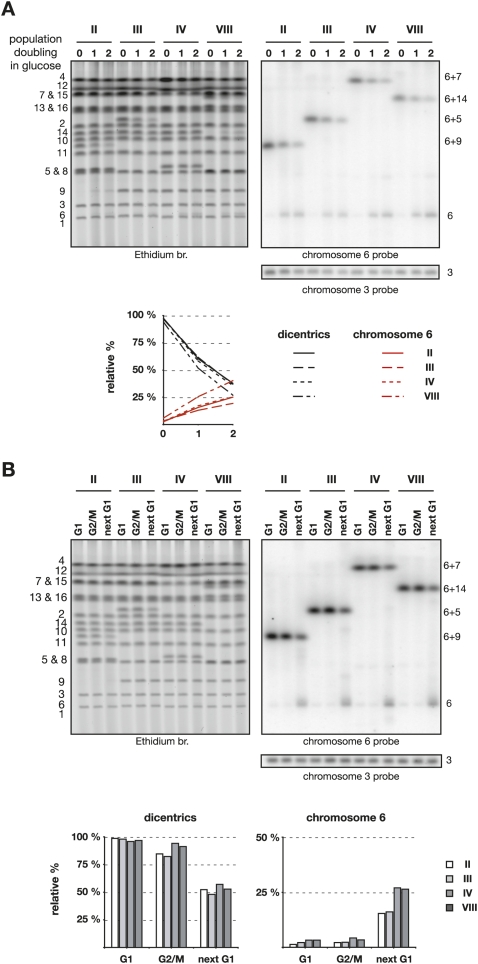
We next asked how the fused chromosomes behave when the function of CEN6 is restored. Cells grown exponentially in the presence of galactose were switched to a glucose-containing medium for one and two population doublings. Karyotype analysis by pulse-field electrophoresis shows two interesting phenomena (Fig. 3A). First, the dicentrics formed by the fused chromosomes fade away when the cells divide in glucose. This is seen by ethidium bromide staining when the dicentric is not overlapping with another chromosome, and by Southern blot. Secondly, a chromosome reappears at the positions of the native chromosomes that were fused together in the dicentrics. This reappearance is visible by ethidium staining at the position of chromosome 6 as well as at the positions of chromosomes 9, 10, and 14 for clones II, VII, and VIII, respectively. Southern blot confirms the reappearance of chromosome 6 in every clone. It also shows the reappearance of chromosome 7 for clones I and IV, and of chromosome 5 for clones III, V, and VI. This indicates that breakage of a dicentric often occurs at or near the telomere fusion joining the two chromosomes. Hence, telomere fusions behave as hot spots particularly prone to breakage.
Figure 3.
Breakage of dicentrics formed by telomere fusion. (A) Cells from clones I–VIII were grown exponentially in galactose-containing synthetic medium (0) and switched to glucose-containing rich medium for one or two population doublings (1 and 2). (Top panel) Chromosomes separated by PFGE were labeled with ethidium bromide and probed successively with fragments from chromosomes 6, 3, 7, and 5. The position of the native and fused chromosomes is labeled on the left. (B) Quantification of the relative signals from the dicentrics and the unfused chromosome 6 seen with chromosome 6 probe and from unfused chromosome 5 (clones III, V, and VI) or chromosome 7 (clones I and IV) seen with chromosome 5 and 7 probes, respectively. The signals were corrected to the signal from chromosome 3 for each lane. For each series, the indicated percentage is relative to the sum of the dicentric and the unfused chromosome 6 in cells grown in galactose.
To quantify the loss of the dicentrics and the reappearance of the chromosomes at their native positions, the Southern blot was rehybridized with a probe from chromosome 3 whose signal was used as an internal loading control (Fig. 3A). The corrected signals were then normalized to the sum of the signals from the dicentric and from the native chromosome in cells growing with galactose (point 0) (Fig. 3B). On average, dicentrics drop from 93% ± 2% in galactose to 57% ± 4% and 33% ± 4% after one and two doublings in glucose, respectively. The proportion of unfused chromosome 6 increases from 7% ± 2% in galactose to 22% ± 4% and 31% ± 7% after one and two doublings in glucose, respectively. The relative signal pooled from unfused chromosome 7 in clones I and IV and from unfused chromosome 5 in clones III, V, and VI increases from 10% ± 3% in galactose to 24% ± 6% and 36% ± 7% after one and two doublings in glucose, respectively. This quantification indicates that ∼40% of the time breakage of the dicentrics restores chromosomes of native size.
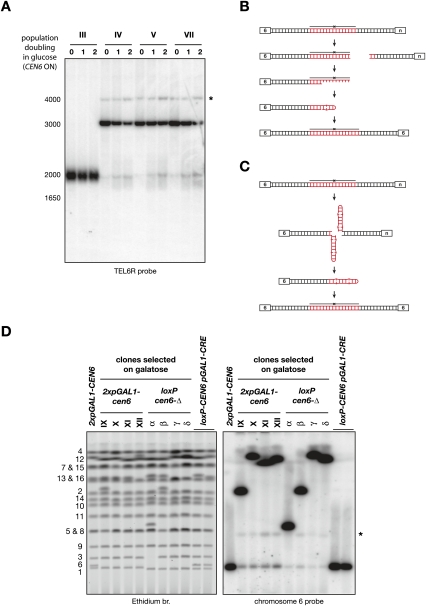
Breakage at TEL 6R
We took advantage of the unique sequence near TEL 6R to have a close-up view at the fusions of clones IV, V, and VII during the first two divisions following the reactivation of CEN6 (Fig. 4A). A loss of the fusion at ∼3 kb is visible, as well as the appearance of a smear around 2 kb. The width of the smear is relatively narrow, suggesting that breakage occurs within the telomeric repeats and not much within the adjacent subtelomeric sequences. A discrete band within the smear also increases in intensity. This band may suggest that breakage often occurs at a preferred position; for example, at the junction of the two telomeres. An alternative explanation is shown in Figure 4B (Bateman 1975; Smith 2008). Breakage may occur randomly within the telomeric repeats, but 5′ degradation may expose the junction as ssDNA, which can fold back on itself, forming the discrete band at ∼2 kb. If this hairpin is filled in and ligated but not opened, replication during the next cell cycle creates a dicentric isochromatid that would explain the XhoI fragment at ∼4 kb as well as the ∼550-kb band detected with a chromosome 6 probe on chromosomes separated by pulse-field electrophoresis (Fig. 2A). Break-induced exchange between chromatids at inverted repeats may also create a dicentric isochromatid (Grossi et al. 2001). We note that the ∼550-kb band migrates slightly faster when the right telomere of chromosome 6 is involved in the fusion than when it is the left telomere. This might be due to different conformations in the gel during pulse-field electrophoresis (Lalande et al. 1987).
Figure 4.
(A) Breakage at TEL 6R. Cells from clones III, IV, V, and VII were grown exponentially in galactose-containing synthetic medium (0) and switched to glucose-containing rich medium for one or two population doublings (1 and 2). Genomic DNA was digested with XhoI, separated in a 0.9% agarose gel, and probed with a TEL 6R-specific fragment. The size marker on the left is in base pairs. An asterisk marks the position of the weak ∼4-kb band. (B) Breakage-and-degradation model for the formation of dicentric isochromatids (Smith 2008). The fused TG1–3 repeats are in red. (C) Cruciform cleavage model for the formation of dicentric isochromatids (Lobachev et al. 2002). (D) Telomere fusion instability in a monocentric context. Chromosomes were separated by PFGE, labeled with ethidium bromide (left panel), and probed with a fragment from chromosome 6 (right panel). The position of the native chromosomes is labeled on the left. An asterisk marks the position of the weak ∼550-kb band.
Telomere fusion instability in a monocentric context
To test whether the residual level of unfused chromosome 6 in galactose was due to a partial activity of CEN6, we constructed a rap1-(Δ) strain with two loxP sites in direct repeats on each side of CEN6. Induction of the CRE recombinase deletes CEN6 from the chromosome and allows the selection of survivors in which chromosome 6 is fused with another chromosome (see the Materials and Methods). Figure 4D shows an example of four cen6-Δ clones (α–δ) next to four other clones where CEN6 is inactivated by transcription from two pGAL1 promoters (IX–XII). Clone δ is a fusion between TEL 6R and a Y′ telomere. The seven others are fusions between TEL 6L and another Y′ telomere (data not shown). Hybridization with a probe from chromosome 6 shows that the residual level of unfused chromosome 6 is reduced by approximately threefold when CEN6 is deleted compared with when it is inactivated by transcription (Fig. 4D). This difference indicates that CEN6 is still sometimes active when transcription is induced from the two pGAL1 promoters. In addition, the steady level of unfused chromosome 6 in the absence of CEN6 despite its permanent counterselection is indicative of a lower but significant instability of the telomere fusions. Interestingly, the band at ∼550 kb is still detectable in the cen6-Δ clones. This instability in a monocentric context may stem from the palindromic structure of telomere fusions (Fig. 4C; Szostak 1983; Murray et al. 1988; Lobachev et al. 2002, 2007; Cote and Lewis 2008; Smith 2008).
Breakage occurs during progression through mitosis
In the previous experiments, breakage was observed in a permissive condition of the rap1-(Δ) mutant. To study dicentrics formed by telomere fusion in a context closer to wild type, we complemented the rap1-(Δ) allele in clones II, III, IV, and VIII with an integrative plasmid carrying a wild-type copy of RAP1. This 8-kb plasmid integrates at the URA3 locus on chromosome 5, sometimes in multiple copies. The dicentrics we studied display one (clones III and VIII), two (clone II), or four (clone IV) copies of RAP1 (data not shown) (also seen in the increased length of chromosome 5 in clones II and IV in Fig. 5A). Complemented cells were grown exponentially in the presence of galactose and switched to a glucose-containing medium for one and two population doublings. In the experiment displayed in Figure 5A, dicentrics in RAP1-complemented cells drop on average from 96% ± 2% in galactose to 58% ± 4% and 34% ± 5% after one and two doublings in glucose, respectively. The proportion of unfused chromosome 6 increases from 4% ± 2% in galactose to 18% ± 5% and 28% ± 9% after one and two doublings in glucose, respectively. This is indistinguishable from the breakage observed previously in rap1-(Δ) cells. Rap1 implication in this process is not ruled out by this result, since its function could still be effective in the permissive condition of the rap1-(Δ) mutant.
Figure 5.
Breakage of dicentrics in RAP1-complemented cells. (A) Clones II, III, IV, and VIII transformed with pRS306-RAP1 were grown exponentially in galactose-containing synthetic medium (0) and switched to glucose-containing rich medium for one or two population doublings (1 and 2). Chromosomes were separated by PFGE, labeled with ethidium bromide (left panel), and probed with a fragment from chromosome 6 (top right panel) and a fragment from chromosome 3 (bottom right panel). The positions of the native and fused chromosomes are labeled on the right. The signals were corrected to the signal from chromosome 3 for each lane. For each series, the indicated percentage is relative to the sum of the dicentric and the unfused chromosome 6 in cells grown in galactose. (B) Cells transformed with pRS306-RAP1 and grown exponentially in galactose-containing synthetic medium were synchronized in G1 with α-factor (G1), released in glucose-containing rich medium, and either blocked in G2/M with nocodazole (G2/M) or allowed to proceed through mitosis to be blocked in the next G1 with α-factor (next G1). Chromosomes were separated by PFGE, labeled with ethidium bromide (left panel), and probed with a fragment from chromosome 6 (top right panel) and a fragment from chromosome 3 (bottom right panel). The positions of the native and fused chromosomes are labeled on the right. The signals were corrected to the signal from chromosome 3 for each lane. For each series, the indicated percentage is relative to the sum of the dicentric and the unfused chromosome 6 in cells in G1 in galactose.
Next, we looked at dicentric breakage in RAP1-complemented cells during a single cell cycle. Cells grown with galactose were synchronized in G1 phase by α-factor and then released from the block in glucose-containing medium. Cells were either blocked at the G2/M transition by nocodazole or allowed to move through mitosis until they were blocked again in the next G1 by α-factor that was added back to the medium. As shown in Figure 5B, transition from G2/M to the next G1 is needed to break the dicentrics and to restore chromosome 6. During this transition, about half of the dicentrics are broken (from 97% ± 1% in G1 and 88% ± 6% in G2/M to 52% ± 4% in the next G1), and a fraction of the signal reappears at the position of chromosome 6 (from 3% ± 1% in G1 and 3% ± 1% in G2/M to 21% ± 6% in the next G1). Thus dicentric breakage at telomere fusions requires progression through mitosis.
Recovery from the fusions
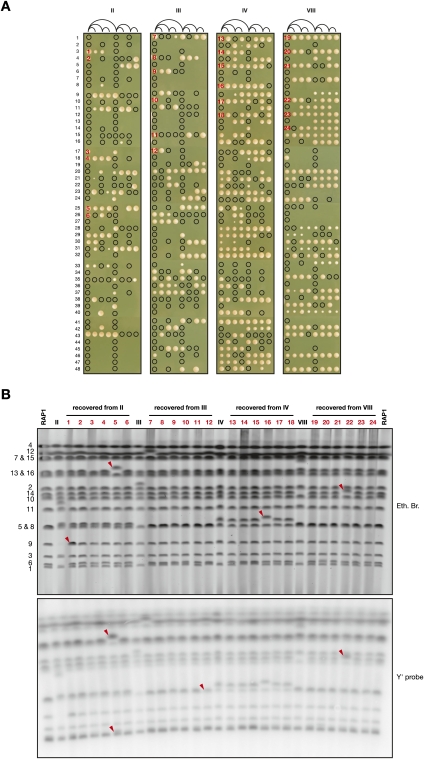
To test their ability to recover from breakage, RAP1-complemented cells containing a dicentric chromosome were grown exponentially in galactose-containing medium and positioned on glucose-containing plates. During their first three generations on glucose, individual cells were separated after each division unless they failed to proceed through cytokinesis. The cells were then left for 3 d at 30°C to form colonies. The result is shown in Figure 6A. A black circle indicates a separated cell that failed to give rise to a colony. Among 48 initial cells from each clone, a majority (26–41) formed at least one colony. However, lethality is very frequent and appears to occur stochastically during the first divisions. Interestingly, the crisis is more acute for clones II and III, and this difference correlates with a lower efficiency at breaking at the fusion during one cell cycle (Fig. 5B). In addition, a few cells failed at the first division, perhaps because the dicentric was already broken and chromosome 6 was missegregated before plating.
Figure 6.
Recovery from the fusions. (A) Clones II, III, IV, and VIII transformed with pRS306-RAP1 were grown exponentially in galactose-containing synthetic medium. (First position from the left) For each clone, 48 individual cells were plated on glucose-containing rich medium. After each of their three first divisions, cells were separated and repositioned on the same line, as indicated in the diagram, unless they failed to proceed through cytokinesis within a few hours. (That is, after a first division, one cell was left at the first position from the left, and the other cell was moved to the fifth position from the left. After a second division, one cell was left at its original position, and the other cell was moved to the third or the seventh position from the left, respectively, etc.). Colonies were photographed after incubation for 3 d at 30°C. A circle indicates the presence of a cell that failed to form a colony. The 24 colonies chosen for PFGE analysis are numbered in red. (B) Chromosomes from clones II, III, IV, and VIII transformed with pRS306-RAP1 grown in galactose-containing synthetic medium and from Lev1212 (RAP1) and the chosen 24 colonies grown in glucose-containing rich medium were separated by PFGE, labeled with ethidium bromide (top panel), and probed with a Y′ fragment (bottom panel). Changes are indicated with red arrows. In colony #1, chromosome 9 appears at two copies. In colony #5, the relative Y′ signal is increased for chromosome 6 or 1 and for chromosome 13 or 16. In colony #11, the Y′ signal is reduced for chromosome 5 or 8. In colony #22, the Y′ signal of chromosome 14 is increased.
The karyotype of 24 colonies obtained from the pedigree was analyzed by pulse-field electrophoresis. We chose six independent colonies for each dicentric (Fig. 6A). In each colony, the chromosomes that formed the dicentrics reappear at or very close to their native positions. The only exception is in colony #22, where chromosome 14 reappears higher than expected (Fig. 6B). Another interesting change is seen in colony #1, where chromosome 9 appears at two copies. In addition, a chromosome not involved in the fusions is increased in size in colonies #5 and #16. Hybridization with a Y′ probe shows that Y′ copy number did not change except in colonies #5, #11, and #22 (Fig. 6B, bottom panel). These rearrangements may originate from breakage within a subtelomeric sequence, followed by break-induced replication with another subtelomere.
Discussion
In this study, we observed that the inactivation of one centromere allows the selection and propagation of head-to-head telomere fusions between chromosomes. When the two centromeres are functional, the dicentrics often break at the telomere fusion during progression through mitosis. This processing restores both chromosomes and allows the cell lineage to recover from the fusion. This shows a remarkable resilience of the genome. It also suggests an unsuspected rescue pathway able to back up NHEJ inhibition at telomeres. This pathway would allow a reversibility of telomere fusions, a common strategy to proofread a biological function. Its existence would imply that, despite several mechanisms ensuring that NHEJ between telomeres remains unlikely, rare fusions sometimes occur in normal cells. Fusion occurrence might be significant during a prolonged quiescent state in spores or in stationary phase, but this remains to be determined.
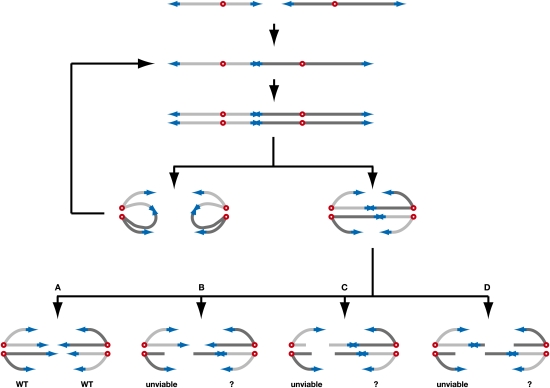
In our assay, ∼40% of the dicentrics regenerate two parental chromosomes after breakage. This partial effectiveness causes lethality among the progeny of the first divisions following dicentric formation, and sometimes genome changes making this process error-prone. In Figure 7, we propose a scenario to explain how a cell with a dicentric either keeps dividing or dies or recovers from the fusion. In anaphase, the two centromeres of each sister chromatid can migrate toward the same pole, thus propagating the dicentric to the next generation in the two offspring. Alternatively, the centromeres of each sister chromatid can migrate toward opposite poles, forming two anaphase bridges. Breakage of the two bridges at the fusion restores a normal karyotype in the two offspring (situation A). Breakage of one bridge at the fusion and breakage of the other bridge away from the fusion creates one progeny missing a chromosome fragment, a lethal situation in haploids (situation B). The other progeny might repair the broken double-strand end by break-induced replication (Lydeard et al. 2007; Smith et al. 2007), recreating the dicentric but with an extra copy of one chromosome (as in seen in colony #1 in Fig. 6B). This outcome would be lethal if chromosome 6 is the duplicated chromosome (Torres et al. 2007). Breakage of the two bridges may also occur away from the fusion, either on the same side relative to the fusion (situation C) or on opposite sides (situation D). In both cases, one progeny misses one or two chromosome fragments and therefore becomes unviable. The other progeny might also repair the two broken double-strand ends by break-induced replication. Fusion by NHEJ might also occur. In situation D, if the two breaks occur nearby, fusion of the two broken ends by single-strand annealing would recreate a single copy of the dicentric.
Figure 7.
Model for the segregation and breakage during one cell cycle of a dicentric formed by a telomere fusion. The telomeres are in blue, and the centromeres are in red. The proposed scenario is described in the second paragraph of the Discussion.
It would be interesting to understand why breakage of the fusions is not more effective. It is possible that the laboratory strain we used had lost some fitness and that other strains will prove more capable of doing it. It is also possible that an inherently error-prone mechanism of breakage comes with a cost on genome stability even in the absence of fusions. A trade-off would set the effectiveness at an intermediate level. A redundancy among the mechanisms established to avoid telomere fusion might also facilitate an evolutionary drift of the rescue pathway. Alternatively, we cannot formally rule out that the function under natural selection is not the breakage of rare telomere fusions, but instead other accidents generating dicentric-like situations that might be more frequent in wild-type cells; for example, entangled telomeres following replication (Miller et al. 2006; Rog et al. 2009).
The mechanism of breakage is unknown except that it occurs in a narrow hot spot (Fig. 4A). This feature will help molecular understanding of the process. Several working models can be imagined. Breakage at the fusion could occur during anaphase when chromatin under tension might be more susceptible to break at a telomeric sequence because of its chromatin structure, a different condensation level, the presence of nicks, or the recruitment of nucleases by the telomeric proteins. Breakage may also be caused by cytokinesis, as observed for entangled chromosomes in Top2-depleted yeast cells (Baxter and Diffley 2008) and for dicentrics created by double-strand break-induced rearrangements in plant cells (Bajer 1964). In this model, the fusion would be recruited to the plane of cleavage during abscission, and would be silent to the NoCut checkpoint that delays cytokinesis in response to lagging chromatin (Norden et al. 2006; Mendoza et al. 2009). It would be interesting to address whether the palindromic structure of the fusion and Rap1 or the factors recruited by Rap1 are important to create a hot spot for breakage. Finally, it is not yet known whether a telomere fusion rescue pathway could exist in other eukaryotes. A recently described temperature-sensitive TRF2 allele might be a tool to address this issue in mammals (Konishi and de Lange 2008). Understanding the mechanism of dicentric breakage at telomere fusions and whether it is conserved in evolution should shed more light on how cells maintain genome stability.
Materials and methods
Strains and plasmids
The strains used in this study are listed in Table 1. The GAL1 promoters were inserted at CEN6 by two successive PCR-mediated transformations using the pFA6a-KanMX6-pGAL1 and pFA6a-skHIS3MX6-pGAL1 plasmids (Longtine et al. 1998). Kan-pGAL1 was inserted 11 bp away from the left side of CEN6. skHIS3-pGAL1 was inserted 81 bp away from the right side of CEN6. The resulting sequence surrounding CEN6 in KANr-pGAL1-CEN6-pGAL1-skHIS3 strains is AAGGAGAAAAAACCCGGATCTCAAAATGATATTTCTTTTCATCACGTGCTATAAAAATAATTATAATTTAAATTTTTTAATATAAATATATAAATTAAAAATAGAAAGTAAAAAAAGAAATTAAAGAAAAAATAGTTTTTGTTTTCCGAAGATGTAAAATAGGTTGAAAGTTAGAAATTAGTATTATAATAGCAAAAAAAATTTAAAGTTAGAAATTAGAATTTAAGGCTCTACACACGCATTTTGAGATCCGGGTTTTTTCTCCTT. The end of the GAL1 promoters is underlined; the CEN6 sequence is in bold.
Table 1.
Yeast strains used in this study
All are from the W303-1a background (ade2-1 trp1-1 ura3-1 leu2-3,112 his3-11,15 can1-100 RAD5). In all listed strains, the UBR1 and ROX1 genes are under the control of the copper-inducible ACE1 promoter. In strains Lev728, Lev730, Lev1125, and Lev1126, rap1 is under the control of a promoter repressible by Rox1 and expresses a protein with an N-terminal tag that makes it a target for Ubr1 and degradation by the N-end rule (Pardo and Marcand 2005).
The loxP sites were inserted at the same positions around CEN6 by two successive PCR-mediated transformations using the pFA6a-klTRP1MX6 and pFA6a-skHIS3MX6 plasmids. The klTRP1 is on the left side of CEN6 within the two loxP sites, and skHIS3 is on the right side of CEN6 outside the two loxP sites. Plasmid pRS315-pGAL1-CRE was transformed into yeast strains Lev1125 and Lev1126.
Plasmid pRS306-RAP1 (Pardo and Marcand 2005) was digested by NdeI prior to transformation into yeast cells.
Amplification of the telomere–telomere fusions by PCR
Genomic DNA was prepared by zymolyase digestion followed by phenol-chloroform extraction. Fusions between Y′ telomeres were amplified with the two following 34-mer: 5′-GTCAGAAAGCCGGGTAAGGTATGACAGCGAGAGT-3′; 5′-GTCAGAAAGCCGGGTAAGGAGTGACAGCGCGAGT-3′.
These primers anneal to the end of the Y′ elements 15–20 bp from the TG1–3 repeats. PCR reactions (30 μL) contained ∼10 ng of genomic DNA, 1× Phusion GC buffer, 3% DMSO, 200 μM each dNTP, 0.5 μM each primer, and 0.6 U of Phusion Polymerase (Finnzymes). The conditions were 30 sec at 98°C, then 28 or 32 cycles of 10 sec at 98°C, 50 sec at 72°C,and 5 min at 72°C. The products (10 μL) were separated through a 1% agarose gel containing 0.1 μg/mL ethidium bromide, and fluorescence was quantified using a Typhoon imager.
Selection of individual fusions
Strains Lev1212 (pGAL1-CEN6-pGAL1), Lev728 [pGAL1-CEN6-pGAL1 rap1-(Δ)], and Lev730 [pGAL1-CEN6-pGAL1 rap1-(Δ) lif1-Δ] were freshly streaked on glucose-containing rich medium (YPD) and then grown exponentially in YPD for 24 h. Parts of the cultures were allowed to reach saturation in 6 d. Cells growing exponentially or in stationary phase were spread at an appropriate dilution on 2% glucose-containing synthetic medium plates and on 2% galactose-containing synthetic medium plates. Colonies were counted after incubation for 4 d at 30°C. Survivors from Lev728 in stationary phase were restreaked on galactose-containing synthetic medium plates and cultured in liquid galactose-containing synthetic medium at 30°C for further analysis. The clones proved to be stable after several restreaks on galactose-containing synthetic medium plates. The steady level of unfused chromosome 6 also remains unchanged in galactose (data not shown).
The karyotypes of a few survivors from Lev1212 (pGAL1-CEN6-pGAL1) and Lev730 [pGAL1-CEN6-pGAL1 rap1-(Δ) lif1-Δ] were studied. Unexpectedly, they display a normal chromosome 6 and two copies of chromosome 2 (data not shown). Chromosome 2 carries the cluster of the GAL7, GAL10, and GAL1 genes whose products process galactose into glucose-phosphate. GAL2, encoding the galactose permease, is on a different chromosome. Introduction into Lev1212 and Lev730 of an extra copy of GAL7, GAL10, and GAL1 on a centromeric plasmid greatly increases the frequency of survivors on galactose (data not shown). This suggests that two copies of chromosome 2 confer resistance to galactose by increasing the dosage of GAL7, GAL10, and GAL1, which may in turn reduce the intracellular concentration of galactose and diminish the rate of transcription from the GAL1 promoters, partially restoring CEN6 function.
Strains Lev1125 and Lev1126 [loxP-CEN6-loxP rap1-(Δ)] transformed with pRS315-pGAL1-CRE were freshly streaked on glucose-containing synthetic medium lacking leucine and then grown to saturation in YPD in 6 d at 30°C. Cells were spread at an appropriate dilution on 2% galactose-containing synthetic medium plates lacking leucine. Survivors were restreaked once on galactose-containing synthetic medium plates lacking leucine and were checked for the loss of the klTRP1 gene linked to the loxP-CEN6 cassette on synthetic medium plates lacking tryptophan. The loss of CEN6 was then confirmed by PCR with primers surrounding the CEN6 locus. The presence of a telomere–telomere fusion in each individual cen6-Δ survivor was determined by PFGE (Fig. 4D) and by Southern blot with XhoI and a Y′ probe (data not shown). The fusion proved to be stable after several restreaks on YPD plates. The steady level of unfused chromosome 6 also remains unchanged (data not shown).
Cloning and sequencing of telomere–telomere fusions
Fusions between telomeres in yeast sometimes generated unique restriction sites at the junction (Pardo and Marcand 2005). The fusion of clones III and VIII displays a HpyCH4V and a RsaI restriction site, respectively. These fusions were amplified by PCR and cleaved at their junction, and each telomere was subcloned and sequenced. The reconstructed sequence of the fusion from clone III displays 147 and 95 bp of TG1–3 repeats: GTCAGAAAGCCGGGTAAGGAGTGACAGCGAGAGTAAAGATAGATGTGAAAAGTGTGGGTGTGGTGTGTGGGTGTGTGGGTGTGGGTGTGGGTGTGGGTGTGGTGTGGGTGTGGTGTGGGTGTGTGTGTGTGGGTGTGGTGTGGGTGTGGTGTGGGTGTGGTGTGGGTGTGGGTGTGGTGTGTGTGTGGGTGTGGTGTGCACCACACCCACACACACACACCCACACACCACACCCACACACCACACCACACACCACACCACACACCACACCACACCCACACACCCACACACACTTTTCACATCTACCTCTACTCTCGCTGTCATACCTTACCCGGCTTTCTGAC.
The reconstructed sequence of the fusion from clone VIII displays 159 and 152 bp of TG1–3 repeats: GTCAGAAAGCCGGGTAAGGAGTGACAGCGCGAGTAAAGGTAAATGTGAAATGTGTGTGTGTGTGGGTGTGTGGGTGTGGGTGTGGGTGTGGTGTGTGGGTGTGTGGGTGTGGTGTGTGTGGTGTGGGTGTGGTGTGTGGTGTGTGTGGGTGTGGTGTGGTGTGGTGTGTGGGTGTGTGTGGGTGTGGGTGTGGTGTGTGTGGGTGTGGTACCCACACACCACACCCACACCCACACACCCACACCCACACCCACACCCACACACCACACCCACACCCACACACCACACCCACACACCACACCCACACACCACACCACACACCACACCACACACCACACCACACCCACACACCCACACACACTTTTCACATCTACCTCTACTCTCGCTGTCATACCTTACCCGGCTTTCTGAC.
The Y′ primers used for amplification are underlined. The HpyCH4V and RsaI sites are in bold.
PFGE
Yeast DNA embedded in agarose plugs was prepared as follows: About 109 cells were washed twice in 1 mL of 50 mM EDTA, 10 mM Tris (pH 7.5) (or 1 mL of 1% Triton, 250 mM EDTA, 10 mM Tris at pH 7.5 for the cells arrested in G1 or G2/M) (Fig. 5B) and resuspended in 150 μL of 50 mM EDTA, 10 mM Tris (pH 7.5). Zymolase (0.6 μL at 20 mg/mL) was added to the suspension, which was quickly warmed to 42°C and mixed with 250 μL of prewarmed 1% agarose LMP and 125 mM EDTA. The suspension was distributed into 80-μL wells placed on a cool surface. The plugs were extruded and incubated for 24 h at 37°C in 1.5 mL of 500 mM EDTA and 10 mM Tris (pH 7.5) followed by 24 h at 55°C in 1.25 mL of 500 mM EDTA, 10 mM Tris (pH 8.0), 1% N-Laurylsarcosyl, and 0.4 mg/mL Proteinase K. Plugs were washed for 1 h three times in 1 mL of 50 mM EDTA, and 10 mM Tris (pH 7.5). Pulse-field electrophoresis was carried out in a 1% agarose gel in 0.5× TBE at 14°C with a CHEF DRII from Bio-Rad with a switch time of 60 sec for 15 h then 90 sec for 9 h at 200 V.
Southern blot
The probes for the Southern blots were PCR products amplified from yeast genomic DNA and were gel-purified. The chromosome 6 probe is a 2.7-kb fragment from coordinates 72471–75177 on the left arm of chromosome 6. The Y′ probe is a 0.85-kb fragment from the distal end of a Y′ element (e.g., coordinates 52–893 of chromosome 8). The TEL 6R probe is a 0.6-kb fragment from coordinates 268971–269589 of chromosome 6. The chromosome 3 probe is a 2.4-kb fragment from coordinates 138842–141231 of chromosome 3. The chromosome 7 probe is 2.3-kb fragment from coordinates 14451–16729 of chromosome 7. The chromosome 5 probe is 2.1-kb fragment from coordinates 113034–115142 of chromosome 5.
Acknowledgments
We thank Julie Cooper for very fruitful discussions on this project, as well as Ofer Rog, Frank Uhlmann, Madalena Tarsounas, Rachel Lescasse, Serge Gangloff, Sylvaine Gasparini, Devanshi Jain, and Laure Sabatier for suggestions and comments. This work is supported by grants from ARC and ANR (ANR-06-BLAN-076).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.571510.
References
- Aihara H, Huang WM, Ellenberger T. An interlocked dimer of the protelomerase TelK distorts DNA structure for the formation of hairpin telomeres. Mol Cell. 2007;27:901–913. doi: 10.1016/j.molcel.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M. Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet. 1992;89:247–249. doi: 10.1007/BF00217134. [DOI] [PubMed] [Google Scholar]
- Bae NS, Baumann P. A RAP1/TRF2 complex inhibits nonhomologous end-joining at human telomeric DNA ends. Mol Cell. 2007;26:323–334. doi: 10.1016/j.molcel.2007.03.023. [DOI] [PubMed] [Google Scholar]
- Bajer A. Cine-micrographic studies on dicentric chromosomes. Chromosoma. 1964;15:630–651. doi: 10.1007/BF00319996. [DOI] [PubMed] [Google Scholar]
- Bateman AJ. Letter: Simplification of palindromic telomere theory. Nature. 1975;253:379–380. doi: 10.1038/253379a0. [DOI] [PubMed] [Google Scholar]
- Baxter J, Diffley JF. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell. 2008;30:790–802. doi: 10.1016/j.molcel.2008.04.019. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, de Villartay JP. Cernunnos interacts with the XRCC4 · DNA–ligase IV complex and is homologous to the yeast nonhomologous end-joining factor Nej1. J Biol Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM. The nature of telomere fusion and a definition of the critical telomere length in human cells. Genes & Dev. 2007;21:2495–2508. doi: 10.1101/gad.439107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli GB, de Lange T. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- Chan SW, Blackburn EH. Telomerase and ATM/Tel1p protect telomeres from nonhomologous end joining. Mol Cell. 2003;11:1379–1387. doi: 10.1016/s1097-2765(03)00174-6. [DOI] [PubMed] [Google Scholar]
- Cote AG, Lewis SM. Mus81-dependent double-strand DNA breaks at in vivo–generated cruciform structures in S. cerevisiae. Mol Cell. 2008;31:800–812. doi: 10.1016/j.molcel.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Daley JM, Palmbos PL, Wu D, Wilson TE. Nonhomologous end joining in yeast. Annu Rev Genet. 2005;39:431–451. doi: 10.1146/annurev.genet.39.073003.113340. [DOI] [PubMed] [Google Scholar]
- de Lange T. How to solve the end-protection problem. Science. 2009;326:948–952. doi: 10.1126/science.1170633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denchi EL, Celli G, de Lange T. Hepatocytes with extensive telomere deprotection and fusion remain viable and regenerate liver mass through endoreduplication. Genes & Dev. 2006;20:2648–2653. doi: 10.1101/gad.1453606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Guo X, Ferguson DO, Chang S. Multiple roles for MRE11 at uncapped telomeres. Nature. 2009;460:914–918. doi: 10.1038/nature08196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MG, Cooper JP. The fission yeast Taz1 protein protects chromosomes from Ku-dependent end-to-end fusions. Mol Cell. 2001;7:55–63. doi: 10.1016/s1097-2765(01)00154-x. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Grossi S, Bianchi A, Damay P, Shore D. Telomere formation by rap1p binding site arrays reveals end-specific length regulation requirements and active telomeric recombination. Mol Cell Biol. 2001;21:8117–8128. doi: 10.1128/MCB.21.23.8117-8128.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Gao Z, Birchler JA. Reactivation of an inactive centromere reveals epigenetic and structural components for centromere specification in maize. Plant Cell. 2009;21:1929–1939. doi: 10.1105/tpc.109.066662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heacock M, Spangler E, Riha K, Puizina J, Shippen DE. Molecular analysis of telomere fusions in Arabidopsis: Multiple pathways for chromosome end-joining. EMBO J. 2004;23:2304–2313. doi: 10.1038/sj.emboj.7600236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Bloom K. Genetic manipulation of centromere function. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill A, Bloom K. Acquisition and processing of a conditional dicentric chromosome in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1368–1370. doi: 10.1128/mcb.9.3.1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA. Origin of human chromosome 2: An ancestral telomere–telomere fusion. Proc Natl Acad Sci. 1991;88:9051–9055. doi: 10.1073/pnas.88.20.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, Kakuma T, Hiraoka Y, Takahashi K. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Jannink G, Duplantier B, Sikorav JL. Forces on chromosomal DNA during anaphase. Biophys J. 1996;71:451–465. doi: 10.1016/S0006-3495(96)79247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz W, Weinstein B, Solomon F. Regulation of tubulin levels and microtubule assembly in Saccharomyces cerevisiae: Consequences of altered tubulin gene copy number. Mol Cell Biol. 1990;10:5286–5294. doi: 10.1128/mcb.10.10.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes & Dev. 2008;22:1221–1230. doi: 10.1101/gad.1634008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D, Rutledge L, Fitzgerald-Hayes M, Hartwell LH. A genetic analysis of dicentric minichromosomes in Saccharomyces cerevisiae. Cell. 1987;48:801–812. doi: 10.1016/0092-8674(87)90077-8. [DOI] [PubMed] [Google Scholar]
- Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalande M, Noolandi J, Turmel C, Rousseau J, Slater GW. Pulsed-field electrophoresis: Application of a computer model to the separation of large DNA molecules. Proc Natl Acad Sci. 1987;84:8011–8015. doi: 10.1073/pnas.84.22.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejeune J, Dutrillaux B, Rethore MO, Prieur M. [Comparison of the structure of chromatids of Homo sapiens and Pan troglodytes (author's transl)] Chromosoma. 1973;43:423–444. doi: 10.1007/BF00406748. [DOI] [PubMed] [Google Scholar]
- Lo AW, Sprung CN, Fouladi B, Pedram M, Sabatier L, Ricoul M, Reynolds GE, Murnane JP. Chromosome instability as a result of double-strand breaks near telomeres in mouse embryonic stem cells. Mol Cell Biol. 2002;22:4836–4850. doi: 10.1128/MCB.22.13.4836-4850.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobachev KS, Gordenin DA, Resnick MA. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- Lobachev KS, Rattray A, Narayanan V. Hairpin- and cruciform-mediated chromosome breakage: Causes and consequences in eukaryotic cells. Front Biosci. 2007;12:4208–4220. doi: 10.2741/2381. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Louis EJ, Haber JE. The structure and evolution of subtelomeric Y′ repeats in Saccharomyces cerevisiae. Genetics. 1992;131:559–574. doi: 10.1093/genetics/131.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowden MR, Meier B, Lee TW, Hall J, Ahmed S. End joining at Caenorhabditis elegans telomeres. Genetics. 2008;180:741–754. doi: 10.1534/genetics.108.089920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydeard JR, Jain S, Yamaguchi M, Haber JE. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature. 2007;448:820–823. doi: 10.1038/nature06047. [DOI] [PubMed] [Google Scholar]
- Mann C, Davis RW. Instability of dicentric plasmids in yeast. Proc Natl Acad Sci. 1983;80:228–232. doi: 10.1073/pnas.80.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcand S, Pardo B, Gratias A, Cahun S, Callebaut I. Multiple pathways inhibit NHEJ at telomeres. Genes & Dev. 2008;22:1153–1158. doi: 10.1101/gad.455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko JF. Micromechanical studies of mitotic chromosomes. Chromosome Res. 2008;16:469–497. doi: 10.1007/s10577-008-1233-7. [DOI] [PubMed] [Google Scholar]
- McClintock B. The stability of broken ends of chromosomes in Zea mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The fusion of broken ends of chromosomes following nuclear fusion. Proc Natl Acad Sci. 1942;28:458–463. doi: 10.1073/pnas.28.11.458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza M, Norden C, Durrer K, Rauter H, Uhlmann F, Barral Y. A mechanism for chromosome segregation sensing by the NoCut checkpoint. Nat Cell Biol. 2009;11:477–483. doi: 10.1038/ncb1855. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Mieczkowska JO, Dominska M, Petes TD. Genetic regulation of telomere-telomere fusions in the yeast Saccharomyces cerevisae. Proc Natl Acad Sci. 2003;100:10854–10859. doi: 10.1073/pnas.1934561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Ferreira MG, Cooper JP. Taz1, Rap1 and Rif1 act both interdependently and independently to maintain telomeres. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- Murray AW, Claus TE, Szostak JW. Characterization of two telomeric DNA processing reactions in Saccharomyces cerevisiae. Mol Cell Biol. 1988;8:4642–4650. doi: 10.1128/mcb.8.11.4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden C, Mendoza M, Dobbelaere J, Kotwaliwale CV, Biggins S, Barral Y. The NoCut pathway links completion of cytokinesis to spindle midzone function to prevent chromosome breakage. Cell. 2006;125:85–98. doi: 10.1016/j.cell.2006.01.045. [DOI] [PubMed] [Google Scholar]
- Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes & Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo B, Marcand S. Rap1 prevents telomere fusions by nonhomologous end joining. EMBO J. 2005;24:3117–3127. doi: 10.1038/sj.emboj.7600778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V, Kolodner RD. Stabilization of dicentric translocations through secondary rearrangements mediated by multiple mechanisms in S. cerevisiae. PLoS One. 2009;4:e6389. doi: 10.1371/journal.pone.0006389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass E, Grabarz A, Plo I, Gautier J, Bertrand P, Lopez BS. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–824. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- Riha K, Heacock ML, Shippen DE. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology. Annu Rev Genet. 2006;40:237–277. doi: 10.1146/annurev.genet.39.110304.095755. [DOI] [PubMed] [Google Scholar]
- Rog O, Miller KM, Ferreira MG, Cooper JP. Sumoylation of RecQ helicase controls the fate of dysfunctional telomeres. Mol Cell. 2009;33:559–569. doi: 10.1016/j.molcel.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Sarthy J, Bae NS, Scrafford J, Baumann P. Human RAP1 inhibits non-homologous end joining at telomeres. EMBO J. 2009;28:3390–3399. doi: 10.1038/emboj.2009.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore D, Bianchi A. Telomere length regulation: Coupling DNA end processing to feedback regulation of telomerase. EMBO J. 2009;28:2309–2322. doi: 10.1038/emboj.2009.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GR. Meeting DNA palindromes head-to-head. Genes & Dev. 2008;22:2612–2620. doi: 10.1101/gad.1724708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CE, Llorente B, Symington LS. Template switching during break-induced replication. Nature. 2007;447:102–105. doi: 10.1038/nature05723. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A, Karlseder J, Holtgreve-Grez H, Jauch A, de Lange T. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2. Curr Biol. 2002;12:1635–1644. doi: 10.1016/s0960-9822(02)01179-x. [DOI] [PubMed] [Google Scholar]
- Surosky RT, Tye BK. Resolution of dicentric chromosomes by Ty-mediated recombination in yeast. Genetics. 1985;110:397–419. doi: 10.1093/genetics/110.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW. Replication and resolution of telomeres in yeast. Cold Spring Harb Symp Quant Biol. 1983;47:1187–1194. doi: 10.1101/sqb.1983.047.01.134. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Therizols P, Duong T, Dujon B, Zimmer C, Fabre E. Chromosome arm length and nuclear constraints determine the dynamic relationship of yeast subtelomeres. Proc Natl Acad Sci. 2010;107:2025–2030. doi: 10.1073/pnas.0914187107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Walmsley RM, Szostak JW, Petes TD. Is there left-handed DNA at the ends of yeast chromosomes? Nature. 1983;302:84–86. doi: 10.1038/302084a0. [DOI] [PubMed] [Google Scholar]
- Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–818. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]