Abstract
We have combined molecular beacon (MB) probes with barcoded metal nanowires to enable no-wash, sealed chamber, multiplexed detection of nucleic acids. Probe design and experimental parameters important in nanowire-based MB assays are discussed. Loop regions of 24 bases and 5 base pair stem regions in the beacon probes gave optimal performance. Our results suggest that thermodynamic predictions for secondary structure stability of solution-phase MB can guide probe design for nanowire-based assays. Dengue virus-specific probes with predicted solution-phase ΔG of folding in 500 mM buffered NaCl of approximately −4 kcal/mol performed better than those with ΔG > −2 or < −6 kcal/mol. Buffered 300–500 mM NaCl was selected after comparison of several buffers previously reported for similar types of assays, and 200–500 mM NaCl was found to be the optimal ionic strength for the hybridization temperatures (25 and 50 °C) and probe designs used here. Target binding to the surface as a function of solution concentration fit a Sips isotherm with Kd = 1.7 ± 0.3 nM. The detection limit was ∼100 pM, limited by incomplete quenching. Single base mismatches could be discriminated from fully complementary targets. Oligonucleotide target sequences specific for human immunodeficiency, hepatitis C, and severe acute respiratory viruses were assayed simultaneously in a no-wash, sealed chamber, multiplexed experiment in which each of three probe sequences was attached to a different pattern of encoded nanowires. Finally, we demonstrated that probe-coated nanowires retain their selectivity and sensitivity in a triplexed assay after storage for over 3 months.
Introduction
Many approaches to nucleic acid detection have appeared, some of which provide exceptional sensitivity1–3 or selectivity.3–6 In addition to these important parameters, ease of use, the ability to simultaneously test for multiple target sequences, and contamination risk can dominate the selection of a particular assay type for a given application. Molecular beacon probes can provide nucleic acid detection under “closed tube” conditions, which simplifies assay performance and greatly reduces contamination risk. Molecular beacons (MBs)7–9 are nucleic acid probe molecules designed with complementarity at their 3′ and 5′ ends such that they fold into a stem-and-loop (hairpin) structure. Traditionally, a fluorophore and a quencher moiety are attached to the opposing ends. When in the hairpin conformation, the quencher is held close to the fluorophore, quenching emission.7,10,11 Binding to target separates the donor and acceptor dyes and results in a fluorescence signal.
Advantages to using MBs for DNA detection include no target labeling, no need to wash after hybridization, and a single hybridization step (as compared to sandwich DNA assays, which require two hybridization steps, with a wash after each one).7 Challenges in multiplexing MB experiments include the requirement for spectrally distinct dyes, each with an efficient quencher.12,13 MB bioassays incorporating four different dyes have been used to enable simultaneous detection of several targets in homogeneous solution.14 Larger numbers of dyes are difficult to spectrally differentiate.
A promising approach to greater levels of multiplexing (i.e., >4) for MB probes is to attach the probe molecules to a surface in an array format.15–18 Tan and co-workers attached biotinylated MBs to fiber optic probes via a layer of streptavidin.19,20 The resulting biosensors provided real-time detection of nucleic acids with ∼1 nM sensitivity and could differentiate single base mismatches from fully complementary targets.19,20 MBs have also been attached to fluorescently encoded microspheres for simultaneous detection of four different sequences.21 Assay performance is influenced by the chemistry at the interface. Lu and co-workers improved quenching in surface-bound MB probes by attaching them to an agarose film on top of glass slides, in an effort to more closely mimic solution conditions.16 Yao and Tan varied the length of a linker between the MB and the surface, and reduced unfavorable electrostatic interactions with the streptavidin layer to optimize assay performance; the increase in emission intensity after target binding rose from 2× the initial (control) value to 5.5×.15 For comparison, the increase in fluorescence for solution-phase MBs can be on the order of 100×.20,22 Indeed, the effect of surface immobilization on MB probes can dominate sensor performance, largely due to high backgrounds caused by inefficient quenching. Recently, Lu, Tan, and co-workers demonstrated a TaqMan probe array, where Taq polymerase nuclease activity results in cleavage of a 5′ quencher to turn on fluorescence at the surface during PCR amplification.23 This approach does not require probe secondary structure on the surface, and thus avoids the problems of ineffective quenching typically observed for immobilized MB probes.
Several groups, including ourselves, have reported MB assays in which the quencher is a metal nanoparticle or metal surface.17,24–27 In this case, the probe strand is attached to the metal particle or surface via one end, and a fluorophore on the other end is quenched by close proximity to the metal surface while the probe maintains its hairpin conformation. Metals can provide extremely efficient quenching; Dubertret et al. showed that 1.4 nm Au clusters gave better performance in molecular beacons than the common molecular quencher, DABCYL.25 Krauss and co-workers bound MB-style probes to planar Au films via 5′ thiol groups. Quenching efficiencies were improved as compared to MBs immobilized on glass.17,24 This could be due to differences in surface chemistry and more efficient quenching by metal surfaces as compared to organic molecules. For molecular quenchers, two mechanisms of quenching are observed: resonant energy transfer, which can occur over several nanometers, and contact quenching, which requires closer approach.28 Fluorescence quenching by metal surfaces is effective out to ∼5 nm, and the large size of the surface means that multiple conformations of the probe molecules can approach to within this separation. Consequently, even linear probes can be quenched in the absence of, and fluorescent in the presence of, target due to the greater conformational flexibility of single-stranded DNA as compared to that of double-stranded DNA.27 This effect is greatest when probe surface densities are low, such that the probes can “lie down” on the metal, interacting not only via the 5′ thiol, but also, transiently, through the 3′ dye and/or the bases; Nie and co-workers demonstrated a homogeneous solution assay based on this effect for 2.5 nm Au nanoparticles.29
We recently reported an assay in which 5′ thiolated MB probes were assembled onto striped metal nanowires (Nanobarcodes Particles, NBCs). Five different nanowire striping patterns were used to identify specific beacon sequences in a five-plex assay for simultaneous detection of nucleic acid signatures for human pathogens.26 The encoded nanowires are several micrometers in length, and ∼300 nm in diameter, and have up to six different stripes of Au or Ag. Many distinguishable “barcode” or metal striping patterns can be encoded into the nanowires during synthesis via templated electrodeposition.30–32 Here, we discuss beacon assembly onto the nanowire surface, as well as the effect of hybridization buffer and of changing the length of the “loop” and “stem” regions of the beacon probes on assay performance. Sensitivity and selectivity of the beacon probe-based nanowire assays are presented. Finally, we demonstrate simultaneous detection of three pathogen-specific DNA oligonucleotides in a sealed chamber, no wash multiplexed assay (Scheme 1), and show that storage of probe-coated wires before use does not negatively impact assay performance. Beyond demonstrating proof-of-principle for a sealed chamber, multiplexed assay of possible future clinical interest, this work provides insight into the effect of surface confinement on molecular beacons. The latter should prove valuable not only for multiplexed MB-based assays such as are described here, but also in the design of experiments in which other structured probes are bound to solid supports, for example, aptamers,33 designed to detect small molecules, ions, and proteins.
Scheme 1.
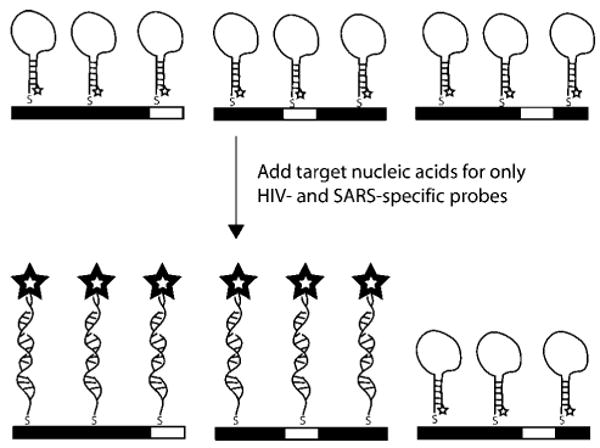
Illustration of Multiplexed Detection of Nucleic Acid Targets by Encoded Nanowires Functionalized with Molecular Beacon Probesa
a In this illustration, wires patterned 00001 (left), 00100 (middle), and 00010 (right) are coated with MB probes SARS, HIV, and HCV, respectively, and complementary target sequences have been added for SARS and HIV only.
Materials and Methods
Materials
The striped nanowires used in this work were commercially available Nanobarcodes Particles (NBCs, Oxonica, Inc.) patterned 000111, 00001, 00010, 00100, and 100100, where 0 and 1 represent ∼0.75 μm segments of Au and Ag, respectively. These particles were synthesized by electrodeposition into aluminum oxide membranes as described previously.30,32,34 Nanowires were stored in ethanol (∼1 × 109 wires per 1 mL of ethanol) and were rinsed three times in water (by centrifugation) to remove the ethanol prior to use. Buffers used in the experiments were: (1) 0.3 M PBS (0.3 M NaCl and 10 mM sodium phosphate, pH 7.0), (2) 40 mM citrate (40 mM citrate in 0.3 M PBS), (3) 0.01 M PBS buffer (0.138 M NaCl; 0.0027 M KCl; 10 mM sodium phosphate, pH 7.4), purchased from Sigma, (4) commercial hybridization buffer (HS114), obtained from Molecular Research Centers, Inc., (5) Tris (100 mM MgCl2, 20 mM Tris-HCl, pH 8.0), and (6) CAC buffer (0.5 M NaCl, 20 mM cacodylic acid, and 0.5 mM EDTA, pH 7.0). Caution! Cacodylic acid is dimethyl arsenate, a toxin and carcinogen; use gloves and a fume hood. All water used in these experiments was purified through a Barnstead Nanopure System to 18 MΩ resistivity. All rinses and washes of samples were done by centrifugation and removal of resulting supernatant. DNA beacons were designed using mfold DNA folding program35 and were purchased from Integrated DNA Technologies, Inc. The sequences used are listed in Table 1. Probes HIV, HCV, SARS, and DENV-2 were designed to detect human immunodeficiency virus, hepatitis C virus, severe acute respiratory syndrome, and strains of Dengue virus subtype 2 (DENV-2), respectively.26,36,37
Table 1.
Probe Sequences Used in This Worka
| name | sequence (5′-3′)b | predicted Tm (°C)c | predicted ΔGc(kcal/mol) | comments |
|---|---|---|---|---|
| HCV | thiol(CH2)6-GCG AGC ATA GTG GTC TGC GGA ACC GGT GAC TCG C-TAMRA | 57.6 | −6.26 | probe specific for a 24-base region of HCV |
| SARS | thiol(CH2)6-GCG AGA GAT GCT GTG GGT ACT AAC CTA CCT CTC GC-TAMRA | 65.1 | −9.77 | probe specific for a 25-base region of SARS, which extends the stem from 5 to 7 bases due to self-complementarity |
| HIV | thiol(CH2)6-GCG AGT GTT AAA AGA GAC CAT CAA TGA GCT CGC-TAMRA | 57.1 | −3.97 | probe specific for a 23-base region of HIV |
| SBM | thiol(CH2)6-GCG AGA TAG TGG TCT GCG GAC TCG C-TAMRA | 60.4 | −4.50 | probe used for mismatch assay; position of mismatch is bold |
| L14 | thiol(CH2)6-GCG AGA TCA ATG AGG AAG CCT CGC-TAMRA | 59.9 | −4.24 | probe with 14-base loop; specific for HIV |
| L24 | thiol(CH2)6-GCG AGG AGA CCA TCA ATG AGG AAG CTG CACT CGC-TAMRA | 53.8 | −3.57 | probe with 24-base loop; specific for HIV |
| L38 | thiol(CH2)6-GCG AGA AAA GAG ACC ATC AAT GAG GAA GCT GCA GAA TGG GAT ACT CGC-TAMRA | 46.8 | −4.95 | probe with 38-base loop; specific for HIV |
| DENV-2(4) | thiol(CH2)6-GCG AGT GTC TGT TAC CAA GGA TCT GTC GC-TAMRA | 44.1 | −1.80 | probe with 4 base pair stem; for DENV-2 |
| DENV-2(5) | thiol(CH2)6-GCG AGG TGT CTG TTA CCA AGG ATC TGC TCG C-TAMRA | 56.6 | −4.00 | probe with 5 base pair stem; for DENV-2 |
| DENV-2(6) | thiol(CH2)6-GCG AGC GTG TCT GTT ACC AAG GAT CTG GCT CGC-TAMRA | 68.6 | −6.27 | probe with 6 base pair stem; for DENV-2 |
| DENV-2(7) | thiol(CH2)6-GCG AGC GGT GTC TGT TAC CAA GGA TCT GCG CTC GC-TAMRA | 74.7 | −8.88 | probe with 7 base pair stem; for DENV-2 |
Targets were synthetic oligonucleotides fully complementary to the loop region of each probe.
The italic portions of the sequences indicate complementary stem regions.
Generated by mfold for most stable secondary structure in 500 mM NaCl at 25 °C.
Disulfide Bond Cleavage
DNA sequences were received as disulfides, which in some experiments were cleaved before use, resulting in a single thiol moiety terminating the sequence. To cleave the disulfide, the DNA was first dissolved in a 100 mM solution of DTT (dithiothreitol) in 1 mL of 0.1 M phosphate buffer (pH 8.3) for 30 min, and then the small thiol fragments were removed using Centri-Spin Separation Columns (Princeton Separations) following the manufacturer protocol. The resulting DNA sequences (terminated with a single –SH group) were diluted in water to a concentration of 10 μM and were stored in the freezer at −80 °C.
Attaching Beacons to Nanowires
(This was used for all experiments unless otherwise noted.) Aliquots of 100 μL of wires were washed and resuspended in 100 μL of 0.01 M PBS. Beacons were attached by adding 500 μL of 5 μM probe in water overnight at room temperature while tumbling. Next, 600 μL of 0.3 M PBS was added and allowed to react for 2 h to assemble a greater number of probes on the surface. The wires were then washed three times with 0.3 M PBS by centrifugation and were resuspended in 100 μL of 0.3 M PBS buffer, for further use.
Pre-cleaved versus Uncleaved Beacons
For this study, HCV beacon was used. One-half of the original batch of the DNA beacon was cleaved by disulfide reduction prior to use (following protocol above), and the other half was not cleaved. Both cleaved and uncleaved beacons were attached to wires patterned 010000 using the attachment protocol previously described (above). For hybridization, 3 μL of beacon derivatized wires was added to 42 μL of CAC buffer and 2 μL of 100 μM DNA target (no target samples simply had the target excluded) and were allowed to hybridize at room temperature for 2 h. Samples were not rinsed prior to imaging.
Effect of Loop Length
Attached to three separate aliquots of wires patterned 000111 were beacon sequences L14, L24, and L28 following the protocol described above. Samples with and without complementary target were prepared by adding 3 μL of probe-coated wires to each of six tubes (six tubes because target and no target sample for each beacon) in 42 μL of CAC buffer and 2 μL of 100 μM DNA target (which was omitted for no target samples). Hybridization was performed at room temperature for 2 h while tumbling. Samples were rinsed two times by centrifugation in 0.3 M PBS buffer prior to imaging. (It is important to note, however, that rinsing is not necessary prior to imaging as discovered in later experiments.)
Effect of Stem Length
Four aliquots of 30 μL of wires patterned 00100 were used as substrates for beacon probes DENV-2(4), DENV-2(5), DENV-2(6), and DENV-2(7). Probe attachment was performed here slightly differently than described above by suspending the wires in 98 μL of 0.01 M PBS buffer, adding 2 μL of respective 100 μM DNA, and allowing the samples to rotate overnight at room temperature. To each sample was added 100 μL of 0.3 M PBS buffer, and the samples were rotated for an additional 2 h at room temperature. Excess DNA was then rinsed out three times with 100 μL aliquots of the 0.3 M PBS buffer. Wires were resuspended in 60 μL of this same buffer and stored at 4 °C until use. Hybridization was performed at room temperature, 40, or 60 °C using 10 μL of probe-coated nanowires added to each target at a final concentration of 5 μM in 30 μL of CAC buffer. Control target samples had the same amount of target added; however, the sequence was noncomplementary to the beacon probe (HCV target was used for the noncomplementary samples). Samples were not rinsed prior to imaging.
Testing Different Hybridization Buffers in Triplex Assays
Triplex assays were performed in four different hybridization buffers in the presence and absence of target. Wires patterned 00100, 00001, and 00010 were coated with beacons HIV, SARS, and HCV, respectively. One microliter of each of the three batches of beacon-coated wires was mixed together in each of the sample tubes such that 3 μL of wires total resided in each tube. Samples were prepared such that all three targets could be added to the triplexed wires in each of four hybridization buffers (PBS, CAC, TRIS, and HS114), and the experiment was duplicated such that no targets were added to separate batches of triplexed wires in each hybridization buffer. Therefore, a total of eight triplexed samples existed (one for each buffer with target and one for each buffer without target). Added to each sample were 47 μL of the specified hybridization buffer and 1 μL of 10 μM of each type of target. (No target samples did not have target added.) This entire protocol was repeated such that hybridization at both room temperature and 50 °C could be studied. All samples were hybridized 1 h at either temperature. The samples were not rinsed before imaging.
Effect of Salt Concentration
Wires patterned 100100 were derivatized with DENV-2(5) beacon following the protocol originally outlined for probe attachment. Hybridization was performed in 20 mM cacodylate buffer to which different amounts of NaCl had been added. Five microliters of the functionalized wires was added to 35 μL each buffer formulation, along with 1 μL of 100 μM DNA target, and were then hybridized for 1 h either at room temperature or 50 °C. Samples were then imaged at room temperature using optical microscopy.
Sensitivity of Nanowire Beacon Assay
Beacon probes (HCV) were attached to nanowires patterned 00010. Final target DNA concentrations 0 to 1 × 10−6 M were prepared in 50 μL of 0.3 M PBS buffer. To 47 μL aliquots of target in 0.3 M PBS buffer was added 3 μL of beacon-coated wires in 0.3 M PBS (yielding a final volume of 50 μL). Beacon targets were allowed to hybridize at 50 °C for 2 h while tumbling. Samples were rinsed three times in 0.3 M PBS before imaging.
Single Base Mismatch Detection
Wires patterned 000010 were derivatized with DNA beacon sequence SBM. Aliquots of 34 μL of 2× TMAC (tetramethyl-ammonium chloride buffer, Sigma), 3 μL of 2 μM oligo target (complementary or containing one of the possible mismatched nucleotides at the location labeled in Table 1 (no target sample simply had target DNA omitted)), and 3 μL of beacon-coated nanowires were mixed, sonicated, and allowed to hybridize at 55 °C for 30 min. The samples were centrifuged and resuspended in 500 μL of 1xSSPE-0.1% SDS buffer (purchased from Promega and added SDS (dodecyl sulfate, sodium salt from Aldrich)) and allowed to mix in this buffer at room temperature for 10 min before removing the buffer by centrifugation. The wires were then suspended in 0.1xSSPE-0.05% SDS for 7 min while rotating at 55 °C, for an additional rinse. The wires were then rinsed three times in 0.5 M CAC buffer and resuspended in 50 μL of CAC buffer before imaging.
Multiplexed Sealed Chamber Assays
To perform sealed chamber assays, single well silicon spacers measuring 20 mm in diameter and 0.5 mm deep were used (Press-to-Seal silicon isolators, Molecular Probes, Eugene, OR). To attach the silicon spacers to coverslips, tape was first applied to the spacer and removed to pull off any lint or particles that would prevent a tight seal; then, they were applied to the coverslip. The tape regimen was then redone on the top side of the spacer before adding reagents. Hybridization buffer was then added (145 μL of 0.3 M PBS), and 3 μL total of beacon-coated wires (in experiments where three types of wires were mixed for multiplexing, 1 μL of each type was used) and 2 μL of 100 μM of each target were added. Beacon sequences HIV, SARS, and HCV were coated onto wires patterned 00100, 00001, and 00010, respectively, using methods previously described. A glass slide was then attached to the top of each sample before placement in an incubator at 50 °C for 10 min. The samples were allowed to cool for 30 min before imaging. Silicon spacers were reused in subsequent experiments by thorough washing in detergent, rinsing in water, air drying, and repeating the tape process to remove any dust prior to use.
Storage in Citrate Buffer
HIV, SARS, and HCV beacons were coated onto wires patterned 00100, 00001, and 00010, respectively, following the attachment protocol outlined above. Once the beacons were attached, the wires were rinsed as described, but instead of storage in 0.3 M PBS buffer, 100 μL of 40 mM citrate buffer was added for storage. After the specified number of days (0, 22, 65, or 110), 1 μL aliquots were removed from each of the three batches, rinsed once in 0.3 M PBS, and mixed in one tube for hybridization. This was done for multiple tubes such that in one tube, all three targets were added, another tube had no targets added, and other tubes contained combinations of certain targets to test multiplexing capabilities. Hybridization was performed with 2 μL of 10 μM target in 47 μL of CAC buffer at 50 °C while tumbling. The samples were not rinsed to remove excess target before imaging.
Optical Microscopy
Brightfield reflectance images were acquired using a Nikon TE-300 inverted microscope equipped with a 12 bit high-resolution Coolsnap HQ camera (Photometrics). A CFI plan fluor 60× oil immersion lens (N.A. = 1.4) was used in conjunction with Image-Pro Plus software (version 4.5) to image the samples. The light source was a 175 W ozone-free Xe lamp, and a Sutter Instruments filter wheel (Lambda 10-2) allowed for wavelength selection. Samples were prepared by first sonicating the tubes of sample to reduce wire clumping and sandwiching a 10 μL aliquot between two coverslips. Wires were allowed to settle onto the bottom slide for 30 s before imaging. All reflectance images were taken at 430 nm, which provides good reflectance contrast between Au and Ag.30,38 Fluorescence images were taken using a filter cube selective for TAMRA fluorophore excitation. All imaging was performed at room temperature.
Probe Surface Coverage Determination
Surface coverage was obtained by adding 5 μL of mercaptoethanol to 200 μL of buffer containing 5 μL of beacon-coated nanowires. Wire concentrations were estimated on the basis of dilutions from the initial 1 × 109 wires/mL stock concentration. The samples were allowed to tumble on a rotator at room temperature overnight. The DNA was displaced into solution and was collected in the supernatant. The fluorescence intensity was determined using a fluorolog-3 fluorimeter, equipped with a 450 W Xe lamp, and double grating excitation spectrometer and a single grating emission spectrometer in a 180 μL volume cuvette. Calibration standards were used to determine the beacon concentrations in each sample.
Results and Discussion
Beacon Attachment Methods
We compared two methods for beacon probe attachment onto the nanowire surface based on 5′ terminal thiol groups. Thiol terminated DNA sequences are purchased as disulfides (DNA5′-S-S-C6H12OH), and generally this disulfide is cleaved using dithiothreitol as a reducing agent, then run down a desalting column to collect the thiolated DNA prior to use. Integrated DNA Technologies recommends cleaving the disulfide bond immediately prior to use to avoid regeneration of the disulfides. This method is routinely used by ourselves and others for preparation of DNA conjugates with colloidal Au nanospheres, which are sensitive to aggregation in the presence of salts and uncharged, short-chain thiols.39–41 However, because adsorption to metal surfaces is known to cleave disulfides,42,43 pre-assembly reduction and separation may be unnecessary for nanowire derivatization. Advantages to allowing the surface to perform the disulfide cleavage reaction include reduced time and effort, and avoiding loss of thiolated DNA during the separation step. To determine whether pre-assembly disulfide cleavage was necessary, we compared mean fluorescence intensities for a 5′ thiolated, 3′ TAMRA molecular beacon probe (HCV) pre-cleaved using DTT before attachment to the nanowires and the same beacon sequence not cleaved prior to adsorption (Figure 1). Beacon probe attachments were performed using identical protocols. In the absence of target, minimal fluorescence intensity is desired, and in the presence of target a great increase in signal is favorable. The “no target” samples should exhibit minimal fluorescence, as beacons should be folded and quenched. Our measured quenching efficiencies (calculated as [1-(signal in absence of target/signal with target)], %) improved from 90% for the DTT cleaved to 96% for the uncleaved samples. However, the overall fluorescence intensity for the uncleaved sample was only about one-third that for the DTT cleaved sample. This corresponded to a substantial difference in beacon probe surface coverage for the two attachment strategies, with 4 × 1012 molecules/cm2 (25 nm2/molecule) for the DTT cleaved probes, and only 7 × 1011 probes/cm2 (140 nm2/molecule) for the uncleaved probes. The large areas occupied by oligonucleotides on surfaces underscore the fact that these molecules are negatively charged and adopt a number of conformations, including dynamic transitions between the hairpin loop and unfolded random coil. We rationalize the difference in surface coverage between the two samples as resulting from more efficient attachment for the cleaved probes, for which the sulfur atoms are less hindered, coupled with the effect of coadsorbed mercaptohexanol molecules in the case of the uncleaved probes (generated by cleavage at the surface). Lower probe coverages would simultaneously provide improved quenching and hybridization efficiencies, due to lower steric and electrostatic repulsions,44–47 as well as lower total fluorescence intensities, due to the smaller number of TAMRA fluorophores on each wire. The coadsorbed mercaptohexanol in the uncleaved samples may also reduce interactions between the DNA bases or backbone and the wire surface. Beacon probes attached via a single point (the 5′ thiol) are more likely to bind to their own stem sequences, allowing them to quench more efficiently, as the stem configuration places the fluorophore in close proximity to the metal surface. The data in Figure 1 show that either pre-cleaved or as-received thiolated oligonucleotides can be used for preparation of probe-coated nanowires. For the remainder of the experiments in this paper, we did not pre-cleave probes prior to assembly.
Figure 1.
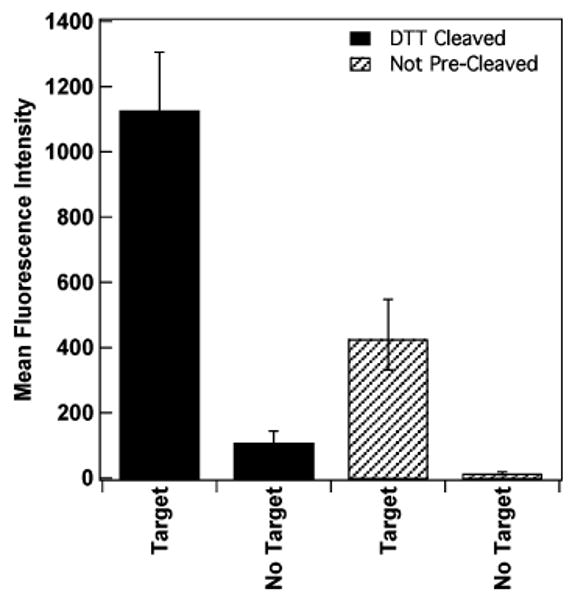
Assays for HCV beacon when pre-cleaved using DTT versus the same beacon not pre-cleaved in the presence and absence of complementary target. Error bars shown are the 95% confidence interval.
Beacon Probe Design
The beacons used here were designed using mfold, a nucleic acid folding program designed by Michael Zuker.35 This program offers insight to MB probe secondary structure and predicts binding energies for the folded structures. These structure predictions, however, do not take into consideration the fact that our beacon probes are attached to a surface, nor the impact of adjacent probe molecules. Therefore, the folding program was used primarily as a guide to avoid the use of beacons that contained a great deal of secondary structure in their loop regions, and to provide a means of comparing the relative thermodynamic stabilities of the probes. Solution-phase molecular beacon probes are typically designed with 15–30 base loop regions and 5–7 base pair stem regions. Shorter loops can provide greater discrimination against single base mismatches, while longer loops can provide greater equilibrium binding constants for target sequences. Stem length dictates the stability of the probe secondary structure; probes having longer stems are more difficult to unfold. We anticipated that these general observations from solution-phase beacons would hold true; nonetheless, the optimal probe design for surface-based experiments could differ substantially from that for solution studies. To identify design rules for surface-bound beacon probes, we compared the performance of nanowire-bound beacon probes as a function of loop length and stem length.
We had previously observed a decrease in beacon probe performance in nanowire-based assays as loop length was increased from 24 to 34 and 44 bases.26 In those experiments, we were unable to measure the probe densities of the nanowire surface, which complicated interpretation. Here, we compare assay performance and probe density for 24-base loops to that for both shorter (14-base) and longer (34-base) loops. Figure 2 compares mean fluorescence intensities for these nanowire-bound beacon probes in the presence and absence of target DNA (target length = loop length). The stem region was 5 base pairs long for each probe. As we reported previously,27 fluorescence intensity in the presence of target increases as probe length increases from 24 to 34 bases (i.e., 14–24 base loops) due to a combination of decreased quenching by the metal surface and increased hybridization efficiency due to the increased stability of the probe–target duplex. For the 38-base loop, intensity decreases despite the fact that quenching should be further reduced for this longer probe, and the thermodynamic stability of the solution-phase analogue of this duplex should be increased. We attribute this to decreased hybridization efficiency for the longer strands, due to increased steric and electrostatic repulsions between probe molecules on the surface.27 The surface coverage was between 1.2 and 2.5 × 1011 probes/cm2 for all three of the probes, with the highest value coming from the 24-base loop probe. Fluorescence signal in the absence of target is lowest for the probe having the 24-base loop, which, combined with the higher signal in the presence of target, gives optimal quenching efficiency for this intermediate length probe.
Figure 2.
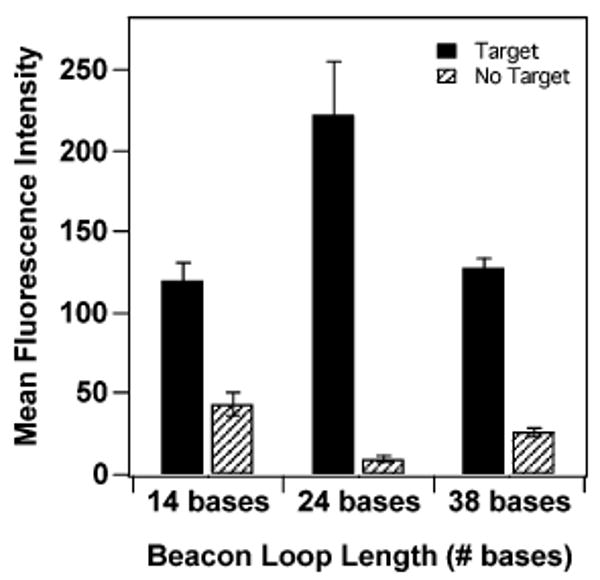
Effect of loop length on fluorescence intensity for molecular beacon probes bound to Ag/Au striped nanowires in the presence and absence of complementary target strands. Stem length was held constant at 5 base pairs. Error bars are the 95% confidence interval.
The effect of stem length was investigated using a series of probes with a 21-base loop region designed to recognize 16 strains of dengue virus subtype 2 (DENV-2), which is the most serious pathogenic variation of DV. Four stem lengths (sequences DENV-2(4), DENV-2(5), DENV-2(6), and DENV-2(7), corresponding to stem lengths of 4, 5, 6, and 7 base pairs, respectively) were predicted by mfold nucleic acid folding software to form secondary structures with ΔG between −2 and −9 kcal/mol in 0.5 M NaCl (the salt concentration used for these experiments). Fluorescence intensities for these probe sequences after incubation at 25 °C in the presence and absence of target oligonucleotides are shown in Figure 3A. As expected, the shorter stems (i.e., least stable hairpin structures) led to higher fluorescence signals both in the presence and in the absence of complementary target DNA strands. This is consistent with less stable hairpin secondary structure formation. Longer stems led to greatly decreased intensity for the complementary target, slightly decreased intensities for the no target samples, but essentially no improvement in the noncomplementary controls. Quenching efficiencies were somewhat similar for all four probes, ranging from ∼90% for the 5 base pair stem to ∼75% for the 7 base pair stem. Repeating the experiment at higher hybridization temperatures (40 and 60 °C) decreased the QE, particularly for the longest stem probes (Figure 3B; fluorescence intensities for the 40 and 60 °C experiments are plotted in Supporting Information Figure 1). We note that QE determination is less accurate for lower intensity samples, and that the apparently anomalous QE for the 7 base pair probe at 40 °C is most likely the result of variability rather than a physical phenomenon unique to this temperature.
Figure 3.
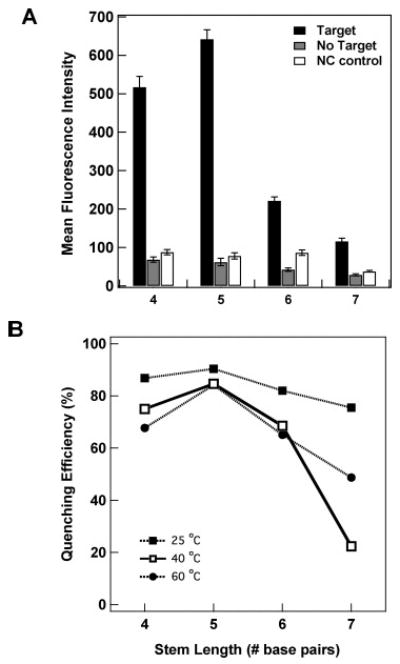
(A) Effect of stem length on fluorescence intensity for DENV-2 molecular beacon probes bound to Ag/Au striped nanowires in the presence and absence of complementary target strands. Hybridization was performed at 25 °C in 500 mM NaCl CAC buffer. Loop length was held constant at 21 bases. Error bars are 95% confidence intervals. (B) Effect of hybridization temperature on quenching efficiency for four stem lengths. Lines connecting the points are present only to guide the eye.
Our results thus far indicate that probes having approximately 24 base loops and 5 base pair stems are optimal under the conditions of these assays (300–500 mM NaCl, 20–60 °C). Performing the hybridizations at room temperature provided the best QE for all four DV stem lengths tested. Changes in either ionic strength or temperature are expected to alter the optimum probe design. For example, if substantially lower ionic strength buffer or higher hybridization temperatures are used, it may be necessary to go to longer stems to maintain quenching efficiency.
Effects of Ionic Strength on MB–Target Duplexes
Salt-dependent electrostatic effects are a major factor in determining the secondary structure and hybridization thermodynamics of nucleic acids. The high density of probe oligonucleotides can be expected to increase the electrostatic repulsions that must be overcome for probe secondary structures or probe–target binding to occur. We compared the performance of the MB-coated nanowires in a triplexed assay format where each of three different MB probes (HIV, SARS, and HCV) was attached to a different nanowire barcode pattern (as shown in Scheme 1, except that all three targets were added). The three probe-coated wires were mixed together, and then either all three targets, or no target (for the negative control), were added for hybridization in one of four different buffers. The four buffers tested were: (1) TRIS (100 mM MgCl2, 20 mM TRIS–HCl, pH 8.0, used by Lu and co-workers for molecular beacons immobilized on agarose films),16 (2) PBS (0.3 M NaCl, 10 mM phosphate, which is commonly used for metal particle-bound DNA hybridization assays),46–50 (3) HS114 (a commercial hybridization buffer, the contents of which are proprietary, which we had previously used for nanowire-bound beacon assays),26 and (4) CAC (0.5 M NaCl, 20 mM cacodylic acid, 0.5 mM EDTA, pH 7.0, which was used by Krauss and co-workers for molecular beacons immobilized on planar Au supports24). Hybridization was performed in each buffer at 25 and 50 °C.
Figure 4 summarizes the results of this experiment. Quenching efficiencies (filled symbols) are plotted on the left axis for all three beacons multiplexed in each buffer formulation at 25 °C (top panel) and 50 °C (bottom panel). The corresponding fluorescence intensities (open symbols) in the presence of target strands are plotted on the right axis. For the 25 °C hybridization, QE varied substantially with the hybridization buffer used, with the more stringent HS114 buffer resulting in QE as low as 24% (for HIV probe), and the highest ionic strength CAC buffer providing QE as high as 88% (for HCV probe). The high salt content of the CAC buffer enables improved performance by screening the electrostatic repulsions due the high density of negatively charged probe molecules at the nanowire surface. The buffers can be ranked in terms of QE as CAC > TRIS > PBS ≈ HS114, and in terms of fluorescence intensity as HS114 > CAC > PBS > TRIS. The best overall performance was observed for CAC, which had the highest QE and the second highest fluorescence intensity. Some differences between the three beacon probes are apparent in Figure 4 (top). For example, the HIV probe is generally the brightest, consistent with the lower thermodynamic stability of this probe's hairpin secondary structure (Table 1).
Figure 4.
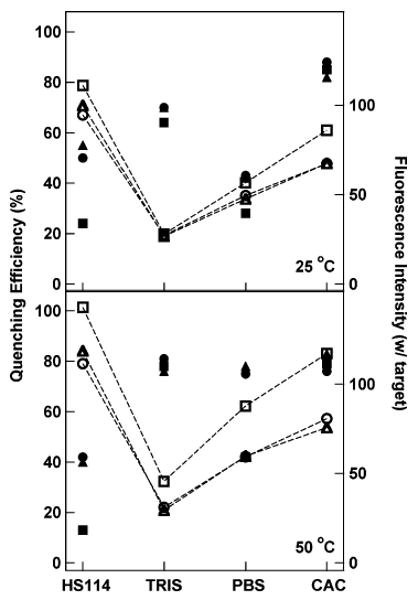
Comparison of assay performance in four different hybridization buffers at 25 °C (top) and 50 °C (bottom). Filled symbols are quenching efficiencies for (■) HIV, (▲) SARS, and (●) HCV. Open symbols are mean fluorescence intensities in the presence of target oligonucleotides for (□) HIV, (△) SARS, and (○) HCV. The lines connecting the fluorescence intensity points are present only to guide the eye.
When hybridization was performed at 50 °C, both fluorescence intensities and QE generally improved (Figure 4, bottom panel). We note, however, that QE for CAC decreased slightly at 50 °C as compared to 25 °C, in agreement with Figure 3B. QE for the four buffers now can be ranked as TRIS ≈ PBS ≈ CAC > HS114, and fluorescence intensity as HS114 > CAC > PBS > TRIS. Although the QE for the HIV probe is anomalously poor for PBS in this data set, good overall performance is achieved with both CAC and PBS buffers at this temperature. The TRIS buffer gave equally good QE, but low fluorescence intensities. HS114, in contrast, gave the highest intensities, but poor quenching. On the basis of these findings, we selected either CAC or PBS buffers for our ongoing studies; when PBS was used, hybridization was performed at 50 °C to avoid the low QE observed at 25 °C (Figure 4, top panel). One advantage of PBS over CAC is avoiding the use of dimethyl arsenate (i.e., cacodylic acid), which is toxic and carcinogenic and therefore must be handled with care.
The most critical aspect of buffer composition is its ability to screen electrostatic repulsions between probe and target DNA molecules as well as between the two ends of the beacon probes. We compared the performance of nanowire-bound DENV-2(5) beacon probes as a function of ionic strength by varying the NaCl concentration between 50 mM and 1.5 M in cacodylate buffer at 25 and 50 °C (Figure 5). At 25 °C, fluorescence intensity in the presence of target increases from 50 mM NaCl to 500 mM NaCl, and then decreases. Fluorescence intensity in the absence of target is lowest for the lower salt samples; however, QE is best for 200 and 500 mM, at 90%. QE drops to 87% at 100 mM, and 82% at 750 mM NaCl. When hybridized at 50 °C, quenching was nearly complete even in the presence of target for ≤100 mM NaCl. As the salt concentration was increased, fluorescence intensity in the presence of target increased substantially to peak at 500 mM, then decreased at higher salt concentrations. Quenching efficiencies were again optimal for 200 and 500 mM NaCl, at 88% for both.
Figure 5.
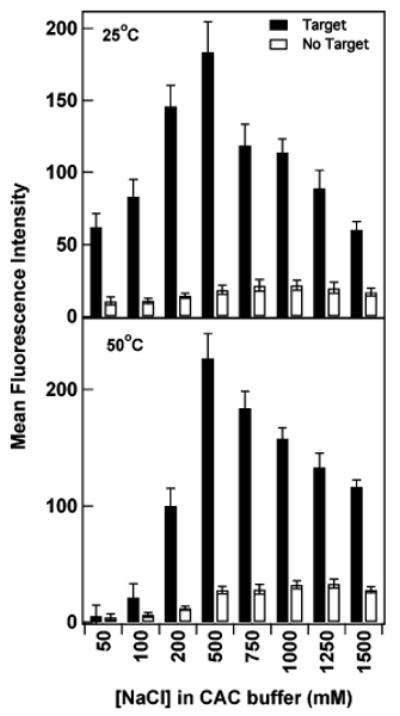
Effect of NaCl concentration on performance of nanowire-bound DENV-2(5) probes. Intensities are shown in the presence (filled bars) and absence (open bars) of complementary target sequence at room temperature and 50 °C. Error bars shown are the 95% confidence interval.
The general trends observed in Figure 5 are consistent with our understanding of MB probe structure at the metal surface. At very low salt, the beacon probes cannot readily bind target molecules and are unable to fold as effectively into secondary structures due to electrostatic repulsions. Thus, the quenching observed for ≤100 mM NaCl in Figure 5, particularly when hybridized at 50 °C, arises not from hairpin formation, but rather from conformational flexibility of the single-stranded probes, which prevents their 3′ dye molecules from extending far enough away from the metal surface to avoid quenching. The very low intensities for low salt samples could also be explained by loss of the probes from the surface due to increased intermolecular electrostatic repulsions. To test for this, we measured the surface coverage of probes after being stored under hybridization conditions in buffer containing either 50 mM or 1 M NaCl, in the absence of target DNA. Surface coverages at 50 mM and 1.0 M NaCl were indistinguishable, at (4 ± 2) × 1011 and (3 ± 1) × 1011 probes/cm2, respectively. Thus, no significant loss of probe DNA occurred at the lower salt concentrations, supporting our interpretation that the low intensities observed in low salt buffers were the result of quenching due to probe flexibility. The surface coverage experiment was performed on samples incubated for 2 h at 40 °C. This is a slightly lower temperature than in the lower panel of Figure 5; however, we also see very low fluorescence intensities in low salt buffers at 40 °C (Supporting Information Figure 2).
The decrease in emission at very high NaCl concentrations in the presence of target observed in Figure 5 presumably arises due to stabilization of the probe's hairpin structure, which must remain fluxional to enable hybridization to complementary target strands. The intensity for the negative control samples is relatively low at all salt concentrations, increasing slightly with NaCl concentration up to 750 or 500 mM, for the 25 and 50 °C data sets, respectively, before leveling off. At both temperatures, the highest intensities in the presence of target are observed in 500 mM NaCl, and optimal QE values are observed in both 200 and 500 mM. For the remainder of the work described here, we used either 300 or 500 mM NaCl. These are relatively high ionic strengths, required due to strong electrostatic repulsions between the adjacent probes on the nanowire surface.49–51
Nanowire MB Assay Sensitivity
The sensitivity of the nanowire beacon assays described here is comparable to that of other surface-bound molecular beacon assays.16,21,22,24,25,29 Figure 6 shows assay response as a function of target oligonucleotide concentration for HCV beacon probes hybridized in 0.3 M PBS at 50 °C. The y-axis is fractional coverage, estimated on the basis of the fluorescence intensity as compared to the maximum intensity. The limit of detection (LOD) for this data was calculated by taking the average fluorescence intensity for the control (background signal) and adding two times its standard deviation.52 We report a LOD = 38.1 + 2(3.0) = 44.1 mean fluorescence units, which translates to a concentration of <100 pM (10 femtomoles in our 100 μL volume), with dynamic range of 3–4 orders of magnitude in concentration. We note that, although the dynamic range is large, this assay is most sensitive to changes in concentration at low concentrations (see Figure 6, inset). The data in Figure 6 could be fit to a Sips isotherm,
Figure 6.
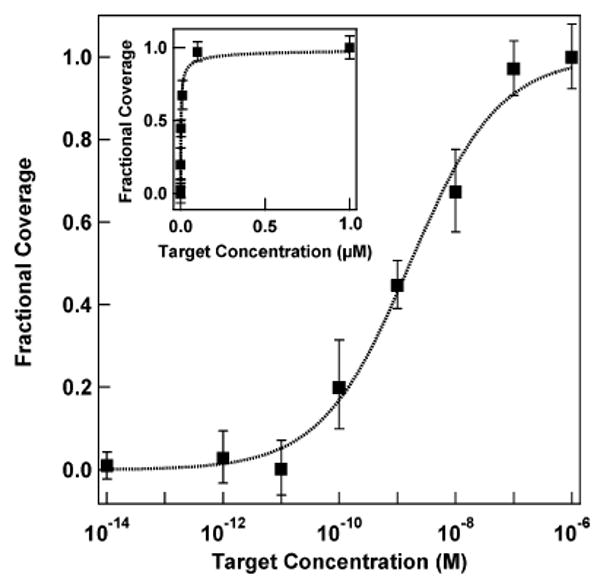
Hybridization adsorption isotherm for target binding to HCV beacons on metal nanowires. Fractional coverage was determined on the basis of fluorescence intensity as compared to intensity at saturation (1 μM). Dotted line is a fit to the Sips isotherm. Inset shows the same data on a linear concentration scale. The error bars are the 95% confidence interval.
| (1) |
where f is the fractional coverage of target binding sites on the beacon probe-coated wire, K is the average equilibrium constant for adsorption, C is the concentration of target in solution, and a is the heterogeneity index.53,54 The Sips isotherm assumes that the heterogeneity in binding sites takes the form of a Gaussian distribution of affinities. The width of this distribution is determined by the magnitude of a, which varies from 0 to 1 (when a = 1, the equation simplifies to the more familiar Langmuir isotherm). Fitting to eq 1 gave a Kd of 1.7 ± 0.3 nM and a heterogeneity index of 0.57 ± 0.06 for the nanowire-bound HCV beacon probes. We hypothesize that the heterogeneity in binding affinities observed in our experiments could arise from: (1) variations in surface probe density between nanowires or at different points on a single nanowire, which would alter steric and electrostatic contributions to the binding affinity; (2) differences in probe conformation, which would affect accessibility; or (3) differences in the signal observed for binding events occurring on Ag versus Au segments of the nanowires, which would not change the affinity but would alter the fluorescence intensity per binding event, which would impact our apparent affinity.
These results differ substantially from those reported for molecular beacons on planar Au surfaces (which fitted a two-state model with a Kd of 0.95 μM and a narrow dynamic range),24 but follow expectations for surface-bound single-stranded probes with no secondary structure, which can generally be fit by Langmuir or related surface adsorption isotherms.55,56 For example, Corn and co-workers fit thermodynamic data for probes on planar Au to a Langmuir isotherm and report similar Kd values (55 nM for an 18-mer probe/target duplex in a 300 mM NaCl, 100 mM urea PBS buffer).57 Peterson et al. found that perfectly matched duplex formation on an Au surface could be fit to a Langmuir isotherm (with Kd =17 nM for a 25-mer duplex in 1 M NaCl buffer), while mismatched duplexes were better modeled by the Sips isotherm.58 The preceding examples both used unstructured probe DNA; our probes are designed to adopt secondary structures, but the population of the folded and unfolded structures in our experiments is not known (the predicted Tm for solution-phase HCV in 0.3 M PBS is 53.6 °C, only slightly above 50 °C hybridization temperature, but far above the 25 °C at which samples were imaged after hybridization). We note that both surface-based immunoassays54 and MB probes in solution also exhibit several orders of magnitude in dynamic range of fluorescence response to target concentration.25
Factors that affect sensitivity include the number of wires in each assay (surface area), surface coverage of beacon probes, hybridization thermodynamics, and beacon probe quenching efficiency. The highest target concentration tested (1 × 10−6 M) provided 3.0 × 1013 target strands/sample, which decreased to 3.0 × 109/sample for 1 × 10−10 M target. For the data shown in Figure 6, ∼3 × 106 nanowires, with 1.3 × 1012 beacon probes/cm2, were used for each sample. This resulted in approximately 2.4 × 1011 total beacon probes per sample. The number of target molecules added as compared to the number of probe molecules present on the nanowires can be estimated at 126% for 10 nM target, 1.3% for 0.1 nM, and 0.13% for 10 pM target. Reducing the number of wires present in the assay, in principle down to a single wire, could improve sensitivity by reducing the volume of ∼0.1 nM target needed to detect a response. However, use of multiple wires simplifies handling and visualization, as well as providing more data points for statistical analysis. Therefore, although it may be possible to improve LOD by reducing the surface available for binding, there are other trade-offs that must be considered. Other approaches to increasing LOD include probe design, to improve Kd, and nanowire surface chemistry, to improve quenching efficiency. Nonetheless, we will ultimately be limited by the binding affinity of the target for the probe strand, and therefore we do not anticipate that this sensing approach will rival ultrasensitive methods such as PCR. Rather, it could offer a route to simple, relatively low sensitivity multiplexing under closed-chamber conditions (i.e., with no target labeling, washing, or other addition of reagents, such that ease of use is increased, and contamination risk is reduced).
Single Base Mismatch Detection
Solution-phase MB probes can provide excellent selectivity, due to their intrinsic secondary structure.4,13 We tested the selectivity of our nanowire-immobilized MBs by comparing their response after exposure to mismatched or fully complementary targets sequences. An HIV-specific probe with a relatively short 14 base loop region, sequence SBM, was used for these experiments to increase the energetic difference between binding the matched versus single base mismatched targets. Figure 7 shows the fluorescence results for each of the target sequences after hybridization at 55 °C for 30 min. We note that this experiment was performed under more stringent hybridization and wash conditions than any other in this paper; poor mismatch discrimination was observed when hybridized in 0.5 M NaCl CAC buffer for 2 h at room temperature.
Figure 7.
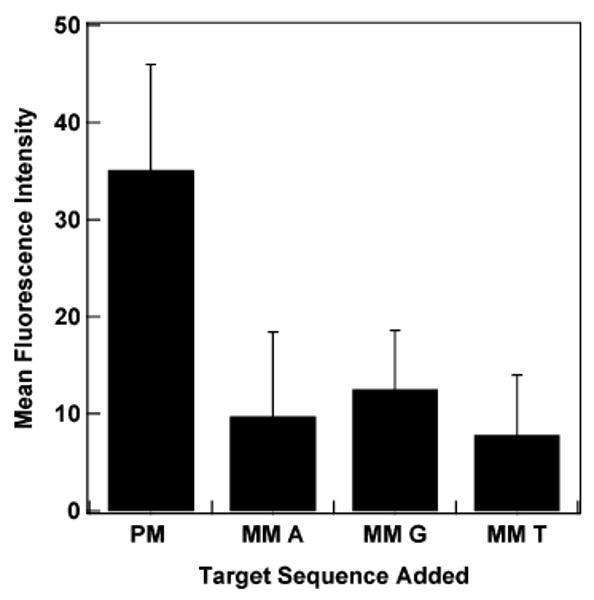
Comparison of response from fully complementary and mismatched target sequences binding to HIV MB probe SBM on the nanowire surface. PM indicates the perfectly matched target; mismatched targets (MM) for each of the bases in place of the C base in the PM are shown. The fluorescence intensity of a sample containing no target was subtracted from each sample. Error bars shown are the 95% confidence interval.
The fully complementary target gave a greater response than any of the single base mismatches (the three mismatches shown correspond to all possibilities for replacing a central C in the target). Discrimination was best for the T, which gave a 4.5-fold difference between the fully complementary and mismatched targets. The A and G mismatch sequences gave 3.6 -and 2.8-fold differences, respectively. Although the nanowire bound beacon probes were able to differentiate the single base mismatches, they did not provide as large of a difference as was observed by Krauss and co-workers for MBs on planar Au (8-fold difference).24 We hypothesize that the greater increase in fluorescence signal in the presence of fully complementary DNA, leading to the larger increase in signal, may be due in part to the longer probe used in ref 24, which would help avoid quenching from the metal surface (36 total bases as compared to 25 bases). We have recently reported that 34 base long sequences exhibit higher fluorescence than 24 base long sequences.27
Multiplexed, No-Wash, Sealed Assays
Diagnostic applications of bioassays under clinical settings must contend with the risk of sample cross-contamination. One advantage of reagent-less approaches such as molecular beacons (whether in solution or surface-bound) is that once the sample is added to the beacon probes, no further manipulation (e.g., addition of reagents, washing) should be required. The ability to perform an assay in a sealed container greatly reduces contamination risk. We therefore considered the performance of a sealed assay, where all reagents were sealed on a microscope slide during both reaction and analysis, as the risk of contamination is greatly reduced. A multiplexed, sealed assay was performed as shown in Scheme 1 by first coating three different MB probes (HIV, SARS, and HCV) on three different patterns of wires (00100, 00001, and 00010, respectively), and then mixing the beacon-coated wires with all hybridizing reagents and target in sealed chamber gaskets on glass slides, which were not opened even for imaging. Representative fluorescence and reflectance microscope images for an assay in which HIV- and SARS-specific targets (but not HCV-specific target) have been added are presented in Figure 8 (the sample was hybridized in PBS at 50 °C). The nanowires that are visible in the fluorescence image correspond to 00100 and 00001 patterns, as evident in the corresponding reflectivity image, indicating that only nanowires coated with HIV- and SARS-specific MB probes gave a positive response.
Figure 8.
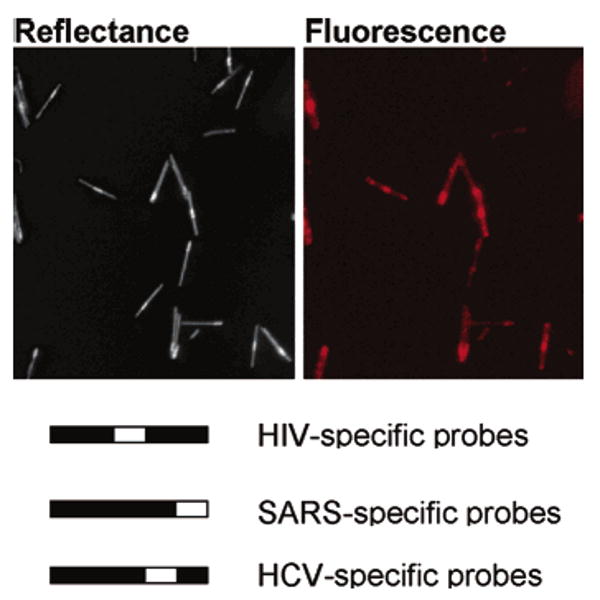
Reflectance and corresponding fluorescence microscopy images of triplexed, sealed chamber assay for HIV, SARS, and HCV target sequences. In this assay, only HIV and SARS targets were added. Thus, while all three nanowire patterns are visible in the reflectance image, only the HIV- and SARS-specific nanowires should be visible in the fluorescence image. Nanowire patterns and corresponding probe specificities are given below the images.
Figure 9 gives quantification for the assay represented in Figure 8 as well as other combinations of targets. Because the individual assays making up this multiplexed experiment exhibited differences in fluorescence response (i.e., the HIV probes were always brighter than the HCV or SARS, as also observed in Figure 4), each probe was normalized independently to simplify interpretation. The quenching efficiencies from this sealed chamber assay were 89%, 87%, and 92% for HIV, HCV, and SARS, respectively. There was good discrimination in triplex samples where only one or two targets are present, considering the assays with all three targets present or no targets present as reference points. We note that the background signal is lowest when no targets have been added, as compared to when one or more targets are added. For example, fluorescence responses for the HCV and SARS assays when only HIV target was added were larger than when no target had been added. This indicates either nonspecific target binding, or incorrect nanowire identification by software, or some combination of the two. This can be improved by beacon probe design and/or optimization of identification software and nanowire electrodeposition. Nonetheless, the multiplexed, sealed assay in its current form unambiguously identified the correct targets in each sample. These results suggest that the elimination of the rinsing step and the reduction on mixing due to the sealed chamber geometry did not negatively affect the ability to perform simultaneous assays for three oligonucleotide targets. Indeed, we see no degradation in assay performance between rinsed and unrinsed samples (data not shown). These results are promising in that sealed chamber assays can help eliminate sample contamination, and personnel exposure, as well as simplifying assay performance. Although our prior demonstration of PCR product detection by nanowire-bound MB was rinsed (i.e., not performed in a sealed chamber),27 the success of solution-phase MB-style probes in the more complex matrices of PCR products, clinical samples, and living cells suggests that the sealed chamber approach used here will be applicable to samples of diagnostic interest.1,59–63
Figure 9.
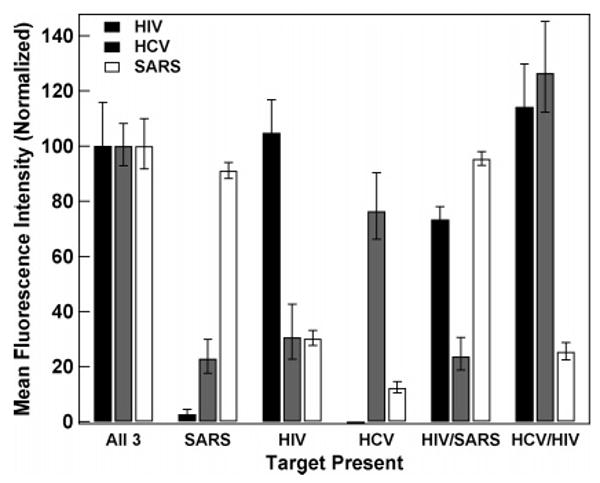
Triplex beacon assays performed and analyzed in a sealed chamber. The labels below the bar graphs indicate which target/targets are present in each assay. Background from a negative control (no targets added) has been subtracted from the data, and the intensity for each probe has been normalized to its intensity in the sample containing all three targets. The error bars shown are the 95% confidence intervals.
Preservation of Assays Using Citrate Buffer
Because future clinical applications of the beacon-coated nanowires would likely involve storage of the bioconjugated wires prior to use, we were interested in determining whether the performance of wires pre-coated in beacon probes deteriorated if not used immediately after preparation. In previous studies, we had found that citrate buffer protected the Ag segments of bare or DNA-coated striped nanowires from Ag degradation and DNA loss in oxygenated, PBS buffers over relatively long periods of time (at least months).64 Therefore, we added 40 mM citrate to the 0.3 M PBS storage buffer for these experiments. Wires were centrifuged and resuspended into 500 mM NaCl CAC buffer prior to hybridization, to avoid any differences in assay performance due to the citrate. Figure 10 shows fluorescence intensities from multiplexed assays in which HIV-, HCV-, and SARS-specific beacon probe sequences are used. In Figure 10A, samples to which all three targets or no target were added are compared after 0, 22, 65, and 110 days of storage. To correct from day-to-day variations in lamp intensity (which were substantial, because between days 22 and 65 we installed a new, brighter lamp), we normalized these data such that the intensity of the brightest nanowire population (in every case, this corresponded to those with the HIV-specific probes) in the presence of target oligonucleotides was defined as 100%. The quenching efficiency for each of the three nanowire populations improved slightly between days 0 and 22, from 78% to 91%, 82% to 88%, and 84% to 93%, for HIV-, HCV-, and SARS-specific probes, respectively. We interpret this improvement as arising from reorganization or loss of some probe strands leading to improved hybridization at the surface. After day 22, essentially no change in QE was observed.
Figure 10.
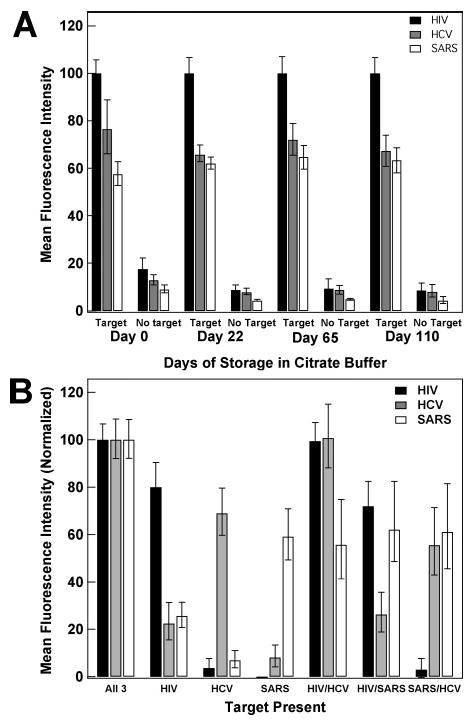
Triplex beacon assay using wires pre-coated in beacons and stored in citrate buffer for various numbers of days. (A) shows target versus no target data for days of storage up to 110 days. Intensities for all three probes on each day have been normalized to the HIV intensity at day 0. (B) shows multiple triplexed assays in the presence of various targets (targets added are indicated under the bars on the graph) after 110 day storage in citrate buffer. Intensities have been normalized for each probe, and the no target background has been subtracted. Error bars shown are the 95% confidence interval.
The relative intensities of the three different beacon probe sequences stayed constant over the 110 day period, with the HIV-specific probes in all cases significantly brighter than the other two probes. This can be understood in light of the greater stability of the hairpin structures for the HCV and SARS probes (Table 1); the HIV probe has the least negative ΔG of the three probes, and its superior performance under the conditions of this assay (500 mM NaCl, 50 °C) is consistent with the results of varying probe ΔG by changing stem length shown in Figure 3B.
Figure 10B shows results for simultaneous assays for HIV-, HCV-, and SARS-specific target oligonucleotides with all permutations of target combinations (i.e., none, all, and mixtures) using the nanowire bioconjugates that had been stored in citrate-containing PBS for 110 days. To simplify interpretation in the multiplexed assay, each probe was normalized independently, and signal from a no-target sample was subtracted from each data point. Despite over 3 months in storage, it remained possible to determine which target sequences were present in this multiplexed assay, and no loss in assay performance was observed.
Conclusions
In this Article, we have focused on initial optimization of MB probe performance on the encoded Au/Ag striped nanowires. Immobilization of the beacon probes leads to a strong electrostatic repulsion within and between probes on the nanowire surface, such that optimal performance requires high salt buffers (300–500 mM NaCl). The length of both the stem and the loop regions of the MB probes impacted performance, and relative thermodynamic stabilities predicted for solution-phase analogies of the probes used here provided useful information despite surface attachment and steric/electrostatic effects. Target binding could be fit to a Sips isotherm, and detection sensitivity for optimum probe stem and loop lengths was on the order of 100 pM. A multiplexed, sealed assay for three viral signature sequences was demonstrated, without reduction in performance as compared to the identical assay performed under nonclosed tube conditions with higher mixing volumes. Beacon-coated nanowires could be prepared ahead of time and stored indefinitely prior to use. No reduction in assay performance was observed after storage in citrate containing buffer for 110 days, the longest time tested. Our results suggest the potential of beacon-coated, barcoded metal nanowires for multiplexed detection of target DNA sequences such as viral signatures. While only three sequences were simultaneously detected in this work, larger numbers of identifiable nanowire patterns have been demonstrated and could be used to increase the level of multiplexing.32,36 No sample manipulations are needed after mixing the molecular beacon probe-coated nanowires with the target DNA, reducing assay complexity and the risk of contamination.
Supplementary Material
Acknowledgments
We thank Willy Valdivia-Granda (Orion Integrated Biosciences) for the Dengue viral genomic signature. This work was funded by the National Institutes of Health (R01 EB000268), the National Science Foundation (NIRT CCR-0303976), and Pennsylvania State University. C.D.K. also acknowledges support from a Beckman Foundation Young Investigator Award, a Sloan Fellowship, and a Dreyfus Teacher-Scholar Award. Oxonica, Inc. acknowledges funding from the National Institute of Standards and Technology Advanced Technology Program Grant (Grant 70NANB1H3028), and from the National Science Foundation (Grant 0418748). S.K.M. acknowledges support from the Penn State Biomaterials and Bionanotechnology Summer Institute.
Footnotes
Supporting Information Available: Effect of stem length on fluorescence intensity for DENV-2 molecular beacon probes bound to Ag/Au striped nanowires at 40 and 60 °C, and effect of NaCl concentration on performance of nanowire-bound DENV-2(5) probes at 40 °C. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Mackay IM, Arden KE, Nitsche A. Nucleic Acids Res. 2002;30:1292–1305. doi: 10.1093/nar/30.6.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Klein D. Trends Mol Med. 2002;8:257–260. doi: 10.1016/s1471-4914(02)02355-9. [DOI] [PubMed] [Google Scholar]
- 2.Nam JM, Thaxton CS, Mirkin CA. Science. 2003;301:1884–1886. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 3.(a) Rosi NL, Mirkin CA. Chem Rev. 2005;105:1547–1562. doi: 10.1021/cr030067f. [DOI] [PubMed] [Google Scholar]; (b) Kricka LJ. Ann Clin Biochem. 2002;39:114–129. doi: 10.1258/0004563021901865. [DOI] [PubMed] [Google Scholar]
- 4.Bonnet G, Tyagi S, Libchaber A, Kramer FR. Proc Natl Acad Sci U S A. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park SJ, Taton TA. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 6.Kwok PY. Annu Rev Genomics Hum Genet. 2001;2:235–258. doi: 10.1146/annurev.genom.2.1.235. [DOI] [PubMed] [Google Scholar]
- 7.Tyagi S, Kramer FR. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 8.Fang X, Li JJ, Perlette J, Tan W, Wang T. Anal Chem. 2000:747A–753A. doi: 10.1021/ac003001i. [DOI] [PubMed] [Google Scholar]
- 9.Broude NE. Trends Biotechnol. 2002;20:249–256. doi: 10.1016/s0167-7799(02)01942-x. [DOI] [PubMed] [Google Scholar]
- 10.Drake TJ, Tan W. Appl Spectrosc. 2004;58:269A–280A. doi: 10.1366/0003702041959406. [DOI] [PubMed] [Google Scholar]
- 11.Tan W, Wang K, Drake TJ. Curr Opin Chem Biol. 2004;8:547–553. doi: 10.1016/j.cbpa.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Tyagi S, Marras SAE, Kramer FR. Nat Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 13.Tyagi S, Bratu DP, Kramer FR. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 14.(a) Vet JAM, Majithia AR, Marras SAE, Tyagi S, Bube S, Poiesz B, Kramer FR. Proc Natl Acad Sci U S A. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sinsimer D, Leekha S, Park S, Marras SAE, Koreen L, Willey B, Naidich S, Musser KA, Kreiswirth BN. J Clin Microbiol. 2005;43:4585–4591. doi: 10.1128/JCM.43.9.4585-4591.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao G, Tan W. Anal Biochem. 2004;331:216–223. doi: 10.1016/j.ab.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 16.Wang H, Li J, Liu H, Liu Q, Mei Q, Wang Y, Zhu J, He N, Lu Z. Nucleic Acids Res. 2002;30:e61. doi: 10.1093/nar/gnf061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du H, Disney MD, Miller BL, Krauss TD. J Am Chem Soc. 2003;125:4012–4013. doi: 10.1021/ja0290781. [DOI] [PubMed] [Google Scholar]
- 18.Palecek E. Trends Biotechnol. 2004;22:55–58. doi: 10.1016/j.tibtech.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Fang X, Tan W. Anal Chem. 1999;71:3101–3105. doi: 10.1021/ac990225w. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Farmerie W, Schuster S, Tan W. Anal Biochem. 2000;283:56–63. doi: 10.1006/abio.2000.4656. [DOI] [PubMed] [Google Scholar]
- 21.Steemers FJ, Ferguson JA, Walt DR. Nat Biotechnol. 2000;18:91–94. doi: 10.1038/72006. [DOI] [PubMed] [Google Scholar]
- 22.Liu X, Tan W. Anal Chem. 1999;71:5054–5059. doi: 10.1021/ac990561c. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Wang H, Shi Z, Wang H, Yang C, Silke S, Tan W, Lu Z. Nucleic Acids Res. 2006;34:e4. [Google Scholar]
- 24.Du H, Strohsahl CM, Camera J, Miller BL, Krauss TD. J Am Chem Soc. 2005;127:7932–7940. doi: 10.1021/ja042482a. [DOI] [PubMed] [Google Scholar]
- 25.Dubertret B, Calame M, Libchaber AJ. Nat Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 26.Sha MY, Yamanaka M, Walton ID, Norton SM, Stoermer RL, Keating CD, Natan MJ, Penn SG. Nanobiotechnology. 2005;1:327–335. doi: 10.1385/NBT:1:4:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoermer RL, Keating CD. J Am Chem Soc. 2006;128:13243–13254. doi: 10.1021/ja0637200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marras SAE, Kramer FR, Tyagi S. Nucleic Acids Res. 2002;30:e122. doi: 10.1093/nar/gnf121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maxwell DJ, Taylor JR, Nie S. J Am Chem Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 30.Nicewarner-Pena SR, Freeman RG, Reiss BD, He L, Pena DJ, Walton ID, Cromer R, Keating CD, Natan MJ. Science. 2001;294:137–141. doi: 10.1126/science.294.5540.137. [DOI] [PubMed] [Google Scholar]
- 31.Keating CD, Natan MJ. Adv Mater. 2003;15:451–454. [Google Scholar]
- 32.Walton ID, Norton SM, Balasingham A, He L, Oviso DF, Jr, Gupta D, Raju PA, Natan MJ, Freeman RG. Anal Chem. 2002;74:2240–2247. doi: 10.1021/ac020073w. [DOI] [PubMed] [Google Scholar]
- 33.(a) Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]; (b) Ellington AD, Szostak JW. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]; (c) Navani NK, Li Y. Curr Opin Chem Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Reiss BD, Freeman RG, Walton ID, Norton SM, Smith PC, Stonas WG, Keating CD, Natan MJ. J Electroanal Chem. 2002;522:95–103. [Google Scholar]
- 35.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrin A, Duracher D, Perret M, Cleuziat P, Mandrand B. Anal Biochem. 2003;322:148–155. doi: 10.1016/j.ab.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 37.The DENV-2 genomic signature is Orion Integrated Biosciences Inc. proprietary information in pending patent applications filed with the U.S. Trademark and Patent Office.
- 38.Nicewarner-Pena SR, Carado AJ, Shale KE, Keating CD. J Phys Chem B. 2003;107:7360. [Google Scholar]
- 39.Enustun BV, Turkevich J. J Am Chem Soc. 1963;85:3317–3328. [Google Scholar]
- 40.Keating CD, Musick MD, Keefe MH, Natan MJ. J Chem Educ. 1999;76:949–955. [Google Scholar]
- 41.Park S, Brown KA, Hamad-Schifferli K. Nano Lett. 2004;4:1925–1926. [Google Scholar]
- 42.Schonherr H, Ringsdorf H. Langmuir. 1996;12:3891–3897. [Google Scholar]
- 43.Gronbeck H, Curioni A, Andreoni W. J Am Chem Soc. 2000;122:3839–3842. [Google Scholar]
- 44.(a) Herne TA, Tarlov MJ. J Am Chem Soc. 1997;119:8916–8920. [Google Scholar]; (b) Steel AB, Levicky RL, Herne TM, Tarlov MJ. Biophys J. 2000;79:975–981. doi: 10.1016/S0006-3495(00)76351-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.(a) Shchepinov MS, Case-Green SC, Southern EM. Nucleic Acids Res. 1997;25:1155–1161. doi: 10.1093/nar/25.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Southern E, Mir K, Shchepinov M. Nat Genet Suppl. 1999;21:5–9. doi: 10.1038/4429. [DOI] [PubMed] [Google Scholar]
- 46.Nicewarner-Peña SR, Raina S, Goodrich GP, Fedoroff NV, Keating CD. J Am Chem Soc. 2002;124:7314–7323. doi: 10.1021/ja0177915. [DOI] [PubMed] [Google Scholar]
- 47.Demers LM, Mirkin CA, Mucic RC, Reynolds RA, III, Letsinger RL, Elghanian R, Viswanadham G. Anal Chem. 2000;72:5535–5541. doi: 10.1021/ac0006627. [DOI] [PubMed] [Google Scholar]
- 48.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. J Am Chem Soc. 1998;120:1959–1964. [Google Scholar]
- 49.Li H, Rothberg L. Proc Natl Acad Sci U S A. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. J Am Chem Soc. 2000;122:9071–9077. [Google Scholar]
- 51.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 52.Harris DC. Quantitative Chemical Analysis. 4th. W H Freeman and Co.; New York: 1995. [Google Scholar]
- 53.Sips R. J Chem Phys. 1948;16:490–495. [Google Scholar]
- 54.Vijayendran RA, Leckband DE. Anal Chem. 2001;73:471–480. doi: 10.1021/ac000523p. [DOI] [PubMed] [Google Scholar]
- 55.(a) Levicky R, Horgan A. Trends Biotechnol. 2006;23:143–149. doi: 10.1016/j.tibtech.2005.01.004. [DOI] [PubMed] [Google Scholar]; (b) Halperin A, Buhot A, Zhulina EB. J Phys: Condens Matter. 2006;18:S463–S490. [Google Scholar]
- 56.Hekstra D, Taussig AR, Magnasco M, Naef F. Nucleic Acids Res. 2003;31:1962–1968. doi: 10.1093/nar/gkg283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nelson BP, Grimsrud TE, Liles MR, Goodman RM, Corn RM. Anal Chem. 2001;73:1–7. doi: 10.1021/ac0010431. [DOI] [PubMed] [Google Scholar]
- 58.Peterson AW, Wolf LK, Georgiadis RM. J Am Chem Soc. 2002;124:14601–14607. doi: 10.1021/ja0279996. [DOI] [PubMed] [Google Scholar]
- 59.Abravaya K, Huff J, Marshall R, Merchant B, Mullen C, Schneider G, Robinson J. Clin Chem Lab Med. 2003;41:468–474. doi: 10.1515/CCLM.2003.070. [DOI] [PubMed] [Google Scholar]
- 60.Tan L, Li Y, Drake TJ, Moroz L, Wang K, Li J, Munteanu A, Yang CJ, Martinez K, Tan W. Analyst. 2005;130:1002–1005. doi: 10.1039/b500308n. [DOI] [PubMed] [Google Scholar]
- 61.Mhlanga MM, Malmberg L. Methods. 2001;25:463–471. doi: 10.1006/meth.2001.1269. [DOI] [PubMed] [Google Scholar]
- 62.Szuhai K, van den Ouweland JM, Dirks RW, Lemaitre M, Truffert JC, Janssen GM, Tanke HJ, Holme E, Maassen JA, Raap AK. Nucleic Acids Res. 2001;29:e13. doi: 10.1093/nar/29.3.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.(a) Sokol D, Zhang X, Lu P, Gewirth AM. Proc Natl Acad Sci U S A. 1998;95:11538–11543. doi: 10.1073/pnas.95.20.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Bratu DP, Cha BJ, Mhlanga MM, Kramer FR, Tyagi S. Proc Natl Acad Sci U S A. 2003;100:13308–13313. doi: 10.1073/pnas.2233244100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoermer RL, Sioss JA, Keating CD. Chem Mater. 2005;17:4356–4361. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


