Abstract
The response of natural microbial communities to environmental change can be assessed by determining DNA- or RNA-targeted changes in relative abundance of 16S rRNA gene sequences by using fingerprinting techniques such as denaturing gradient gel electrophoresis (DNA-DGGE and RNA-DGGE, respectively) or by stable isotope probing (SIP) of 16S rRNA genes following incubation with a 13C-labeled substrate (DNA-SIP-DGGE). The sensitivities of these three approaches were compared during batch growth of communities containing two or three Nitrosospira pure or enriched cultures with different tolerances to a high ammonia concentration. Cultures were supplied with low, intermediate, or high initial ammonia concentrations and with 13C-labeled carbon dioxide. DNA-SIP-DGGE provided the most direct evidence for growth and was the most sensitive, with changes in DGGE profiles evident before changes in DNA- and RNA-DGGE profiles and before detectable increases in nitrite and nitrate production. RNA-DGGE provided intermediate sensitivity. In addition, the three molecular methods were used to follow growth of individual strains within communities. In general, changes in relative activities of individual strains within communities could be predicted from monoculture growth characteristics. Ammonia-tolerant Nitrosospira cluster 3b strains dominated mixed communities at all ammonia concentrations, and ammonia-sensitive strains were outcompeted at an intermediate ammonia concentration. However, coexistence of ammonia-tolerant and ammonia-sensitive strains occurred at the lowest ammonia concentration, and, under some conditions, strains inhibited at high ammonia in monoculture were active at high ammonia in mixed cultures, where they coexisted with ammonia-tolerant strains. The results therefore demonstrate the sensitivity of SIP for detection of activity of organisms with relatively low yield and low activity and its ability to follow changes in the structure of interacting microbial communities.
Molecular characterization of natural microbial communities has demonstrated the existence of novel high-level taxonomic groups with no cultured representatives and with significant diversity within phylogenetic and functional groups already established through analysis of organisms in laboratory culture. Autotrophic ammonia-oxidizing bacteria (AOB) exemplify the latter situation. Their low growth rates and the limited number of readily measured phenotypic characteristics available for identification of these organisms necessitate the use of molecular techniques for characterization of their diversity in natural environments. Phylogenetic analysis of 16S rRNA gene sequences places the majority of cultivated autotrophic bacterial ammonia oxidizers in a monophyletic group within the Betaproteobacteria (8, 26). Amplification and phylogenetic analysis of 16S rRNA gene sequences from enrichment cultures of ammonia oxidizers and sequences of environmental clones (31) suggest the existence of novel groups with no cultivated representative and considerable diversity within those represented by pure cultures.
Increased awareness of microbial diversity has raised questions regarding links between species diversity and functional diversity, functional redundancy, and the influence of environmental conditions on the activities of representatives of different phylotypes. For ammonia-oxidizing bacteria, relationships exist between broad phylogenetic groups and the environments from which laboratory isolates were obtained, which are linked, in some cases, to differences in physiological characteristics (11). There is also evidence of links between the relative abundance of different ammonia oxidizer groups and environmental conditions (1, 13, 14, 18, 21, 23, 34), suggesting selection for organisms with particular physiological characteristics. In one study (36), a combination of molecular and physiological studies has demonstrated links between species diversity, functional diversity, and soil nitrification kinetics. However, for ammonia oxidizers and other groups, there is little direct evidence about which strains within diverse communities are active under particular conditions or the extent of competition for substrates.
Stable isotope probing (SIP) (24, 27) of nucleic acids provides direct evidence of which members of mixed communities are active. This involves addition of substrates labeled with a stable isotope (most commonly 13C), extraction of nucleic acids, separation of 12C- and 13C-labeled nucleic acids by density gradient centrifugation, and subsequent molecular analysis. Sequences amplified from 13C-labeled DNA or RNA are derived from organisms actively assimilating the substrate. This approach has been used to identify organisms that utilize methane or methanol (4, 19), organic compounds (15, 20), or CO2 (6, 9) in microcosms and those that assimilate plant root exudates in the field (28). SIP therefore links phylogeny to ecosystem function and has identified established and novel groups by utilizing labeled compounds in complex soil communities. The technique also enables in situ physiological studies and investigation of interactions between organisms in mixed cultures belonging to the same functional group. For autotrophic betaproteobacterial ammonia oxidizers, amplification of 16S rRNA genes from 13C-labeled DNA during incubation with [13C]CO2 has the potential for discriminating which strains are active under specific conditions. Assessment of the discriminatory ability of this approach in complex natural environments requires studies under controlled and well-characterized conditions. The first aim of this study was, therefore, to assess the ability of SIP to discriminate activities of different members of simple mixed communities in comparison with direct measurement of product concentration and DNA- and RNA-denaturing gradient gel electrophoresis (DGGE). The second was to determine whether the activities of members of mixed communities of ammonia-oxidizing bacteria, in particular, their ability to grow at high ammonia concentrations, could be predicted from their physiological characteristics in monoculture. Of particular interest was whether strains with low ammonia tolerance are competitive at low ammonia concentrations. Mixed cultures were assembled from pure culture representatives of Nitrosospira clusters 0, 3a, and 3b (26, 36), which are frequently found in soil environments, and from enrichment cultures containing representatives of these clusters with heterotrophic contaminants. Other criteria for choice of community members were similarities in specific growth rate and cultivation conditions to enable meaningful competition experiments.
MATERIALS AND METHODS
Bacterial strains and cultivation conditions.
Pure and enrichment cultures used in growth and interaction experiments are listed in Table 1. Nitrosospira briensis strain C-128 was kindly provided by Woods Hole Oceanographic Institution (Woods Hole), and Nitrosospira strains 40KI and L115 were provided by Lars Bakken (Agricultural University of Norway). Other cultures were obtained in previous studies at Aberdeen (Scotland). All enrichment cultures contained a single ammonia oxidizer, as detected by DGGE analysis of 16S rRNA genes (see below). All cultures were maintained in inorganic medium (30), as modified by Powell and Prosser (22), containing 3.6 mM NH4+ as ammonium sulfate. Phylogenetic association of strains was according to Purkhold et al. (25). Additionally, phylogenetic analysis of full-length 16S rRNA gene sequences was carried out as described by Webster et al. (36) for cultivated representatives of the major AOB clusters, including Nitrosospira sp. Wyke2, Nitrosospira sp. Wyke8, and Nitrosospira sp. EnI299.
TABLE 1.
Pure and enrichment cultures of ammonia-oxidizing bacteria used in mixed communities and assessment of their growth in monoculture at different ammonium concentrations
| Community and straina | Sample type and source | Culture type | 16S rRNA gene accession no(s).b | Reference(s) or source | Phylogenetic group | Growth at initial ammonium concn (mM) ofc: |
||
|---|---|---|---|---|---|---|---|---|
| 3.6 | 14.3 | 71.4 | ||||||
| Community A | ||||||||
| Nitrosospira sp. En13 | Craibstone soil, pH 4.5; Scotland | Pure | AY856079 | 29 | 3a | + | + | − |
| N. briensis C-128 | Soil isolate; Woods Hole Oceanographic Institution (USA) | Pure | L35505 | 33 | 3b | + | + | + |
| Community B | ||||||||
| Nitrosospira sp. 40KI | Cultivated clay loam soil, pH 6.5; Ås, Norway | Pure | X84565 | 35 | 0 | + | + | − |
| Nitrosospira sp. L115 | Ombrotrophic peat, pH 4.0; Lakkasuo area, Finland | Pure | X84662, AY123796 | 25, 35 | 3a | + | + | − |
| Nitrosospira sp. EnU4a* | Natural (unimproved) grassland soil; Sourhope, Scotland | Enrichment | AY727034 | 36 | 3b | + | + | + |
| Community C | ||||||||
| Nitrosospira sp. Wyke2* | Grassland soil; Exeter, UK | Enrichment | EF175095 | This study | 0 | + | + | − |
| Nitrosospira sp. Wyke8 | Grassland soil; Exeter, UK | Enrichment | EF175096 | This study | 3a | + | + | − |
| Nitrosospira sp. EnI299* | Fertilized grassland soil; Sourhope, Scotland | Enrichment | EF175101 | This study | 3b | + | + | + |
All cultures contained a single ammonia oxidizer strain, but cultures marked with an asterisk also contained a nitrite oxidizer.
GenBank database.
Growth was determined after incubation for 30 days in inorganic medium with the indicated initial ammonia concentration. +, growth; −, no growth or a significant lag phase.
Assessment of sensitivity to high ammonia concentrations.
To assess sensitivity to ammonia concentration, each ammonia oxidizer strain was grown in triplicate monocultures in 150 ml of inorganic growth medium with three initial ammonium concentrations (3.6, 14.3, and 71.4 mM NH4+) in 250-ml Erlenmeyer flasks inoculated with 1 ml of a fresh stationary phase culture. Cultures were incubated without shaking at 28°C in the dark, and pH was maintained at values in the range of 7 to 8.5 by addition of 5% (wt/vol) sodium carbonate when the pH of the medium fell below 7. Samples (0.7 ml) were taken immediately after inoculation and after 4, 8, 10, 13, 16, 20, 24, and 29 days for assessment of growth by colorimetric quantitative analysis for combined nitrite-N plus nitrate-N concentrations using an Alpkem RFA Autoanalyser (Alpkem Corporation, Clackamas, OR).
Growth of mixed cultures.
Growth of three mixed communities (Table 1) was investigated in 150 ml of inorganic growth medium (described above) at three initial ammonium concentrations (3.6, 14.3, and 71.4 mM NH4+) in 250-ml Erlenmeyer flasks inoculated with 5 ml of a fresh stationary phase culture of each strain. Flasks were sealed with butyl rubber septum stoppers, and the headspace was purged with CO2-free air. 13C-labeled CO2 was provided by dropwise addition of 1 ml of 5% (wt/vol) Na213CO3 (99% atom enriched; CEH-Gas, Cambridge, United Kingdom), and an equimolar solution of HCl was used simultaneously for titration. The pH of the medium was then adjusted to approximately 7.5 using 13C-labeled Na2CO3 and was maintained at values in the range of 7 to 8.5 by regular addition of the same solution. Flasks were incubated at 28°C in the dark, and samples (5 ml) were taken immediately after inoculation and after 2, 5, 10, 20, 30, 53, and 81 days. A 0.5-ml volume of sample was used for assessment of growth by analysis of nitrite-N plus nitrate-N concentrations, as described for monocultures, and 4.5-ml volumes of samples taken at 0, 5, 10 and 20 days were subjected to molecular analysis.
Nucleic acid extraction and cDNA synthesis.
Cells from 4.5-ml samples were centrifuged (8,400 × g for 10 min) and then diluted in 0.5 ml of extraction buffer [0.7 M NaCl, 0.1 M Na2(SO3), 0.1 M Tris-HCl (pH 7.5), 0.05 M EDTA (pH 8), and 1% SDS]. The mixture was transferred in 2-ml screw-cap Blue Matrix Ribolyser tubes (Hybaid, Ltd., Ashford, United Kingdom) and, after addition of 0.5 ml of phenol-chloroform-isoamyl alcohol (25:24:1), cells were disrupted with a ThermoHybaid Ribolyser Cell Disruptor (Hybaid) (at speed 4 for 30 s). Further extraction and precipitation of nucleic acids were carried out as described by Griffiths et al. (7). For RNA-targeted analysis of ammonia-oxidizing bacteria, cDNA synthesis was performed as previously described (34).
16S rRNA genes/rRNA-DGGE.
Betaproteobacterial AOB strains were targeted using 16S rRNA genes, which provide better discrimination of closely related Nitrosospira strains during DGGE than functional genes. The relative abundance of different ammonia oxidizer strains targeting either 16S rRNA genes or cDNA was determined using a nested PCR approach, with first-round amplification using primer set CTO189f-CTO654r (12), which generated products that acted as templates for second-round amplification using primers P2/P3 (17). Prior to secondary amplification, PCR products were diluted 1:10 for samples taken after 0, 2, and 5 days and 1:20 for samples taken after 10 and 20 days. PCR conditions were as previously described (5). Amplification products were analyzed using DGGE as described previously (31) using a DCode Universal Mutation Detection System (Bio-Rad Laboratories, Hemel Hempstead, United Kingdom) and 1-mm-thick 8% polyacrylamide gels containing a denaturant gradient of 38 to 62%. Gels were electrophoresed for 16 h at 75 V and 60°C and were silver stained and analyzed as previously described (16).
Stable isotope probing.
12C- and 13C-labeled DNA were fractionated by density gradient centrifugation as described by Freitag et al. (6). Briefly, a cesium chloride solution was prepared in TE (Tris-EDTA) buffer, pH 8 (approximately 1.27 g ml−1), and the average density was measured with an Atago-R-5000 hand refractometer (Uni-IT Inc., Tokyo, Japan) and adjusted to a refractive index of 1.398 (1.683 g ml−1 buoyant density) by adding small amounts of CsCl or TE buffer, if necessary. Centrifugation medium was prepared by mixing 0.5 μg of DNA with 0.75 μl of ethidium bromide (0.1 mg ml−1) and 200 μl of the CsCl solution. The mixture was added to 4.8 ml of CsCl solution in 5.1-ml Quick-Seal polyallomer tubes (Beckman Coulter, Palo Alto, CA), and 100 μl of mineral oil was added to the upper surface. Tubes were centrifuged in a VTI 65.2 vertical rotor (Beckman Coulter) at 227,640 × g for 18 h at 20°C. Twelve fractions of equal volume (approximately 250 μl) were sampled from the base of the centrifuge tube using an LKB Bromma 2112 Redirac fraction collector (Pharmacia LKB, Cambridge, United Kingdom) with displacement of the cesium chloride solution by water added to the top of the tube using a Gilson MiniPuls 3 peristaltic pump (Anachem, Luton, United Kingdom). The density of each fraction collected was determined by measurement of the refractive index. DNA in each fraction was precipitated overnight using 2 volumes of 30% polyethylene glycol and resuspended in 30 μl of sterile water after being washed with 70% ethanol. PCR amplification of ammonia oxidizer 16S rRNA gene fragments and DGGE analysis were performed as for DNA-DGGE.
Nucleotide sequence accession numbers.
The sequences of the 16S rRNA genes of Nitrospira sp. strains Wyke2, Wyke8, and EnI299 have been deposited in the GenBank database under accession numbers EF175095, EF175096, and EF175101, respectively.
RESULTS
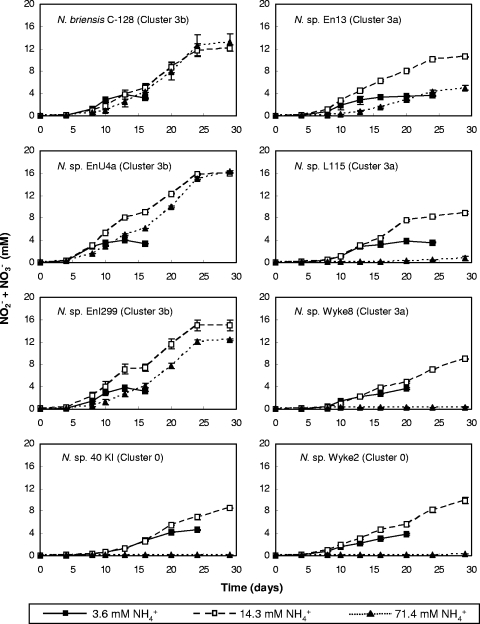
Several Nitrosospira pure cultures and enrichment cultures containing a single ammonia oxidizer were tested for sensitivity to high ammonia concentrations by growth in liquid batch culture at initial ammonium concentrations of 3.6, 14.3, and 71.4 mM NH4+ (subsequently referred to as low, intermediate, and high ammonia concentrations). Nitrite/nitrate production in all cultures was preceded by a short lag phase of at least 4 days, after which the concentration increased approximately linearly before either complete conversion of ammonia or entry into stationary phase. The three Nitrosospira cluster 3b strains grew at all ammonia concentrations (Fig. 1 and Table 1). Two of the three Nitrosospira cluster 3a strains (Nitrosospira sp. L115 and Nitrosospira sp. Wyke8) and both cluster 0 strains grew at the low and intermediate ammonia concentrations but were inhibited at the high ammonia concentration (Fig. 1). The cluster 3a strain Nitrosospira sp. En13 was the only noncluster 3b strain that grew at all ammonia concentrations; but growth at the high ammonia concentration was preceded by an extended lag phase of approximately 10 days, and there was little evidence of growth after 25 days (Fig. 1). This strain is therefore considered, for the purposes of this study, to be ammonia sensitive.
FIG. 1.
Changes in nitrite/nitrate concentration during growth of eight Nitrosospira strains on inorganic medium with initial ammonium concentrations of 3.6 (▪), 14.3 (□), or 71.4 (▴) mM NH4+. Growth was assessed by production of nitrite/nitrate, and plotted data represent means of measurements from triplicate flasks. Error bars represent standard errors.
Information on ammonia sensitivity of the eight Nitrosospira strains was used to construct three communities of two or three ammonia oxidizers with similar nitrite/nitrate production rates at low ammonia but with different responses to intermediate and high ammonia. Each community was represented by strains belonging to different phylogenetic clusters, enabling discrimination by DGGE analysis. Each was grown at low, intermediate, and high initial ammonia to determine whether activities and competition reflected growth in monoculture. DNA-DGGE and RNA-DGGE were used to assess the activities of different community members, reflected in changes in the relative abundance of community members and indicated by changes in relative band intensities. Activity was also assessed using DGGE-DNA-SIP by the appearance of bands in 13C-labeled DNA fractions, indicating active assimilation of 13CO2 during ammonia oxidation.
Interactions between one ammonia-tolerant and one ammonia-sensitive strain (community A).
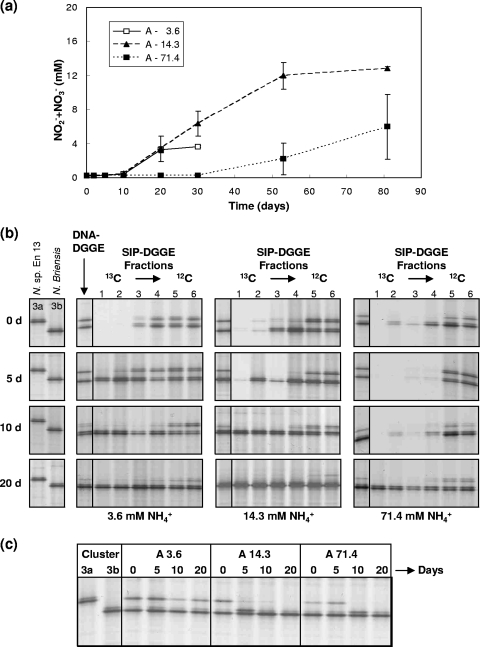
Community A contained an ammonia-tolerant strain, N. briensis, and an ammonia-sensitive strain, Nitrosospira sp. En13, with similar growth kinetics at low and intermediate ammonia concentrations. Growth in monoculture suggested that both would grow at low and intermediate ammonia concentrations but that N. briensis would dominate at a high ammonia concentration. Growth rates of the mixed culture (Fig. 2a) at low and intermediate ammonia were similar, with a lag phase of 10 days and complete conversion of ammonia after 20 and 53 days, respectively. There was a long lag period of 30 days at the high ammonia concentration; subsequent growth was substantial, but ammonia was not completely oxidized during the incubation period.
FIG. 2.
Changes in nitrite/nitrate concentration (a) and changes in community structure (b and c) of community A containing an ammonia-sensitive (Nitrospira sp. En 13, cluster 3a) and an ammonia-tolerant strain (N. briensis, cluster 3b) on inorganic medium with initial ammonium concentrations of 3.6, 14.3, or 71.4 mM NH4+, and supplied with [13C]CO2. In panel a, data represent means calculated from triplicate flasks, and error bars represent standard errors. Community structure was assessed by DGGE analysis of 16S rRNA gene fragments amplified from extracted DNA (b) or RNA (c). In panel b, for each sampling interval (0 to 20 days; top to bottom), a panel containing DGGE profiles of each monoculture is followed by three panels (left to right), one for each initial ammonia concentration. The first lane is a DNA-DGGE profile generated from extracted DNA. The second and third lanes are generated from fractions containing 12C- or 13C-labeled DNA following density gradient centrifugation of extracted DNA. Six sequential fractions are presented (labeled 1 to 6) from which the first two and the last two are considered the most representative 13C and 12C fractions, respectively, while the middle fractions (3 and 4) are considered intermediate.
Relative band intensities of DNA- and RNA-DGGE profiles determined and compared visually in samples taken immediately after inoculation were similar and reflected inoculation of similar cell numbers of each strain. Evidence of differential activity was first observed at 10 days, with an increase in relative intensity of N. briensis, which subsequently dominated DNA-DGGE profiles (Fig. 2b). Similar changes were observed in RNA-DGGE profiles but generally occurred earlier and were more marked (Fig. 2c). In particular, the relative intensity of the Nitrosospira sp. En13 band decreased more rapidly and to a greater extent in RNA-DGGE profiles at all ammonia concentrations. DNA- and RNA-DGGE data therefore indicate eventual domination by the ammonia-tolerant strain at all initial ammonia concentrations, with community changes first detectable after at least 10 days in DNA-DGGE profiles and after at least 5 days in RNA-DGGE profiles.
Immediately after inoculation and after 5 days of incubation (Fig. 2b), DNA-DGGE and 12C-labeled DNA-SIP-DGGE profiles were indistinguishable at all initial ammonia concentrations. No bands were detectable visually in 13C-labeled DNA-SIP-DGGE profiles in day 0 samples, as expected, with the exception of faint bands in the 13C-labeled DNA fractions at intermediate and high ammonia. Analysis of DGGE profiles from all gradient fractions enables clear distinction between true 13C enrichment and 12C contamination and suggests that these faint bands were due to imperfect fractionation. 13C-labeled DNA-SIP-DGGE bands indicative of the ammonia-tolerant N. briensis were observed after incubation of cultures for 5 days in low and intermediate ammonium concentrations and in subsequent samples (Fig. 2b). N. briensis bands were also observed in 13C-derived DGGE profiles from cultures exposed to high ammonia after 20 days. A band of low relative intensity corresponding to the ammonia-sensitive strain, En13, was also detected in the 13C fractions at day 5 and in subsequent samples at low ammonia but not at intermediate or high ammonia. The DNA-SIP-DGGE data therefore indicate that N. briensis was the only strain that grew and incorporated [13C]CO2 at intermediate and high ammonia. The strain also dominated at the low ammonia concentration, but strain En13 was also detected in 13C fractions, indicating some growth of this strain at the lowest initial ammonia concentration.
DNA-DGGE profiles therefore indicated an increase in relative abundance of N. briensis, but effects were observed later than in RNA-DGGE and DNA-SIP-DGGE and did not distinguish growth and death of either strain. DNA-DGGE was therefore not sufficiently sensitive to detect differences in the activities of N. briensis and En13. Incorporation of [13C]CO2 was detected at all initial ammonia concentrations before detectable changes in nitrite/nitrate production. At low and intermediate initial ammonia concentrations, [13C]CO2 incorporation was detected 5 days before detectable nitrite/nitrate production, while at high initial ammonia concentrations it was detected 10 days prior to detectable nitrite/nitrate increases. In general, reverse transcription-PCR (RT-PCR)-DGGE profiles obtained at later time points (10 or 20 days) were similar to the 13C-labeled DNA-SIP-DGGE profiles and also allowed detection of changes in relative activities before DNA-DGGE and before detectable nitrite/nitrate production.
Interactions between one ammonia-tolerant strain and two ammonia-sensitive strains (community B).
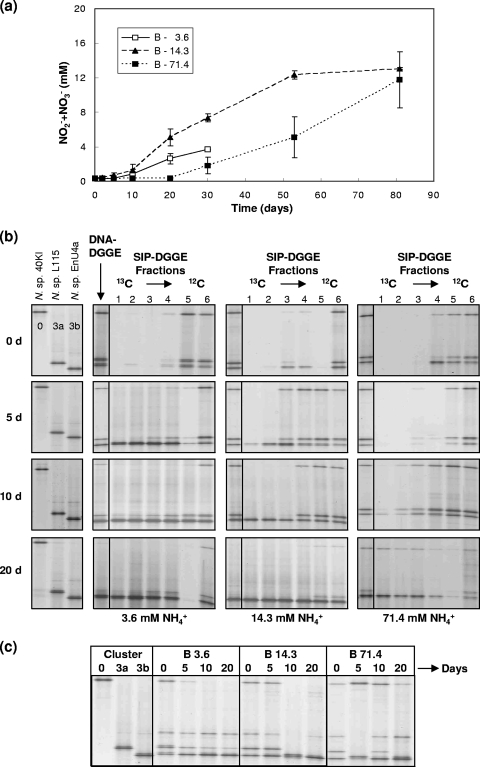
Community B contained one ammonia-tolerant, fast-growing strain, Nitrosospira EnU4a, and two ammonia-sensitive Nitrosospira strains, 40KI and L115. Growth in monoculture suggested that EnU4a would dominate, even at low and intermediate ammonia. Growth of the mixed culture occurred at all initial ammonia concentrations but with a lag phase of at least 20 days at high ammonia (Fig. 3a).
FIG. 3.
Changes in nitrite/nitrate concentration and in community structure during growth of community B containing an ammonia-tolerant strain (Nitrospira sp. EnU4a, cluster 3b) and two ammonia-sensitive strains (Nitrospira sp. L115 and Nitrospira sp. 40KI, representing clusters 3a and 0, respectively). See the legend of Fig. 1 for further information.
All three strains were detected in DNA-DGGE profiles by similar band intensities for all ammonia concentrations until day 10 (Fig. 3b). At day 20, strain EnU4a was the only band detectable at intermediate ammonia. At low ammonia, strain L115 was detectable only as a faint band, and 40KI was not detectable while the converse situation was observed at high ammonia. Similar but faster changes were observed in RNA-DGGE profiles. Again, strain EnU4a dominated profiles (Fig. 3c), and strains L115 and 40KI were detected at low and high ammonia, respectively, after incubation for 20 days. In addition, strain 40KI could not be detected at low ammonia after incubation for 10 days, and both strains were not detectable at intermediate ammonia after 10 days.
DNA-DGGE and 12C-labeled DNA-SIP-DGGE profiles (Fig. 3c) were similar until day 10, after which DNA-DGGE profiles were more similar to DGGE profiles of fractions representing 13C-labeled DNA. No band was detectable in 13C-labeled DNA-SIP-DGGE profiles (Fig. 3b) at day 0, but EnU4a was detectable at low and intermediate ammonia at 5 days, or 15 days prior to detection of changes in DNA-DGGE profiles and detectable increases in nitrite/nitrate concentration. At high ammonia, Nitrosospira EnU4a was first detected in day 10 samples; it dominated all 13C-labeled DNA-SIP-DGGE profiles and was the only strain detected at intermediate ammonia. Strains L115 and 40KI, however, were detected in 13C-labeled DNA-SIP-DGGE profiles at low and high ammonia after 10 and 20 days, respectively.
Interactions between one ammonia-tolerant strain and two ammonia-sensitive strains in enrichment cultures (community C).
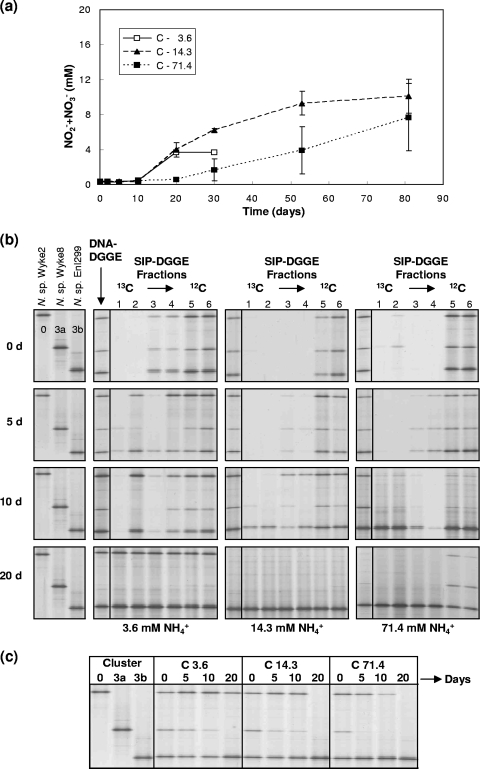
The third community investigated, community C, contained enrichments with two ammonia-sensitive Nitrosospira strains, Wyke2 and Wyke8, and an ammonia-tolerant enrichment, Nitrosospira EnI299. In monoculture, all strains had similar growth rates at low ammonia, suggesting that all would be active in the mixed community at this concentration, but only strain EnI299 grew at high ammonia. Nitrite/nitrate production by the mixed culture was similar at low and intermediate ammonia after an apparent lag of at least 10 days. Growth at high ammonia was inhibited, and nitrite/nitrate production, although detected at day 20, was lower than at other ammonia concentrations (Fig. 4a).
FIG. 4.
Changes in nitrite/nitrate concentration and in community structure during growth of community C containing an ammonia-tolerant strain (Nitrospira sp. EnI299, cluster 3b) and two ammonia-sensitive strains (Nitrospira sp. Wyke2 and Nitrospira sp. Wyke8, representing clusters 0 and 3a, respectively) in enrichment cultures. See the legend of Fig. 1 for further information.
DNA-DGGE remained unchanged until day 20, when bands representing strains EnI299 and Wyke2 dominated profiles at low ammonia, and only strain EnI299 could be detected at high ammonia (Fig. 4b). Similar changes were observed in RNA-DGGE profiles, but strain Wyke2 was no longer detectable after incubation for 20 days at intermediate and high ammonia. Wyke8 bands were not detectable after 20 days at all ammonia concentrations and disappeared earlier at high ammonia (Fig. 4c).
DNA-DGGE and 12C-labeled DNA-SIP-DGGE profiles were again similar, with the exception of samples taken at days 10 and 20 at high ammonia. At low ammonia, 13C-labeled DNA-SIP-DGGE profiles were dominated by Wyke2 and EnI299, which were first detected after incubation for 5 days, before detectable nitrite/nitrate production. At intermediate and high ammonia, only Nitrosospira strain EnI299 was detected in 13C-labeled DNA-SIP-DGGE profiles, with initial detection at 10 days, again before detectable nitrite/nitrate production. The data therefore indicate that the community was dominated by a single strain at intermediate and high ammonia, but two members of the community grew at low ammonia.
DISCUSSION
Inhibition by ammonia.
Phylogenetic analysis of AOB sequences from laboratory isolates and enrichments and from environmental clones provides support for the existence of several Nitrosospira clusters linked to environmental origin and physiology (11). Of particular relevance to this study is sensitivity to high ammonia concentrations of members of Nitrosospira cluster 3a and tolerance within cluster 3b (36). Growth of ammonia oxidizers follows Monod kinetics at noninhibitory concentrations (2, 10) and Haldane kinetics at high, inhibitory concentrations (32). Differences in growth and inhibition kinetics, as a function of ammonia, will influence the outcome of competition between ammonia oxidizers. For example, Bollmann et al. (3) showed that a Nitrosomonas cluster 6a strain outcompeted Nitrosomonas europaea at limiting ammonia concentrations during continuous culture. However, ammonia concentrations investigated in this study were higher than reported saturation constants for ammonia (10), and competition will have resulted from differences in inhibition; in addition, the authors investigated nitrosomonads rather than nitrosospiras, which may have different levels of “within-genus” physiological diversity.
Responses of strains to ammonia concentration varied, as observed in previous studies. All Nitrosospira cluster 3b strains were tolerant to the high ammonia concentration, while all but one Nitrosospira cluster 3a and cluster 0 strains were sensitive to high ammonia. The exception, Nitrosospira sp. En13, grew at all ammonia concentrations but exhibited a significant lag phase at high ammonia. The possibility of growth of ammonia-sensitive strains with prolonged incubation cannot be excluded. For example, a small increase in nitrite/nitrate production was detected after incubation of strain L115 for 30 days (Fig. 1). However, in many environmental situations, the competitive advantage to be gained from ammonia tolerance will be of greatest value in the short term, with rapid response required before ammonia is utilized by other microorganisms or plants. In this respect, the lag phase observed for Nitrosospira sp. En13 is likely to have a significant influence on its competitiveness during transient increases in ammonia concentration, and it was therefore considered to be ammonia sensitive.
Methods for assessment of activity.
One aim of this study was to compare the abilities of four approaches—measurement of product formation, traditional DNA- and RNA-DGGE, and SIP—in detecting activities of ammonia-oxidizing bacteria. Detection of differences by DNA-DGGE requires sufficient growth for detectable changes in relative abundance, as indicated by relative band intensity. RNA-DGGE might be expected to increase sensitivity through earlier changes as increased ribosome synthesis will precede DNA replication, and changes may be accentuated through greater ribosome copy number. SIP, however, showed significant advantages over other methods for a number of reasons. First, it was capable of detecting ammonia-oxidizing activity through incorporation of [13C]CO2 into ammonia oxidizer 16S rRNA gene sequences before detection of significant changes in nitrite/nitrate concentrations. Second, the data demonstrate the relatively poor resolution of traditional DNA-DGGE approaches. Although changes were observed in the relative intensities of DNA-DGGE bands, corresponding to relative abundance of different community members, these occurred later than the appearance of bands in 13C-labeled DNA-SIP-DGGE profiles. More importantly, DNA-DGGE profiles do not distinguish between dormant and active strains or between increased band relative intensities due to growth of a particular strain or death of others. In contrast, SIP provides direct evidence of incorporation of labeled substrates. RNA-DGGE gave intermediate sensitivity, with detection of changes earlier than DNA-DGGE but later than SIP. RNA-DGGE, however, like DNA-DGGE, assesses only relative abundance and is therefore unable to distinguish between the increased activity of some strains and decreased activity of others.
The benefits of SIP must be balanced against the disadvantages associated with all molecular techniques, including biases associated with DNA extraction and PCR amplification, and the technical difficulties associated with SIP should not be underestimated. In addition, some applications of SIP require consideration of utilization of compounds produced as by-products or end products by primary utilizers of labeled substrates (24). However, for autotrophic ammonia oxidizers, uptake of organic by-products is likely to be negligible, and [13C]CO2 released through respiration will be assimilated only by active ammonia oxidizers, by nitrite oxidizers (if present), and, to a minor extent, by heterotrophic assimilation. Cross-feeding is therefore unlikely to be significant. There is also considerable potential for increased sensitivity of SIP analysis of ammonia oxidizers. DNA was diluted by at least one order of magnitude prior to fractionation, and sensitivity could be increased by use of RNA-SIP, greatly increasing target copy number. This suggests that, despite the additional problems associated with analysis of complex communities in environmental samples, the approach may enable determination of which ammonia oxidizers are active under different environmental conditions. The sensitivity achieved may also be sufficient for assessment of activity of ammonia oxidizers and other functional groups at substrate concentrations similar to those experienced in natural environments rather than at the high concentrations typically used for laboratory culture.
Interactions within communities.
The three communities investigated were designed to analyze interactions between closely related ammonia oxidizers, representative of those found in terrestrial environments. In general, relative activities within communities at high ammonia were predicted by those in monoculture, in that ammonia-tolerant cluster 3b strains always dominated, and, in communities A and C, activity of ammonia-sensitive strains could not be detected at the high initial ammonia concentration. In community B, activity of Nitrosospira 40KI (cluster 0) was detected, and this strain incorporated 13C at high ammonia, which inhibited its growth in monoculture. However, incorporation was not detected until incubation for 20 days and might have been detected in monoculture if SIP had been employed. Nevertheless, this suggests that there are potential interactions between these two strains and that strains within natural communities may behave differently from monocultures.
Cluster 3b strains also dominated activity at low and intermediate ammonia concentrations. Although activity of ammonia-sensitive strains could generally be detected at low ammonia, they never dominated mixed cultures. This was not predicted by growth in monoculture. For example, growth rates of monocultures En13 and N. briensis were similar at low and intermediate ammonia. However, in mixed culture (community A), N. briensis dominated both DNA-SIP and RT-PCR profiles at all ammonia concentrations. En13 was detected at low ammonia, but it was present at much lower relative abundance than N. briensis, and there was no evidence of growth or activity of En13 at intermediate ammonia. Similarly, for community C, monoculture growth suggested that strains Wyke2 (cluster 0) and Wyke8 (cluster 3a) would not compete well at the high ammonia concentration, as was observed, but they were outcompeted by strain EnI299 (cluster 3b) at intermediate ammonia even though monoculture growth characteristics suggested that they would be active. At low ammonia, although all strains within community C had similar growth rates in monoculture, only two strains, Wyke2 and EnI299, showed activity in SIP-DGGE and RT-PCR-DGGE profiles. These results indicate possible interspecies interactions between ammonia-oxidizing bacterial communities, perhaps through production of metabolites or growth factors that result in cooperation or competition.
In summary, results show that cluster 3b strains that grow rapidly at high ammonia concentrations in monocultures dominated all mixed communities at all ammonia concentration treatments. Cluster 3a and cluster 0 strains, characterized as ammonia sensitive when grown in monocultures, were inhibited at high ammonia concentrations in mixed communities and were probably outcompeted at intermediate ammonia concentrations by cluster 3b strains. However, coexistence of these strains with cluster 3b strains occurred at the lowest ammonia concentrations although they never outcompeted cluster 3b strains. High diversity in communities of soil AOB is often regarded as an indication of functional redundancy. However, the observed differences in competitiveness of the investigated AOB strains suggests that the cooccurrence of ammonia-sensitive Nitrosospira cluster 3a strains and more competitive cluster 3b strains in soil AOB results from different ecological strategies of the former strains, such as the occupation of different spatial or temporal niches, or from effects of factors other than ammonia concentration on ecological fitness. In addition, under some conditions, strains that were inhibited by high ammonia in monoculture were active at high ammonia in mixed cultures, where they coexisted with ammonia-tolerant strains.
Acknowledgments
M.T. acknowledges support from the Greek State Scholarship Foundation (IKY).
Footnotes
Published ahead of print on 12 February 2010.
REFERENCES
- 1.Avrahami, S., and R. Conrad. 2005. Cold-temperate climate: A factor for selection of ammonia oxidizers in upland soil? Can. J. Microbiol. 51:709-714. [DOI] [PubMed] [Google Scholar]
- 2.Belser, L. W., and E. L. Schmidt. 1980. Growth and oxidation kinetics of three genera of ammonia oxidizing nitrifiers. FEMS Microbiol. Lett. 7:213-216. [Google Scholar]
- 3.Bollmann, A., M. J. Bar-Gilissen, and H. J. Laanbroek. 2002. Growth at low ammonium concentrations and starvation response as potential factors involved in niche differentiation among ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 68:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cébron, A., L. Bodrossy, Y. Chen, A. C. Singer, I. P. Thompson, J. I. Prosser, and J. C. Murrell. 2007. Identity of active methanotrophs in landfill cover soil as revealed by DNA-stable isotope probing. FEMS Microbiol. Ecol. 62:12-23. [DOI] [PubMed] [Google Scholar]
- 5.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Freitag, T. E., L. Chang, and J. I. Prosser. 2006. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684-696. [DOI] [PubMed] [Google Scholar]
- 7.Griffiths, R. I., A. S. Whiteley, A. G. O'Donnell, and M. J. Bailey. 2000. Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66:5488-5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 9.Jia, Z., and R. Conrad. 2009. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ. Microbiol. 11:1658-1671. [DOI] [PubMed] [Google Scholar]
- 10.Keen, G. A., and J. I. Prosser. 1987. Steady-state and transient growth of autotrophic nitrifying bacteria. Arch. Microbiol. 147:73-79. [Google Scholar]
- 11.Koops, H. P., and A. Pommerening-Röser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 12.Kowalchuk, G. A., J. R. Stephen, W. De Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Roux, X., F. Poly, P. Currey, C. Commeaux, B. Hai, G. W. Nicol, J. I. Prosser, M. Schloter, E. Attard, and K. Klumpp. 2008. Effects of aboveground grazing on coupling among nitrifier activity, abundance and community structure. ISME J. 2:221-232. [DOI] [PubMed] [Google Scholar]
- 14.Mahmood, S., and J. I. Prosser. 2006. The influence of synthetic sheep urine on ammonia oxidizing bacterial communities in grassland soil. FEMS Microbiol. Ecol. 56:444-454. [DOI] [PubMed] [Google Scholar]
- 15.Mahmood, S., J. I. Prosser, and G. I. Paton. 2005. Cultivation-independent in situ molecular analysis of bacteria involved in degradation of pentachlorophenol in soil. Environ. Microbiol. 7:1349-1360. [DOI] [PubMed] [Google Scholar]
- 16.McCaig, A. E., L. A. Glover, and J. I. Prosser. 2001. Numerical analysis of grassland bacterial community structure under different land management regimens by using 16S ribosomal DNA sequence data and denaturing gradient gel electrophoresis banding patterns. Appl. Environ. Microbiol. 67:4554-4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muyzer, G., E. C. Dewaal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S ribosomal RNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicol, G. W., S. Leininger, C. Schleper, and J. I. Prosser. 2008. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10:2966-2978. [DOI] [PubMed] [Google Scholar]
- 19.Noll, M., P. Frenzel, and R. Conrad. 2008. Selective stimulation of type I methanotrophs in a rice paddy soil by urea fertilization revealed by RNA-based stable isotope probing. FEMS Microbiol. Ecol. 65:125-132. [DOI] [PubMed] [Google Scholar]
- 20.Padmanabhan, P., S. Padmanabhan, C. DeRito, A. Gray, D. Gannon, J. R. Snape, C. S. Tsai, W. Park, C. Jeon, and E. L. Madsen. 2003. Respiration of 13C-labeled substrates added to soil in the field and subsequent 16S rRNA gene analysis of 13C-labeled soil DNA. Appl. Environ. Microbiol. 69:1614-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patra, A. K., L. Abbadie, A. Clays-Josserand, V. Degrange, S. J. Grayston, N. Guillaumaud, P. Loiseau, F. Louault, S. Mahmood, S. Nazaret, L. Philippot, F. Poly, J. I. Prosser, and X. Le Roux. 2006. Effects of management regime and plant species on the enzyme activity and genetic structure of N-fixing, denitrifying and nitrifying bacterial communities in grassland soils. Environ. Microbiol. 8:1005-1016. [DOI] [PubMed] [Google Scholar]
- 22.Powell, S. J., and J. I. Prosser. 1986. Effect of copper on inhibition by nitrapyrin of growth of Nitrosomonas europaea. Curr. Microbiol. 14:177-179. [Google Scholar]
- 23.Prosser, J. I., and T. M. Embley. 2002. Cultivation-based and molecular approaches to characterisation of terrestrial and aquatic nitrifiers. Antonie Van Leeuwenhoek 81:165-179. [DOI] [PubMed] [Google Scholar]
- 24.Prosser, J. I., J. I. Rangel-Castro, and K. Killham. 2006. Studying plant-microbe interactions using stable isotope technologies. Curr. Opin. Biotechnol. 17:98-102. [DOI] [PubMed] [Google Scholar]
- 25.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 26.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: Implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radajewski, S., I. R. McDonald, and J. C. Murrell. 2003. Stable-isotope probing of nucleic acids: a window to the function of uncultured microorganisms. Curr. Opin. Biotechnol. 14:296-302. [DOI] [PubMed] [Google Scholar]
- 28.Rangel-Castro, J. I., J. I. Prosser, C. M. Scrimgeour, P. Smith, N. Ostle, P. Ineson, A. Meharg, and K. Killham. 2004. Carbon flow in an upland grassland: effect of liming on the flux of recently photosynthesized carbon to rhizosphere soil. Global Change Biol. 10:2100-2108. [Google Scholar]
- 29.Shaw, L. J., G. W. Nicol, Z. Smith, J. Fear, J. I. Prosser, and E. M. Baggs. 2006. Nitrosospira spp. can produce nitrous oxide via a nitrifier denitrification pathway. Environ. Microbiol. 8:214-222. [DOI] [PubMed] [Google Scholar]
- 30.Skinner, F. A., and N. Walker. 1961. Growth of Nitrosomonas europaea in batch and continuous culture. Arch. Mikrobiol. 38:339-349. [DOI] [PubMed] [Google Scholar]
- 31.Stephen, J. R., G. A. Kowalchuk, M. A. V. Bruns, A. E. McCaig, C. J. Phillips, T. M. Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suwa, Y., Y. Imamura, T. Suzuki, T. Tashiro, and Y. Urushigawa. 1994. Ammonia-oxidizing bacteria with different sensitivities to (N4)2SO4 in activated sludges. Water Res. 28:1523-1532. [Google Scholar]
- 33.Teske, A., E. Alm, J. M. Regan, S. Toze, B. E. Rittmann, and D. A. Stahl. 1994. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J. Bacteriol. 176:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tourna, M., T. E. Freitag, G. W. Nicol, and J. I. Prosser. 2008. Growth, activity and temperature responses of ammonia-oxidizing archaea and bacteria in soil microcosms. Environ. Microbiol. 10:1357-1364. [DOI] [PubMed] [Google Scholar]
- 35.Utåker, J. B., L. Bakken, Q. Q. Jiang, and I. F. Nes. 1996. Phylogenetic analysis of seven new isolates of ammonia-oxidizing bacteria based on 16S rRNA gene sequences. Syst. Appl. Microbiol. 18:549-559. [Google Scholar]
- 36.Webster, G., T. M. Embley, T. E. Freitag, Z. Smith, and J. I. Prosser. 2005. Links between ammonia oxidiser species composition, functional diversity and nitrification kinetics in grassland soils. Environ. Microbiol. 7:676-684. [DOI] [PubMed] [Google Scholar]