Abstract
The I-TASSER server is an integrated platform for automated protein structure and function prediction based on the sequence-to-structure-to-function paradigm. Starting from an amino acid sequence, I-TASSER first generates three-dimensional atomic models from multiple threading alignments and iterative structural assembly simulations. The function of the protein is then inferred by structurally matching the 3D models with other known proteins. The output from a typical server run contains full-length secondary and tertiary structure predictions, and functional annotations on ligand-binding sites, Enzyme Commission numbers and Gene Ontology terms. An estimate of accuracy of the predictions is provided based on the confidence score of the modeling. This protocol provides new insights and guidelines for designing of on-line server systems for the state-of-the-art protein structure and function predictions. The server is available at http://zhang.bioinformatics.ku.edu/I-TASSER.
Keywords: I-TASSER, protein structure prediction, protein function prediction
INTRODUCTION
“I have a protein of interest but don’t know its structure/function” is one of the most common problems that many molecular and cell biologists face in their research. This impediment has been aggravated in recent years due to the fact that the percentage of protein sequences in UniProtKB/TrEMBL1 with a solved protein structure in the PDB library2 plunged to 0.6% by the end of 2009; this number was 2% in 2004 and 1.2% in 2007. Recent advances in computer algorithms for predicting protein structure and function have alleviated this problem, and provides biologists with valuable information about their proteins of interest3.
Computational methods for predicting three dimensional (3D) protein structures have been historically divided into three categories, based on the availability of template structures in the PDB library. In comparative modeling (CM)4, evolutionarily-related homologous templates are identified by sequence or sequence profile comparisons5, and high-resolution models can be generated by simply copying the framework of the template structures or by satisfying the spatial restraints collected from the template structures. Since proteins from different evolutionary origins may have similar structure, threading methods6–7 are designed to match the query sequence directly onto the 3D structures of other solved proteins with the goal of recognizing folds similar as the query, even when there is no evolutionary relationship between the query and the template protein. Finally, for query proteins that have no structurally related protein in the PDB library, the structure must be built from scratch by ab initio modeling8–10. This is the hardest case and success is limited to small proteins with <120 amino acids3, 11.
As a general trend, the boundaries between the conventional categories of protein structure prediction methods have become increasingly blurred. While both comparative modeling and threading methods use sequence-profile and profile-profile alignments for identifying the templates, many ab initio modeling algorithms use evolutionary or knowledge-based information for collecting spatial restraints10, 12 or for identifying local structural building blocks13. Recent community-wide CASP experiments11, 14–16 have demonstrated significant advantages of composite approaches in protein structure prediction, which combine various techniques from threading, ab initio modeling, and atomic-level structure refinement approaches3, 17–19. I-TASSER (Iterative Threading ASSEmbly Refinement)10 is one example of the composite approaches, and has been ranked as the best method for the automated protein structure prediction in the last two CASP experiments14, 18, 20–21.
The biological usefulness of the predicted protein models relies on the accuracy of the structure prediction22. For example, high-resolution models with RMSD in 1–2 Å, typically generated by CM using close homologous templates, usually meet the highest structural requirements and are sometime suitable for computational ligand-binding studies and virtual compound screening23–25. Medium-resolution models, roughly in the RMSD range of 2~5 Å and typically generated by threading and CM from distantly homologous templates, can be used for identifying the spatial locations of functionally important residues, such as active sites and the sites of disease-associated mutations26–29. However, many of the functionally important sites are located on loop regions which show large structural variability although the scaffold of the protein structures is conserved. Thus, accurately modeling of the loop regions is still an important yet unsolved problem in template-based modeling30–31. Finally, even models with the lowest resolution, from an otherwise meaningful prediction, i.e. models with an approximately correct topology, predicted using either ab initio approaches or based on weak hits from threading, have a number of uses including protein domain boundary identification32–33, topology recognition, and family/superfamily assignment34–35.
As the biological function of protein molecules is determined by their 3D shape (which dictates how the protein interacts with ligands or other protein molecules)36, one of the most common motivations for predicting the protein structure is to use the structural information to gain insight into the protein’s biological function. A convenient approach to the structure-based functional assignment involves global structural comparison of protein pairs for fold recognition and family assignment34–35, which in many cases can be directly used to infer function22, 37. However, it is increasingly recognized that the relationship between structure and function is not always straightforward, as many protein folds/families are known to be functionally promiscuous38, and different folds can perform the same function39. When the global structures are not similar, functional similarity may arise due to the conserved local structural motifs which perform the same biochemical function, although in different global structural frameworks. In a recent development of I-TASSER40, the methodology was extended for annotating the biological function using the predicted protein structures, based on a combination of local and global structural similarities with proteins of known function. Using this method, the biological functions (including ligand binding sites, Enzyme Commission numbers and Gene Ontology terms) of a substantial number of protein targets were correctly identified based on similarities to non-homologous proteins, which otherwise could not have been inferred from sequence or profile-based searches5.
The success of the I-TASSER methods in the blind CASP experiments18, 20 and the large-scale benchmarking tests10, 35, 41–42 makes it a useful tool for automated protein structure and function annotation. In the past 24 months, the online I-TASSER server has generated >30,000 full-length structure and function predictions for over 6,000 registered biologists from 82 countries. Compared to a number of other useful on-line structure predictions tools43–51, the uniqueness of the I-TASSER sever is in the significant accuracy and reliability of full-length structure prediction for protein targets of varying difficulty and the comprehensive structure-based function predictions. Especially, the inherent template fragment reassembly procedure has the power to consistently drive the initial template structures closer to the native structure10, 14, 16. For example, in CASP8, the final models generated by the I-TASSER server had a lower RMSD to the native structure than the best threading template for 139 out of 164 domains, with an overall RMSD reduction by 1.2 Å (on average from 5.45 Å in templates to 4.24 Å in the final models)20. Here, one purpose of this protocol is to provide detailed guidelines to help the biologists to use the I-TASSER server in designing their online structure and function prediction experiments. Meanwhile, since the I-TASSER system is based on the general sequence-to-structure-to-function paradigm, the described protocol can be valuable to the developers of other similar bioinformatics systems.
I-TASSER Server
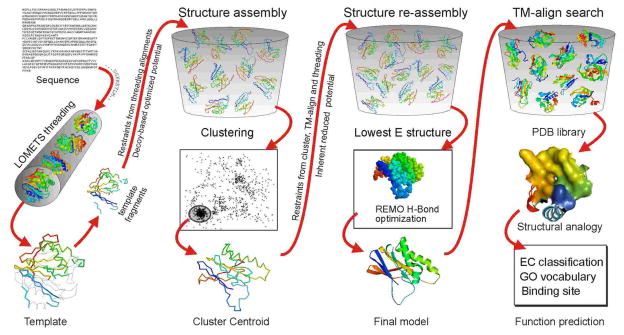
Detailed descriptions of the I-TASSER methodology for protein structure and function prediction have been provided elsewhere10, 20, 40. For the sake of completeness, here we give a brief outline of the method, which is divided into four general stages (Figure 1).
Figure 1.
A schematic representation of the I-TASSER protocol for protein structure and function predictions. The protein chains are colored from blue at N-terminus to red at the C-terminus.
Stage 1: threading
Threading refers to a bioinformatics procedure for identifying template proteins from solved structure databases which have a similar structure or similar structural motif as the query sequence. In the first stage of I-TASSER, the query sequence is matched against a non-redundant sequence database by PSI-BLAST5, to identify evolutionary relatives. A sequence profile is then created based on a multiple alignment of the sequence homologs, which is also used to predict the secondary structure using PSIPRED52. Assisted by the sequence profile and the predicted secondary structure, the query sequence is then threaded through a representative PDB structure library using LOMETS53, a locally installed meta-threading server combining 7 state-of-the-art threading programs (FUGUE54, HHSEARCH46, MUSTER55, PROSPECT56, PPA10, SP357 and SPARKS58). In the individual threading programs, the templates are ranked by a variety of sequence-based and structure-based scores. The top template hits from each threading program are then selected for further consideration. The quality of the template alignments (and therefore the difficulty of modeling the targets) is judged based on the statistical significance of the best threading alignment, i.e. the Z-score which is defined as the energy score in standard deviation units relative to the statistical mean of all alignments.
Stage 2: structural assembly
In the second stage, continuous fragments in threading alignments are excised from the template structures, and are used to assemble structural conformations of the sections that aligned well, with the unaligned regions (mainly loops/tails) built by ab initio modeling10, 12.To improve the efficiency of conformational search, I-TASSER adopts a reduced model to represent the protein chain, with each residue described by its Cα atom and side-chain center of mass. Because the regions not aligned during the threading process usually have a lower modeling accuracy, the structure modeling in these regions is confined to a lattice system of grid size 0.87 Å12, which helps to reduce the entropy of conformational search. Although this grid size may introduce considerable uncertainty of conformational representations in comparative modeling (which has usually an error range of 1–2 Å), it does not generate observable effect in the ab initio modeling, as it often has an error level of 4–6 Å. The threading aligned regions usually have a higher accuracy. The modeling in these regions is therefore off lattice and the template fragments are kept rigid during the simulations, which helps to maintain the fidelity of the high-resolution structures in these regions. The fragment assembly is performed using a modified replica-exchange Monte Carlo simulation technique,59 which implements several replica simulations in parallel at different temperatures, with the temperatures periodically exchanged between the replicas; the energy barriers are flattened by a hyperbolic function to speed up the jumps of simulations between different energy basins. The overall simulation is guided by a composite knowledge-based force field, which includes: (1) general statistical terms derived from the PDB (C-alpha/side-chain correlations12, H-bonds60 and hydrophobicity61); (2) spatial restraints from threading templates53; and (3) sequence-based contact predictions from SVMSEQ62. Partly because of the consideration of the hydrophobic interactions and the bias towards radius of gyration in the energy force field, the current I-TASSER procedure is designed to best fold single-domain globular proteins (the procedure for modeling multiple-domain proteins using I-TASSER will be discussed in the next section). The conformations generated in the low-temperature replicas during the refinement simulation are clustered by SPICKER63 with the purpose of identifying low free-energy states. Cluster centroids are then obtained by averaging the 3D coordinates of all the clustered structural decoys. For further details of the structural assembly procedure, the readers are advised to read Refs.10, 42 for the on-and-off lattice system, Refs.12, 18, 20 for the force field development, and Ref.59 for the MC search engine.
Stage 3: model selection and refinement
In the third stage, the fragment assembly simulation is performed again starting from the selected cluster centroids. While the inherent I-TASSER potential remains unchanged in the second run, external constraints are pooled from the LOMETS threading alignments and the PDB structures that are structurally closest to the cluster centroids, as identified by TM-align64. The purpose of the second iteration is to remove steric clashes as well as to refine the global topology of the cluster centroids. The decoys generated during the second round of simulations are clustered again, and the lowest energy structures are selected as input for REMO65, which generates the final structural models by building all-atom models from Cα traces through the optimization of hydrogen-bonding networks.
Stage 4: structure-based functional annotation
In the last stage, the function of the query protein is inferred by structurally matching the predicted 3D models against the proteins of known structure and function in the PDB. For this purpose, three protein structure/function libraries have been constructed independently and bi-weekly updated; at present, these include a library of 5,798 non-redundant entries with known Enzyme Commission (EC) numbers66, a library of 26,045 non-redundant entries with known Gene Ontology (GO) terms67, and a library of 19,658 non-redundant entries with known ligand-binding sites. The structural analogs of the query protein in the GO library are mainly matched based on the global topology using TM-align64, and a consensus is derived based on the frequency of occurrence of the GO terms. The structural analogs in the EC and binding site libraries are matched based on both global and local structural similarity40. While the global structural similarity search is used for recognizing proteins with similar global fold, the local similarity search provides a complementary method, identifying analogs that have a different fold but perform similar function due to the conservation of active/binding sites. The functional analogs from the global search results are ranked based on the conserved structural patterns present in the model, measured using a scoring scheme that combines TM-score68, RMSD, sequence identity, and coverage of the structure alignment40. Here, TM-score (template modeling score) is defined to assess the topological similarity of protein structure pairs with a value in the range of (0, 1], a higher score indicating better structural match. Statistically, a TM-score <0.17 means a randomly selected protein pair with the gapless alignment taken from PDB; TM-score >0.5 corresponds to the protein pairs of similar folds69. The statistical meaning of TM-score is independent of protein size68. The local similarity search looks for conserved spatial motifs in the predicted I-TASSER model, with the candidates ranked based on their structure and sequence similarity to functional cavities (binding pockets) in known structures. Finally, the results from the global and local search are combined to present a comprehensive list of functional analogs.
Estimation of prediction accuracy
Assessing the quality of a prediction is important because this assessment eventually determines how biologists will use the prediction in their research. For estimating the accuracy of the structure predictions, a confidence score named C-score is defined based on the quality of the threading alignments and the convergence of the I-TASSER’s structural assembly refinement simulations, i.e.
| (1) |
where M is the number of structure decoys in the cluster and Mtot is the total number of decoys generated during the I-TASSER simulations. 〈RMSD〉 is the average root-mean-squared deviation of the decoys to the cluster centroid. Z(i) is the Z-score of the best template generated by ith threading in the seven LOMETS programs and Z0(i) is a program-specified Z-score cutoff for distinguishing between good and bad templates. The C-score scheme has been extensively tested in large-scale benchmarking tests40, 42, 70. When tested on predicted structures, the Pearson correlation between C-score and the TM-score (the absolute difference between model to the native structure) was found to be 0.91, which is a significantly high value, having in mind that the mathematic range of the Pearson correlation is between 0 (for random variables) and 1 (for identical variables). When a C-score cutoff of -1.5 is used to select models of correct topology, both the false positive and the false negative rate are below 0.1, which means that more than 90% of the quality predictions are correct. Combining C-score and protein length, the accuracy of the I-TASSER models can be predicted with an average error of 0.08 for the TM-score and 2 Å for the RMSD (root mean square deviation)70. Again, considering the big quality variations of protein structure predictions (i.e. TM-score in 0–1 and RMSD in 0~30Å), these estimation errors are very low and the assessments should provide quantitative guidance of model quality for the users.
For the function predictions, the confidence score is defined based on the C-score of the structure prediction and the global and/or local structural similarity between the predicted models and their structural analogs in the PDB40. For the EC numbers, using an EC-score cutoff of 1.1, the first three EC digits can be correctly assigned in 72.4% cases. Similarly for the GO terms, using a GO-score cutoff of 0.5, 85.1% of molecular functions, 76.9% of biological processes, and 74.6% of cellular locations can be correctly assigned. These values are consistently higher than traditional sequence-based methods, while the PSI-BLAST search (using E-value <0.001) results in an overall precision of 56.2% for EC number, 81.1% for molecular function, 64.8% for biological process, and 68.2% for the cellular component Gene Ontology terms40. The predicted binding sites in the modeled structure are evaluated based on the BS-score, which measures the fitness of the ligand-model complex. Using a BS-score cutoff of 0.5, the success rate for identifying the correct binding site in the predicted model is 72.3%, which is also higher than that by PSI-BLAST search (with an overall success rate of 62.2%) in our benchmark test.
It needs to be mentioned that despite extensive benchmark tests18, 20, 40, 70, there can be considerable uncertainty and error in the automated estimation of the quality of structure and function predictions. The final and essential validation of the predictions should therefore be made based on the experimental data collected by the users. Before the entire structure becomes available, other indirect structural information from the data like mutagenesis experiments, affinity labeling, NMR dipolar coupling, cryo-electron microscopy, and circular dichroism and dual polarization interferometry experiments, can provide important information for validating the predicted models and help in deciding whether the predictions can be useful for further experimental design and study.
Experimental Design
Modeling of multi-domain proteins
Since the I-TASSER force field has been designed for modeling single-domain proteins, the procedure for modeling multiple-domain protein is slightly different from that of single-domain proteins, but is fully automated. First, the domain boundaries are defined based on the LOMETS threading programs, i.e. if a segment of query sequence of >80 residues have no alignment with template proteins in top two threading hits, the target is treated as a multiple-domain protein and the domain boundary is defined at the borders of the aligned/unaligned sections. Next, two types of assembly simulations are implemented: one simulation is conducted for modeling the whole-chain structure which provides a guide for domain orientations; another simulation is carried out for modeling the single-domain structures individually. Finally, to obtain the full-length model, the models of individual domains are docked together using the whole-chain I-TASSER model as a template. The docking simulation is performed using a quick Metropolis Monte Carlo simulation where the energy is defined as the RMSD of the individual domain models to the whole-chain I-TASSER template plus the reciprocal of the number of inter-domain steric clashes. The purpose is to generate a global model which has a similar domain orientation to the whole-chain I-TASSER model but with the minimum number of steric clashes. This procedure is applied only to proteins that have some domains that are not aligned in the top-scoring templates. If multi-domain templates are available and all domains of query protein are aligned, the whole chain will be modeled in I-TASSER using the full-chain template.
If the domain boundary information is available to the user, e.g. from some experimental data, it is recommended that the user should first split the sequence into individual domains and then submit each domain individually to the server. This will not only speed up the I-TASSER prediction process but also result in a more reliable structure and function prediction, since the current pipeline of the I-TASSER methodology has been optimized for modeling single-domain proteins20. Domain boundaries in protein sequences can also be predicted by using freely available external online programs such as NCBI CDD (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml) or PFAM (http://pfam.sanger.ac.uk/).
Specifying external restraints
Spatial information, including residue-residue contacts and distances, can be used as restraints to guide the I-TASSER structure assembly simulations. I-TASSER normally collects restraints from the threading templates, but these often contain errors because of the uncertainty of templates and alignments. Nevertheless, threading-based restraints have been proven to be essential for the I-TASSER structure assembly18, 20. The new version of the I-TASSER server allows the user to specify additional restraints based on experimental evidence or biological insights. Because restraints from experiments normally have a higher accuracy than those derived from threading alignments, user-specified spatial information can be very useful for improving the quality of the structure assembly, especially for the non-homologous protein targets. Our benchmark test shows that by using as few as N/8 NOE restraints, obtained from the NMR experiments (where N is the length of the protein), the current simulation procedure is able to successfully fold 75% of the proteins of up to 200 residues, which could not be folded without using spatial restraints because of the lack of appropriate templates71.
There are two methods by which the user can input restraints to the I-TASSER server.
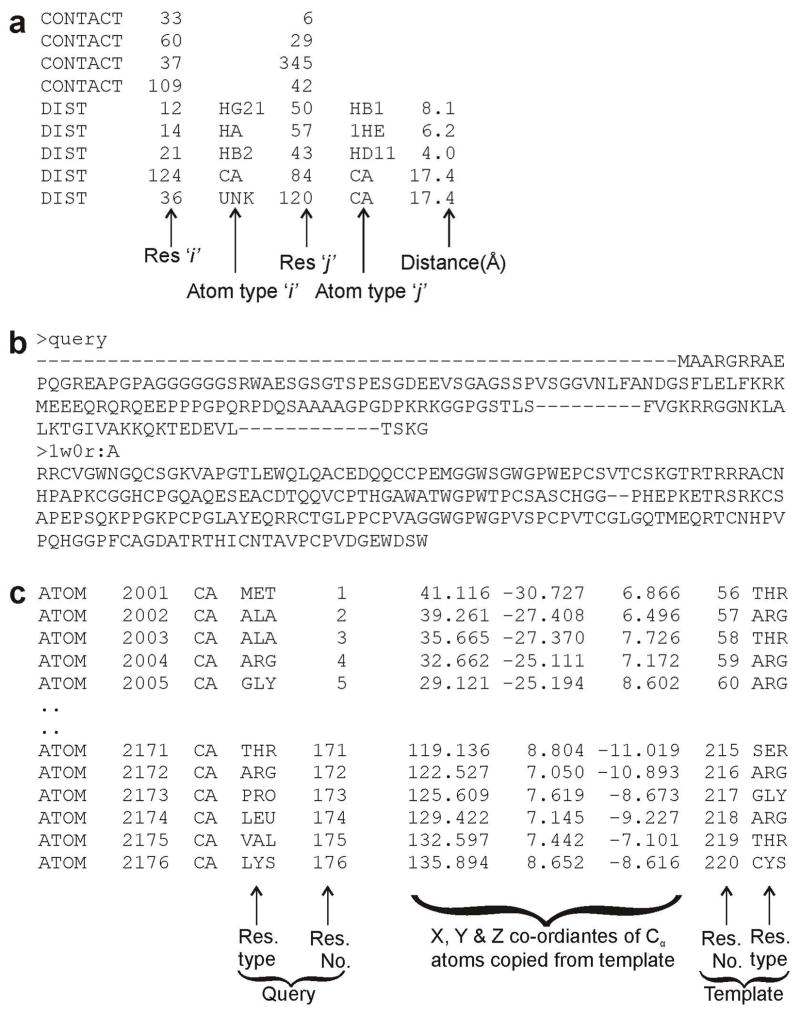
A. Specifying a set of atomic distances and contacts
Restraint data from NMR or cross-linking experiments can be specified by uploading a restraint file. A typical example is shown in Figure 2a. Column 1 specifies the type of restraint, i.e. “DIST” or “CONTACT”. For distance restraints (DIST), columns 2 and 4 contain the residue positions (i, j) and columns 3 and 5 contain atom names in the residues. Column 6 contains the distance between the two atoms in Angstrom (Å). I-TASSER will try to bring these atom pairs close to the specified distance during the structure refinement simulations. For contact restraints (CONTACT), columns 2 and 3 contain the positions (i, j) of residues which should be in contact. I-TASSER will try to draw the side-chain centers of mass of these two residues into contact during the simulations.
Figure 2.
Example of external restraint files that users can use to specify (a) residue-residue contact/distance restraints; (b) query-template alignment in FASTA format; and (c) query-template alignment in 3D format.
B. Designating a specific PDB structure as template
I-TASSER normally starts with a set of protein templates identified by the LOMETS threading programs, where the template library consists of a representative PDB subset at a pair-wise sequence identity cutoff of 70%. Users can specify a solved protein structure as the template, as the desired template may not be included in our library or the desired template may not be identified by LOMETS even though it is in the library. To specify a template, users can either upload a PDB formatted structure file or input a PDB ID and the I-TASSER server will obtain the structure from the PDB library. Once a template is specified, the I-TASSER simulation will start from the template with restraints mainly collected from it; but the simulation will also use the threading-based LOMETS restraints with the purpose to model the unaligned regions as well as adjust the reassembly of aligned regions.
The weight of the LOMETS restraints varies depends on the target type. Here, the query proteins are categorized into easy or hard targets based on the statistical significance of the threading alignments (see Step 17 for detail). The templates for easy targets are usually from homologous proteins and the alignments have a higher accuracy, while templates for hard targets are mostly from non-homologous proteins and the alignments have a lower accuracy. Because the accuracy of the LOMETS restraints is different for different targets, the weight of implementing the LOMETS restraints is stronger for easy targets than that in the case of hard targets, which have been systematically tuned based on large-scale benchmark training53.
When I-TASSER uses a template, it needs to know both the 3D structure and the alignment between the query and the template sequence. If users upload a template structure without specifying the alignment, I-TASSER will generate the query-template alignment using the MUSTER program55, an algorithm to align protein sequences to structures based on multiple information sources including secondary structure, sequence profile, solvent accessibility, and structure fragment profiles.
Users can also specify their own query-template alignments. The I-TASSER server accepts alignment in two formats: the FASTA format (Figure 2b) and the 3D format (Figure 2c). The FASTA format is standard and described at http://www.ncbi.nlm.nih.gov/blast/fasta.shtml. The 3D format is similar to the standard PDB format (http://www.wwpdb.org/documentation/format32/sect9.html), but two columns derived from the templates are added to the ATOM records (see Figure 2c):
Columns 1–30: Atom (C-alpha only) and residue names for the query sequence.
Columns 31–54: Coordinates of C-alpha atoms of the query copied from the corresponding atoms in the template.
Columns 55–59: Corresponding residue number in the template based on alignment
Columns 60–64: Corresponding residue name in the template
MATERIALS
Equipment
A personal computer with an Internet connection and a web browser.
Data
Amino acid sequence of the protein of interest in FASTA format.
Software
A molecular visualizing software, like RASMOL (http://www.openrasmol.org) or PYMOL (http://pymol.sourceforge.net), for viewing the 3D structure of the modeled protein and the predicted functional sites.
PROCEDURE
Sequence submission and restraint specification
To submit a protein sequence, visit the I-TASSER web page (http://zhang.bioinformatics.ku.edu/I-TASSER).
Copy and paste the amino acid sequence of a single protein chain into the provided form or directly upload the sequence from the computer by clicking the “Browse” button. At present, the I-TASSER server accepts protein sequences with a length between 10–1500 amino acids. ?TROUBLESHOOTING
Provide an e-mail address. The results will be mailed to the user at this address once the job is completed.
Provide a name for the protein. This is optional and is provided for user’s convenience. If no name is provided, a default name (“your protein”) will be assigned to the submitted sequence.
To add external residue contact/distance restraints or to specify a solved protein structure as a template, prepare a restraint file and upload it using the corresponding button or provide the PDB ID and chain ID in the provided form. Read about adding restraints in the Experimental Design section of this protocol or click on “More explanation on how to add restraints”.
-
To eventually submit the sequence with/without additional restraints, click the “Run I-TASSER” button. Upon clicking the button, the browser will be directed to an acknowledgement page which will display a confirmation of the submitted sequence, a job identification number, restraint information, and a link to the page that will contain the detailed results once the job is completed. The user may choose to bookmark this link for future reference.
PAUSE POINT: Once the sequence is successfully submitted, it is stored in a database until all other sequences in the server queue are processed. However, if the same sequence was submitted earlier on the server by another user within the last 30 days, the result will be copied from the previous run and sent to the user without rerunning the protein. If the previous result was generated more than 30 days ago, a new run will be initiated because new template proteins may have become available since the last run.
Upon completion of structure and function predictions for the submitted query sequence, an e-mail notification containing image of the predicted structures and a link to the result page on the server is sent to the user.
Availability of the results
7 Track the modeling status (optional). Users can track the status of all the submitted jobs on the I-TASSER server by visiting the queue page (http://zhang.bioinformatics.ku.edu/I-TASSER/queue.php). An estimated time for completion of the running jobs is also displayed in this page, where the time is counted from the time point when the visitor opens the page and is updated every 10 minutes. ?TROUBLESHOOTING
8 Search submitted/completed targets (optional). Users can click on the “Search” tab, displayed in the navigation bar of the queue page, to visit a new webpage at http://zhang.bioinformatics.ku.edu/I-TASSER/search.html. The page allows the user to search through the I-TASSER server using: (a) a job ID (e.g. ‘S12345’, the search will return a link to the result page of this target); or (b) a query sequence (the search will returns all homologous proteins with the sequence identity to the query protein >40%); or (c) an email address (the search will return all the targets that have been submitted by the user within the last one year). The search for homologs of the query protein is helpful for the users to make comparisons between the modeling result of the query protein and homologous targets. It can also save user’s time, if the same protein is found to have been modeled previously. To maintain the privacy and confidentiality of users, searching by email requires a password, which can be easily obtained at http://zhang.bioinformatics.ku.edu/I-TASSER/registration.html.
-
9 Visit the page containing the prediction results by clicking on the link provided in the email-notification or open the link bookmarked in Step 6. An example result page is available at http://zhang.bioinformatics.ku.edu/I-TASSER/output/example.
CAUTION To maintain sufficient free space on the server, the results will be deleted 365 days after they are made available to the user. The user can keep a copy of the result page locally in his/her computer by saving the complete web page.
Sequence and predicted secondary structure
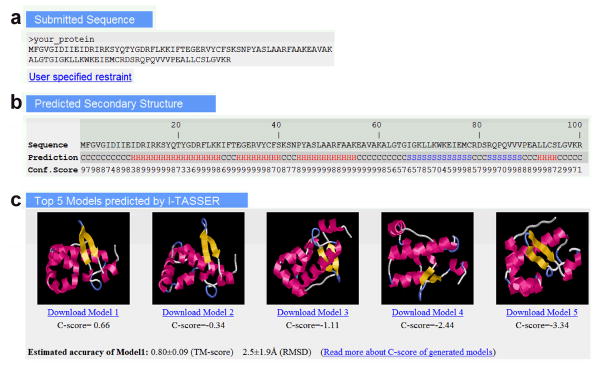
10 View the top of the result page to check the submitted amino acid sequence in FASTA format (Figure 3a) and the predicted secondary structures (Figure 3b). If the user has specified experimental restraints, a link is provided to the page containing user-specified restraint information. A typical secondary structure prediction contains three states: alpha helix (H), beta strand (S) and coil (C), with confidence scores for each residue. The secondary structure shown here is the state with the highest confidence score. The confidence score for each residue is shown in the next row with values ranging between 0–9, where a higher score indicates a prediction with higher confidence52. The predicted secondary structure can be used for estimating the number of secondary structure elements and the tertiary structure class of the query protein.
Figure 3.
An illustrative example of the I-TASSER result page showing (a) query sequence in FASTA format and a link to the user-specified restraints; (b) predicted secondary structure of the query protein; and (c) image of the top-five predicted models and links for downloading the PDB formatted structure files. The confidence score for estimating the model quality is reported as C-score. The secondary structures in the model are highlighted in red (for α-helices) and yellow (for β-strands).
Predicted 3D structures
11 Scroll down further to see GIF images of up to five predicted models, with highlighted regular secondary structures (see Figure 3c). This will help in quickly ascertaining the tertiary structure class and topology of the query protein from the modeled structure(s). ?TROUBLESHOOTING
12 Download the coordinate file (in PDB format) of the predicted structures by clicking on the “Download Model” link below the image of each individual model. To interactively view the modeled structure on the computer, open these files by any molecular visualization program. Some of the freely available programs commonly used for the visualization of protein structures are listed in the MATERIALS section.
13 View the confidence score of the structure modeling, shown as C-scores displayed below the download link for each model. As described in the I-TASSER Server section, C-score is an estimate of the quality of the predicted models, and is calculated based on the significance (i.e. Z-score) of the threading alignments in LOMETS and the convergence (i.e. cluster density) of the I-TASSER simulations (see Eq. 1). C-score is typically in the range [-5, 2], where a higher score reflects a model of better quality. In general, models with C-score > -1.5 have a correct fold. Here, the C-score of the model should not be confused with the TM-score68. While TM-score is a measure of structural similarity between the predicted model and the native structure, C-score is an estimate of the confidence of structure prediction. ?TROUBLESHOOTING
14 View the estimated TM-score and RMSD to the native structure for the first model, shown as “Estimated accuracy of the first model”. ?TROUBLESHOOTING
15 Click on the “more about C-score of generated models” link to open a new page containing further information about the estimated TM-score and RMSD values for the first model, as well as the cluster size and cluster density for all the predicted models. The estimated TM-score and RMSD values reported here are values calculated based on the correlation of C-score with TM-score and RMSD in the benchmark test70. Nevertheless, C-score is listed for all the models as a reference. The quality of the lower-ranked models can be assessed partially based on their cluster density and the cluster size, where the models associated with clusters of larger size and higher density are on average closer to the native structure. ?TROUBLESHOOTING
Threading templates
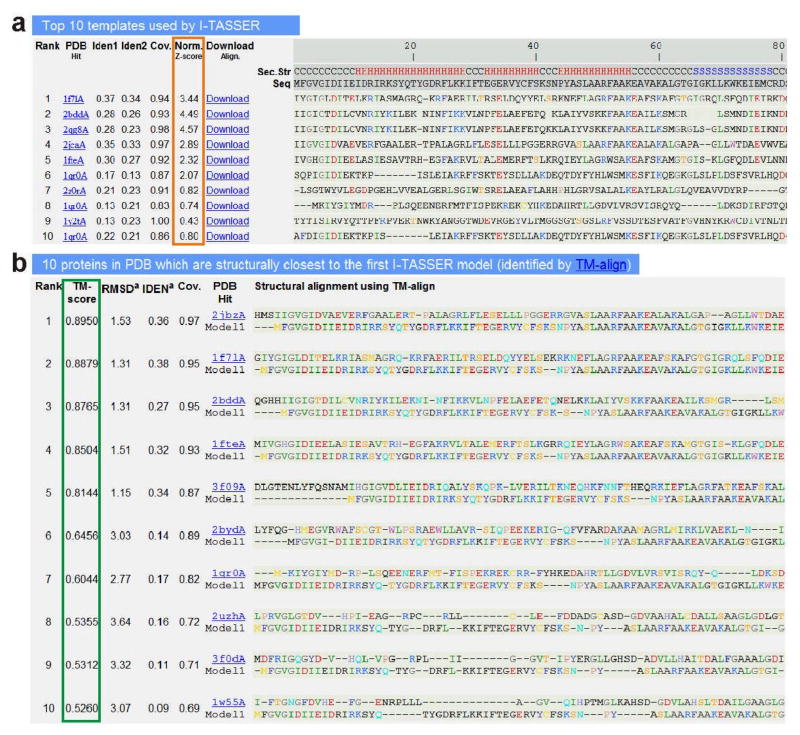
16 View the next section of the result page to analyze the top 10 threading templates for the query protein sequence, as identified by the LOMETS threading program (an example is shown in Figure 4a). The threading-aligned regions of these templates provide the building blocks and the spatial restraints in the I-TASSER fragment assembly simulations. ?TROUBLESHOOTING
-
17 View the “Norm. Z-score” column of the table to analyze the quality of threading alignments (highlighted in orange rectangle in Figure 4a). The quality of the threading alignment is usually estimated based on the Z-score of the alignment, which reflects how significant the alignment is as compared to the average. However, in LOMETS meta-server, the Z-score scale is different in different threading programs, which renders the comparison of Z-scores among different threading programs based on their absolute values meaningless. Instead, a normalized Z-score is presented in this column, and is equal to the Z-score of the alignment divided by a program-specific cutoff Z0, where Z0 has been determined based on large-scale threading benchmark tests53 for differentiating ‘good’ and ‘bad’ templates, i.e. a template with a Z-score greater than Z0 usually implies that the alignment corresponds to a correct fold. Similarly, an alignment with a normalized Z-score >1 reflects a confident alignment.
CRITICAL STEP: If most of the top threading alignments have a normalized Z-score >1, the accuracy of the final model is usually high. However, if the coverage of the top threading alignments is low and the alignments are confined to only a small region of query protein, then a high normalized Z-score is not a good indicator of modeling accuracy for the full-length model. In these cases, the query protein usually contains more than one domain and it is recommended to split the sequence into individual domains based on predicted domain boundaries and then submit each domain individually to the server. ?TROUBLESHOOTING
18 View the percentage sequence identity in the threading-aligned region (column “Iden. 1”) and in the whole chain (column “Iden. 2”) to judge the homology level between the query and the template proteins. A higher sequence identity is an indicator of evolutionary relatedness between the query and template proteins. A sequence identity that is high in the threading-aligned region but low for the whole-chain alignment indicates a conserved structural motif/domain present in the query and the template protein.
19 View the threading alignments to identify conserved residues/motifs/regions in the query and the template proteins. The aligned residues in the template that are identical to the corresponding query residues are colored based on their amino-acid property in the alignment. In many cases these regions/residues are of functional significance.
20 Click on the PDB code and chain identifier of the templates in the “PDB Hit” column. The browser will be directed to the RCSB website showing information about the template protein. On the RCSB web page, scroll down and click the links shown in the “Derived Data” field to see SCOP, CATH and PFAM classifications of the template protein and the associated Gene Ontology (GO) terms for analyzing the function.
21 Download the threading alignment (optional). Users can download PDB formatted threading alignment file by clicking on the “Download Align” link. The alignment file can be opened in any molecular visualization program listed in the Materials section, and can also be used for adding additional restraints during the structure modeling, as described in step 5 and repeating steps 1–18.
Figure 4.
An illustrative example of the I-TASSER result page showing (a) top ten threading templates and the alignments for the query protein identified by LOMETS; and (b) structural analogs and their alignment with the I-TASSER model, as identified by TM-align from the PDB library. The quality of the threading alignment in (a) is evaluated based on their normalized Z-score (highlighted in orange), where a normalized Z-score >1 reflects a good alignment. The ranking of the analogs shown in (b) is based on the TM-score (highlighted in green) of the structural alignment.
Structural analogs of the predicted model
22 View the structural analogs of the top-scoring I-TASSER model in the PDB library as identified by the structural alignment program TM-align64 (an example is shown in Figure 4b). The structural analogs are ranked based on the TM-score (highlighted in green rectangle) between the I-TASSER model and the TM-align templates. Detected structural analogs with a TM-score >0.5 can be used for determining the structure class/protein family of the predicted query protein structure69.
23 View the ‘RMSD’, ‘IDEN’ and ‘Cov.’ columns in the table to analyze theparameters derived from the structural alignment. RMSD and IDEN are the RMSD and the sequence identity in the regions structurally aligned by TM-align, and reflects the conservation of spatial motifs in the model and the structural analog.
24 Analyze the structural alignment obtained from TM-align64 to identify the structural conservation/variations that are present in the query protein and the structural analogs. Structurally aligned residue pairs in the alignment are highlighted in color based on their amino-acid property, while the unaligned regions are indicated by “-”.
Function prediction based on the predicted structure
25 View the identified functional analogs of the query protein and the confidence scores of the predictions based on the predicted 3D model. The function prediction result (an example is shown in Figure 5) is divided into three subsections: Enzyme Classification (EC) numbers, Gene Ontology (GO) terms, and ligand binding sites.
26 View the ‘TM-score’, ‘RMSD’, ‘IDEN’ and ‘Cov.’ columns in each subsection to quantitatively evaluate the global structure similarity and conservation of spatial patterns between the I-TASSER model and the functional analogs.
Figure 5.
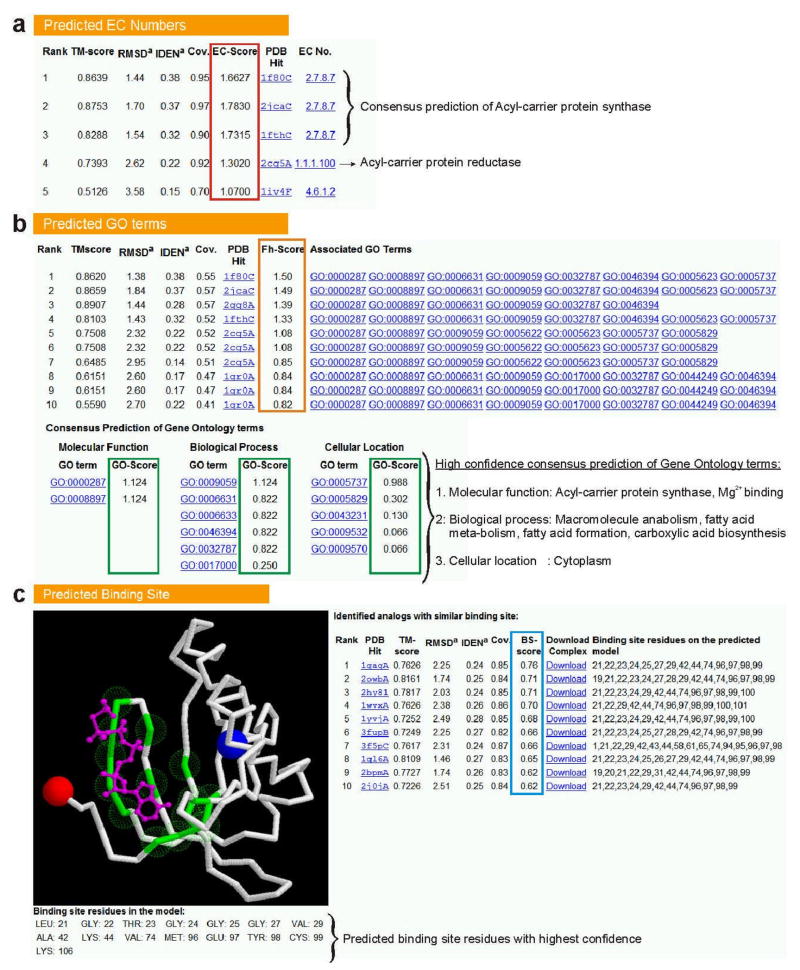
Illustrative examples of the I-TASSER function predictions on (a) Enzyme Commission numbers; (b) Gene Ontology terms; and (c) ligand-binding sites. The confidence level of the function prediction on EC number and binding site is shown in EC-score and BS-score columns (highlighted in red and blue rectangles, respectively). For GO, the analogs are first sorted based on Fh-score (in orange rectangle) and then a consensus of the predictions is derived from the top-scoring analogs and the confidence score of the GO prediction is defined as the ‘GO-score’ shown in green. The image in (c) shows the top-scoring binding site prediction in 3D model along with the bound ligand (in magenta), where the binding residues in protein are shown as transparent green spheres. The N- and C-terminus residues are marked by blue and red spheres, respectively.
Function subsection 1: Enzyme Commission (EC) number predictions
27 View the top five potential enzyme analogs along with their EC numbers which are displayed in the “Predicted EC numbers” table. An example is shown in Figure 5a.
-
28 View the confidence score shown in “EC-Score” column to determine whether the EC number of the analog can be used for functional annotation of the query protein. Based on a large-scale benchmarking test on the predicted models for 97 enzymes with unique functions, where homologous templates were excluded from both the threading and the enzyme library, we found40 that the first three digits of EC numbers can be correctly predicted from the first identified functional analog for 51 proteins; 38 of these analogs had an EC-Score >1.1.
CRITICAL STEP: Although an EC-Score >1.1 is a good indicator of the functional similarity between the query and the identified enzyme analogs, the users are advised to consult both the EC-Score and the consensus of the EC numbers associated with the analogs of the similar fold (i.e. TM-score >0.5). For example, if most of the identified functional analogs with similar folds have the same first 3 EC number digits (shown as an example in Figure 5a), and the EC-Score is higher than 1.1, the likelihood of the prediction to be correct is very high. On the contrary, if the EC-Score is high but there is no consensus of the EC numbers among the identified analogs, the prediction will become less reliable and users are advised to consult the GO term predictions, presented in the next subsection, because retooling of active sites can cause drastic shifts in function as expressed by the EC number, even in very closely related enzymes38. In most of these cases, the proteins usually bind a similar ligand or are part of the same biological pathway40. For cases when all EC-Scores are <1.1, it is possible that the query protein is either not an enzyme or the confidence level of the structure prediction and therefore the predictability of the function is low.
29 Click on the Enzyme Commission (EC) numbers to visit the ExPASy Enzyme database. This database provides a detailed description of the enzyme families, namely, the reactions catalyzed by the enzyme, the co-factor required, and the metabolic pathway.
Function subsection 2: Gene Ontology term predictions
30 View the functional analogs and their associated Gene Ontology terms in the table describing “Predicted GO terms” (an example is shown in Figure 5b). Most of the analogs are associated with multiple GO terms, which describe their highest level of molecular function, biological process and cellular location in the GO hierarchy. Click on each of these terms to visit the Amigo website (http://amigo.geneontology.org/) for analyzing the definition and lineage of each term.
-
31 View the Fh-score (Functional homology score) associated with the analogs to get a partial estimation of the confidence level of transferring functional annotation from these analogs. Based on our benchmarking study of 218 modeled protein structures using non-homologous templates40, we found that 50% of the native GO terms could be correctly identified from the first identified analogs for 122 test proteins using an Fh-score cutoff of 0.8, achieving an overall accuracy of 56%.
CRITICAL STEP: In the benchmarking study40, we found that Fh-score is a strong indicator of functional similarity between the predicted structure and detected analogs. However, because the function of proteins is multi-faceted and the unanimity of functionalities of the identified analogs usually yields a more reliable prediction, a consensus between a GO term and its ancestor terms in the ontology has been proven to be a more reliable indicator of the GO terms, which is therefore provided in the next table. It is recommended that the user analyses the consensus prediction shown in the table.
32 View the “Consensus prediction of GO terms” table for the consensus prediction of GO terms. This table is collected from the functional analogs having an Fh-score >1.0. If no analogs have an Fh-score greater than the cutoff score, the consensus prediction is derived from top 10 analogs regardless of the Fh-score. The table contains the GO terms and the associated confidence scores for the predicted molecular function, biological process and cellular localization. The confidence score (GO-score) for each term is derived based on weighted frequencies of occurrence of each term, where the weights are taken from the Fh-score of the templates from which the function is derived. Based on the benchmarking test, the best false positive and false negative rates are obtained for the GO terms associated with a GO-score cutoff=0.5, with decreasing coverage of prediction at deeper ontology levels40.
Function subsection 3: Ligand-binding site predictions
33 View the best identified ligand-binding site in the predicted structure, shown as a GIF image at the bottom of the result page (illustrated in Figure 5c). The backbone of the model in the image is shown as white solid lines, while the binding site residues of the query protein are highlighted as transparent green spheres in the image. Ligand atoms are shown as “ball and stick” in magenta. The N- and C-terminus residues are depicted by blue and red spheres, respectively. The residue number and amino acid type of the highlighted binding site residues are shown beneath the image.
34 View the list of the top 10 identified functional analogs and the derived binding site residues on the model in the table (right of the image). The bound ligands in these structures can be tracked by clicking on the PDB links.
35 Analyze the confidence level of the predictions based on the BS-score. The BS-score is based on the local structural similarity of the ligand binding sites and the sequence identity between the I-TASSER model and the structural analogs40. When the BS-score of the analogs is >0.5 and the predicted binding site residues are clustered together, the confidence level of the prediction is usually high.
36 Download ligand-protein complex (optional). Users can download the PDB formatted file containing the model and the bound ligand by clicking on the “Download” link. The binding site and the docked ligand on the model can be viewed interactively by opening these files in any molecular visualization program. Rendering the ligand as a space-filling model or depicting predicted binding site residues as a surface will aid in visualizing the binding site cleft and help with analyzing the ligand-protein interactions.
TIMING
The procedure of structure and function prediction by the I-TASSER server takes 6–10 hours for a typical medium-size protein (~200–400 residues), although larger proteins require a longer Monte Carlo simulation and hence longer waiting time. When a user submits a sequence, however, the actual processing time also depends on the number of jobs in the queue. We have currently devoted a cluster of 2000 HP DL1000h (Nehalem) processors to the I-TASSER server, and in most cases users can receive the results within 1–2 days. For getting an even faster response on structure modeling, see TROUBLESHOOTING (Step 7).
TROUBLESHOOTING
How to handle large/multi-domain proteins? (Step 2)
At present, the I-TASSER server accepts protein sequences of length up to 1,500 amino acids. For sequences longer than 1,500 amino acids, since these are predominantly multi-domain proteins, the user should split the sequence into domains and separately submit the sequence of the individual domains. See the Experimental Design section for tips on how to identify domain boundaries in multi-domain protein sequences.
How to speed up the modeling procedure? (Step 7)
The major time-consuming part of the I-TASSER modeling process is the Monte Carlo simulation for structure refinement assembly. If the user needs a quicker response and/or the structural refinement is unnecessary, we provide two threading servers, the LOMETS meta-threading server (http://zhang.bioinformatics.ku.edu/LOMETS) and the MUSTER single threading server (http://zhang.bioinformatics.ku.edu/MUSTER), from which results can be obtained within about an hour. These threading servers build full-length models by MODELLER72, which aims at constructing a 3D model close to the template without performing extensive structure refinement. For an even faster structure prediction, users are recommended to use the PSI-BLAST servers (http://blast.ncbi.nlm.nih.gov or http://www.ebi.ac.uk/Tools/psiblast). Although PSI-BLAST is less sensitive than the state-of-art threading algorithms in detecting distantly-homologous templates, it is very robust in identifying highly accurate alignment if close homologous protein exist in the databases.
Why is the number of generated models less than 5? (Step 11)
The I-TASSER server normally outputs top five structure models as ranked on the confidence score. There are some cases where the number of final models is less than 5. This is because the top templates identified by LOMETS are very similar to each other, and the I-TASSER simulations converge. Therefore, the number of structure clusters is <5, even when the RMSD cutoff in SPICKER63 is set to the minimum (3.5 Å). In these cases, the C-score is usually high, which indicates a high-quality structure prediction.
What can I do if the C-score of my model is low? (Step 13)
Like the majority of template-based methods in the field, the quality of the predicted model from I-TASSER relies on the availability and quality of the threading templates as identified by LOMETS. In CASP8, for example, the correlation between the RMSD of the final models by I-TASSER and the initial templates by LOMETS is around 0.8920. Therefore, a prediction with low C-score values usually indicates lack of good templates in the protein structure library, while ab initio modeling of medium-to-large size proteins without using templates is the major challenge in the field. For these cases, we suggest the users to seek for other sources of structural information, such as data from mutagenesis or cross-linking experiments on the target protein, which can provide residue-residue contact and distance information. The information from these sources can be specified as external restraints to improve the modeling quality, while using the convenient interface of I-TASSER server (see Experimental Design and Step 5 of PROCEDURE).
Despite of dependence of modeling results on templates, we note significant advantage of I-TASSER methodology in both template structure refinement and ab initio structure modeling for small proteins, which makes the I-TASSER server unique and different from many of other widely-used homology modeling tools like PSI-Blast5 and MODELLER72. For example, in CASP820, I-TASSER drew the best threading templates closer to the native structure in 139 out of 164 cases. In CASP718, the ab initio procedure of I-TASSER generated correct models with RMSD in the range of 3–6 Å, for 7/19 New Fold targets with sequence up to 132 residues long. These data demonstrate the significant ability of the I-TASSER server in modeling protein targets in the “twilight zone” which have no or weakly homologous templates.
Why does only the first model have quality estimation? (Step 14)
TM-score and RMSD to native of the first model are predicted based on their strong correlation with the C-score, as observed in the large-scale benchmark test70. However, the correlation of C-score and quality of lower-ranked models (i.e. 2nd-5th models) is much weaker than that for the first model, which cannot result in a meaningful estimation of the absolute quality for the lower rank models. This is understandable because the conformational space covered by the I-TASSER simulations is limited. For easy targets, almost all decoys are near-native and the structures are mainly aggregated in the first cluster. After removing the structures in the first cluster, the size of the lower-ranked clusters will be much smaller than the first one and comparable to that of hard targets. But the quality of the lower-rank clusters from the easy targets is still on average better than that from hard targets because most of the decoys generated for hard targets are incorrect. This makes the overall correlation between C-score and the lower-ranked clusters very weak when combining data from both easy and hard targets.
Nevertheless, there is a correlation between the relative rank and the quality of the clusters for the same target. In the large-scale benchmark test70, for example, the average TM-score (RMSD) of the top-five models are 0.501 (9.6 Å), 0.468 (10.6 Å), 0.466 (10.7 Å), 0.461 (11.1 Å), and 0.454 (11.3 Å), respectively. Thus, having the expected quality of the first model and the C-score of each model, the users can estimate the relative quality of lower rank models based on the relative rank and their C-score information.
Should I trust TM-score or RMSD in the model quality estimation? (Step 15)
It can be professedly confusing when the server’s quality estimation shows a high RMSD value but with a good or reasonable TM-score. This often happens when the protein is big. In these situations, we suggest that the users should judge the quality of the predicted model based on the expected TM-score rather than the expected RMSD.
First, it is well-known that RMSD is not a good measure for the protein structural similarity especially when RMSD is high, because RMSD weights all residue pairs equally and a local structural change or tail disorientation can result in a big RMSD, although the topology of the core structure is the same. Also, the average RMSD of random structure pairs depends on the length of proteins73, which renders the absolute value of RMSD less meaningful for comparing proteins of different size. TM-score68 has been designed to specifically alleviate this issues by weighting small distance stronger than the large distance. Therefore, TM-score is more sensitive to the topology change than RMSD. Since TM-score adopts a length-dependent scale to normalize the distance, the average TM-score of random protein pairs does not depend on the protein size, with TM-score <0.17 meaning random predictions and TM-score >0.5 meaning correct topology for all sizes of proteins69.
Second, as a consequence of the sensitivity of TM-score on structural topology, we found in our benchmark test68 that the correlation coefficient of C-score and TM-score (0.91) is much higher than that of C-score and RMSD (0.75). Therefore, the estimation of TM-score is usually more reliable than that of RMSD for the I-TASSER models, i.e. TM-score estimation has usually a much smaller systematic error than RMSD in our estimation.
What can I do if no significant templates are identified? (Step 16)
In most cases, this means that there is no good template which has been solved so far for this target. We therefore suggest the users to seek for other experimental studies which can provide information for collecting additional spatial restraints and use it to improve the I-TASSER modeling results. Since the threading programs in LOMETS use representative PDB libraries to build template alignments for the query protein, in some rare cases, it is possible that a good template for the query protein may not have been included in the library or the template may not have been correctly identified by LOMETS threading programs, even though it is present in the library. In these cases, the users are encouraged to try the possibility of identifying templates on their own using specific tools or functional analysis. I-TASSER allows users to specify templates with or without alignment. Read more about adding external spatial restraints or specifying protein structure as a template in the Experimental Design section of this protocol.
Should I trust Z-score or C-score for judging final modeling result? (Step 17)
Z-score measures the statistical significance of the threading alignment and is strongly correlated with the quality of the threading template. C-score is a combination of the Z-score of threading templates and the structural density of SPICKER cluster that reflects the convergence of the I-TASSER simulations. C-score is therefore more strongly correlated with the quality of the final model than the Z-score of the templates. Accordingly, TM-score and RMSD of the model is estimated based on the C-score and the length of the query sequence.
ANTICIPATED RESULTS
Once the job is completed, the user is notified by an email message which contains the images of the predicted structures and a link to the I-TASSER website where the complete result is deposited. The result page contains:
Predicted secondary structures.
Up to five full-length atomic models along with the estimated accuracy (TM-score and RMSD to the native) of the models.
The top ten templates and alignments that have been identified by LOMETS and used in the assembly of the full-length model.
The top ten structural analogs which are structurally closest to the predicted 3D model.
Functional predictions for the query protein in terms of Enzyme Commission numbers, Gene Ontology terms, and ligand-binding sites, with a confidence estimate provided for each prediction.
Acknowledgments
We thank Dr. A. Szilagyi for reading the manuscript. This work was supported in part by the Alfred P. Sloan Foundation; and the National Science Foundation (Career Award 0746198); and the National Institute of General Medical Sciences (GM083107, GM084222).
Footnotes
AUTHOR CONTRIBUTIONS STATEMENT
Y.Z conceived and supervised the project. A.R, A.K and Y.Z designed and performed the experiments. A.R and Y.Z wrote the manuscript.
COMPETING FINANCIAL INTEREST
The authors declare that they have no competing financial interests.
Contributor Information
Ambrish Roy, Email: ambroy@umich.edu.
Alper Kucukural, Email: alper@ku.edu.
Yang Zhang, Email: zhng@umich.edu.
References
- 1.The UniProt, C. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2008 [Google Scholar]
- 2.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Y. Progress and challenges in protein structure prediction. Curr Opin Struct Biol. 2008;18:342–8. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marti-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 5.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowie JU, Luthy R, Eisenberg D. A method to identify protein sequences that fold into a known three-dimensional structure. Science. 1991;253:164–170. doi: 10.1126/science.1853201. [DOI] [PubMed] [Google Scholar]
- 7.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–9. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 8.Liwo A, Lee J, Ripoll DR, Pillardy J, Scheraga HA. Protein structure prediction by global optimization of a potential energy function. Proc Natl Acad Sci U S A. 1999;96:5482–5. doi: 10.1073/pnas.96.10.5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–25. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 10.Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jauch R, Yeo HC, Kolatkar PR, Clarke ND. Assessment of CASP7 structure predictions for template free targets. Proteins. 2007;69:57–67. doi: 10.1002/prot.21771. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Y, Kolinski A, Skolnick J. TOUCHSTONE II: A new approach to ab initio protein structure prediction. Biophys J. 2003;85:1145–1164. doi: 10.1016/S0006-3495(03)74551-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J Mol Biol. 1997;268:209–25. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 14.Battey JN, et al. Automated server predictions in CASP7. Proteins. 2007;69:68–82. doi: 10.1002/prot.21761. [DOI] [PubMed] [Google Scholar]
- 15.Moult J, et al. Critical assessment of methods of protein structure prediction-Round VII. Proteins. 2007;69 (Suppl 8):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp J, Bordoli L, Battey JN, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69:38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 17.Das R, et al. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@home. Proteins. 2007;69:118–128. doi: 10.1002/prot.21636. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y. Template-based modeling and free modeling by I-TASSER in CASP7. Proteins. 2007;69:108–117. doi: 10.1002/prot.21702. [DOI] [PubMed] [Google Scholar]
- 19.Zhou H, et al. Analysis of TASSER-based CASP7 protein structure prediction results. Proteins. 2007;69 (Suppl 8):90–7. doi: 10.1002/prot.21649. [DOI] [PubMed] [Google Scholar]
- 20.Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cozzetto D, et al. Evaluation of template-based models in CASP8 with standard measures. Proteins. 2009;77 (Suppl 9):18–28. doi: 10.1002/prot.22561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y. Protein structure prediction: when is it useful? Curr Opin Struct Biol. 2009;19:145–55. doi: 10.1016/j.sbi.2009.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ekins S, Mestres J, Testa B. In silico pharmacology for drug discovery: applications to targets and beyond. Br J Pharmacol. 2007;152:21–37. doi: 10.1038/sj.bjp.0707306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becker OM, et al. An integrated in silico 3D model-driven discovery of a novel, potent, and selective amidosulfonamide 5-HT1A agonist (PRX-00023) for the treatment of anxiety and depression. J Med Chem. 2006;49:3116–35. doi: 10.1021/jm0508641. [DOI] [PubMed] [Google Scholar]
- 25.Brylinski M, Skolnick J. Q-Dock: Low-resolution flexible ligand docking with pocket-specific threading restraints. J Comput Chem. 2008;29:1574–88. doi: 10.1002/jcc.20917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arakaki AK, Zhang Y, Skolnick J. Large-scale assessment of the utility of low-resolution protein structures for biochemical function assignment. Bioinformatics. 2004;20:1087–96. doi: 10.1093/bioinformatics/bth044. [DOI] [PubMed] [Google Scholar]
- 27.Yue P, Moult J. Identification and analysis of deleterious human SNPs. J Mol Biol. 2006;356:1263–74. doi: 10.1016/j.jmb.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 28.Boyd A, et al. A random mutagenesis approach to isolate dominant-negative yeast sec1 mutants reveals a functional role for domain 3a in yeast and mammalian Sec1/Munc18 proteins. Genetics. 2008;180:165–78. doi: 10.1534/genetics.108.090423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Y, Li Z, Godzik A. Modeling and analyzing three-dimensional structures of human disease proteins. Pac Symp Biocomput. 2006:439–50. [PubMed] [Google Scholar]
- 30.Keedy DA, et al. The other 90% of the protein: assessment beyond the Calphas for CASP8 template-based and high-accuracy models. Proteins. 2009;77 (Suppl 9):29–49. doi: 10.1002/prot.22551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tress M, Ezkurdia I, Grana O, Lopez G, Valencia A. Assessment of predictions submitted for the CASP6 comparative modeling category. Proteins. 2005;61 (Suppl 7):27–45. doi: 10.1002/prot.20720. [DOI] [PubMed] [Google Scholar]
- 32.Moult J. Comparative modeling in structural genomics. Structure. 2008;16:14–6. doi: 10.1016/j.str.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Tress M, et al. Assessment of predictions submitted for the CASP7 domain prediction category. Proteins. 2007;69 (Suppl 8):137–51. doi: 10.1002/prot.21675. [DOI] [PubMed] [Google Scholar]
- 34.Malmstrom L, et al. Superfamily assignments for the yeast proteome through integration of structure prediction with the gene ontology. PLoS Biol. 2007;5:e76. doi: 10.1371/journal.pbio.0050076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Devries ME, Skolnick J. Structure modeling of all identified G protein-coupled receptors in the human genome. PLoS Comput Biol. 2006;2:e13. doi: 10.1371/journal.pcbi.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lopez G, Rojas A, Tress M, Valencia A. Assessment of predictions submitted for the CASP7 function prediction category. Proteins. 2007;69 (Suppl 8):165–74. doi: 10.1002/prot.21651. [DOI] [PubMed] [Google Scholar]
- 37.Brylinski M, Skolnick J. A threading-based method (FINDSITE) for ligand-binding site prediction and functional annotation. Proc Natl Acad Sci U S A. 2008;105:129–34. doi: 10.1073/pnas.0707684105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roy A, Srinivasan N, Gowri VS. Molecular and structural basis of drift in the functions of closely-related homologous enzyme domains: implications for function annotation based on homology searches and structural genomics. In Silico Biol. 2009;9:S41–55. [PubMed] [Google Scholar]
- 39.Bork P, Sander C, Valencia A. Convergent evolution of similar enzymatic function on different protein folds: the hexokinase, ribokinase, and galactokinase families of sugar kinases. Protein Sci. 1993;2:31–40. doi: 10.1002/pro.5560020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy A, Kucukural A, Mukherjee S, Hefty PS, Zhang Y. Large scale benchmarking of protein function prediction using predicted protein structures. 2009 Submitted. [Google Scholar]
- 41.Zhang Y, Skolnick J. Tertiary structure predictions on a comprehensive benchmark of medium to large size proteins. Biophys J. 2004;87:2647–55. doi: 10.1529/biophysj.104.045385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc Natl Acad Sci U S A. 2004;101:7594–9. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karplus K, Barrett C, Hughey R. Hidden Markov models for detecting remote protein homologies. Bioinformatics. 1998;14:846–856. doi: 10.1093/bioinformatics/14.10.846. [DOI] [PubMed] [Google Scholar]
- 44.McGuffin LJ, Jones DT. Improvement of the GenTHREADER method for genomic fold recognition. Bioinformatics. 2003;19:874–81. doi: 10.1093/bioinformatics/btg097. [DOI] [PubMed] [Google Scholar]
- 45.Wallner B, Elofsson A. Pcons5: combining consensus, structural evaluation and fold recognition scores. Bioinformatics. 2005;21:4248–54. doi: 10.1093/bioinformatics/bti702. [DOI] [PubMed] [Google Scholar]
- 46.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–60. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 47.Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ginalski K, Elofsson A, Fischer D, Rychlewski L. 3D-Jury: a simple approach to improve protein structure predictions. Bioinformatics. 2003;19:1015–8. doi: 10.1093/bioinformatics/btg124. [DOI] [PubMed] [Google Scholar]
- 49.Fischer D. 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51:434–41. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- 50.Kim DE, Chivian D, Baker D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004;32:W526131. doi: 10.1093/nar/gkh468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–71. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 52.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 53.Wu S, Zhang Y. LOMETS: a local meta-threading-server for protein structure prediction. Nucleic Acids Res. 2007;35:3375–82. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–57. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 55.Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72:547–56. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu Y, Xu D. Protein threading using PROSPECT: design and evaluation. Proteins. 2000;40:343–54. [PubMed] [Google Scholar]
- 57.Zhou H, Zhou Y. Fold recognition by combining sequence profiles derived from evolution and from depth-dependent structural alignment of fragments. Proteins. 2005;58:321–8. doi: 10.1002/prot.20308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H, Zhou Y. Single-body residue-level knowledge-based energy score combined with sequence-profile and secondary structure information for fold recognition. Proteins. 2004;55:1005–13. doi: 10.1002/prot.20007. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Kihara D, Skolnick J. Local energy landscape flattening: Parallel hyperbolic Monte Carlo sampling of protein folding. Proteins. 2002;48:192–201. doi: 10.1002/prot.10141. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Hubner I, Arakaki A, Shakhnovich E, Skolnick J. On the origin and completeness of highly likely single domain protein structures. Proc Natl Acad Sci USA. 2006;103:2605–10. doi: 10.1073/pnas.0509379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen H, Zhou HX. Prediction of solvent accessibility and sites of deleterious mutations from protein sequence. Nucleic Acids Res. 2005;33:3193–9. doi: 10.1093/nar/gki633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu S, Zhang Y. A comprehensive assessment of sequence-based and template-based methods for protein contact prediction. Bioinformatics. 2008;24:924–31. doi: 10.1093/bioinformatics/btn069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Skolnick J. SPICKER: a clustering approach to identify near-native protein folds. J Comput Chem. 2004;25:865–71. doi: 10.1002/jcc.20011. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Skolnick J. TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–9. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Zhang Y. REMO: A new protocol to refine full atomic protein models from C-alpha traces by optimizing hydrogen-bonding networks. Proteins. 2009;76:665–76. doi: 10.1002/prot.22380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barrett AJ. Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). Enzyme Nomenclature. Recommendations 1992. Supplement 4: corrections and additions (1997) Eur J Biochem. 1997;250:1–6. doi: 10.1111/j.1432-1033.1997.001_1.x. [DOI] [PubMed] [Google Scholar]
- 67.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–10. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- 69.Xu JR, Zhang Y. How significant is a protein structure similarity with TM-core=0.5? 2009 doi: 10.1093/bioinformatics/btq066. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li W, Zhang Y, Skolnick J. Application of sparse NMR restraints to large-scale protein structure prediction. Biophys J. 2004;87:1241–8. doi: 10.1529/biophysj.104.044750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 73.Betancourt MR, Skolnick J. Universal similarity measure for comparing protein structures. Biopolymers. 2001;59:305–9. doi: 10.1002/1097-0282(20011015)59:5<305::AID-BIP1027>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]