Abstract
Magnetosome biomineralization and magnetotaxis in magnetotactic bacteria are controlled by numerous, mostly unknown gene functions that are predominantly encoded by several operons located within the genomic magnetosome island (MAI). Genetic analysis of magnetotactic bacteria has remained difficult and requires the development of novel tools. We established a Cre-lox-based deletion method which allows the excision of large genomic fragments in Magnetospirillum gryphiswaldense. Two conjugative suicide plasmids harboring lox sites that flanked the target region were subsequently inserted into the chromosome by homologous recombination, requiring only one single-crossover event, respectively, and resulting in a double cointegrate. Excision of the targeted chromosomal segment that included the inserted plasmids and their resistance markers was induced by trans expression of Cre recombinase, which leaves behind a scar of only a single loxP site. The Cre helper plasmid was then cured from the deletant strain by relief of antibiotic selection. We have used this method for the deletion of 16.3-kb, 61-kb, and 67.3-kb fragments from the genomic MAI, either in a single round or in subsequent rounds of deletion, covering a region of approximately 87 kb that comprises the mamAB, mms6, and mamGFDC operons. As expected, all mutants were Mag− and some were Mot−; otherwise, they showed normal growth patterns, which indicates that the deleted region is not essential for viability in the laboratory. The method will facilitate future functional analysis of magnetosome genes and also can be utilized for large-scale genome engineering in magnetotactic bacteria.
Magnetosomes are unique membrane-enveloped organelles that are formed by magnetotactic bacteria (MTB) for magnetic navigation (2, 37). The mechanism of magnetosome formation is within the focus of a multidisciplinary interest and has relevance for biotechnological applications (5). It has been recognized that the biomineralization of inorganic magnetite crystals and their assembly into highly ordered magnetosome chains are under strict genetic control. Recent studies combining proteomic and bioinformatic approaches suggested that the genetic determination of magnetosome formation is complex and may potentially involve 25 to 50 gene functions (15), with unknown numbers of accessory genes and those controlling signal transduction and motility to achieve effective magnetotaxis (8, 9, 12, 26, 27, 29). However, the functional characterization of these candidate genes has been lagging behind. This is due to technical difficulties and the lack of facile tools for genetic manipulation of MTB. Allelic replacement systems have been established for Magnetospirillum magneticum (18) and Magnetospirillum gryphiswaldense (39, 40), but so far, there are only few examples of these for magnetosome genes that were functionally characterized because of the tedious and cumbersome procedures required for mutant generation (11, 19, 28, 31-32). Most genes controlling magnetosome formation in these and other MTB are located within a genomic magnetosome island (MAI) (34), which is genetically instable during stationary growth (47) and more or less conserved in other MTB (12, 13, 35). Most known magnetosome genes are organized within several conserved operons, which are interspersed with large, poorly conserved genome sections of unknown functions that have been speculated to represent genetic junk irrelevant for magnetotaxis but to cause genetic instability by their high content of repeats and transposable elements (34, 47). Thus, for large-scale functional genome analysis and rearrangements of the MAI, there is a great need for additional and more efficient genetic methods.
Artificial genome recombination systems have been described for a number of bacteria. Many of them are based on the Cre-loxP system of the P1 phage (42). The Cre-loxP recombination system is a simple two-component system that is recognized as a powerful genetic tool in a multitude of eukaryotic and prokaryotic organisms (4, 6, 48). The Cre protein belongs to the integrase family of site-specific recombinases and catalyzes reciprocal site-specific recombination of DNA at 34-bp loxP sites, resulting in either excision or inversion, depending on the parallel or antiparallel orientation of the loxP sites, respectively (21). It does not require any host cofactors or accessory proteins (7). Cre-lox deletion has several advantages over other methods, such as a high efficiency and the independency of the length of DNA located between the two lox sites. The utility of Cre-lox systems has been demonstrated in a wide variety of Gram-positive and Gram-negative bacteria (17, 22-23). In several studies, it was applied for the generation of large-scale deletions, as in for example, the Gram-positive Corynebacterium glutamicum (43-46) and Bacillus subtilis (49).
In M. gryphiswaldense, the functionality of a Cre-loxP antibiotic marker recycling system (25) has been previously demonstrated by deletion of a single gene based on double-crossover insertion of two loxP sites, followed by subsequent Cre-mediated excision (31). In this study, we describe a novel strategy for Cre-loxP-mediated deletion of large genomic fragments which requires only two single crossovers. The system has been validated by the generation of three large deletions, two single and one combination within the MAI, which demonstrated that the total deleted region of approximately 87 kb is not essential for viability and growth in the laboratory.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All strains used in this study are presented in Table 1. Liquid cultures of all Magnetospirillum strains were grown in modified flask standard medium (FSM) (10). Colonies of M. gryphiswaldense were obtained on activated charcoal agar medium (ACAM) that was incubated at 30°C (41). For growth of Escherichia coli strains, lysogeny broth (3) was supplemented with 1 mM dl-α,ɛ-diaminopimelic acid (Sigma-Aldrich, Switzerland). Culture conditions for E. coli strains were as previously described (30). Antibiotics were used at the following concentrations for E. coli: 25 μg/ml kanamycin (Km), 12 μg/ml tetracycline (Tet), and 15 μg/ml gentamicin (Gm). Antibiotics were used at the following concentrations for M. gryphiswaldense strains: 5 μg/ml kanamycin, 5 μg/ml tetracycline, and 20 μg/ml gentamicin. The pKmobGII plasmid contains a chromogenic marker, the gusA gene, which encodes the β-glucoronidase (GUS) enzyme. The concentration of the substrate X-Gluc (5-bromo-4-chloro-3-indoxyl-β-d-glucuronidase) (AppliChem GmbH, Germany) was 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) or source |
|---|---|---|
| M. gryphiswaldense strains | ||
| MSR-1 R3/S1 | Rifr Smr, spontaneous mutant | 40 |
| MSR-1B | Spontaneous mutant, lacking 40.385 kb genomic region | 34 and 47 |
| MSR-1_SU1 | MSR-1::pSU12 | This study |
| MSR-1_SU4 | MSR-1::pSU12::pSU13 | This study |
| ΔmamAB#K7 | ΔmamAB | This study |
| ΔmamAB#K7_pSU28 | ΔmamAB::pSU28 | This study |
| MSR-1_SU12 | ΔmamAB with deletion to mgr4029 | This study |
| MSR-1B_SU13 | MSR-1B::pSU25 | This study |
| MSR-1B_SU14 | MSR-1B::pSU25::pSU37 | This study |
| MSR-1BΔmgr4058tomgr4146 | MSR-1B extension of deletion from mgr4058 to mgr4146 | This study |
| E. coli strain BW29427 | dap auxotroph derivative of E. coli strain B2155 | B. Wanner |
| Plasmids | ||
| pJet1.2 | Apr, eco47IR (lethal restriction enzyme gene), rep (pMB-1) | Fermentas |
| pGEM-T Easy | Apr, lacZα, PCR cloning vector | Promega |
| pKmobGII | Knr, pMB-1 replicon, gusA, lacZα | 16 |
| pAS200 | Gmr, COLE1 ori, sacB of Bacillus subtilis | This study |
| pCM184 | Kmr Apr Tcr | 25 |
| pCM157 | Tcr, Cre expression vector | 25 |
| pSU12 | pKmobGII with upstream mamAB-flanking sequence in EcoRI | This study |
| pSU13 | pAS200 with downstream mamAB-flanking region | This study |
| pSU25 | pSU12 cut with XbaI and HincII, blunted, and ligated with upstream sequence of mgr4058 | This study |
| pSU37 | pCM184 cut with BglII and BsaXI, blunted, self-ligated, then cut with SalI and NheI, blunted, and self-ligated, resulted in pCM184 derivative with downstream mgr4146-flanking sequence in AgeI | This study |
| pSU28 | pSU12 cut with XbaI and HincII and ligated sticky and blunt end with upstream sequence of mgr4029 | This study |
The optical density and magnetic response (Cmag) of M. gryphiswaldense R3/S1 cultures were measured turbidimetrically at 565 nm as previously described (38). For conjugation experiments, E. coli strain BW29427 was used as a donor and cultivated (K. Datsenko and B. L. Wanner, unpublished data) as previously described (33). Conjugative transfers of plasmids were performed as described by Schultheiss et al. (40), with slight modifications. For selection of homologous recombination events, up to 5 × 109 cells were mixed and incubated microaerobically on ACAM for 8 h. Cells were flushed from the agar surface into sterile medium. To increase the ratio of homologous recombination events, the cells were incubated in this medium for 2 h before they were plated onto ACAM with the appropriate antibiotics for plasmid selection.
DNA techniques.
Total DNA from M. gryphiswaldense strains used in this study was isolated as described previously (24). Genetic constructs used in this work were generated using standard PCR procedures. Primer sequences for amplification of DNA fragments from M. gryphiswaldense MSR-1 were derived from GenBank sequence deposition no. CU459003. For sequencing, we used BigDye Terminator version 3.1 chemistry on an ABI 3700 capillary sequencer (Applied Biosystems, Darmstadt, Germany). Sequence data were analyzed with Lasergene 6 (DNAStar Inc., Madison, WI). Primers were purchased from Sigma-Aldrich (Steinheim, Germany) and MWG Biotech (Ebersberg, Germany). For Southern blot hybridization, DNA was isolated, digested with restriction enzymes, electrophoresed, and blotted on a Hybond-N membrane (Amersham). Probe DNA was labeled with radioactive [α-32P]dATP by using the HexaLabel kit (Fermentas, St. Leon-Rot, Germany) and the primers SondemamABfw and SondemamABrw. Prehybridization and hybridization were carried out at 65°C. Signals were detected with a PhosphorImager Typhoon 9400 scanner (Amersham Pharmacia).
Construction of the loxP site plasmids.
For the construction of the inserted loxP plasmids we amplified flanked regions upstream and downstream of the deletion targets. The PCR products were cloned into pGEM-T Easy or pJET1.2 plasmids, sequenced, and subcloned into suicide plasmids. For the construction of the mamAB deletion mutant, a 1.453-kb fragment was amplified by PCR by using primers 5′mamABfw_SU and 5′mamAB_SUloxP (see Table S1 in the supplemental material), which included the loxP sequence, and then subcloned into the pGEM-T Easy vector (Promega, Mannheim, Germany). The upstream fragment of the mamAB operon was sequenced and excised with EcoRI and ligated with the pKmobGII vector containing the gusA gene (16), resulting in pSU12. pKmobGII carries, besides kanamycin resistance, the chromogenic gusA marker. The downstream construct of the mamAB operon was PCR amplified using the primers 3′mamABfw_SUloxP and 3′mamABrw_SU. The resulting 953-bp fragment was sequenced and subcloned into pAS200 (constructed from pBBRMCS5 by replacing oriV with the COLE1 origin of replication [ori] and inserting a sacB gene) to yield pSU13. pSU12 was cut with XbaI and HincII to delete the 5′-end mamAB fragment without the loxP site. pKmobGII containing one loxP site was used for cloning of further upstream fragments. pSU25 was generated from a 2,539-bp fragment localized 345 bp from the spontaneous deletion of the MSR-1B strain and cloned sticky and blunt into pKmobGII::loxP using XbaI and HincII. The same cloning steps were used to create the pSU28 plasmid containing a 2,391-bp sequenced fragment. pSU37 contained a downstream fragment of mgr4146 of 2,337 bp, localized approximately 27 kb downstream of the spontaneous deletion. This fragment was amplified by PCR by using primers 3′MGR4146SUfw and 3′MGR4146SUrw and then subcloned into pGEM-T Easy vector. The fragment was excised with NotI, blunted, and ligated blunt end into the AgeI cleavage site of a pCM184 derivate containing one loxP site and a gentamicin cassette. The gentamicin cassette was subcloned blunt from the pBBRMCS5 plasmid with PstI.
RESULTS
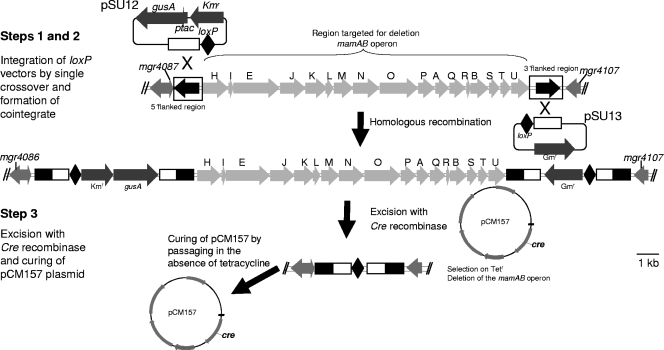
Our strategy for the construction of large deletions relies on a combination of homologous and site-specific recombination, with the latter mediated by the Cre-lox system (Fig. 1). First, homologous regions (1 to 2.5 kb) flanking the 5′ and 3′ ends of the targeted regions, respectively, are cloned while each is adjacent to loxP sites on two different suicide vectors marked differentially by a kanamycin and a gentamicin resistance cassette, respectively. The vectors are then conjugated into the recipient and inserted via homologous recombination by single crossovers into the recipient chromosomes to form a double cointegrate. The relative orientations of the lox sites with respect to the homologous fragments are chosen such that they flank as direct repeats the entire target region encompassing the two inserted plasmids in the double cointegrate to allow subsequent excision of the vector sequences, including the resistance markers, leaving behind only a scar of one 34-bp loxP site. Thus, chromosome integration requires only two single crossovers, which are easier to select for than double crossovers that occur at low frequencies and have remained tedious to screen for in M. gryphiswaldense (14, 40). The efficiencies of single insertions were between 1.34 × 10−5 and 1 × 10−8. Excision is then induced by conjugational transfer of the replicative plasmid pCM157 (25), from which the Cre recombinase is expressed in trans, resulting in the deletion of the targeted chromosomal segment (Fig. 1). The helper plasmid pCM157 is then cured from the deletant strain by relief of antibiotic selection, which had previously been observed to occur at a frequency of 10−1 after one transfer (31).
FIG. 1.
Schematic representation of steps applied for the generation of a Cre-loxP-mediated 16.3-kb genomic deletion within the genomic magnetosome island (MAI) comprising the entire mamAB operon. Other deletions were generated in an analogous way. See the text for details.
We first tested this strategy to delete the mamAB operon that was already known to be not essential for growth (34, 47) but was expected to have a nonmagnetic phenotype (Mag−), as shorter spontaneous deletions mapping within the mamAB operon resulted in cells lacking magnetite crystals (47). The mamAB operon is localized within the MAI, extends over 16.4 kb, and comprises 17 genes of mostly unknown function that are cotranscribed from a single promoter (36). In addition to genes implicated in magnetite biomineralization, genes with functions in magnetosome chain assembly (mamK and mamJ) are carried within the operon (18, 32). For the generation of a defined 16.362-kb mamAB deletion, flanking homologous regions of 1.460 kb upstream and 0.952 kb downstream were cloned into the lox destination vectors derived from pKmobGII containing a gusA marker gene (16), resulting in pSU12 and pSU13, respectively. Conjugation of pSU12 yielded 54 Km-resistant colonies that were blue on X-Gluc plates due to the presence of β-glucuronidase encoded by gusA. After verification of proper chromosomal insertion in all clones by PCR (data not shown), one single-insertant clone named MSR-1_SU1 was selected. Subsequent conjugation of pSU13, which was derived from a pBBRMCS-5 backbone (20) by elimination of the origin of replication, into the single-insertant strain MSR-1_SU1 resulted in 10−6 to 1.2 × 10−8 double cointegrants (MSR-1_SU4) per recipient that were resistant to both kanamycin and gentamicin and formed blue colonies (16) (see Fig. S1A in the supplemental material). Growth and magnetite formation were not affected in MSR-1_SU4.
Lox-mediated excision of the targeted region was initiated by conjugational introduction of pCM157 carrying the Cre recombinase into double-cointegrate strain MSR-1_SU4. After several transfers, we used replica plating to identify 10 out of approximately 300 clones, sensitive to both kanamycin and gentamicin, that had lost their blue color due to loss of the plasmid-borne gusA gene, and hence had lost the targeted chromosomal segment (see Fig. S1B in the supplemental material). Precise excision was verified by PCR amplification and sequencing of a 1.5-kb fragment spanning the excision site and Southern blotting (see Fig. S1C in the supplemental material). To cure pCM157 from this strain, which was named ΔmamAB#K7, it was serially transferred in the absence of tetracycline. Clones that had lost the plasmid were identified after five consecutive passages by replica plating. Light microscopy, transmission electron microscopy (TEM), and Cmag results revealed that strain ΔmamAB#K7 had lost its capability to align in magnetic fields and to form magnetite crystals (Mag−), whereas morphology, growth, and motility were unaffected, confirming that the mamAB operon is essential for magnetosome formation but is not required for growth.
Generation of multiple deletions.
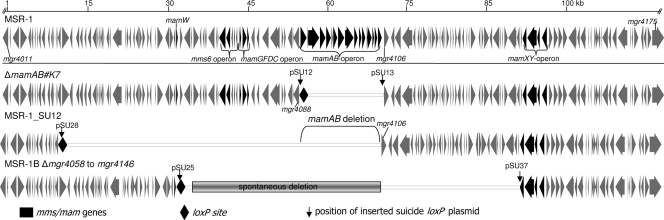
In the next experiments, we wanted to use the technique for genome engineering using the generation of multiple large deletions within the MAI. The target regions were selected because they were predicted to be involved in magnetosome synthesis but not essential for growth (47). As the Cre-mediated excision of mamAB left behind an intact loxP site in strain ΔmamAB#K7, we reused this site for another round of deletion (Fig. 2). Therefore, pSU28 harboring a second loxP site and a 2.5-kb fragment that is homologous to a chromosomal region 45 kb upstream of the left boundary of the mamAB operon was recombined via conjugation into deletion strain ΔmamAB#K7. Upon introduction of pCM157 harboring cre, the targeted fragment of 45 kb was precisely excised, as confirmed by PCR analysis, yielding strain MSR-1_SU12. Thus, a contiguous chromosomal region of 61 kb was eliminated in total by the two consecutive rounds of deletion. Like ΔmamAB#K7, strain MSR-1_SU12 displayed no phenotype other than loss of magnetosomes (Mag−); that is, its morphology, growth characteristics, and motility were indistinguishable from those of the wild type.
FIG. 2.
Molecular organization of the MAIs of mutant strains after Cre-loxP deletion of three different genomic sections.
In a further example, we used strain MSR-1B as a parent, which is Mag− due to a spontaneous deletion of 40 kb comprising a part of the MAI (34, 47). We wanted to further extend this deletion by excision of an additional 27-kb fragment adjacent to the deletion site (Fig. 2). Homologous fragments flanking the targets of 2.539 and 2.337 kb were cloned into lox-pKmobGII and a pCM184 derivate, yielding pSU25 and pSU37, respectively. Consecutive insertion and subsequent Cre excision yielded 4 out of 26 Tetr Kans Gens clones in which the target region was properly excised, as verified by PCR, resulting in strain MSR-1BΔmgr4058tomgr4146. In total, the extent of the chromosomal deletion in strain MSR-1BΔmgr4058tomgr4146 was 67.345 kb. Strain MSR-1BΔmgr4058tomgr4146 showed a phenotype identical to that of its parental strain MSR-1B, i.e., it was Mag− and Mot− (nonmotile) but displayed otherwise normal growth and morphology.
DISCUSSION
We describe a strategy for the generation of large-scale deletions by site-specific recombination in magnetotactic bacteria. Compared to conventional deletion, the Cre-loxP-based method provides several advantages. First, it is very efficient on large fragments, and marker recycling by the site-specific Cre recombinase enables the construction of strains bearing multiple genetic modifications. We have also demonstrated that the single loxP site that remains inserted can be recycled by reusing it in consecutive rounds of deletion. For example, by the combination of a single loxP site with different loxP insertions, the generation of a range of genome deletions with variable lengths has become straightforward. Similar methods were first described for use in E. coli by vector-mediated excisions (VEX) (1) and later applied during genome engineering in C. glutamicum (46). Our method has been specifically adapted for application in magnetospirilla by the modification of a pair of conjugative suicide vectors for lox introduction. In addition, the presence of the gusA gene (16) encoding β-glucoronidase on one of the vectors allows it to monitor vector insertion and subsequent excision. Another specific advantage of our strategy is that it solely relies on single crossovers, which are easier to enforce than double-crossover events (31, 40). Frequencies of RecA-mediated single-crossover insertions in M. gryphiswaldense were previously reported to be 10−6 compared to frequencies of 10−8 for double crossovers (40). In the presented examples and further unpublished mutagenesis experiments, we observed single insertion frequencies between 2 × 10−3 and 1 × 10−6. Subsequent Cre-mediated deletion of the target fragment could usually be detected at approximately 10−3 after 3 to 7 passages. This was significantly less frequent compared to a frequency of 1.0 × 10−1 after only one passage, as previously described (31), and seemed to vary in an unpredictable manner depending on the particular target and the distance between the lox sites. Precise excision of the target fragments was found in about 4 to 15% of the tested Kans Gens GusA− clones. The total time requirement to generate a mutant using this method depended on the particular region, but generation could be usually achieved within several weeks. This is generally faster and more efficient than using conventional allelic replacement techniques for construction of unmarked deletion methods.
We have demonstrated the usefulness of this method for the construction of 16.3-kb, 61-kb, and 67.3-kb deletions within the genomic MAI, either alone or in combination. In total, a region of approximately 87 kb was covered by overlapping deletions, comprising the mamAB, mms6, and mamGFDC operons. As was expected from previous genetic analysis (47), all mutants were nonmagnetic (Mag−) and some were nonmotile (Mot−), but otherwise they showed normal growth patterns under the tested conditions. This indicates that the deleted regions are not essential for viability in the laboratory, which further corroborates our previous assumption that the regions neighboring the known magnetosome operons within the MAI rather comprise accessory genes that are dispensable depending on environmental conditions and might be lost or acquired horizontally (12).
As vectors and techniques are compatible with the closely related magnetotactic bacterium M. magneticum, the described strategy probably can also be used in the latter strain and potentially other MTB. It can be further envisioned that this method can be used for several applications in future approaches. In addition to a systematic functional analysis of the genomic MAI, it could be utilized for targeted genome engineering in magnetotactic bacteria. For example, genomic regions which are not required for growth and magnetosome formation but which contain a large number of repeats and transposons that cause instability of the MAI could be eliminated. This would result in genetically stable host strains synthesizing magnetosomes, which would facilitate the production and application of the biogenic magnetic nanoparticles in biotechnology.
Supplementary Material
Acknowledgments
This study was supported by Deutsche Forschungsgemeinschaft and the Max Planck Society.
We are grateful to Emanuel Katzmann for electron microscopy of mutant strains and André Scheffel for provision of pAS200 and helpful suggestions in the initial phase of the project.
Footnotes
Published ahead of print on 19 February 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ayres, E. K., V. J. Thomson, G. Merino, D. Balderes, and D. H. Figurski. 1993. Precise deletions in large bacterial genomes by vector-mediated excision (VEX). The trfA gene of promiscuous plasmid RK2 is essential for replication in several gram-negative hosts. J. Mol. Biol. 230:174-185. [DOI] [PubMed] [Google Scholar]
- 2.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 3.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bucholtz, F. 2008. Principles of site-specific recombinase (SSR) technology. J. Vis. Exp. pii:718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faivre, D., and D. Schüler. 2008. Magnetotactic bacteria and magnetosomes. Chem. Rev. 108:4875-4898. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh, K., and G. D. Van Duyne. 2002. Cre-loxP biochemistry. Methods 28:374-383. [DOI] [PubMed] [Google Scholar]
- 7.Gopaul, D. N., F. Guo, and G. D. Van Duyne. 1998. Structure of the Holliday junction intermediate in Cre-loxP site-specific recombination. EMBO J. 17:4175-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grünberg, K., E. C. Müller, A. Otto, R. Reszka, D. Linder, M. Kube, R. Reinhardt, and D. Schüler. 2004. Biochemical and proteomic analysis of the magnetosome membrane in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 70:1040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grünberg, K., C. Wawer, B. M. Tebo, and D. Schüler. 2001. A large gene cluster encoding several magnetosome proteins is conserved in different species of magnetotactic bacteria. Appl. Environ. Microbiol. 67:4573-4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heyen, U., and D. Schüler. 2003. Growth and magnetosome formation by microaerophilic Magnetospirillum strains in an oxygen-controlled fermentor. Appl. Microbiol. Biotechnol. 61:536-544. [DOI] [PubMed] [Google Scholar]
- 11.Huang, Y., W. Zhang, W. Jiang, C. Rong, and Y. Li. 2007. Disruption of a fur-like gene inhibits magnetosome formation in Magnetospirillum gryphiswaldense MSR-1. Biochemistry (Moscow) 72:1247-1253. [DOI] [PubMed] [Google Scholar]
- 12.Jogler, C., M. Kube, S. Schübbe, S. Ullrich, H. Teeling, D. A. Bazylinski, R. Reinhardt, and D. Schüler. 2009. Comparative analysis of magnetosome gene clusters in magnetotactic bacteria provides further evidence for horizontal gene transfer. Environ. Microbiol. 11:1267-1277. [DOI] [PubMed] [Google Scholar]
- 13.Jogler, C., W. Lin, A. Meyerdierks, M. Kube, E. Katzmann, C. Flies, Y. Pan, R. Amann, R. Reinhardt, and D. Schüler. 2009. Towards cloning the magnetotactic metagenome: identification of magnetosome island gene clusters in uncultivated magnetotactic bacteria from different aquatic sediments. Appl. Environ. Microbiol. 75:3972-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jogler, C., and D. Schüler. 2006. Genetic analysis of magnetosome biomineralization. In D. Schüler (ed.), Magnetoreception and magnetosomes in bacteria. Springer, Heidelberg, Germany.
- 15.Jogler, C., and D. Schüler. 2009. Genomics, genetics, and cell biology of magnetosome formation. Annu. Rev. Microbiol. 63:501-521. [DOI] [PubMed] [Google Scholar]
- 16.Katzen, F., A. Becker, M. V. Ielmini, C. G. Oddo, and L. Ielpi. 1999. New mobilizable vectors suitable for gene replacement in gram-negative bacteria and their use in mapping of the 3′ end of the Xanthomonas campestris pv. campestris gum operon. Appl. Environ. Microbiol. 65:278-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, T. Y., H. U. Kim, J. M. Park, H. Song, J. S. Kim, and S. Y. Lee. 2007. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol. Bioeng. 97:657-671. [DOI] [PubMed] [Google Scholar]
- 18.Komeili, A., Z. Li, D. K. Newman, and G. J. Jensen. 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242-245. [DOI] [PubMed] [Google Scholar]
- 19.Komeili, A., H. Vali, T. J. Beveridge, and D. Newman. 2004. Magnetosome vesicles are present prior to magnetite formation and MamA is required for their activation. Proc. Nat. Acad. Sci. U. S. A. 101:3839-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovach, M. E., R. W. Phillips, P. H. Elzer, R. M. Roop II, and K. M. Peterson. 1994. pBBR1MCS: a broad-host-range cloning vector. Biotechniques 16:800-802. [PubMed] [Google Scholar]
- 21.Kuhn, R., and R. M. Torres. 2002. Cre/loxP recombination system and gene targeting. Methods Mol. Biol. 180:175-204. [DOI] [PubMed] [Google Scholar]
- 22.Lambert, J. M., R. S. Bongers, and M. Kleerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 73:1126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leibig, M., B. Krismer, M. Kolb, A. Friede, F. Gotz, and R. Bertram. 2008. Marker removal in staphylococci via Cre recombinase and different lox sites. Appl. Environ. Microbiol. 74:1316-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marmur, J. 1961. A procedure for the isolation of deoxiribonucleic acid from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 25.Marx, C. J., and M. E. Lidstrom. 2002. Broad-host-range Cre-lox system for antibiotic marker recycling in gram-negative bacteria. Biotechniques 33:1062-1067. [DOI] [PubMed] [Google Scholar]
- 26.Matsunaga, T., M. Nemoto, A. Arakaki, and M. Tanaka. 2009. Proteomic analysis of irregular, bullet-shaped magnetosomes in the sulphate-reducing magnetotactic bacterium Desulfovibrio magneticus RS-1. Proteomics 9:3341-3352. [DOI] [PubMed] [Google Scholar]
- 27.Matsunaga, T., Y. Okamura, Y. Fukuda, A. T. Wahyudi, Y. Murase, and H. Takeyama. 2005. Complete genome sequence of the facultative anaerobic magnetotactic bacterium Magnetospirillum sp. strain AMB-1. DNA Res. 12:157-166. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura, C., J. G. Burgess, K. Sode, and T. Matsunaga. 1995. An iron-regulated gene, magA, encoding an iron transport protein of Magnetospirillum sp. strain AMB-1. J. Biol. Chem. 270:28392-28396. [DOI] [PubMed] [Google Scholar]
- 29.Richter, M., M. Kube, D. A. Bazylinski, T. Lombardot, F. O. Glöckner, R. Reinhardt, and D. Schüler. 2007. Comparative genome analysis of four magnetotactic bacteria reveals a complex set of group-specific genes implicated in magnetosome biomineralization and function. J. Bacteriol. 189:4899-4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Scheffel, A., A. Gärdes, K. Grünberg, G. Wanner, and D. Schüler. 2008. The major magnetosome proteins MamGFDC are not essential for magnetite biomineralization in Magnetospirillum gryphiswaldense but regulate the size of magnetosome crystals. J. Bacteriol. 190:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheffel, A., M. Gruska, D. Faivre, P. Graumann, A. Linaroudis, J. M. Plitzko, and D. Schüler. 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110-115. [DOI] [PubMed] [Google Scholar]
- 33.Scheffel, A., and D. Schüler. 2007. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J. Bacteriol. 189:6437-6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schübbe, S., M. Kube, A. Scheffel, C. Wawer, U. Heyen, A. Meyerdierks, M. H. Madkour, F. Mayer, R. Reinhardt, and D. Schüler. 2003. Characterization of a spontaneous nonmagnetic mutant of Magnetospirillum gryphiswaldense reveals a large deletion comprising a putative magnetosome island. J. Bacteriol. 185:5779-5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schübbe, S., T. J. Williams, G. Xie, H. E. Kiss, T. S. Brettin, D. Martinez, C. A. Ross, D. Schüler, B. L. Cox, K. H. Nealson, and D. A. Bazylinski. 2009. Complete genome sequence of the chemolithoautotrophic marine magnetotactic coccus strain MC-1. Appl. Environ. Microbiol. 75:4835-4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schübbe, S., C. Würdemann, J. Peplies, U. Heyen, C. Wawer, F. O. Glöckner, and D. Schüler. 2006. Transcriptional organization and regulation of magnetosome operons in Magnetospirillum gryphiswaldense. Appl. Environ. Microbiol. 72:5757-5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schüler, D. 2008. Genetics and cell biology of magnetosome formation in magnetotactic bacteria. FEMS Microbiol. Rev. 32:654-672. [DOI] [PubMed] [Google Scholar]
- 38.Schüler, D., R. Uhl, and E. Baeuerlein. 1995. A simple light-scattering method to assay magnetism in Magnetospirillum gryphiswaldense. FEMS Microbiol. Lett. 132:139-145. [Google Scholar]
- 39.Schultheiss, D., R. Handrick, D. Jendrossek, M. Hanzlik, and D. Schüler. 2005. The presumptive magnetosome protein Mms16 is a PHB-granule bound protein (phasin) in Magnetospirillum gryphiswaldense. J. Bacteriol. 187:2416-2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schultheiss, D., M. Kube, and D. Schüler. 2004. Inactivation of the flagellin gene flaA in Magnetospirillum gryphiswaldense results in non-magnetotactic mutants lacking flagellar filaments. Appl. Environ. Microbiol. 70:3624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultheiss, D., and D. Schüler. 2003. Development of a genetic system for Magnetospirillum gryphiswaldense. Arch. Microbiol. 179:89-94. [DOI] [PubMed] [Google Scholar]
- 42.Sternberg, N., and D. Hamilton. 1981. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J. Mol. Biol. 150:467-486. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki, N., H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2005. New multiple-deletion method for the Corynebacterium glutamicum genome, using a mutant lox sequence. Appl. Environ. Microbiol. 71:8472-8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, N., H. Nonaka, Y. Tsuge, S. Okayama, I. Masayuki, and H. Yukawa. 2005. Multiple large segment deletion method for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 69:151-161. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, N., S. Okayama, H. Nonaka, Y. Tsuge, M. Inui, and H. Yukawa. 2005. Large-scale engineering of the Corynebacterium glutamicum genome. Appl. Environ. Microbiol. 71:3369-3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, N., Y. Tsuge, M. Inui, and H. Yukawa. 2005. Cre/loxP-mediated deletion system for large genome rearrangements in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 67:225-233. [DOI] [PubMed] [Google Scholar]
- 47.Ullrich, S., M. Kube, S. Schübbe, R. Reinhardt, and D. Schüler. 2005. A hypervariable 130-kilobase genomic region of Magnetospirillum gryphiswaldense comprises a magnetosome island which undergoes frequent rearrangements during stationary growth. J. Bacteriol. 187:7176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Duyne, G. D. 2001. A structural view of Cre-loxP site-specific recombination. Annu. Rev. Biophys. Biomol. Struct. 30:87-104. [DOI] [PubMed] [Google Scholar]
- 49.Yan, X., H. J. Yu, Q. Hong, and S. P. Li. 2008. Cre/lox system and PCR based genome engineering in Bacillus subtilis. Appl. Environ. Microbiol. 74:5556-5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.