Abstract
The epiphyte Pseudomonas syringae pv. syringae 22d/93 (Pss22d) produces the rare amino acid 3-methylarginine (MeArg), which is highly active against the closely related soybean pathogen Pseudomonas syringae pv. glycinea. Since these pathogens compete for the same habitat, Pss22d is a promising candidate for biocontrol of P. syringae pv. glycinea. The MeArg biosynthesis gene cluster codes for the S-adenosylmethionine (SAM)-dependent methyltransferase MrsA, the putative aminotransferase MrsB, and the amino acid exporter MrsC. Transfer of the whole gene cluster into Escherichia coli resulted in heterologous production of MeArg. The methyltransferase MrsA was overexpressed in E. coli as a His-tagged protein and functionally characterized (Km, 7 mM; kcat, 85 min−1). The highly selective methyltransferase MrsA transfers the methyl group from SAM into 5-guanidino-2-oxo-pentanoic acid to yield 5-guanidino-3-methyl-2-oxo-pentanoic acid, which then only needs to be transaminated to result in the antibiotic MeArg.
Microbial plant pathogens cause severe losses in agriculture each year (1). For example, the plant pathogen Pseudomonas syringae pv. glycinea is responsible for bacterial blight of soybean, a leaf spot disease of great economic impact. Besides chemical treatment, biocontrol agents that antagonize microbial plant pathogens are gaining increasing importance in fighting plant diseases (6, 11, 27). In a screening for possible biocontrol strains, an epiphytic bacterium showing a strong and selective activity against the pathogen P. syringae pv. glycinea was isolated from soybean leaves (29). The strain was characterized as Pseudomonas syringae pv. syringae 22d/93 (Pss22d). The antagonism of Pss22d against P. syringae pv. glycinea has been demonstrated successfully in vitro and in planta under greenhouse and field conditions (19, 29). In order to identify the molecular basis of the antagonism of Pss22d against P. syringae pv. glycinea, we focused on its secondary metabolites. Besides the well-known lipodepsipeptides syringomycin and syringopeptin (3), Pss22d produces the rare amino acid 3-methylarginine (MeArg) (5). As little as 20 nmol of MeArg strongly and selectively inhibits P. syringae pv. glycinea but no other pseudomonads in vitro (29). Since the inhibition can be compensated for by l-arginine supplementation but not by any other essential amino acid, it is likely that the toxin acts as an inhibitor of the arginine biosynthesis pathway or an arginine-dependent pathway, such as nitric oxide formation (13, 16). Feeding experiments and Tn5 transposon mutagenesis suggested that MeArg is produced by an S-adenosyl methionine (SAM)-dependent methyltransferase (5) converting the enol of 5-guanidino-2-oxo-pentanoic acid to 5-guanidino-3-methyl-2-oxo-pentanoic acid. An analogous reaction is known to occur with the methyltransferases GlmT, DptI, and LptI, which form 3-methylglutamate from α-ketoglutarate (18). On the way to MeArg, only a transaminase catalyzing the formation of MeArg from 5-guanidino-3-methyl-2-oxo-pentanoic acid and an amino acid exporter to secrete the toxin would be needed.
Here, we describe the identification and functional characterization of the MeArg biosynthesis gene cluster from the epiphyte Pss22d.
MATERIALS AND METHODS
Strains, plasmids, and cultivation conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. syringae strains were cultured and maintained on King's B (KB) medium (17). The Pss22d mutants were cultivated on selective KB medium containing spectinomycin (25 μg/ml). The complemented strain P. syringae pv. syringae 22d/93.1C (Pss22d.1C) was grown on KB agar containing kanamycin (25 μg/ml). Escherichia coli strains were maintained on Standard 1 medium (Merck). The E. coli strains 3150 (Ec3150) and 2795 (Ec2795) were cultivated on Standard 1 agar containing ampicillin (25 μg/ml).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Pseudomonas syringae pv. syringae strains | ||
| Pss22d | Wild type, isolated from soybean | 46 |
| Pss22d.1 | Spr, transposon mutant of Pss22d, Tn5 insertion in methyltransferase gene mrsA | 6 |
| Pss22d.1C | Spr Kmr, Pss22d.1 harboring plasmid pB3150 for complementation of MeArg production | This study |
| Pseudomonas syringae pv. glycinea strain | ||
| Psg1a | Wild type, isolated from soybean | 46 |
| Escherichia coli strains | ||
| Ec2795 | DH5α containing plasmid pG2795 | This study |
| Ec3150 | DH5α containing plasmid pG3150 | This study |
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 17 |
| BL21(DE3) | F−dcm ompT lon hsdSB(rB− mB−) gal λ (DE3) | Novagen |
| S17-1λpir | recA thi pro hsdR M+, RP4:2-Tc:Mu:Km Tn7, λpir, Tpr Smr | 48 |
| Plasmids | ||
| pMKm | Source of Kmr cassette | 33 |
| pGEM-T Easy | Cloning vector, Apr | Promega |
| pJet 1.2 | Cloning vector, Apr | Fermentas |
| pBBR1MCS | Cloning vector, Cmr | 27 |
| pET28b(+) | Cloning vector, Kmr | Novagen |
| pUT/mini-Tn5 Sm/Sp | Smr Spr Apr, contains mini-Tn5 | 10 |
| pG2795 | Apr, contains a 2,795-bp fragment carrying the MeArg biosynthesis cluster of Pss22d without the exporter gene mrsC in pGEM-T Easy | This study |
| pJ2795 | Apr, contains a 2,795-bp fragment carrying the MeArg biosynthesis cluster without the exporter gene mrsC in pJet 1.2 | This study |
| pG1342 | Apr, contains a 1,343-bp adaptor PCR fragment carrying a part of the exporter gene mrsC of Pss22d in pGEM-T Easy | This study |
| pJ3150 | Apr, contains a 3,150-bp PCR fragment carrying the MeArg biosynthesis cluster of Pss22d in pJet 1.2 | This study |
| pG3150 | Apr, contains a 3,150-bp PCR fragment carrying the MeArg biosynthesis cluster of Pss22d in pGEM-T Easy | This study |
| pB3150 | Cmr, contains a 3,150-bp PCR fragment carrying the MeArg biosynthesis cluster of Pss22d in pBBR1MCS | This study |
| pET28b(+)-MrsA | Kmr, contains a 1,053-bp PCR fragment carrying the methyltransferase gene mrsA of Pss22d | This study |
Agar diffusion assay.
In order to test for MeArg production by P. syringae pv. syringae strains and Pss22d mutants, agar diffusion assays with P. syringae pv. glycinea 1a/96 (Psg1a) as the indicator strain were performed (5). Briefly, Psg1a was cultured on KB agar plates overnight at 28°C. Single colonies of Psg1a were used to prepare a suspension of approximately 4 × 108 CFU/ml in sterile water. For preparation of test plates (diameter, 135 mm), 50 ml of 5b agar medium (10) was warmed to 50°C and supplemented with 2 ml of the Psg1a suspension. The strains to be tested were inoculated onto the agar plates, and the plates were incubated at 28°C for 24 h. To determine the relative toxin concentration, a standard curve using purified MeArg was generated (4). Levels of syringomycin and syringopeptin production were determined using agar diffusion assays against Geotrichum candidum and Bacillus megaterium as indicator strains, respectively (26).
Tn5 mutagenesis.
Transposon mutagenesis of Pss22d was performed by two-parent matings using E. coli S17-1λpir containing the plasmid pUT/mini-Tn5 Sm/Sp (30) as the donor strain. Plate matings were carried out on Standard 1 agar at 28°C overnight. Pss22d mutants were selected on MG agar (15) containing spectinomycin (25 μg/ml) as the selection agent. The methyltransferase mrsA transposon mutant P. syringae pv. syringae 22d/93.1 (Pss22d.1) (Table 2) was identified by screening for the loss of antibiotic activity against P. syringae pv. glycinea in the agar diffusion assay, followed by liquid chromatography-mass spectrometry (LC-MS) analysis (5). The growth rates of Pss22d and the mutant Pss22d.1 in HSC medium (21) at 28°C were determined by measuring the optical density at 578 nm (OD578).
TABLE 2.
Characterization of Pss22d, Pss22d.1, and the complemented mutant Pss22d.1C as well as E. coli strains Ec2795 and Ec3150 containing 3-methylarginine biosynthesis genese
| Strain | 3-Methylarginine concn (μg/ml) at 24 ha |
Growth (OD578) at 24 h | SMc | SPd | |
|---|---|---|---|---|---|
| Without arginine | With arginineb | ||||
| Pseudomonas syringae pv. syringae strains | |||||
| Pss22d (wild type) | 74.7 ± 13.0 | 0.0 ± 0.0 | 7.3 ± 0.1 | + | + |
| Pss22d.1f | 0.0 ± 0.0 | 0.0 ± 0.0 | 7.3 ± 0.1 | + | + |
| Pss22d.1C | 61.1 ± 5.2 | 0.0 ± 0.0 | 8.2 ± 0.1 | + | + |
| Escherichia coli strains | |||||
| Ec2795 | 0.0 ± 0.0 | 0.0 ± 0.0 | 2.9 ± 0.1 | − | − |
| Ec3150 | 4.3 ± 0.0 | 0.0 ± 0.0 | 3.0 ± 0.1 | − | − |
Concentrations were estimated by a standard curve (6).
5b agar medium (10) supplemented with 0.1 mM l-arginine.
SM, syringomycin halo (+, like that of the wild type; −, no production).
SP, syringopeptin halo (+, like that of the wild type; −, no production).
Values for 3-methylarginine concentrations and growth levels are given as means ± standard deviations.
The predicted function of the disrupted gene of mutant Pss22d.1 was that of a SAM-dependent methyltransferase.
Cloning and sequencing of the 3-methylarginine biosynthesis cluster.
Genomic DNA was isolated following standard procedures (24). Preparation of plasmid DNA was performed using a Qiagen plasmid mini kit (Qiagen, Hilden, Germany). The insertion sites of Tn5 in the Pss22d mutants were determined by cloning the insertion loci into the plasmid pBBR1MCS and sequencing of the insert (5). A 265-bp fragment flanking the Tn5 insertion in the MeArg-deficient mutant Pss22d.1 showed high homology to SAM-dependent methyltransferases and was identified as part of the MeArg biosynthesis gene cluster (5). The noncoding region upstream of the methyltransferase gene was used to design the reverse primer MTrev (5′-ATCAGGAATGCGGCACTACA-3′). Analysis of the sequences surrounding the methyltransferase gene in the genome sequence of P. syringae pv. syringae B728a (PssB728a) revealed the presence of two open reading frames (ORFs) showing high similarity to genes coding for an aminotransferase and an amino acid exporter. The genome sequence of PssB728a was used to design the forward primer LTfwd (5′-TTGGATTCGCGGTTTCGA-3′), binding a sequence in the amino acid exporter gene. A 2,795-bp fragment was PCR amplified from genomic DNA of Pss22d using the primers LTfwd and MTrev and cloned into pJET 1.2 (Fermentas, St. Leon-Rot, Germany), yielding plasmid pJ2795, which was sequenced using standard primers from the pJET 1.2 kit. Additionally, the 2,795-bp fragment was cloned into pGEM T-easy (Promega, Mannheim, Germany), yielding plasmid pG2795. To clone the complete biosynthesis cluster, an adaptor PCR was performed (22). Genomic DNA of Pss22d was digested with the restriction enzyme BclI and subsequently ligated with the BclI adaptor (5′-GATCCCCTATAGTGAGTCGTATTAAC-3′ [restriction site underlined]) overnight. Using the adaptor primer (5′-CCCTATAGTGAGTCGTATTAAC-3′) and primer LTrev (5′-ATTCCGTGGCTCTGAAGT-3′), derived from the sequence of the 2,795-bp fragment in pJ2795, a 1,342-bp fragment was PCR amplified from genomic DNA of Pss22d and cloned into pGEM T-easy, yielding plasmid pG1342. The primers MeArgfwd (5′-GTGAATCACGCCTCGAAG-3′) and MeArgrev (5′-GGAGAGTCTGAATTTTTGCG-3′) were deduced from the sequences of pJ2795 and pG1342 and used to PCR amplify the complete MeArg biosynthesis gene cluster of Pss22d. The amplified 3,150-bp fragment was cloned into pJET 1.2, yielding plasmid pJ3150, which was sequenced using standard primers from the pJET 1.2 kit. For complementation of MeArg-deficient mutants, the 3,150-bp fragment was first cloned into pGEM T-Easy, yielding plasmid pG3150. Subsequently, a 3,150-bp XhoI-XbaI fragment cut from pJ3150 was ligated into XhoI-XbaI-digested pBBR1MCS, yielding plasmid pB3150.
In silico analysis of the 3-methylarginine biosynthetic genes.
The nucleotide sequence of the putative MeArg biosynthetic gene cluster was translated into the corresponding amino acid sequences and analyzed using the BLASTP program from the National Center for Biotechnology Information. The fragment cloned in pJ3150 was compared to the genome sequence of PssB728a using VectorNTI software (Invitrogen, Karlsruhe, Germany), ClustalX2 (28), and TreeViewX (20). Promoter site prediction was performed with BPROM software (Softberry Inc., Mount Kisco, NY).
Cloning of the SAM-dependent methyltransferase MrsA.
The SAM-dependent methyltransferase MrsA was PCR amplified from genomic DNA of Pss22d using the primers MTfwd (5′-CATATGAATCTGCTTGACTCTATAAA-3′) and MTrev (5′-CTCGAGTCATGCCCTCCGCAGCAGATA-3′) (the start and stop codons are shown in bold, and the NdeI and XhoI restriction sites are underlined). The amplified 1,060-bp fragment was digested with NdeI and XhoI and cloned into NdeI-XhoI-digested pET28b(+) (Novagen), yielding plasmid pET28b(+)-MrsA. The identity of the insert was confirmed by sequencing (JenaGen, Jena, Germany).
Overproduction and purification of MrsA.
To overproduce the SAM-dependent methyltransferase MrsA, the plasmid pET28b(+)-MrsA was transformed into E. coli BL21(DE3). One liter of Standard 1 medium containing kanamycin (25 μg/ml) was inoculated with 20 ml of an overnight culture and incubated at 180 rpm at 37°C. At an OD578 of 0.9 to 1.3, cultures were induced with 0.2 ml of 1 M isopropyl-β-d-thiogalactopyranoside (IPTG) and then incubated at 16°C to an OD578 of 1.8 to 2.0. The cells were harvested by centrifugation. After resuspension of the cells in 50 ml binding buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.9, 5 mM imidazole, 10% glycerol), the cells were broken using a French pressure cell press (SIM-AMINCO; Spectronic Instruments Inc., Rochester, NY). The cell lysate was centrifuged (8,000 × g, 4°C, 40 min), and the supernatant was loaded onto Ni2+-nitrilotriacetate resin (Qiagen, Hilden, Germany) preequilibrated with binding buffer. The column was washed with 10 ml binding buffer and 4 ml wash buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.9, 60 mM imidazole, 10% glycerol). To elute the methyltransferase, 6 ml of elution buffer (20 mM Tris-HCl, 500 mM NaCl, pH 7.9, 200 mM imidazole, 10% glycerol) was added. Imidazole was removed by buffer exchange to 100 mM NaCl, 50 mM HEPES, pH 7.5, using Vivaspin-6 concentrators (30,000 molecular weight cutoff; Vivascience, Hannover, Germany) (6,000 × g, 4°C, 60 min). The purity of the methyltransferase MrsA was determined by SDS-PAGE and electrospray ionization mass spectrometry (ESI-MS).
Generation of 5-guanidino-2-oxo-pentanoic acid.
Fifty milligrams of d-arginine in 5 ml phosphate buffer (30 mM, pH 8.3) was incubated with 46 U of d-amino acid oxidase from porcine kidney (Sigma, Deisenhofen, Germany) and 29,500 U of catalase from bovine liver (Sigma, Deisenhofen, Germany) for 24 h. After precipitation of the enzyme by acidification (pH 1.0) and vortexing with 5 ml CH2Cl2, the aqueous layer was concentrated and subjected to medium-pressure liquid chromatography (MPLC) (Büchi, Flawil, Switzerland) purification using RP-18 resin and gradient elution (solvent A, H2O; solvent B, methanol [MeOH]). For LC-MS analysis, an Agilent HP1100 high-pressure liquid chromatography (HPLC) system (Agilent, Waldbronn, Germany) hooked to an LTQ mass spectrometer (Thermo Fisher, Egelsbach, Germany) fitted with a Luna NH2-HPLC column (150 mm by 2 mm, 5 μm; Phenomenex, Aschaffenburg, Germany) under hydrophobic interaction chromatography (HILIC) (5) conditions was used. The details of the HPLC program are as follows: 3 min at 100% solvent A (MeCN, 0.1% acetic acid [AcOH]), followed by 27 min at 100% solvent B (H2O, 0.1% AcOH) (flow rate, 0.2 ml/min). High-resolution electrospray ionization tandem mass spectrometry (HR-ESI-MS-MS) experiments were performed by direct injection of purified samples into an LTQ Orbitrap (Thermo Fisher, Egelsbach, Germany).
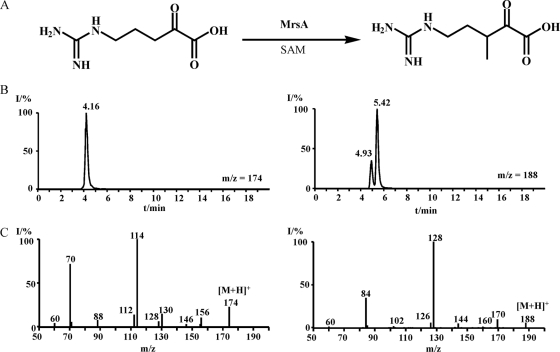
5-Guanidino-2-oxo-pentanoic acid eluted at 23.0 min; its mass spectral data are as follows: HR-ESI-MS [M+H]+ measured 174.08748, calculated 174.08787; ESI-MS-MS of [M+H]+ 60 (2), 70.06532 (41), 114.05496 (100), 130.09769 (8), 156.07690 (12).
Functional characterization of MrsA.
In order to determine the methyltransferase activity required for the conversion of 2-oxo acids (5-guanidino-2-oxo-pentanoic acid, pyruvate, phenylpyruvate, and α-ketoglutarate) to 3-methyl-2-oxo acids, approximately 0.6 μM of the methyltransferase was mixed with 2-oxo acids (1.6 mM) and S-adenosylmethionine (SAM) (1.6 mM), with 100 μl reaction buffer (glycine buffer, 30 mM, pH 8.5) (145-μl final volume). The reaction mixture was incubated at 28°C for 40 min. Control reactions were performed without enzyme, SAM, or 2-oxo acids. All incubations were stopped by addition of 10 μl HCl (10 M) and 50 μl CH2Cl2 and vortexing. The conversion of 5-guanidino-2-oxo-pentanoic acid to 5-guanidino-3-methyl-2-oxo-pentanoic acid at different pH and temperature values was analyzed by LC-ESI-MS-MS comparing the peak ratios of 2-oxo acid to those of 3-methyl-oxo acid (for HPLC conditions, see above). The pH optimum of the methyltransferase (3.5 μg) was studied over a pH range of 1.0 to 11.0 using 100 μl citrate buffer (30 mM, pH 1 to 5), 100 μl phosphate buffer (30 mM, pH 5 to 8.5), and 100 μl glycine buffer (30 mM, pH 8.5 to 11) (145-μl final volume). The temperature dependence of the enzyme was analyzed from 10°C to 60°C (30 mM phosphate buffer, pH 8.0).
For nuclear magnetic resonance (NMR) analysis of 5-guanidino-3-methyl-2-oxo-pentanoic acid, 4 mg 5-guanidino-2-oxo-pentanoic acid (23 mmol), 10 mg SAM (23 mmol), and 60 μg enzyme were incubated in phosphate buffer (30 mM, pH 8.3) overnight. After precipitation of the enzyme by vortexing with CHCl3, the sample was centrifuged, and the aqueous layer was concentrated in vacuo and subjected to MPLC separation followed by HPLC purification (see above). NMR measurements were performed with a Bruker DRX 500 spectrometer (Bruker, Rheinstetten, Germany) using the D2O signal for calibration (D2O, 4.68 ppm).
The product of the MrsA reaction eluted at 22.0 min; its mass spectral and NMR data are as follows: HR-ESI-MS [M+H]+ measured 188.10320, calculated 188.10352; ESI-MS-MS of [M+H]+ 60 (0.2), 84.08094 (11), 128.07075 (100), 144.11314 (1.5), 170.09250 (7.7); 1H NMR (500 MHz, D2O) δ 0.85 (d, J = 6.85, 3H), 1.51 to 1.61 (m, 1H), 1.93 to 2.01 (m, 1H), 2.25 to 2.34 (m, 1H), 3.28 to 3.36 (m, 1H), 3.44 to 3.51 (m, 2H).
Kinetics of MrsA.
Kinetic data for MrsA were determined by LC-MS using the peak area of the base peak ions of the MS-MS spectra of 5-guanidino-2-oxo-pentanoic acid (m/z = 114) and 5-guanidino-3-methyl-2-oxo-pentanoic acid (m/z = 128) to determine their amounts at different time points (0.5, 1, 3, 5, 10, 15, 30, 45, and 60 min) using 3.3 μM MrsA and the substrates 5-guanidino-2-oxo-pentanoic acid and SAM both at the same concentration (0.4, 1.2, 2.0, 4.6, or 7.7 mM) per assay, with 100 μl phosphate buffer (30 mM, pH 9) at 45.0°C (150-μl final volume). Km was determined after calculation of the reaction rates at the different substrate concentrations by use of the Lineweaver-Burk plot.
RESULTS
Characterization of Pss22d Tn5 mutants with altered 3-methylarginine production.
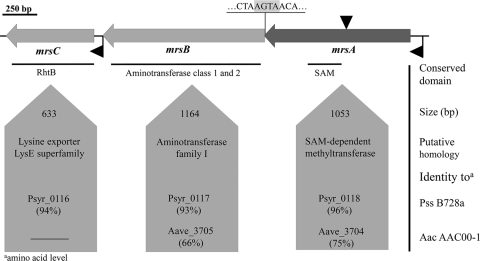
To identify the genes involved in MeArg biosynthesis, mini-Tn5 mutagenesis was carried out. Out of 2,468 Tn5 mutants, 21 were affected in MeArg production, but only the mutant Pss22d.1 did not produce MeArg although its lipodepsipeptide formation was comparable to that of the wild type. Sequencing of the DNA flanking the mini-Tn5 insert from Pss22d.1 revealed that the insertion was located in a gene with high similarity to genes coding for SAM-dependent methyltransferase from PssB728a (Psyr_0118; 96% identity, GenBank accession no. AAY35192) and Acidovorax avenae subsp. citrulli (Aave_3704; 75% identity, GenBank accession no. ABM34252) (Fig. 1). Due to the high homology of the disrupted gene to SAM-dependent methyltransferases, the mutant Pss22d.1 was a promising candidate for a MeArg biosynthesis mutant because this enzyme function is necessary to produce MeArg (5).
FIG. 1.
3-Methylarginine biosynthesis gene cluster from P. syringae pv. syringae 22d/93, i.e., mrsA (GenBank accession no. FJ788104), mrsB (GenBank accession no. FJ788104), and mrsC (GenBank accession no. FJ788104) genes, coding for a SAM-dependent methyltransferase, an aminotransferase, and a 3-methylarginine exporter, respectively. The lines below the physical map represent conserved domains identified by a BLASTP search (Psyr_0116, AAY35190; Psyr_0117, AAY35191; Psyr_0118, AAY35192; Aave_3705, ABM34253; Aave_3704, ABM34252). The insertion site of the mini-Tn5 in the mrsA gene of the Pss22d.1 mutant is indicated (▾). Predicted sites for putative promoters were analyzed with BPROM software and are also indicated (◂). The sizes and homologies of the ORFs are given.
Analysis of the 3-methylarginine biosynthesis gene cluster.
Analysis of the sequences surrounding the methyltransferase gene in PssB728a revealed the presence of two ORFs showing high similarity to genes coding for aminotransferases and amino acid exporters. Thus, the sequences of PssB728a were used to design primers to PCR amplify the putative MeArg biosynthesis gene cluster from genomic DNA of Pss22d. Because no significant similarities were found in the sequences downstream of the exporter gene, adaptor PCR was used to identify the downstream sequence in Pss22d. Finally, two primers, MeArgfwd and MeArgrev, were designed to amplify a 3,150-bp fragment from Pss22d containing the complete MeArg biosynthesis cluster. The three ORFs were designated mrsA, mrsB, and mrsC (for 3-methylarginine synthesis). Analysis of the deduced amino acid sequences indicated high degrees of similarity between the proteins from Pss22d and PssB728a (93 to 96% identity) (Fig. 1). Moreover, MrsA and MrsB showed significant sequence homology to a putative methyltransferase (Aave_3704; 75% identity, GenBank accession no. ABM34252) and a putative aminotransferase (Aave_3705; 66% identity, GenBank accession no. ABM34253), respectively, from Acidovorax avenae subsp. citrulli AAC00-1. However, no homologue of MrsC was found in A. avenae subsp. citrulli (Fig. 1).
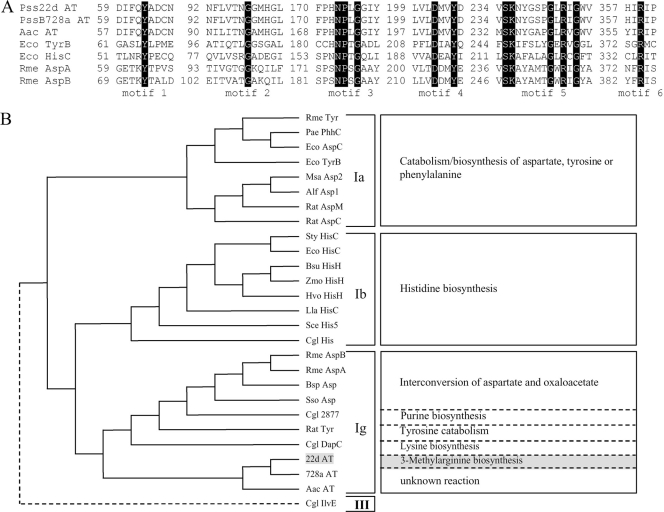
MrsA was predicted to be a SAM-dependent methyltransferase with a conserved SAM domain belonging to the type 12 family (6). MrsB contains the motifs typical of an aminotransferase of family I (15) (Fig. 2A). From the multiple sequence alignment with members of aminotransferase family I, we deduced that MrsB belongs to the subfamily Iγ (14). This subfamily contains aminotransferases catalyzing the formation of different amino acids, e.g., lysine and tyrosine (Fig. 2B) (9, 14). MrsC shows high similarity to a putative lysine transporter from PssB728a (8) and belongs to the LysE superfamily. A detailed motif analysis revealed that MrsC groups to the RhtB subfamily (2) (Fig. 3).
FIG. 2.
(A) Sequence alignment for members of aminotransferase superfamily I. Abbreviations (GenBank or SwissProt accession numbers): Pss22d AT, P. syringae pv. syringae 22d/93 aminotransferase MrsB (FJ788104); PssB728a AT, P. syringae pv. syringae B728a aminotransferase Psyr_117 (YP_233229); Aac AT, A. avenae subsp. citrulli aminotransferase (YP_972027); Eco TyrB, E. coli aromatic aminotransferase (X03628); Eco HisC, E. coli imidazole acetol phosphate (IAP) aminotransferase (P06986); Rme AspA, Rhizobium meliloti aspartate aminotransferase A (L05064); Rme AspB, Rhizobium meliloti aspartate aminotransferase B (L12149). (B) Dendrogram of the class I aminotransferase superfamily, generated with the ClustalX multiple-alignment program. The primary function(s) of each aminotransferase is indicated (right). Abbreviations (GenBank or SwissProt accession numbers): Rme Tyr, Rhizobium meliloti aromatic aminotransferase (L05065); Pae PhhC, Pseudomonas aeruginosa aromatic aminotransferase (M88627); Eco AspC, E. coli aspartate aminotransferase (X03629); Eco TyrB, E. coli aromatic aminotransferase (X03628); Msa Asp2, Medicago sativa aspartate aminotransferase (AAB46611); Alf Asp1, alfalfa aspartate aminotransferase 1 (P28011); Rat AspM, rat mitochondrial aspartate aminotransferase (M18467); Rat AspC, rat cytosolic aspartate aminotransferase (J04171); Sty HisC, Salmonella enterica serovar Typhimurium HisC (J01804); Eco HisC, E. coli IAP aminotransferase (P06986); Bsu HisH, Bacillus subtilis IAP aminotransferase (P17731); Zmo HisH, Zymomonas mobilis IAP aminotransferase (L36343); Hvo HisH, Haloferax volcanii IAP aminotransferase (M33161); Lla HisC, Lactococcus lactis IAP aminotransferase (M90760); Sce His5, Saccharomyces cerevisiae IAP aminotransferase (X05650); Cgl His, Corynebacterium glutamicum histidine biosynthesis (AY238320); Rme AspA, Rhizobium meliloti aspartate aminotransferase A (L05064); Rme AspB, Rhizobium meliloti aspartate aminotransferase B (L12149); Bsp Asp, Bacillus sp. aspartate aminotransferase (P23034); Sso Asp, Sulfolobus solfataricus aspartate aminotransferase (X16505); Cgl 2877, Corynebacterium glutamicum purine synthesis (AY238316); Rat Tyr, rat tyrosine aminotransferase (X02741); Cgl DapC, Corynebacterium glutamicum lysine biosynthesis (AY170830); Cgl IlvE, Corynebacterium glutamicum IlvE (AF424637).
FIG. 3.
Multiple sequence alignment for RhtB and LysE amino acid transporters. Both subfamilies are members of the LysE superfamily and differ in the shown motifs. Abbreviations of the exporters (GenBank accession numbers): Pss22d Exp, P. syringae pv. syringae 22d/93 MrsC (FJ788104); PssB728a Exp, P. syringae pv. syringae B728a lysine exporter protein LysE/YggA (YP233228); Ec YeaS, E. coli neutral amino acid efflux system protein (NP416312); Pa YcaR, Pseudomonas aeruginosa leucine export protein (NP253445); Ec RhtC, E. coli threonine efflux system protein (YP026264); Ps CmaU, Pseudomonas syringae biosynthesis of coronamic acid protein (AAC46034); Ah YggA, Aeromonas hydrophila putative amino acid transporter YggA (SwissProt no. P52047); Ec YggA, E. coli arginine exporter protein (NP289490).
A precise analysis of the sequences of mrsA and mrsB revealed an overlapping region in the start/stop sequences (mrsA-ATGA-mrsB) (Fig. 1). This suggests that mrsA and mrsB are translationally coupled, which was supported by the finding of only one putative promoter region (Fig. 1). Downstream of the MeArg biosynthesis cluster, a gene with high similarity to a putative transposase from PssB728a was found. Upstream of the MeArg biosynthesis cluster, an ORF with similarity to a gene coding for a mannuronan C-5 epimerase from Pseudomonas syringae pv. tomato (41% amino acid identity, GenBank accession no. AAO57541) was identified.
Complementation of the mutant Pss22d.1 and heterologous expression of the entire MeArg biosynthesis gene cluster in E. coli.
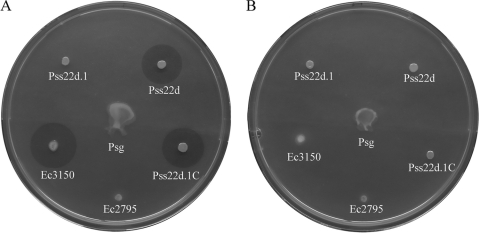
The MeArg-deficient mutant Pss22d.1 was complemented with the plasmid pB3150, harboring the genes mrsA, mrsB, and mrsC. This complementation completely restored MeArg production in Pss22d.1, resulting in a growth inhibition of Psg1a in the agar diffusion assay similar to that of the wild-type strain Pss22d (Table 2; Fig. 4). Moreover, the high-copy-number plasmid pG3150, containing the MeArg biosynthesis genes mrsA, mrsB, and mrsC from Pss22d, was introduced into E. coli DH5α. The resulting strain, Ec3150, produced MeArg. In HSC liquid medium Ec3150 generated only small amounts of MeArg (Table 2), but on 5b agar plates Ec3150 produced amounts of MeArg similar to those produced by the parent strain Pss22d, causing comparable inhibition zones in the agar diffusion assay against P. syringae pv. glycinea (Fig. 4). The inhibition zones in the agar diffusion assay caused by Ec3150 were completely reversed after addition of 0.1 mM arginine (Fig. 4). Furthermore, the production of MeArg was verified by LC-MS-MS.
FIG. 4.
Agar diffusion bioassays (5b agar medium [10]) performed with the indicator strain Psg1a (sensitive to 3-methylarginine) without l-arginine (A) and with 0.1 mM l-arginine (B), which compensates for the toxicity of 3-methylarginine (5). Pss22d, Pseudomonas syringae pv. syringae wild-type strain; Pss22d.1, methyltransferase Tn5 mutant; Pss22d.1C, methyltransferase Tn5 mutant complemented with the plasmid pB3150 harboring the genes mrsA, mrsB, and mrsC; Ec2795, Escherichia coli DH5α complemented with the plasmid pG2795 harboring the genes mrsA and mrsB; Ec3150, E. coli DH5α complemented with the plasmid pG3150 harboring the genes mrsA, mrsB, and mrsC; Psg, Pseudomonas syringae pv. glycinea wild-type strain.
Heterologous expression and functional characterization of the SAM-dependent methyltransferase MrsA.
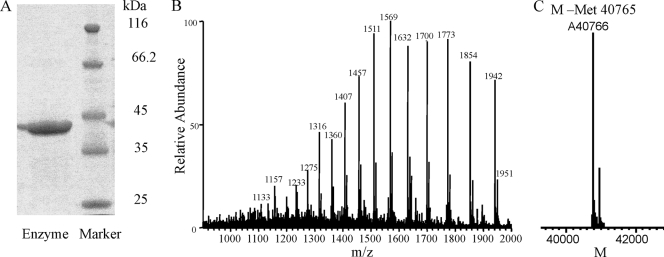
In order to characterize the key enzyme in the biosynthesis of 3-methylarginine, we overexpressed the SAM-dependent methyltransferase MrsA as a His-tagged protein in E. coli BL21(DE3) using plasmid pET28b(+)-MrsA. Overexpression and enzyme purification yielded 7 mg/ml pure protein, as can be seen by SDS-PAGE (Fig. 5A). ESI-MS analysis confirmed the identity of the protein, with the determined protein mass minus methionine (M-Met) of 40,766 Da being in very good accordance with the calculated mass of 40,765 Da (Fig. 5B).
FIG. 5.
SDS-PAGE (A), ESI-MS (B), and deconvoluted mass (C) of the SAM-dependent methyltransferase MrsA, involved in 3-methylarginine biosynthesis.
The methyltransferase MrsA efficiently converts 5-guanidino-2-oxo-pentanoic acid to 5-guanidino-3-methyl-2-oxo-pentanoic acid in the presence of SAM (Fig. 6). Control reactions without enzyme or without one of the substrates did not show any product formation. The enzyme has its pH optimum at pH 9. The catalytic activity decreases 3-fold at pH 7 and 10-fold at pH 10. At 45°C maximal conversion was achieved, whereas at the physiological temperature of 28°C the enzyme activity decreased 3-fold compared to the optimum. The methylation of 5-guanidino-2-oxo-pentanoic acid is fast, with a Km of 7 mM and a kcat of 85 min−1.
FIG. 6.
Conversion of 5-guanidino-2-oxo-pentanoic acid into 5-guanidino-3-methyl-2-oxo-pentanoic acid by the SAM-dependent methyltransferase MrsA. Reaction scheme (A), LC-MS ion traces at m/z 174 and m/z 188 (B), and ESI-MS-MS (C) for 5-guanidino-2-oxo-pentanoic acid and 5-guanidino-3-methyl-2-oxo-pentanoic acid. I/%, relative intensity (percent).
Use of other 2-oxo acids, such as pyruvate, α-ketoglutarate, and phenylpyruvate, as the substrate did not result in any 3-methyl-2-oxo acid formation.
DISCUSSION
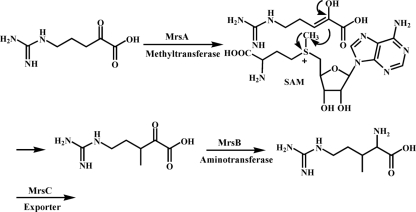
By use of Tn5 transposon mutagenesis, the 3-methylarginine-deficient mutant Pss22d.1 was obtained and served to identify a 3-kb gene cluster responsible for the biosynthesis of MeArg. The gene cluster contained three ORFs designated mrsA, mrsB, and mrsC. MrsA showed high homology to SAM-dependent methyltransferases, MrsB was identified as a putative aminotransferase belonging to the subfamily Iγ, and MrsC was assigned to code for a 3-methylarginine exporter due to its similarity to amino acid exporters (2) (Fig. 1). The SAM-dependent methyltransferase MrsA methylates 5-guanidino-2-oxo-pentanoic acid to 5-guanidino-3-methyl-2-oxo-pentanoic acid. Subsequently, the putative aminotransferase MrsB transaminates 5-guanidino-3-methyl-2-oxo-pentanoic acid to result in 3-methylarginine, which is secreted via the MeArg exporter MrsC (Fig. 7).
FIG. 7.
Suggested biosynthetic pathway of 3-methylarginine formation in Pss22d.
The involvement of the three genes in MeArg biosynthesis was proven by complementation of the mutant Pss22d.1 with the plasmid pB3150 harboring the entire MeArg biosynthesis gene cluster (mrsA, mrsB, and mrsC). The complemented mutant showed a clear inhibition of P. syringae pv. glycinea in the agar diffusion assay. In addition, E. coli DH5α bearing the plasmid pG3150 was shown to produce MeArg by use of the agar diffusion assay and LC-MS for detection. In contrast, E. coli DH5α bearing plasmid pG2795, which harbors only the genes mrsA and mrsB, did not cause an inhibition zone in the agar diffusion assay, which suggests the function of MrsC as a MeArg exporter (Fig. 4). The complementation experiments are in line with the high homology of MrsC to LysE exporters, which serve to export the basic amino acids l-lysine and l-arginine (7).
The deduced genes from the MeArg biosynthesis cluster of Pss22d showed a high similarity to genes from PssB728a. However, in spite of PssB728a containing a similar set of genes, this strain did not produce MeArg. A possible reason for this is a lack of function of the genes (96% identity to mrsA, 93% identity to mrsB, and 94% identity to mrsC at the protein level) or the possibility that PssB728a does not express the corresponding proteins under the growth conditions used. Alternatively, another related product may be produced by PssB728a.
In order to study the MeArg biosynthesis cluster of Pss22d in more detail, we heterologously expressed its key enzyme, the methyltransferase MrsA, as a His-tagged protein in E. coli. The purified enzyme converted 5-guanidino-2-oxo-pentanoic acid to 5-guanidino-3-methyl-2-oxo-pentanoic acid in the presence of SAM as the methyl group donor (Km, 7 mM; kcat, 85 min−1) (Fig. 6). The Km of MrsA is about 100 times higher than that of the related GlmT, a SAM-dependent methyltransaminase that produces (2S,3R)-3-methylglutamate, and the kcat of MrsA is 850 times higher than that of GlmT (0.11 min−1) (18). Similarly to other related methyltransferases, such as GlmT, DptI, and LptI, which convert α-ketoglutarate to 3-methylglutamate in a highly selective manner (18), MrsA is specific for its substrate and did not accept any other substrates (pyruvate, α-ketoglutarate, or phenylpyruvate). Unfortunately, our attempts to overexpress the aminotransferase MrsB in E. coli were not successful, so far preventing us from producing larger amounts of MeArg in vitro.
The trio of mrs genes is sufficient for the epiphyte Pss22d to produce and export 3-methylarginine. Thus, Pss22d exhibits a remarkable efficiency to generate with such a small set of genes a potent toxin against the plant pathogen P. syringae pv. glycinea. Most other natural amino acid toxins require more enzymatic steps for their formation, e.g., the well-known arginine analogue canavanine, which is produced by the jack bean (Canavalia ensiformis) (23). Even though natural products from microorganisms make use of rare amino acids as constituents of non-ribosomally produced peptides, only a few free nonproteinogenic amino acids, such as MeArg, are known to come from microorganisms (25). Free rare amino acids appear to be more common among plants and often serve as defense compounds (12).
Although the mode of action of MeArg still has to be investigated, we suspect MeArg to act as an inhibitor of the arginine biosynthesis pathway or an arginine-dependent pathway (5). MeArg probably also interferes with the formation of the signal compound nitric oxide, because 5-(2-methylisothioureido)-2-amino-3-methylpentanoic acid, a synthetic analogue closely related to MeArg, strongly inhibited mammalian nitric oxide synthases, which are important targets for treating diseases such as diabetes, septic shock, and various neurodegenerative diseases (13).
In summary, the genes responsible for the production of MeArg by the epiphyte Pss22d were identified. MeArg is synthesized from 5-guanidino-2-oxo-pentanoic acid, which is methylated by the SAM-dependent methyltransferase MrsA. The resulting 5-guanidino-3-methyl-2-oxo-pentanoic acid is likely transaminated by the putative aminotransferase MrsB to result in 3-methylarginine. The toxin is secreted via the amino acid exporter MrsC (Fig. 7). Identification of the MeArg biosynthesis gene cluster may provide the basis for its large-scale biotechnological production in order to test its potential for control of the soybean pathogen P. syringae pv. glycinea or its potential for pharmacological applications.
Acknowledgments
S.D.B. and B.V. are grateful for financial support from the Deutsche Forschungsgemeinschaft (VO 558/6-3). J.H. and D.S. are thankful for funding from the Jena School for Microbial Communication (JSMC) of the Deutsche Forschungsgemeinschaft. D.S. is grateful for financial support from the Deutsche Forschungsgemeinschaft by means of an Emmy Noether fellowship (SP 1106/3-1) and for funding from the Verband der Chemischen Industrie and the Max Planck Society.
Footnotes
Published ahead of print on 26 February 2010.
REFERENCES
- 1.Agrios, G. N. 2005. Plant pathology, 5th ed. Elsevier Academic Press, London, United Kingdom.
- 2.Aleshin, V. V., N. P. Zakataeva, and V. A. Livshits. 1999. A new family of amino-acid-efflux proteins. Trends Biochem. Sci. 24:133-135. [DOI] [PubMed] [Google Scholar]
- 3.Bender, C., D. Palmer, A. Penaloza-Vázquez, V. Rangaswamy, and M. Ullrich. 1996. Biosynthesis of coronatine, a thermoregulated phytotoxin produced by the phytopathogen Pseudomonas syringae. Arch. Microbiol. 166:71-75. [Google Scholar]
- 4.Braun, S. D., J. Hofmann, A. Wensing, H. Weingart, M. Ullrich, D. Spiteller, and B. Völksch. 2010. In vitro antibiosis by Pseudomonas syringae Pss22d, acting against the bacterial blight pathogen of soybean plants, does not influence in planta biocontrol. J. Phytopathol. 158:288-295. [Google Scholar]
- 5.Braun, S. D., B. Völksch, J. Nüske, and D. Spiteller. 2008. 3-Methylarginine from Pseudomonas syringae pv. syringae 22d/93 suppresses the bacterial blight caused by its close relative Pseudomonas syringae pv. glycinea. Chembiochem 9:1913-1920. [DOI] [PubMed] [Google Scholar]
- 6.Cirvilleri, G., A. Bonaccorsi, G. Scuderi, and M. Scortichini. 2005. Potential biological control activity and genetic diversity of Pseudomonas syringae pv. syringae strains. J. Phytopathol. 153:654-666. [Google Scholar]
- 7.Eggeling, L., and H. Sahm. 2003. New ubiquitous translocators: amino acid export by Corynebacterium glutamicum and Escherichia coli. Arch. Microbiol. 180:155-160. [DOI] [PubMed] [Google Scholar]
- 8.Feil, H., W. S. Feil, P. Chain, F. Larimer, G. DiBartolo, A. Copeland, A. Lykidis, S. Trong, M. Nolan, E. Goltsman, J. Thiel, S. Malfatti, J. E. Loper, A. Lapidus, J. C. Detter, M. Land, P. M. Richardson, N. C. Kyrpides, N. Ivanova, and S. E. Lindow. 2005. Comparison of the complete genome sequences of Pseudomonas syringae pv. syringae B728a and pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 102:11064-11069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu, W., G. Zhao, C. Eddy, and R. A. Jensen. 1995. Imidazole acetol phosphate aminotransferase in Zymomonas mobilis: molecular genetic, biochemical, and evolutionary analyses. J. Bacteriol. 177:1576-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guthke, R., J. Nüske, R. Schorcht, W. Fritsche, and W. Knorre. 1984. Dynamic model of discontinuous and continuous phaseolotoxin production of Pseudomonas syringae pv. phaseolicola. Z. Allg. Mikrobiol. 24:427-435. [PubMed] [Google Scholar]
- 11.Handelsman, J., and E. V. Stabb. 1996. Biocontrol of soilborne plant pathogens. Plant Cell 8:1855-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hylin, J. W. 1969. Toxic peptides and amino acids in foods and feeds. J. Agric. Food Chem. 17:492-496. [Google Scholar]
- 13.Ijuin, R., N. Umezawa, S. Nagai, and T. Higuchi. 2005. Evaluation of 3-substituted arginine analogs as selective inhibitors of human nitric oxide synthase isozymes. Bioorg. Med. Chem. Lett. 15:2881-2885. [DOI] [PubMed] [Google Scholar]
- 14.Jensen, R. A., and W. Gu. 1996. Evolutionary recruitment of biochemically specialized subdivisions of family I within the protein superfamily of aminotransferases. J. Bacteriol. 178:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keane, P. J., A. Kerr, and P. B. New. 1970. Crown gall of stone fruit. 2. Identification and nomenclature of Agrobacterium isolates. Aust. J. Biol. Sci. 23:585-590. [Google Scholar]
- 16.Kers, J. A., M. J. Wach, S. B. Krasnoff, J. Widom, K. D. Cameron, R. A. Bukhalid, D. M. Gibson, B. R. Crane, and R. Loria. 2004. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429:79-82. [DOI] [PubMed] [Google Scholar]
- 17.King, E. O., M. S. K. Ward, and D. E. Raney. 1954. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 44:301-307. [PubMed] [Google Scholar]
- 18.Mahlert, C., F. Kopp, J. Thirlway, J. Micklefield, and M. A. Marahiel. 2007. Stereospecific enzymatic transformation of alpha-ketoglutarate to (2S,3R)-3-methyl glutamate during acidic lipopeptide biosynthesis. J. Am. Chem. Soc. 129:12011-12018. [DOI] [PubMed] [Google Scholar]
- 19.May, R., B. Völksch, and G. Kampmann. 1997. Antagonistic activities of epiphytic bacteria from soybean leaves against Pseudomonas syringae pv. glycinea in vitro and in planta. Microb. Ecol. 34:118-124. [DOI] [PubMed] [Google Scholar]
- 20.Page, R. D. 2002. Visualizing phylogenetic trees using TreeView. Curr. Protoc. Bioinformatics, chapter 6, unit 6.2. [DOI] [PubMed]
- 21.Palmer, D. A., and C. L. Bender. 1993. Effects of environmental and nutritional factors on production of the polyketide phytotoxin coronatine by Pseudomonas syringae pv. glycinea. Appl. Environ. Microbiol. 59:1619-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers, Y. C., A. C. Munk, L. J. Meincke, and C. S. Han. 2005. Closing bacterial genomic sequence gaps with adaptor-PCR. Biotechniques 39:31-34. [DOI] [PubMed] [Google Scholar]
- 23.Rosenthal, G. A. 1982. L-Canavanine metabolism in jack bean, Canavalia ensiformis (L) Dc (Leguminosae). Plant Physiol. 69:1066-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Sanada, M., T. Miyano, S. Iwadare, J. M. Williamson, B. H. Arison, J. L. Smith, A. W. Douglas, J. M. Liesch, and E. Inamine. 1986. Biosynthesis of fluorothreonine and fluoroacetic acid by the thienamycin producer, Streptomyces cattleya. J. Antibiot. 39:259-265. [DOI] [PubMed] [Google Scholar]
- 26.Scholz-Schroeder, B. K., M. L. Hutchison, I. Grgurina, and D. C. Gross. 2001. The contribution of syringopeptin and syringomycin to virulence of Pseudomonas syringae pv. syringae strain B301D on the basis of sypA and syrB1 biosynthesis mutant analysis. Mol. Plant Microbe Interact. 14:336-348. [DOI] [PubMed] [Google Scholar]
- 27.Shoda, M. 2000. Bacterial control of plant diseases. J. Biosci. Bioeng. 89:515-521. [DOI] [PubMed] [Google Scholar]
- 28.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Völksch, B., J. Nüske, and R. May. 1996. Characterization of two epiphytic bacteria from soybean leaves with antagonistic activities against Pseudomonas syringae pv. glycinea. J. Basic Microbiol. 36:453-462. [DOI] [PubMed] [Google Scholar]
- 30.Wilson, K. J., A. Sessitsch, J. C. Corbo, K. E. Giller, A. D. L. Akkermans, and R. A. Jefferson. 1995. Beta-glucuronidase (GUS) tansposons for ecological and genetic-studies of rhizobia and other Gram-negative bacteria. Microbiology 141:1691-1705. [DOI] [PubMed] [Google Scholar]