SUMMARY
Previous structures of Arp2/3 complex, determined in the absence of a nucleation-promoting factor and actin, reveal its inactive conformation. The study of the activated structure has been hampered by uncontrollable polymerization. We have engineered a stable activated complex consisting of Arp2/3 complex, the WCA activator region of N-WASP and one actin monomer, and studied its structure in solution by small angle X-ray scattering. The scattering data support a model in which the first actin subunit binds at the barbed end of Arp2, and disqualify an alternative model that places the first actin subunit at the barbed end of Arp3. This location of the first actin and bound W motif constraints the binding site of the C motif to subunits Arp2 and ARPC1, from where the A motif can reach subunits Arp3 and ARPC3. The results support a model of activation that is consistent with most of the biochemical observations.
Keywords: Actin filament nucleation, cytoskeleton dynamics, Arp2/3 complex
INTRODUCTION
The initiation of actin polymerization in cells requires filament nucleation factors, including Arp2/3 complex and formins (Pollard, 2007). These molecules stabilize actin polymerization seeds, whose formation is rate-limiting during actin assembly (Sept and McCammon, 2001). Arp2/3 complex is targeted by multiple signaling pathways that result in actin cytoskeleton remodeling during processes such cell migration, endocytosis and the maintenance of cell morphology.
Arp2/3 complex consists of seven proteins, including the actin-related proteins Arp2 and Arp3. By itself, Arp2/3 complex has low nucleation activity. The complex is activated by nucleation-promoting factors (NPFs), among which the most prominent are members of the WASP/WAVE family of proteins (Goley and Welch, 2006; Pollard, 2007). The C-terminal region of WASP/WAVE proteins, known as the WCA (or VCA) region, constitutes the shortest fragment necessary for Arp2/3 complex activation (Machesky et al., 1999). The W (WASP-Homology 2 or WH2) motif within this region binds the first actin monomer of the new filament branch, whereas the C (central or connecting) and A (acidic) motifs interact with various subunits of Arp2/3 complex, helping to stabilize the activated conformation. The actin monomer bound to the W motif, together with Arp2 and Arp3, are thought to form a trimeric seed for the nucleation of a filament branch, which emerges at a 70° angle from the side of a preexisting filament. According to this model, Arp2 and Arp3 are the first two subunits at the pointed end of the new filament branch.
The crystal structure of Arp2/3 complex was first determined in the absence of nucleotide and NPF (Robinson et al., 2001). In the structure, Arp2 and Arp3 are separated and the nucleotide cleft of Arp3 is wide open, whereas subdomains 1 and 2 of Arp2 are disordered. Therefore, this structure was described as the inactive conformation of the complex (Robinson et al., 2001). Subsequently Arp2/3 complex was crystallized in the presence of nucleotide and nucleotide analogs (Nolen and Pollard, 2007). Nucleotide binding favors closure of the nucleotide cleft of Arp3 and marginally stabilizes subdomains 1 and 2 of Arp2. However, the relative position of Arp2 and Arp3 was unchanged, indicating that ATP binding is not sufficient to activate Arp2/3 complex. Indeed, it is generally believed that the binding of nucleotide and WCA are thermodynamically coupled and that these two factors contribute together to activate Arp2/3 complex (Dayel et al., 2001; Goley et al., 2004; Le Clainche et al., 2001).
The structure of Arp2/3 complex in the branch and with bound WASP has been studied using electron microscopy (Egile et al., 2005; Rodal et al., 2005). These structures consistently indicate that a major conformational change takes place upon activation, bringing Arp2 and Arp3 into an actin filament-like arrangement at the pointed end of the new filament branch. However, none of the existing structures resolves the location and interactions of the WCA activator region with subunits of Arp2/3 complex. This question has mainly been addressed by cross-linking experiments, showing that CA can be cross-linked to subunits Arp2, ARPC1, Arp3 and ARPC3 (Kelly et al., 2006; Kreishman-Deitrick et al., 2005; Weaver et al., 2002; Zalevsky et al., 2001). Because of the short length of the WCA polypeptide (73 aa), and considering that both the C (Panchal et al., 2003) and W motifs comprise helical segments, with W bound at the barbed end of the first actin monomer (Chereau et al., 2005), it is difficult to rationalize how CA can span subunits Arp2, ARPC1, Arp3 and ARPC3 in the complex. This question cannot be fully addressed without knowing the binding site of the first actin subunit in the activated complex. Here we present a solution study by small angle X-ray scattering (SAXS) of an engineered activated Arp2/3 complex containing the WCA region of N-WASP and the first actin subunit of the new filament branch. The study reveals that the first actin subunit binds at the barbed end of Arp2, which also sheds light on the location and interactions of the CA region of N-WASP with other subunits of Arp2/3 complex. Based on the results we propose a general model of activation of Arp2/3 complex by NPFs.
RESULTS
Engineering of a stable activated actin-WCA-Arp2/3 complex
A major limitation in the structural investigation of actin polymerization seeds is their intrinsic dynamic character. Arp2/3 complex polymerization seeds evolve quickly into filaments, and the complex itself is believed to exist in multiple states (Goley et al., 2004; Rodal et al., 2005). To overcome these limitations, we devised a strategy to block the polymerization seed into a single activated species by structure-based cross-linking of WCA to actin. From the high-resolution crystal structures of various W motifs bound to actin (Chereau et al., 2005), we determined that N-WASP residue Ser 426 binds in close proximity to and facing actin Cys 374. Therefore, we expressed a WCA mutant in which Ser 426 was replaced by Cys and the endogenous Cys 427 by Ala. Note that another W motif precedes WCA in N-WASP, but this feature is not conserved among NPFs. The Cys-WCA construct was cross-linked to actin with nearly 100% efficiency and the cross-linked complex was purified to homogeneity and characterized by analytical ultracentrifugation (AUC) (Figure 1a and 1d).
Figure 1. Design and characterization of activated actin-WCA-Arp2/3 complex.
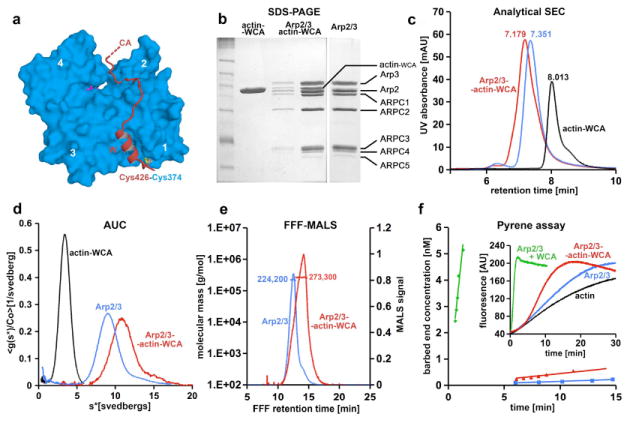
(a) From the structure of actin complexes with the W motifs of various cytoskeletal proteins (Chereau et al., 2005), we determined that N-WASP residue Ser 426 could be mutated to Cys and cross-linked to actin Cys 374 with minimal structural disturbance. The actin subdomains are numbered 1 to 4.
(b) Non-reducing coomassie-stained gel showing cross-linked actin-WCA, purified Arp2/3 complex, and Arp2/3 complex with bound actin-WCA.
(c) Arp2/3 complex with bound actin-WCA (red) has shorter retention time on analytical size exclusion chromatography than Arp2/3 complex alone (blue). Actin-WCA is also shown for reference (black). Retention times are indicated above each peak.
(d) The heavier actin-WCA-Arp2/3 complex sediments faster by analytical ultracentrifugation than Arp2/3 complex alone, with sedimentation coefficients of ~11 S and ~8.8 S, respectively.
(e) Determination of the molecular masses of Arp2/3 complex with and without bound actin-WCA by FFF-MALS. The measured masses were 224,200 Da for Arp2/3 complex alone (expected value from sequence 223,600 Da) and 273,300 for actin-WCA-Arp2/3 complex (expected 274,060 Da).
(f) Time course of barbed end formation of 2 μM Mg-ATP-actin (6% pyrene-labeled) alone (black) or in the presence of 25 nM Arp2/3 complex and 250 nM native WCA (green) or 25 nM actin-WCA-Arp2/3 complex (red). The inset shows the time course of fluorescence increase upon polymerization. Nucleation rates were determined from the polymerization rate, the concentration of actin monomers remaining, and the rate constant for barbed-end elongation (10 μM-1s-1).
Cross-linked actin-WCA was mixed with Arp2/3 complex at a 1.2:1 ratio. The resulting complex was separated from excess actin-WCA by size exclusion chromatography (Figure 1b). Analytical size exclusion chromatography shows that the complex has lower retention time than Arp2/3 complex alone, indicating that actin-WCA is bound (Figure 1c). Dissociation of actin-WCA was not observed, suggesting that the nine-protein complex is stable. To confirm this finding, we compared the sedimentation velocities of Arp2/3 complex with and without actin-WCA. Both complexes formed single boundaries, which are distinguishable from each other and from actin-WCA (Figure 1d). In agreement with a previous study (Mullins et al., 1997), the sedimentation coefficient of Arp2/3 complex alone was ~8.8 S. Binding of actin-WCA increased the sedimentation coefficient to ~11 S. The molecular masses of the complexes were estimated using flow-field fractionation combined with multi-angle light scattering (FFF-MALS) (Figure 1e). The measured mass of Arp2/3 complex alone was 224,200 Da, which is strikingly similar to the expected value from sequence (223,600 Da). The observed mass of the complex upon addition of actin-WCA was 273,300 Da, also in excellent agreement with the expected value (274,060 Da).
Combined, these results suggested that actin-WCA forms a stable 1:1 complex with Arp2/3 complex. This is in sharp contrast with previous studies that generally used excess WCA in order to ensure saturated binding, which is due to WCA’s low intrinsic affinity (2 μM) for Arp2/3 complex (Panchal et al., 2003). Based on a dilution series AUC experiment, we estimated that the binding affinity of actin-WCA for Arp2/3 complex is ~70 nM (Figure S1). Because similarly cross-linked actin-W (lacking the C and A motifs) did not bind Arp2/3 complex (Figure S2), we conclude that the increased affinity results from the combined affinities of actin and the CA region. In other words, actin, even when bound to W, has very low affinity for inactive Ap2/3 complex, which probably contributes to the low nucleation activity of the complex in the absence of NPFs.
The resulting nine-protein complex contains ATP, actin, and WCA, three main factors known to cooperate in the activation of Arp2/3 complex (Pollard, 2007). However, because 1) the complex cannot release WCA, as it normally occurs during branch formation (Egile et al., 2005), and 2) covalently bound W at the barbed end of actin sterically hinders the addition of actin monomers, the cross-linked complex was expected to have low nucleation activity. To test this assumption we used the pyrene-actin polymerization assay (Figure 1f). We compared the nucleation activity of actin-WCA bound Arp2/3 complex with that of Arp2/3 complex in the presence of a 10-fold molar excess of native WCA. As expected, the nucleation rate (change in the number of barbed ends over time) of actin-WCA bound Arp2/3 complex (0.064 ± 0.006 nM min-1) was much lower than that of Arp2/3 complex with free WCA (4.37 ± 0.6 nM min-1), and only marginally higher than that of Arp2/3 complex alone (0.015 ± 0.002 nM min-1).
X-ray scattering study of activated actin-WCA-Arp2/3 complex
The conformation of Arp2/3 complex with bound actin-WCA was investigated by SAXS, a method that yields information about the overall shape of macromolecules (Koch et al., 2003; Petoukhov and Svergun, 2007; Svergun and Koch, 2002). To validate the applicability of SAXS to this particular system, the solution structure of Arp2/3 complex alone was analyzed first and compared to the crystal structure of the inactive complex (Nolen and Pollard, 2007; Robinson et al., 2001). The radius of gyration (Rg) derived from the scattering curve and representing the average size of the particle in solution was 47±0.1 Å, which closely matches the value calculated from the crystal structure of Arp2/3 complex (46.4 Å). Furthermore, the scattering curve calculated from the crystal structure with the program CRYSOL (Svergun et al., 1995) fits closely the experimental data, as characterized by a discrepancy value χ2 of 1.55 (Figure 2). While the χ2 is the most reliable parameter to evaluate a scattering model, we also used a model-independent approach to reconstruct the solution shape of Arp2/3 complex ab initio from the scattering data with the program DAMMIN (Svergun, 1999). The final ab initio envelope consisted of the average, calculated with the program DAMAVER (Volkov and Svergun, 2003), of fifteen dummy atom models. This envelope fits the crystal structure well, both visually and quantitatively, as reflected by a normalized spatial discrepancy (NSD) value of 1.42 (Figure 2). These results indicate that the crystal structure of inactive Arp2/3 complex is preserved in solution and suggest that SAXS can be reliably used to reconstruct the structure of the complex.
Figure 2. Structure of inactive Arp2/3 complex.
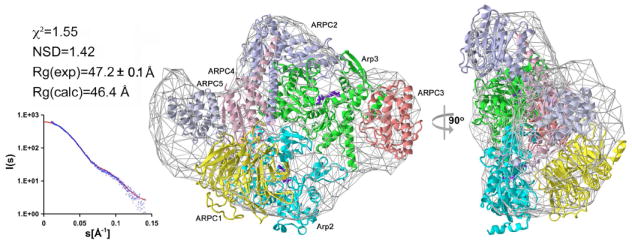
Fitting statistics of the crystal structure of inactive Arp2/3 complex to the scattering data (left). The experimental and computed scattering intensities are displayed as a function of momentum transfer s = 4π sin(θ)/λ, where 2θ is the scattering angle and λ = 1.5 Å is the X-ray wavelength. The scattering data were analyzed in the range 0.01 < s < 0.15 Å-1, which yields information about the overall shape of the molecule. Higher resolution data were discarded in this analysis due to increased noise. Two orientations of the structure, rotated by 90° and docked into the ab initio SAXS envelope (right). Individual proteins are shown as ribbon diagrams (color scheme: Arp2, cyan; Arp3, green, ARPC1, yellow; ARPC2, purple; ARPC3, maroon; ARPC4, pink; ARPC5, gray).
The same approach was then applied to Arp2/3 complex with bound actin-WCA. The experimental scattering profile in this case was compared to theoretical curves calculated from alternative models of activation. Other than the addition of actin and WCA, activation is thought to involve conformational changes within Arp2/3 complex itself (Pollard, 2007). While some of these changes may be minor, significant and well-documented changes involve repositioning of Arp2 into a filament-like conformation with Arp3 (Egile et al., 2005) and nucleotide-dependent closure of their clefts (Goley et al., 2004; Nolen and Pollard, 2007). We adopted the structure of nucleotide bound inactive Arp2/3 complex and rebuilt subdomains 1 and 2 of Arp2 by analogy with actin (Figure S3a and S3b). Arp2 was then moved alone into an actin filament-like conformation with Arp3 (Figure S3d), as previously proposed (Aguda et al., 2005). This mode of activation minimizes the magnitude of the structural changes, produces no steric clashes, and is consistent with the fact that Arp2 is flexible and partially disordered in the inactive structure (a different mode of activation is discussed below). The crystal structure of actin-W (Chereau et al., 2005) was then added at the barbed end of either Arp2 or Arp3 (Figures S3e and S3f), according to the arrangement of actin subunits in the filament (Holmes et al., 1990). These two models test two radically different pathways of Arp2/3 complex activation previously proposed (Aguda et al., 2005; Beltzner and Pollard, 2004; Chereau et al., 2005).
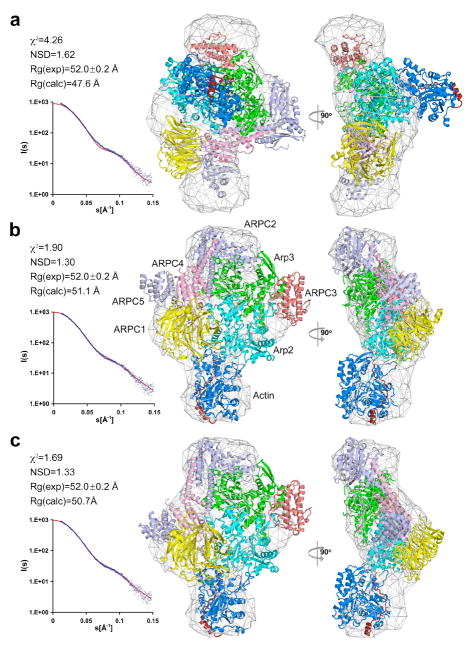
Comparison of the experimental and calculated scattering curves and radiuses of gyration and model docking into the averaged ab initio envelope, all clearly disqualified the model with actin at the barbed end of Arp3 (Figure 3a) and gave strong support to the model with actin at the barbed end of Arp2 (Figure 3b). Indeed, in the range of scattering vectors up to S=1.5 nm-1, a range that accounts for the overall shape of a particle (Svergun et al., 2001), the discrepancy between the experimental and model-based curves for these two models is χ2=4.26 (indicating major systematic deviations and poor fit) and χ2=1.90 (considered a good fit), respectively. Docking into the ab initio envelope is also significantly better when actin is bound at the barbed end of Arp2 (NSD=1.30) than Arp3 (NSD=1.62). Finally, the Rg value derived from the scattering curve (52.0±0.2 Å) is in close agreement with the value calculated for the model with actin at the barbed end of Arp2 (51.1 Å), but significantly larger than that of the model with actin at the barbed end of Arp3 (47.6 Å).
Figure 3. Scattering statistics for different models of actin-WCA-Arp2/3 complex.
(a) Fitting of the model with actin bound at the barbed end of Arp3 to the scattering data (left). In this model Arp2 was moved alone into a filament-like conformation with Arp3 (see also Figure S3f). Two orientations of the model, rotated by 90°, are shown inside the averaged grid envelope resulting from the ab initio SAXS reconstruction (right). A poor fit to the scattering data disqualifies this model. The individual proteins are shown as ribbon diagrams and colored according to other figures of the paper (W motif, red; actin, blue; Arp2, cyan; Arp3, green, ARPC1, yellow; ARPC2, purple; ARPC3, maroon; ARPC4, pink; ARPC5, gray).
(b) Fitting of the model with actin bound at the barbed end of Arp2 after activation of Arp2/3 complex by moving Arp2 alone alongside Arp3 (see also Figure S3). The orientation is as in Figure 2.
(c) Fitting of the model with actin bound at the barbed end of Arp2 after activation of Arp2/3 complex by moving together Arp2, ARPC1, ARPC4 and ARPC5 (see also Figure S3).
The two models considered above are based on the generally accepted assumption that Arp2 occupies a filament-like position alongside Arp3 in the activated structure (Aguda et al., 2005; Egile et al., 2005; Goley and Welch, 2006; Pollard, 2007; Robinson et al., 2001). Two different pathways have been proposed to accomplish the realignment of Arp2 with respect to Arp3 during activation. In the two models considered above we moved subunit Arp2 alone (Figure S3), which in the structure of the inactive complex appears to be loosely connected to other subunits of the complex. However, a more substantial rearrangement of the complex, involving a rotation of subunits Arp2, ARPC1, ARPC4 and ARPC5 relative to subunits Arp3, ARPC2 and ARPC3 was originally proposed (Robinson et al., 2001) (Figure S4). We do not favor this pathway because it involves a comparatively large structural change and the breakage of multiple hydrophobic interactions along a large interface between the two halves of the complex. However, at the resolution of this study, we cannot distinguish between the two pathways (i.e. moving Arp2 alone vs. moving Arp2, ARPC1, ARPC4 and ARPC5 as a block). In fact, after addition of the first actin subunit at the barbed end of Arp2 (Figure S4c) the second pathway yields a somewhat better χ2 value (1.69), which may suggest that changes other than the movement of Arp2 occur during activation. Note that independent of the pathway of activation, the models with actin at the barbed end of Arp3 are clearly excluded by the scattering data (Figures S3f and S4d).
The first actin subunit had been proposed to occupy the next available space in the short-pitch filament helix at the barbed end of Arp3 (Beltzner and Pollard, 2004), which due to steric clashes only becomes accessible if Arp2 moves first. In other words, if this proposal is correct, activation of the complex must precede the addition of the first actin subunit. In contrast, we found here that the first actin subunit binds at the barbed end of Arp2, which can either precede or follow activation, since both models are steric clash free (Figure S3c and S3e). Therefore, we also tested a model in which actin is bound at the barbed end of Arp2 in the inactive conformation of the original crystal structure (Figure S3c). Fitting of this model to the experimental data was considerably worse (χ2=2.58, NSD=1.47, Rgcalc=52.6 Å) than fitting of the pre-activated models with actin at the barbed end of Arp2 (Figure 3b and 3c). However, the differences in fitting statistics are not large enough to totally exclude this model. Therefore, the main conclusion from the scattering experiments is that the first actin monomer bound to the W motif of NPFs interacts at the barbed end of Arp2.
DISCUSSION
Crystal structures of Arp2/3 complex have been determined in the inactive conformation (Nolen and Pollard, 2007; Robinson et al., 2001), while the structure of the complex in the branch has been studied using electron microscopy (Egile et al., 2005). However, important questions remain concerning the mechanism of activation and the role of WCA in this process. Structural methods require monodisperse species which, owing to the dynamic nature of actin polymerization seeds, has been a major limitation in the study of activated Arp2/3 complex. We have found a solution to this problem by structure-based cross-linking actin and WCA prior to binding to Arp2/3 complex. This approach resulted in a homogeneous and stable polymerization seed suitable for structural investigation (Figure 1).
The structure of actin-WCA-Arp2/3 complex in solution was reconstructed by SAXS (Figure 3). We tested four models of activation that fall into two major categories: actin at the barbed end of Arp3 (Beltzner and Pollard, 2004), which can only occur if Arp2 moves first into an activated conformation, and actin at the barbed end of Arp2 (Aguda et al., 2005; Chereau et al., 2005), which can either precede or follow the movement of Arp2. While the scattering data clearly disqualifies the model with actin at the barbed end of Arp3, we cannot reliably exclude any of the models with actin at the barbed end of Arp2 (Figure 3). It is possible that the complex populates multiple intermediate states with actin at the barbed end of Arp2, and that binding to the side of a pre-existing filament fixes the complex into a unique activated conformation (Figure 4). As previously proposed (Aguda et al., 2005; Egile et al., 2005; Pollard, 2007; Robinson et al., 2001), this unique activated conformation is most likely one in which Arp2 occupies a filament-like conformation alongside Arp3, which consistently gave the best fit to the scattering data (Figures 3b and 3c). It remains unclear how Arp2 moves into the activated state; alone (Aguda et al., 2005), together with subunits ARPC1, ARPC4 and ARPC5 (Robinson et al., 2001), or a combination of the two. Although moving Arp2, ARPC1, ARPC4 and ARPC5 as a group results in a slightly better fit to the scattering data, we favor moving Arp2 alone because this subunit appears to be in an unstable position in the original structure of the inactive complex and because this pathway of activation involves fewer structural changes. Slight changes to this model, the existence of which has been suggested by a previous study (Goley et al., 2004), may result in even better fit to the scattering data. However, such alterations of the original crystal structure are not supported at low resolution.
Figure 4. Model of activated Arp2/3 complex.
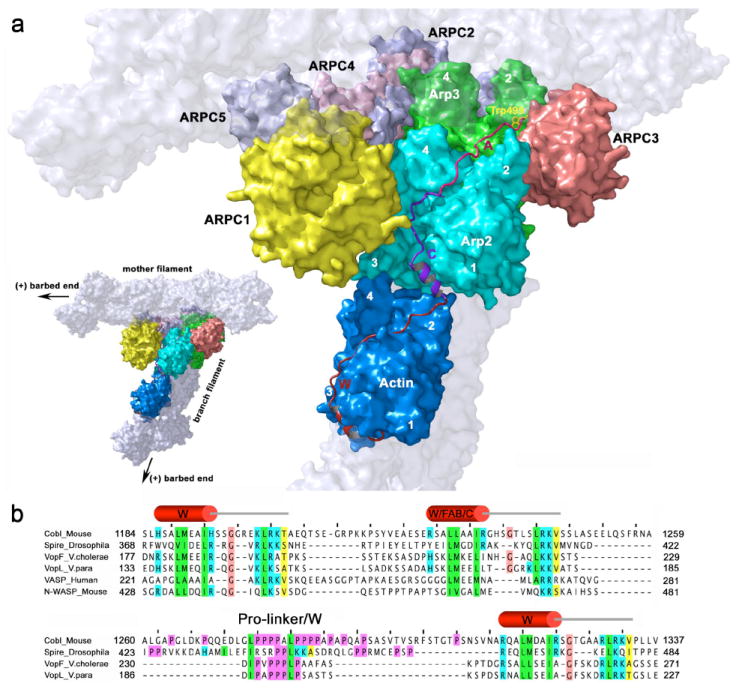
(a) Model of activated Arp2/3 complex with actin bound at the barbed end of Arp2 (as in Figure 3b), showing the most likely path of the WCA polypeptide. The mother and daughter filaments and their respective barbed (or +) ends are tentatively shown for reference (light gray, semi-transparent), but note that the precise organization of the branch is not addressed by this study. Subdomains 1 to 4 of actin and Arp2 and subdomains 2 and 4 of Arp3 are numbered. In the activated structure, Arp2 moves to occupy a filament-like conformation next to Arp3. The first actin subunit, bound to the W motif of N-WASP (red), binds at the barbed end of Arp2. The position of the W motif in the activated complex is derived from the crystal structure of actin-W (Chereau et al., 2005). This location of the first actin and bound W motif imposes constraints on the location of the C and A motifs (magenta and pink). Thus, the hydrophobic cleft between subdomains 1 and 3 of Arp2 is right in the path of the CA polypeptide as it progresses toward Arp2/3 complex. We predict that the helical portion of the C motif (Panchal et al., 2003) binds in this cleft. This idea is also supported by sequence similarity with the W motif and by the fact that tandem W motifs constitute a common architecture among actin filament nucleators (see part b). The C-terminal portion of the C motif runs near the interface between Arp2 and ARPC1, from where the A motif can reach a hydrophobic pocket at the interface between Arp3 and ARPC3, which is consistent with existing binding and cross-linking studies (Kelly et al., 2006; Kreishman-Deitrick et al., 2005; Pan et al., 2004; Weaver et al., 2002; Zalevsky et al., 2001). The conserved Trp 499 of the A motif (yellow) may bind in a hydrophobic pocket at the interface between Arp3 and ARPC3, formed by amino acids Tyr202, Phe 203, Leu 207, Leu 277, Phe 287 of Arp3 and Phe 54 and Phe 163 of ARPC3.
(b) Alignment of the WC motifs of N-WASP with tandem W motifs of proteins involved in actin assembly. Conserved amino acids are colored according to their chemical characteristics (green, hydrophobic; blue, basic; magenta, prolines; brown, glycines; yellow, small conserved amino acid). The WC motifs of N-WASP align particularly well with the G- and F-actin binding (GAB and FAB) motifs of vasodilator-stimulated phosphoprotein (VASP) (Ferron et al., 2007). Since FAB binds F-actin, it appears likely that the C motif binds Arp2 within Arp2/3 complex (see text). Tandem W motifs are emerging as a common architecture in actin filament nucleation. Spire (Quinlan et al., 2005), Cobl (Ahuja et al., 2007) and the Vibrio proteins VopL (Liverman et al., 2007) and VopF (Tam et al., 2007) are recently discovered filament nucleators that utilize tandem W motifs to assemble actin monomers into polymerization seeds. Of note, Cobl, VopL and VopF contain only three W motifs and present a long Pro-rich linker between the second and third W motifs. Although this linker in spire presents all the signature features of a W motif, it is also rich in Pro residues. By analogy with these proteins, we predict that WC constitutes a specialized form of tandem W motifs, where C has evolved specificity for Arp2, allowing for the combination of the nucleation and branching functions by Arp2/3 complex. Sequence accession numbers are: Cobl (Mouse, CAI36023); Spire (Drosophila melanogaster, Q9U4F1); VopF (Vibrio cholerae, AAZ32252); VopL (Vibrio parahaemolyticus, NP_800881); VASP (Human, P50552); N-WASP (Mouse, CAC69994).
The models analyzed here are all based on the generally accepted idea that the two Arps and the actin subunit associated with the W motif of NPFs form a trimeric seed for nucleation of a filament branch that emerges at a 70° angle from the side of a preexisting filament. One group, however, sustains that Arp2/3 complex binds to the barbed end of the mother filament, where one of the Arps is incorporated into the mother filament while the other Arp becomes part of the branch (Carlier et al., 2003; Pantaloni et al., 2000). This proposal implies a profound reorganization of Arp2/3 complex, which is incompatible with existing structural data, including this work.
Defining the location of the first actin subunit to the barbed end of Arp2 imposes considerable constraints on the way WCA binds to and activates Arp2/3 complex. We know from the crystal structure of actin-W that the N-terminal half of the W motif forms an amphiphilic helix that binds in the hydrophobic cleft between actin subdomains 1 and 3 (Chereau et al., 2005). The C-terminal half of the W motif climbs along the actin surface toward the pointed end of the actin monomer. The ensuing C motif must therefore bind Arp2, ARPC1 or both (Figure 4a and legend). Specifically, the hydrophobic cleft of Arp2 is in the path of the CA polypetide as it progresses toward Arp2/3 complex (Figure 5a). Like the W motif, the N-terminal portion of the C motif forms an amphiphilic helix (Panchal et al., 2003). We (Chereau and Dominguez, 2006; Chereau et al., 2005) and others (Aguda et al., 2005; Hertzog et al., 2004) have pointed out that the C motif presents sequence similarity with the W motif and that it may bind to Arp2 in a way analogous to the actin-W interaction. The same cannot be proposed for Arp3, because its hydrophobic cleft is occupied by a C-terminal extension of the polypeptide chain. The WC motifs of WASP family proteins also present clear sequence similarity with the G- and F-actin binding motifs (GAB-FAB) of Ena/VASP proteins (Figure 4b). We have recently shown that the GAB and W motifs bind similarly to actin (Chereau et al., 2005; Ferron et al., 2007). Since the FAB motif of Ena/VASP binds F-actin, it appears likely that the closely related C motif of N-WASP binds one of the actin-related subunits of Arp2/3 complex. There is one additional consideration in support of this model. Other than Arp2/3 complex and formin (Pollard, 2007), three filament nucleators have recently been identified: Spire (Quinlan et al., 2005), Cobl (Ahuja et al., 2007) and the related Vibrio proteins VopL (Liverman et al., 2007) and VopF (Tarn et al., 2007). With the exception of formin, all known filament nucleators use tandem W motifs to assemble actin monomers into a polymerization seed (Figure 4b). By analogy with these proteins we predict that the WC of N-WASP constitutes a specialized form of tandem W motifs, where C has evolved specificity for Arp2. In other words, N-WASP and other NPFs can be conceptually viewed as filament nucleators that bring together one actin monomer with the actin-related proteins of Arp2/3 complex. The association of the Arps with five other proteins in the larger Arp2/3 complex probably emerged from a need to integrate nucleation and branching within a single system. Together, these considerations and structural constraints resulting from the SAXS analysis allow us to propose that the N-terminal helical region of the C motif binds in the hydrophobic cleft of Arp2, which is in close proximity to subunit ARPC1 (Figure 4a).
A recent proposal that the role of the C motif is to first activate Arp2/3 complex and then deliver the first actin subunit to the complex by binding to a different region in the actin monomer than the W motif (Kelly et al., 2006), is not supported by our results. Although we also find that actin binds the C motif (albeit with weak affinity), our results suggest that this interaction also takes place through the hydrophobic cleft between actin subdomains 1 and 3. Indeed, cross-linked actin-W, in which this cleft is permanently occupied by the W motif, does not bind WCA (Figure S5), suggesting that there is no second binding site on the actin monomer for any part of WCA.
The CA region has been cross-linked to subunits Arp2, ARPC1, Arp3 and ARPC3 of Arp2/3 complex (Kelly et al., 2006; Kreishman-Deitrick et al., 2005; Weaver et al., 2002; Zalevsky et al., 2001). In particular, NMR, cross-linking, and cortactin competition studies place the N-terminal helical portion of the C motif in contact with subunits Arp2 and ARPC1 and the C-terminus of the A motif in contact with subunits Arp3 and ARPC3 (Kreishman-Deitrick et al., 2005; Weaver et al., 2002). Interpretation of these results has led to the proposal that CA connects these four subunits in the activated complex, bridging the pointed ends of Arp2 and Arp3 (Pollard, 2007). Constraints imposed by the length of the WCA polypeptide, in particular considering that both W (Chereau et al., 2005) and C (Panchal et al., 2003) contain helical segments, mean that these interactions cannot be achieved if the first actin subunit binds at the barbed end of Arp3 as previously suggested (Beltzner and Pollard, 2004). In contrast, the model of activated Arp2/3 complex resulting from the SAXS study is fully consistent with the binding data (Figure 4a). Indeed, as it emerges from the cleft in Arp2, the C-terminal portion of the C motif runs near the interface between subunits Arp2 and ARPC1. This path takes the A motif to the interface between subunits Arp3 and ARPC3, where the conserved Trp 499 may bind in a hydrophobic pocket formed by amino acids Tyr202, Phe 203, Leu 207, Leu 277, Phe 287 of Arp3 and Phe 54 and Phe 163 of ARPC3. Another study implicates this Trp in direct interaction with ARPC1, suggesting a different binding site for the A motif (Pan et al., 2004). A proposal that reconciles this result with the cross-linking studies is that the A motif binds to different subunits of Arp2/3 complex during different stages of activation (Aguda et al., 2005). The scattering model of activated Arp2/3 complex would be consistent with both modes of interaction.
In conclusion, in the model of activated Arp2/3 complex proposed here (Figure 4a), the location of the W motif is crystallographically determined to the hydrophobic cleft of the first actin (Chereau et al., 2005), which binds at the barbed end of Arp2, the location of the C motif is strongly constrained to the hydrophobic cleft in Arp2, whereas the position of the A motif is consistent with existing binding studies (Kelly et al., 2006; Kreishman-Deitrick et al., 2005; Pan et al., 2004; Weaver et al., 2002; Zalevsky et al., 2001).
EXPERIMENTAL PROCEDURES
Purification of Arp2/3 complex
Arp2/3 complex was isolated from bovine brain with modifications to a previously published protocol (Higgs et al., 1999). Briefly, 400 g of frozen brain was homogenized in 800 ml of Breaking Buffer (20 mM Tris pH 8.0, 120 mM NaCl, 5 mM MgCl2, 5mM EGTA, 1 mM DTT, 1 mM PMSF, 20 μg/ml leupeptin, 6 μg/ml aprotinin, 6 μg/ml pepstatin, 20 μg/ml soy bean trypsin inhibitor and 0.1 mM benzamidine) and clarified by centrifugation at 12,000 g for 30 min. The supernatant was loaded onto a Macro-Prep High Q column (Bio-Rad, Melville, NY) pre-equilibrated with Breaking Buffer. The flow-through, containing Arp2/3 complex, was applied onto a WCA affinity column equilibrated with Breaking Buffer supplemented with 0.1 mM ATP. Arp2/3 complex was eluted in 20 mM Tris pH 8.0, 25 mM KCl, 400 mM MgCl2, 1 mM EGTA, 1 mM DTT, 0.1 mM ATP, dialyzed against 20 mM MES pH 6.4, 20 mM KCl, 2 mM MgCl2, 1 mM EGTA and 0.1 mM ATP and further purified through a Mono S column (Amersham Biosciences, Piscataway, NJ). The complex was then dialyzed against 20 mM HEPES pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA and 0.1 mM ATP and concentrated using an ultrafiltration cell (Millipore, Billerica, MA). The concentration of the purified complex was determined spectrophotometrically using a theoretically calculated extinction coefficient of 234,080 M-1 cm-1.
Cloning and expression of Cys-WCA
The cDNA encoding for mouse N-WASP was obtained from ATCC (clone number 3169027). The WCA fragment corresponding to amino acids 426-501 was amplified by PCR, with a forward primer containing mutations Ser 426 to Cys and Cys 427 to Ala, and cloned between the Ndel and EcoRI sites of vector pTYB12 (New England BioLabs, Ipswich, MA). This vector comprises a chitin-binding domain (for affinity purification) and an intein (for self-cleavage after purification). The Cys-WCA construct was expressed in BL21(DE3) cells (Invitrogen, Carlsbad, CA). Cells were grown in Terrific Broth medium at 37°C until the OD at 600 nm reached a value of 1.0-1.2. The expression was induced by addition of 0.5 mM isopropylthio-β-D-galactoside (IPTG) and carried out for 5 hours at 20°C. Cells were harvested by centrifugation and resuspended in chitin- affinity-column equilibration buffer (20 mM Hepes pH 7.5, 500 mM NaCl, 1 mM EDTA and 100 μM PMSF). Standard purification on a chitin affinity column (New England Biolabs) was followed by HPLC purification on a reverse phase C12 column using a gradient of CH3CN (0-90%) with addition of 0.1% TFA. The purified Cys-WCA was dialyzed against 2 mM Tris pH 7.5, 0.2 mM CaCl2 and its concentration was determined spectrophotometrically using a theoretically calculated extinction coefficient of 5500 M-1 cm-1.
Cross-linking of Cys-WCA to actin and binding to Arp2/3 complex
Actin was purified from rabbit skeletal muscle (Pardee and Spudich, 1982) and mixed with Cys-WCA at a 1.2:1 ratio, using 5,5’-dithiobis(2-nitrobenzoic acid) (Sigma-Aldrich) to facilitate the cross-linking reaction as described (Graceffa and Jancso, 1991). Excess actin was then removed on a S100 column (Pfizer-Pharmacia) in 20 mM HEPES pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA and 0.1 mM ATP. Actin was similarly cross-linked to a synthetic peptide corresponding to the W motif of mouse N-WASP (amino acids 425-448), which also contains the two mutations Ser 426 to Cys and Cys 427 to Ala. Actin-WCA was mixed with Arp2/3 complex at a 1.2:1 ratio and excess actin-WCA was removed on a S200 column (Pfizer-Pharmacia). The concentration of the nine-protein complex was determined spectrophotometrically using a theoretical extinction coefficient of 283,170 M-1 cm-1, derived from the amino acid sequences.
Actin polymerization assay
Pyrene-actin polymerization assays were carried out using a Cary Eclipse fluorescence spectrophotometer (Varian, Palo Alto, CA) and analyzed as described (Harris and Higgs, 2006). Prior to data acquisition, 2 μM Mg-ATP-actin (6% pyrene-labeled) was mixed with 25 nM Arp2/3 complex alone, 25 nM Arp2/3 complex with 250 nM WCA peptide, and 25 nM Arp2/3-actin-WCA complex in F-buffer (10 mM Tris pH 7.5, 1 mM MgCl2, 50 mM KCl, 1 mM EGTA, 0.1 mM NaN3, 0.02 mg/mL BSA, 0.2 mM ATP).
Analytical Ultracentrifugation
Sedimentation velocity experiments were conducted in a Beckman Optima XL-I ultracentrifuge (Beckman, Palo Alto, CA) using an An50 Ti eight-hole rotor. Data were acquired with the interference optics system using sapphire windows and meniscus-matching 12-mm aluminum-filled Epon centerpieces with interference slit window holders on the top window (Biomolecular Interaction Technology Center, Durham, NH). Prior to spinning, samples were dialyzed against 20 mM HEPES pH 7.5, 100 mM KCl, 2 mM MgCl2, 1 mM EGTA, 0.1 mM ATP for at least 16 hours. Velocity experiments were conducted at 50,000 rpm and 20°C.
Flow field fractionation and multi-angle light scattering
The stability of Arp2/3 complex with bound actin-WCA was first characterized by analytical ultracentrifugation (SI). The molecular masses and monodispersity of Arp2/3 complex with and without actin-WCA were then analyzed using an Eclipse Asymmetric Flow Field Fractionation (FFF) system coupled with a DAWN HELEOS Multi-Angle Light Scattering (MALS) detector and an Optilab rEX refractometer (Wyatt Technology Corparation, Santa Barbara, CA). This configuration allows for the determination of the absolute molar mass distribution of the complexes separated by the FFF unit. The FFF trapezoidal flow channel was fit with a 350 μm teflon spacer over a membrane made of regenerated cellulose with either 5 or 10 kDa molecular weight cutoff (depending on sample). For each sample, a volume of 100 μl at a the concentration of 1 mg/ml was injected using an Agilent 1100 HPLC system (Agilent Technologies, Palo Alto, CA), which also delivered the mobile phase to the FFF unit. Samples were first focused for 6 min at the head of the channel with two opposing lateral flow fields of 1 ml/min and a vertical cross-flow field of 2 ml/min, and then eluted with a lateral flow rate of 1.0 ml/min and a cross-flow rate of 2 ml/min. The experiments were performed in the normal elution mode, in which particles elute in order of increasing size. The eluting samples were detected through the refractometer and the 18-angle MALS detector, equipped with a 50 mV solid-state laser operating at 658 nm. Baseline scattering, peak boundaries and molecular masses of the samples were determined using the Astra software (Wyatt Technology Corp., Santa Barbara, CA).
SAXS data collection and analysis
Small angle X-ray scattering (SAXS) data from solutions of Arp2/3 complex with and without actin-WCA were collected at the X33 beamline of the European Molecular Biology Laboratory (DESY, Hamburg) using a MAR345 image plate detector (Roessle et al., 2007). Data were collected at a sample-detector distance of 2.7 m and in the range of scattering vectors 0.1 < s < 5 Å. The measurements were performed at 10°C for two different concentrations (0.45 mg/ml and 0.9 mg/ml for Arp2/3 complex and 0.55 mg/ml and 1.1 mg/ml for Arp2/3-complex-actin-WCA). No systematic differences were detected between the two concentrations, indicating the absence of inter-particle interference at these concentrations. To monitor for radiation damage, two 2-minute exposures of protein samples were collected and no damage was observed. The average background scattering of the buffer, determined from two independent measurements (before and after each sample measurement), was subtracted from the average scattering profile of the protein. The data were processed by standard procedures with the program PRIMUS (Konarev et al., 2003). The forward scattering I(0) and the radii of gyration Rg were estimated from the Guinier approximation (Guinier, 1939), assuming that at very small angles (s < 1.3/Rg) the intensity is represented as I(s) = I(0) exp(-(sRg)2/3). Maximum particle dimensions Dmax were computed using the indirect transform package GNOM (Svergun, 1992), which also gives the distance distribution functions p(r). The ab initio shape reconstructions were performed with the program DAMMIN (Svergun, 1999), which represents a macromolecule as an assembly of densely packed beads. Simulated annealing is then used to build a compact interconnected configuration of beads inside a sphere with diameter Dmax that fits the experimental data Iexp(s) to minimize discrepancy:
where N is the number of experimental points, c is a scaling factor and Icalc(sj) and σ(sj) are the calculated intensity and experimental error at the momentum transfer, sj, respectively. For each of the complexes, fifteen ab initio models were generated and averaged using the program DAMAVER (Volkov and Svergun, 2003). The final envelopes were obtained by averaging the models and removing low occupancy beads. Atomic models were fit into the ab initio envelopes with the program SUPCOMB (Kosik and Svergun, 2001). This program finds the best alignment between two models by minimizing the normalized spatial discrepancy (NSD), which is a normalized average of minimal distances between every point s1i of structure 1 and all points s2i of structure 2 (ρ(s1i,2)), and every point s2i of structure 2 and all points S1i of structure 1 (ρ(s2i,1)):
where Ni is the number of points (either atoms or dummy atoms) in a structure and di is the average distance between adjacent points in a structure. The scattering curves of atomic models were calculated and compared with the experimental curves using the program CRYSOL (Svergun et al., 1995), which either predicts theoretical scattering patterns or fits the experimental data by adjusting the excluded volume and the contrast of the hydration layer.
Supplementary Material
Acknowledgments
This work was supported by NIH grants GM073791. MVP and DIS were supported by the European Union design study SAXIER (contract RIDS-011934). We thank Paul Leavis and Elizabeth Gowell for the synthesis of the W motif of N-WASP.
References
- Aguda AH, Burtnick LD, Robinson RC. The state of the filament. EMBO Rep. 2005;6:220–226. doi: 10.1038/sj.embor.7400363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-Bleu Is an Actin Nucleation Factor and Controls Neuronal Morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltzner CC, Pollard TD. Identification of functionally important residues of Arp2/3 complex by analysis of homology models from diverse species. J Mol Biol. 2004;336:551–565. doi: 10.1016/j.jmb.2003.12.017. [DOI] [PubMed] [Google Scholar]
- Carlier MF, Le Clainche C, Wiesner S, Pantaloni D. Actin-based motility: from molecules to movement. Bioessays. 2003;25:336–345. doi: 10.1002/bies.10257. [DOI] [PubMed] [Google Scholar]
- Chereau D, Dominguez R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J Struct Biol. 2006;155:195–201. doi: 10.1016/j.jsb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc Natl Acad Sci U S A. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Holleran EA, Mullins RD. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc Natl Acad Sci U S A. 2001;98:14871–14876. doi: 10.1073/pnas.261419298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Rouiller I, Xu XP, Volkmann N, Li R, Hanein D. Mechanism of filament nucleation and branch stability revealed by the structure of the Arp2/3 complex at actin branch junctions. PLoS Biol. 2005;3:e383. doi: 10.1371/journal.pbio.0030383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F, Rebowski G, Lee SH, Dominguez R. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP. EMBO J. 2007 doi: 10.1038/sj.emboj.7601874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Rodenbusch SE, Martin AC, Welch MD. Critical conformational changes in the Arp2/3 complex are induced by nucleotide and nucleation promoting factor. Mol Cell. 2004;16:269–279. doi: 10.1016/j.molcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Goley ED, Welch MD. The ARP2/3 complex: an actin nucleator comes of age. Nat Rev Mol Cell Biol. 2006;7:713–726. doi: 10.1038/nrm2026. [DOI] [PubMed] [Google Scholar]
- Graceffa P, Jancso A. Disulfide cross-linking of caldesmon to actin. J Biol Chem. 1991;266:20305–20310. [PubMed] [Google Scholar]
- Guinier A. La diffraction des rayons X aux petits angles; application a I’etude de phenomenes ultramicroscopiques. Ann Phys (Paris) 1939;12:161–231. [Google Scholar]
- Harris ES, Higgs HN. Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 2006;406:190–214. doi: 10.1016/S0076-6879(06)06015-0. [DOI] [PubMed] [Google Scholar]
- Hertzog M, van Heijenoort C, Didry D, Gaudier M, Coutant J, Gigant B, Didelot G, Preat T, Knossow M, Guittet E, Carlier MF. The beta-thymosin/WH2 domain; structural basis for the switch from inhibition to promotion of actin assembly. Cell. 2004;117:611–623. doi: 10.1016/s0092-8674(04)00403-9. [DOI] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry. 1999;38:15212–15222. doi: 10.1021/bi991843+. [DOI] [PubMed] [Google Scholar]
- Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Kranitz H, Dotsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MH, Vachette P, Svergun DI. Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys. 2003;36:147–227. doi: 10.1017/s0033583503003871. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS - a Windows-PC based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- Kosik KS, Svergun DI. Automated matching of high- and low-resolution structural models. J Appl Crystallog. 2001;34:33–41. [Google Scholar]
- Kreishman-Deitrick M, Goley ED, Burdine L, Denison C, Egile C, Li R, Murali N, Kodadek TJ, Welch MD, Rosen MK. NMR analyses of the activation of the Arp2/3 complex by neuronal Wiskott-Aldrich syndrome protein. Biochemistry. 2005;44:15247–15256. doi: 10.1021/bi051065n. [DOI] [PubMed] [Google Scholar]
- Le Clainche C, Didry D, Carlier MF, Pantaloni D. Activation of Arp2/3 complex by Wiskott-Aldrich Syndrome protein is linked to enhanced binding of ATP to Arp2. J Biol Chem. 2001;276:46689–46692. doi: 10.1074/jbc.C100476200. [DOI] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, Leung DW, Yarbrough ML, Burdette DL, Rosen MK, Orth K. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc Natl Acad Sci U S A. 2007;104:17117–17122. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Mullins RD, Higgs HN, Kaiser DA, Blanchoin L, May RC, Hall ME, Pollard TD. Scar, a WASp-related protein, activates nucleation of actin filaments by the Arp2/3 complex. Proc Natl Acad Sci U S A. 1999;96:3739–3744. doi: 10.1073/pnas.96.7.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins RD, Stafford WF, Pollard TD. Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J Cell Biol. 1997;136:331–343. doi: 10.1083/jcb.136.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen BJ, Pollard TD. Insights into the influence of nucleotides on actin family proteins from seven structures of Arp2/3 complex. Mol Cell. 2007;26:449–457. doi: 10.1016/j.molcel.2007.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Egile C, Lipkin T, Li R. ARPC1/Arc40 mediates the interaction of the actin-related protein 2 and 3 complex with Wiskott-Aldrich syndrome protein family activators. J Biol Chem. 2004;279:54629–54636. doi: 10.1074/jbc.M402357200. [DOI] [PubMed] [Google Scholar]
- Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK. A conserved amphipathic helix in WASP/Scar proteins is essential for activation of Arp2/3 complex. Nat Struct Biol. 2003;10:591–598. doi: 10.1038/nsb952. [DOI] [PubMed] [Google Scholar]
- Pantaloni D, Boujemaa R, Didry D, Gounon P, Carlier MF. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat Cell Biol. 2000;2:385–391. doi: 10.1038/35017011. [DOI] [PubMed] [Google Scholar]
- Pardee JD, Spudich JA. Purification of muscle actin. Methods Enzymol. 1982;85(Pt B):164–181. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr Opin Struct Biol. 2007 doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Pollard TD. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu Rev Biophys Biomol Struct. 2007;36:451–477. doi: 10.1146/annurev.biophys.35.040405.101936. [DOI] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Robinson RC, Turbedsky K, Kaiser DA, Marchand JB, Higgs HN, Choe S, Pollard TD. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Sokolova O, Robins DB, Daugherty KM, Hippenmeyer S, Riezman H, Grigorieff N, Goode BL. Conformational changes in the Arp2/3 complex leading to actin nucleation. Nat Struct Mol Biol. 2005;12:26–31. doi: 10.1038/nsmb870. [DOI] [PubMed] [Google Scholar]
- Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, Svergun DI. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Crystallogr. 2007;40:s190–s194. [Google Scholar]
- Sept D, McCammon JA. Thermodynamics and kinetics of actin filament nucleation. Biophys J. 2001;81:667–674. doi: 10.1016/S0006-3495(01)75731-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Barberato C, Koch MH. CRYSOL: a program to evaluate X-ray solution scattering of biological macro-molecules from atomic coordinates. J Appl Crystallog. 1995;28:768–773. [Google Scholar]
- Svergun DI, Koch MH. Advances in structure analysis using small-angle scattering in solution. Curr Opin Struct Biol. 2002;12:654–660. doi: 10.1016/s0959-440x(02)00363-9. [DOI] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarn VC, Serruto D, Dziejman M, Brieher W, Mekalanos JJ. A Type III Secretion System in Vibrio cholerae Translocates a Formin/Spire Hybrid-like Actin Nucleator to Promote Intestinal Colonization. Cell Host and Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Crystallog. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AM, Heuser JE, Karginov AV, Lee WL, Parsons JT, Cooper JA. Interaction of cortactin and N-WASp with Arp2/3 complex. Curr Biol. 2002;12:1270–1278. doi: 10.1016/s0960-9822(02)01035-7. [DOI] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria acta protein. Acta binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–3475. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.