Abstract
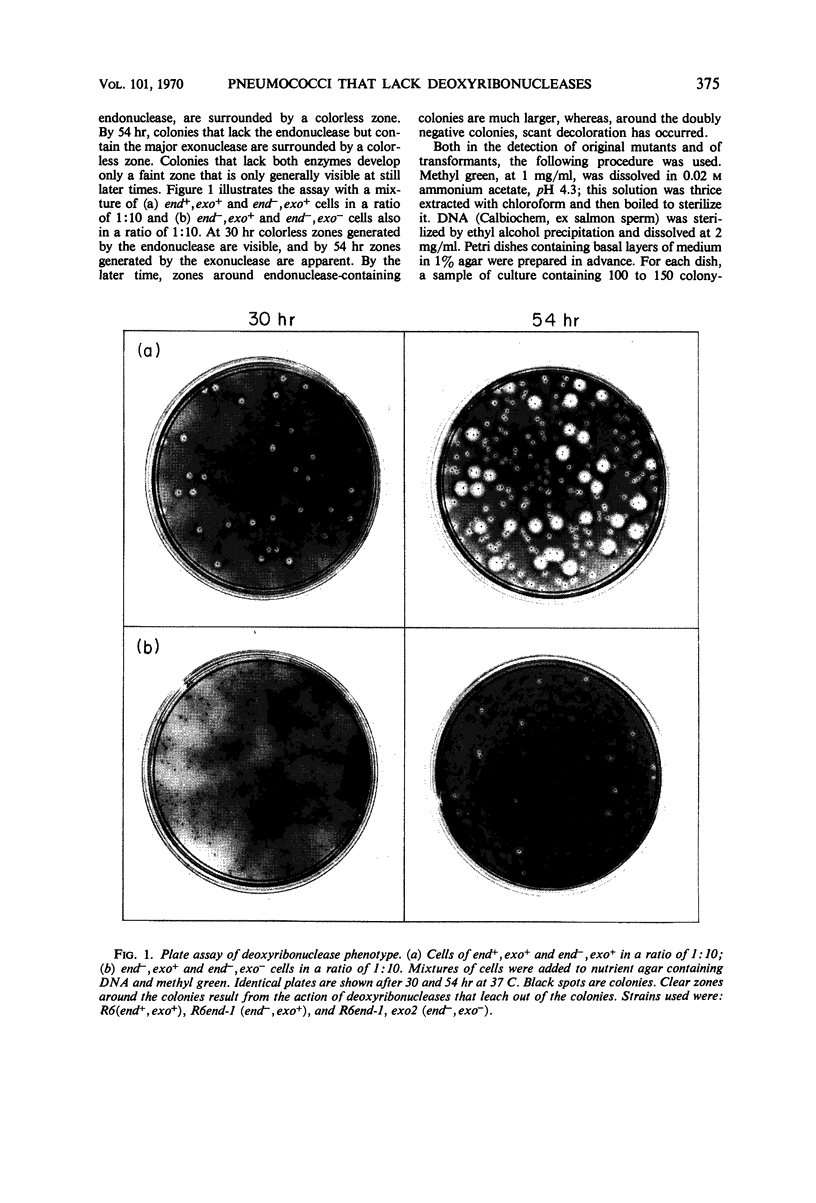
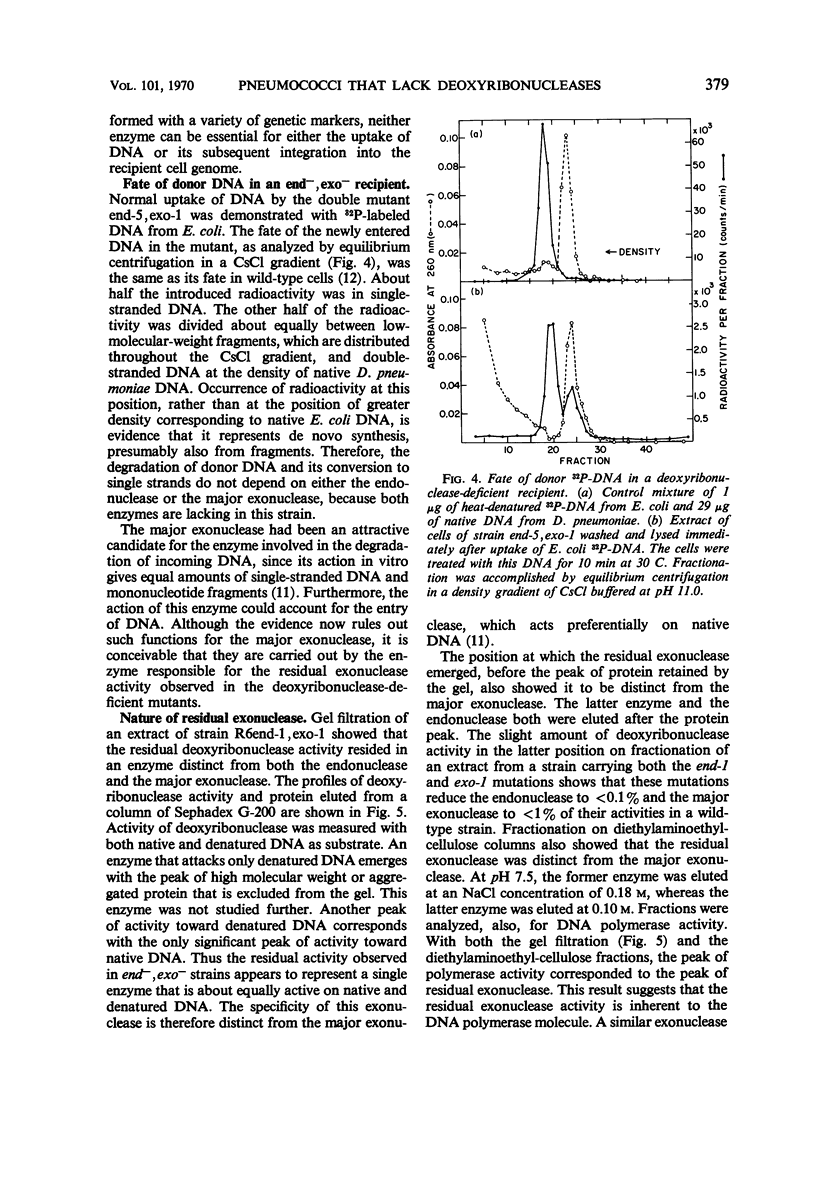
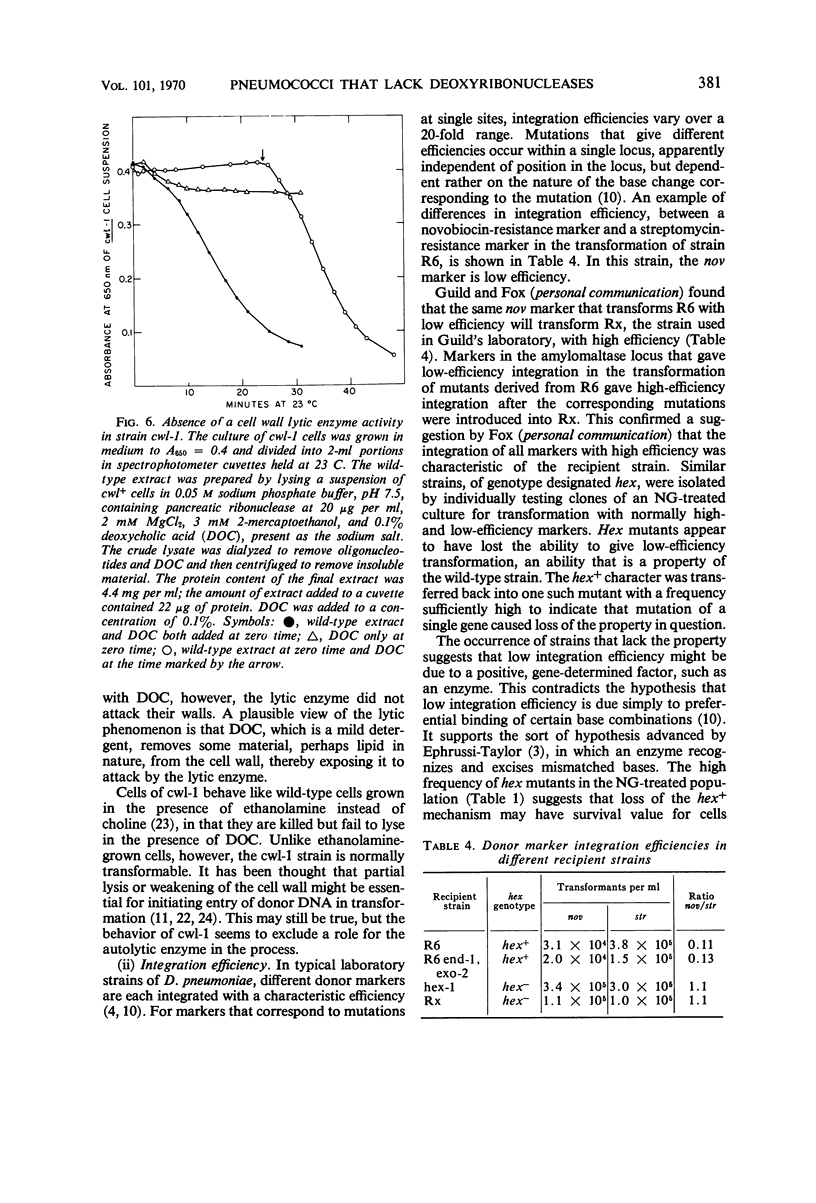
Mutants of Diplococcus pneumoniae that lacked the two major deoxyribonucleases of the cell—one an endonuclease, the other an exonuclease preferentially active on native deoxyribonucleic acid (DNA)—were obtained. The development of a method for detecting mutant colonies, based on the binding of methyl green to DNA, facilitated isolation of the mutants. Neither enzyme was essential for growth of the cells, for repair of ultraviolet damage, or for any phase of DNA-mediated transformation. Residual deoxyribonuclease activity in the double mutant corresponded to an exonuclease, approximately one-fifth as active as the major exonuclease, that attacked native and denatured DNA equally well. This activity appeared to be associated with the DNA-polymerase enzyme. A mutant that apparently lacked a cell wall lytic enzyme was also fully transformable. A mutant strain that was four times more sensitive to ultraviolet light than the wild type also transformed normally. Recipient cells of this strain were deficient in the repair of ultraviolet-irradiated transforming DNA. Mutants were found which, unlike the wild type, integrated donor markers only with high efficiency, thereby indicating that a particular cellular component that is susceptible to loss by mutation, such as an enzyme, is responsible for low integration efficiency.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dürwald H., Hoffmann-Berling H. Endonuclease-I-deficient and ribonuclease I-deficient Escherichia coli mutants. J Mol Biol. 1968 Jul 14;34(2):331–346. doi: 10.1016/0022-2836(68)90257-x. [DOI] [PubMed] [Google Scholar]
- Ephrussi-Taylor H., Gray T. C. Genetic studies of recombining DNA in pneumococcal transformation. J Gen Physiol. 1966 Jul;49(6):211–231. doi: 10.1085/jgp.49.6.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi-Taylor H., Sicard A. M., Kamen R. Genetic Recombination in DNA-Induced Transformation of Pneumococcus. I. the Problem of Relative Efficiency of Transforming Factors. Genetics. 1965 Mar;51(3):455–475. doi: 10.1093/genetics/51.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREEN D. M. A host-specific variation affecting relative frequency of transformation of two markers in pneumococcus. Exp Cell Res. 1959 Nov;18:466–480. doi: 10.1016/0014-4827(59)90312-x. [DOI] [PubMed] [Google Scholar]
- Ghei O. K., Lacks S. A. Recovery of donor deoxyribonucleic acid marker activity from eclipse in pneumococcal transformation. J Bacteriol. 1967 Mar;93(3):816–829. doi: 10.1128/jb.93.3.816-829.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KURNICK N. B. The determination of desoxyribonuclease activity by methyl green; application to serum. Arch Biochem. 1950 Nov;29(1):41–53. [PubMed] [Google Scholar]
- LACKS S., HOTCHKISS R. D. A study of the genetic material determining an enzyme in Pneumococcus. Biochim Biophys Acta. 1960 Apr 22;39:508–518. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lacks S., Greenberg B. Deoxyribonucleases of Pneumococcus. J Biol Chem. 1967 Jul 10;242(13):3108–3120. [PubMed] [Google Scholar]
- Lacks S. Integration efficiency and genetic recombination in pneumococcal transformation. Genetics. 1966 Jan;53(1):207–235. doi: 10.1093/genetics/53.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKAZAKI T., KORNBERG A. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID. XV. PURIFICATION AND PROPERTIES OF A POLYMERASE FROM BACILLUS SUBTILIS. J Biol Chem. 1964 Jan;239:259–268. [PubMed] [Google Scholar]
- Patrick M. H., Rupert C. S. The effects of host-cell reactivation on assay of U.V.-irradiated Haemophilus influenzae transforming DNA. Photochem Photobiol. 1967 Jan;6(1):1–20. doi: 10.1111/j.1751-1097.1967.tb08785.x. [DOI] [PubMed] [Google Scholar]
- RICHARDSON C. C., SCHILDKRAUT C. L., APOSHIAN H. V., KORNBERG A. ENZYMATIC SYNTHESIS OF DEOXYRIBONUCLEIC ACID. XIV. FURTHER PURIFICATION AND PROPERTIES OF DEOXYRIBONUCLEIC ACID POLYMERASE OF ESCHERICHIA COLI. J Biol Chem. 1964 Jan;239:222–232. [PubMed] [Google Scholar]
- Setlow J. K., Brown D. C., Boling M. E., Mattingly A., Gordon M. P. Repair of deoxyribonucleic acid in Haemophilus influenzae. I. X-ray sensitivity of ultraviolet-sensitive mutants and their behavior as hosts to ultraviolet-irradiated bacteriophage and transforming deoxyribonucleic acid. J Bacteriol. 1968 Feb;95(2):546–558. doi: 10.1128/jb.95.2.546-558.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow R. B. The photochemistry, photobiology, and repair of polynucleotides. Prog Nucleic Acid Res Mol Biol. 1968;8:257–295. doi: 10.1016/s0079-6603(08)60548-6. [DOI] [PubMed] [Google Scholar]
- Tichy P., Landman O. E. Transformation in quasi spheroplasts of Bacillus subtilis. J Bacteriol. 1969 Jan;97(1):42–51. doi: 10.1128/jb.97.1.42-51.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasz A. Biological consequences of the replacement of choline by ethanolamine in the cell wall of Pneumococcus: chanin formation, loss of transformability, and loss of autolysis. Proc Natl Acad Sci U S A. 1968 Jan;59(1):86–93. doi: 10.1073/pnas.59.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOUNG F. E., SPIZIZEN J. BIOCHEMICAL ASPECTS OF COMPETENCE IN THE BACILLUS SUBTILIS TRANSFORMATION SYSTEM. II. AUTOLYTIC ENZYME ACTIVITY OF CELL WALLS. J Biol Chem. 1963 Sep;238:3126–3130. [PubMed] [Google Scholar]




